To the Editor:
Mouse Peg1/Mest encodes a protein with sequence homology to the alpha/beta-hydrolase (Sado et al. 1993). The gene maps to an imprinted region of mouse chromosome 6 and is expressed monoallelically from the paternal allele (Kaneko-Ishino et al. 1995). When the null allele is paternally transmitted, the offspring exhibit severe intrauterine growth retardation (Lefebvre et al. 1998). Uniparental disomy of mouse chromosome 6 is associated with a similar phenotype, presumably as a result of lack of expression of Peg1/Mest (Ferguson-Smith et al. 1991). The human homologue, PEG1/MEST, has been mapped to 7q31.3, within a region of conserved synteny corresponding to mouse chromosome 6, and is monoallelically expressed from the paternal allele in a wide variety of tissues during prenatal and postnatal development. Uniparental disomy of chromosome 7 in humans is associated with phenotypic features of Russell-Silver syndrome (MIM 180860), characterized by intrauterine growth retardation with dysmorphic features such as triangular facies. PEG1/MEST, as the only known imprinted gene on chromosome 7, has been considered a candidate gene for the syndrome (Kobayashi et al. 1997; Lefebvre et al. 1997; Riesewijk et al. 1997).
Imprinting of PEG1/MEST is apparently lost in lymphocytes and transformed lymphoblastoid cell lines. In these tissues, PEG1/MEST is apparently expressed from both the paternal allele and the maternal allele (Riesewijk et al. 1997). Furthermore, PEG1/MEST is transcribed in lymphoblastoid cell lines from patients with maternal uniparental disomy of chromosome 7, or “upd(7)mat” (Cuisset et al. 1997; Riesewijk et al. 1997). Because upd(7)mat cells lack a paternal allele of PEG1/MEST, the transcript must derive from the maternal allele.
The purpose of this report is to delineate the underlying mechanism of apparent loss of imprinting in lymphocytes,to better understand the control of imprinting of the human PEG1/MEST gene. In general, loss of imprinting may be accounted for by several mechanisms. First, imprinting can be regulated in a tissue-specific way. Relaxation of imprinting or biallelic expression of imprinted genes is observed in some tissues. Examples include insulin (Ins) 1 and Ins2 (Giddings et al. 1994; Deltour et al. 1995), and Ube3a (Albrecht et al. 1997). Second, imprinting may be controlled in a promoter-specific manner. Such promoter-specific imprinting was first identified in the IGF2 gene (Vu and Hoffman 1994; Ekstroem et al. 1995). In liver and chondrocytes, the IGF2 transcript from the P1 promoter is always derived from both the paternal allele and the maternal allele, whereas transcripts from other promoters (P2–P4) are expressed solely from the paternal allele. This finding demonstrated that both imprinting and a lack of imprinting could occur within a single gene in a single tissue, suggesting that regional imprinting factors might be important. Third, imprinting can be governed in an isoform-specific way when a single transcription unit encodes different proteins. Maternally derived (e.g., from NESP55), paternally derived (e.g., from XLALPHAS), and biallelically derived (e.g., from GSALPHA) proteins are produced by different patterns of promoter use and alternative splicing of a single transcription unit, GNAS1 (Hayward et al. 1998; Peters et al. 1999).
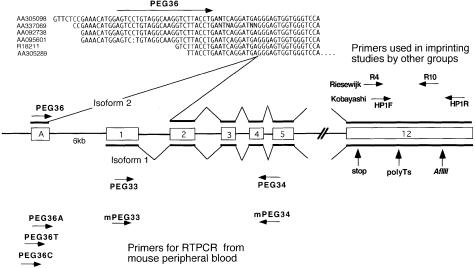
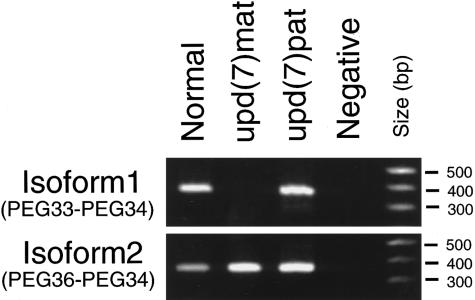
Comparison of the 5′ end of the expressed-sequence tag (EST) sequences assembled as the PEG1/MEST UniGene cluster (Hs. 79284) revealed that six EST clones—AA305098 and AA305289 (colon carcinoma), AA337069 (endometrial tumor), R18211 (infant brain), AA095601 (8-wk fetal heart), and AA092738 (10-wk fetal heart)—share a novel sequence joined to exon 2 of PEG1/MEST (Cuisset et al. 1997), suggesting transcription of an alternative isoform (fig. 1). We first characterized the alternative isoform of PEG1/MEST and examined expression of each of the original and novel isoforms independently. In the following discussion, the original isoform and the alternative one will be referred to as “isoform 1” and “isoform 2,” respectively. To delineate the genomic structure of the PEG1/MEST transcription unit containing the two isoforms, finished genomic-sequence contigs of 7q31.3, deposited at the University of Washington Genome Center were surveyed and aligned against the isoform 1–specific and the isoform 2–specific cDNA sequences, by means of Sequencher software (Gene Codes). Because a mapping study had indicated that genetic distance between PEG1/MEST and D7S649 (also known as “sWSS1203”) was <1 cM (Kobayashi et al. 1997), sequence contigs flanking the PAC clone djs213 containing D7S649 were analyzed (GenBank accession number AC007938). Comparison of the genomic sequence of PAC djs201 and cDNA sequences of isoform 1 and isoform 2 revealed the following: (1) the two isoforms have distinctive first exons (the first exon of isoform 2 will be referred to as “exon A,” the first exon of isoform 1 as “exon 1”); and (2) exon A is located 6 kb upstream of exon 1. Exon A contains a stop codon only 6 bases 5′ of the exon-intron boundary. It is likely that the start codon of isoform 2 is within exon 2 and that exon A comprises the 5′ UTR of isoform 2. Exon A is ⩾57 bp in length. Expression of isoform 1 and isoform 2 in lymphoblastoid cells was detected by means of reverse transcription–coupled PCR (RT-PCR) assay. Either the forward PCR primer (PEG36 [5′-agtcctgtaggcaaggtcttacctg]), based on the isoform 2–specific sequence in exon A, or the forward primer specific for the exon 1 of the isoform 1 (PEG33 [5′-atgggataacgcggccatggtg-3′]), was used with the reverse primer that anneals to the portion of the cDNA sequence shared between the two isoforms (PEG34 [5′-atagtgatgtggtctcggtttgtcactg-3′]) (fig. 1). A upd(7)mat lymphoblastoid cell line (GM11496) (Spence et al. 1988) and a paternal uniparental disomy of chromosome 7, or “upd(7)pat,” lymphoblastoid cell line (Pan et al. 1998) were obtained from the National Institute of General Medical Sciences (NIGMS Coriell Cell Repositories) and from the tissue culture core at Baylor College of Medicine, respectively. The cells were cultured under standard conditions, and total RNA was extracted by means of an RNA purification kit (QIAGEN). One microgram of total RNA was used to synthesize cDNA with the Superscript preamplification system (GIBCO/BRL), and 1% of the resulting material was used for RT-PCR. The cycling conditions were 94°C for 10 min (1 cycle); 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min (40 cycles); and 72°C for 10 min (1 cycle). RT-PCR revealed that the upd(7)mat cell line expressed isoform 2 but not isoform 1, whereas normal lymphocytes and the upd(7)pat cell line expressed both isoform 1 and isoform 2 (fig. 2).
Figure 1.
Alternative splicing of human PEG1/MEST. Arrows indicate primers used for expression studies. RT was followed by PCR amplification by use of exon specific primers. Top, Human EST sequences that matched with the first exon of the alternative isoform. Middle, Exon organization of the PEG1/MEST transcription unit that transcribes two isoforms. In the present study, RT-PCR was done either with PEG36 and PEG34 or with PEG33-PEG34, to detect isoform 2 and isoform 1, respectively. Poly T's and AflII correspond to polymorphic sites in the 3′ UTR that have been described elsewhere. Riesewijk et al. (1997) and Kobayashi et al. (1997) used primer pair R4 and R10 and primer pair HP1F and HP1R, respectively. Bottom, Primers used for RT-PCR, from mouse peripheral blood.
Figure 2.
Expression patterns of the PEG1/MEST isoforms. Expression of isoforms 1 and 2 in normal, upd(7)mat, and upd(7)pat lymphoblastoid cell lines were analyzed by means of RT-PCR. Isoform 1 is expressed in upd(7)pat but not in upd(7)mat. Isoform 2 is expressed in both cell lines. Hence, isoform 1 is imprinted, whereas isoform 2 is not.
In this study, we have demonstrated that (1) an alternative isoform of PEG1/MEST is expressed concurrently with the original isoform in adult lymphocytes and lymphoblastoid cell lines and (2) isoform 1 (the original isoform) is expressed only from the paternal allele, whereas isoform 2 (the alternative isoform) is expressed from both the paternal allele and the maternal allele. These results are discordant with the results of previous studies, which support biallelic expression of the PEG1/MEST in lymphocytes. In retrospect, it is understandable why the previous studies failed to identify such differential imprinting: the primers used for RT-PCR in other studies would not have allowed discrimination between the imprinted isoform and the nonimprinted isoform (fig. 1). In lymphocytes, recognition of an imprinted isoform (isoform 1) was masked by the presence of the nonimprinted form (isoform 2).
Other studies have demonstrated that, in upd(7)mat lymphocytes, only the methylated allele is present at the promoter of the isoform 1 of PEG1/MEST, whereas both methylated and unmethylated alleles are present in normal lymphocytes (Riesewijk et al. 1997). We now conclude, on the basis of findings from the present study, that parental-of-origin–specific loss of isoform 1 expression is strictly correlated with the methylation of the promoter of isoform 1. Documentation of this tight correlation validates the use of methylation analysis of PEG1/MEST gene in lymphocytes as a diagnostic assay for upd(7)mat.
Identification of isoform-specific imprinting illustrates several important issues with respect to imprinting studies in general. First, effort should be made to identify isoforms when one is evaluating new potentially imprinted genes. A potentially imprinted gene could be mistakenly disregarded if isoform-specific imprinting is overlooked. As shown in this study, use of the EST database can be very helpful in the identification of alternative isoforms. Second, imprinted genes that are allegedly subject to tissue-specific imprinting may need further evaluation. As shown with PEG1/MEST and GNAS1, nonimprinted or reciprocally imprinted isoforms may be expressed in tissues in which imprinting is apparently lost (Hayward et al. 1998; Peters et al. 1999). Third, the concept of leaky expression needs to be challenged. Examples of leaky expression include p57kip2 (Reik and Maher 1997) and IMPT1/ORCTL2 (Cooper et al. 1998; Dao et al. 1998). With respect to PEG1/MEST, a minimal but detectable level of expression from the maternal allele was observed in early (6–9 wk) human embryos, and this was considered to be leaky expression from the imprinted inactive maternal allele (Kobayashi et al. 1997). It is probable that these leaky transcripts from the maternal allele represent isoform 2, in light of the fact that isoform 2 is expressed as early as 8–10 wk in fetal heart (EST sequences AA092738 and AA095601). Similarly, the concept of interspecific imprinting differences may need revision. In contrast to the human PEG1/MEST gene, the mouse gene is not expressed from the paternal allele in lymphocytes (Riesewijk et al. 1997), nor is leaky expression from the maternal allele observed in mouse embryos (Kaneko-Ishino et al. 1995). Hence, a difference, in imprinting patterns, between mice and humans may simply reflect absence of isoform 2 in the mice. In fact, evaluation of the mouse Peg1/Mest UniGene cluster (Mm. 1089), consisting of 181 mouse EST sequences, revealed no evidence of alternative splicing: all 40 ESTs that contained exon 2 were flanked by exon 1 sequence, not by exon A–like sequence. Furthermore, RT-PCR, done with cDNA obtained from mouse peripheral blood by means of mouse-specific primer positioned within exon 4/5 of mouse Peg1/Mest gene (mPEG34 [5′-atgtggtctcggcttgtcactg-3′]), in combination with any of the four human-specific primers positioned within exon A (PEG36, PEG36A [5′-agtcctgtaggcaaggtcttacctga-3′], PEG36T [5′-gagtcctgtaggcaaggtcttacct-3′], and PEG36C [5′-gagtcctgtaggcaaggtcttacc-3′]) failed to amplify, whereas primer mPEG33 (5′-gggataatgcggccatggtg-3′), designed on the basis of mouse exon 1 sequence (the exon unique to isoform 1), yielded a specific PCR product when used with mPEG34 (fig. 1). These observations support the contention that isoform 2 may not be expressed in mouse peripheral blood and/or lymphocytes. In summary, human PEG1/MEST is imprinted in an isoform-specific manner rather than in a tissue-specific manner in lymphocytes.
Acknowledgments
We thank Mr. Taichi Suzuki, from Tokyo Technical College, for excellent laboratory assistance. This work was supported, in part, by a grant from the Pharmacia-Upjohn Fund for Growth & Development Research.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Web/Genbank [Google Scholar]
- NIGMS Coriell Cell Repository, http://locus.umdnj.edu/nigms [Google Scholar]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for Russell-Silver syndrome [MIM 180860])
- UniGene, http://www.ncbi.nlm.nih.gov/UniGene/ [Google Scholar]
- University of Washington Genome Center, http://www.genome.washington.edu/UWGC/chr-7/c7project.htm [Google Scholar]
References
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV (1997) Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet 17:75–78 [DOI] [PubMed]
- Cooper PR, Smilinich NJ, Day CD, Nowak NJ, Reid LH, Pearsall RS, Reece M, et al (1998) Divergently transcribed overlapping genes expressed in liver and kidney and located in the 11p15.5 imprinted domain. Genomics 49:38–51 [DOI] [PubMed]
- Cuisset L, Le Stunff C, Dupont JM, Vasseur C, Cartigny M, Despert F, Delpech M, et al (1997) PEG1 expression in maternal uniparental disomy 7. Ann Genet 40:211–215 [PubMed]
- Dao D, Frank D, Qian N, O'Keefe D, Vosatka RJ, Walsh CP, Tycko B (1998) IMPT1, an imprinted gene similar to polyspecific transporter and multi-drug resistance genes. Hum Mol Genet 7:597–608 [DOI] [PubMed]
- Deltour L, Montagutelli X, Guenet JL, Jami J, Paldi A (1995) Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev Biol 168:686–688 [DOI] [PubMed]
- Ekstroem TJ, Cui H, Li X, Ohlsson R (1995) Promoter specific IGF2 imprinting status and its plasticity during human liver development. Development 121:309–316 [DOI] [PubMed]
- Ferguson-Smith AC, Cattanach BM, Barton SC, Beechey CV, Surani MA (1991) Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature 351:667–670 [DOI] [PubMed]
- Giddings SJ, King CD, Harman KW, Flood JF, Carnaghi LR (1994) Allele specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat Genet 6:310–313 [DOI] [PubMed]
- Hayward BE, Moran V, Strain L, Bonthron DT (1998) Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci USA 95:15475–15480 [DOI] [PMC free article] [PubMed]
- Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, et al (1995) Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet 11:52–59 [DOI] [PubMed]
- Kobayashi S, Kohda T, Miyoshi N, Kuroiwa Y, Aisaka K, Tsutsumi O, Kaneko-Ishino T, et al (1997) Human PEG1/MEST, an imprinted gene on chromosome 7. Hum Mol Genet 6:781–786 [DOI] [PubMed]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA (1998) Abnormal maternal behaviour and growth retardation associated with loss of imprinted gene Mest. Nat Genet 20:163–169 [DOI] [PubMed]
- Lefebvre L, Viville S, Barton SC, Ishino F, Surani MA (1997) Genomic structure and parent-of-origin-specific methylation of Peg1. Hum Mol Genet 6:1907–1915 [DOI] [PubMed]
- Pan Y, McCaskill CD, Thompson KH, Hicks J, Casey B, Shaffer LG, Craigen WJ (1998) Paternal isodisomy of chromosome 7 associated with complete situs inversus and immotile cilia. Am J Hum Genet 62:1551–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Wroe SF, Wells CA, Miller HJ, Bodle D, Beechey CV, Williamson CM, et al (1999) A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA 96:3830–3835 [DOI] [PMC free article] [PubMed]
- Reik W, Maher ER (1997) Imprinting in clusters: lessons from Beckwith-Wiedemann syndrome. Trends Genet 13:330–334 [DOI] [PubMed]
- Riesewijk AM, Hu L, Schulz U, Tariverdian G, Hoglund P, Kere J, Ropers HH, et al (1997) Monoallelic expression of human PEG1/MEST is paralleled by parent-specific methylation in fetuses. Genomics 42:236–244 [DOI] [PubMed]
- Sado T, Nakajima N, Tada M, Takagi N (1993) A novel mesoderm-specific cDNA isolated from a mouse embryonal carcinoma cell line. Dev Growth Differ 35:551–560 [DOI] [PubMed] [Google Scholar]
- Spence JE, Perciaccante RG, Greig GM, Willard HF, Ledbetter DH, Hejtmancik JF, Pollack MS, et al (1988) Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet 42:217–226 [PMC free article] [PubMed]
- Vu TH, Hoffman AR (1994) Promoter-specific imprinting of the human insulin-like growth factor-II gene. Nature 371:714–717 [DOI] [PubMed]