Summary
Lysinuric protein intolerance (LPI) is a rare autosomal recessive defect of cationic amino acid transport caused by mutations in the SLC7A7 gene. We report the genomic structure of the gene and the results of the mutational analysis in Italian, Tunisian, and Japanese patients. The SLC7A7 gene consists of 10 exons; sequences of all of the exon-intron boundaries are reported here. All of the mutant alleles were characterized and eight novel mutations were detected, including two missense mutations, 242A→C (M1L) and 1399C→A (S386R); a nonsense mutation 967G→A (W242X); two splice mutations IVS3 +1G→A and IVS6 +1G→T; a single-base insertion, 786insT; and two 4-bp deletions, 455delCTCT and 1425delTTCT. In addition, a previously reported mutation, 1625insATCA, was found in one patient. It is noteworthy that 242A→C causes the change of Met1 to Leu, a rare mutational event previously found in a few inherited conditions. We failed to establish a genotype/phenotype correlation. In fact, both intrafamilial and interfamilial phenotypic variability were observed in homozygotes for the same mutation. The DNA-based tests are now easily accessible for molecular diagnosis, genetic counseling, and prenatal diagnosis of LPI.
Introduction
Lysinuric protein intolerance (LPI; MIM 222700) is a rare autosomal recessive disease that is relatively common in Finland (Simell 1995) and Italy (Incerti et al. 1993). Clinical findings of LPI include vomiting, diarrhea, failure to thrive, episodes of coma, hepatosplenomegaly, and osteoporosis. A life-threatening lung involvement, mainly alveolar proteinosis, and severe renal involvement have also been reported (Parenti et al. 1995; Simell 1995; Santamaria et al. 1996a). Metabolic derangement is characterized by reduced intestinal absorption of cationic amino acid (lysine, ornithine, arginine [CAA]), increased renal excretion of CAA, orotic aciduria, and dysfunction of urea cycle leading to hyperammonemia. The disease is considered relatively benign when appropriately treated with citrulline supplementation and a low-protein diet, but this treatment may be insufficient to prevent severe complications. LPI is caused by a defective transport of CAA at the basolateral membrane of the epithelial cells of intestine and kidney (Simell 1995). This transport is mediated by systems y+ and y+L, the former exerted by human CAA transporters 1, 2, and 4 (CAT1-4) (Palacin et al. 1998). The latter is induced by heterodimers consisting of the 4F2 heavy chain (4F2hc) and the light chain of either the solute carrier family 7A, member 6 (SLC7A6) or member 7 (SLC7A7) (Torrents et al. 1998, 1999). Previous studies excluded human CAT1-4 and 4F2hc as candidate genes of LPI. Recently, the LPI gene SLC7A7, located on chromosome 14q11.2, has been identified (Borsani et al. 1999; Torrents et al. 1999). SLC7A7 protein forms an ∼135-kDa disulfide bond–dependent heterodimer with 4F2hc in oocytes, which, on reduction, results in two peptides of ∼85 kDa (4F2hc) and ∼40 kDa (SLC7A7). A hydrophobicity plot of SLC7A7 predicts 12 transmembrane (TM) domains with both C- and N-terminal segments in the cytoplasm. Five mutations of the SLC7A7 gene were reported in Italian, Finnish, and Spanish patients with LPI (Borsani et al. 1999; Torrents et al. 1999). A splice-acceptor mutation (IVS5-2A→T, previously reported as 1136-2A→T) accounts for the founder LPI allele in Finland (Borsani et al. 1999; Torrents et al. 1999). In the same study, the 1625insATCA mutation was found in homozygosity in three unrelated Italian pedigrees, all originating from a restricted geographical area and all sharing a common haplotype linked to the LPI locus. Surprisingly, a different mutation, 197-740del, was found in homozygosity in another patient originating from the same restricted area. This result suggested a possible mutational heterogeneity of the SLC7A7 gene in Italian patients with LPI.
In the present study, we report the structure of the human SLC7A7 gene, which enables a rapid screening of mutations in patients with LPI. In addition, we report the identification of eight novel mutations, definitely indicating an unusually high degree of mutational heterogeneity of LPI, at least in Italy.
Subjects, Material, and Methods
Patients
Fourteen patients with LPI from 11 independent families were investigated for the presence of mutations of the SLC7A7 gene. Clinical findings of the Italian and Tunisian patients were reported elsewhere (Parini et al. 1991; Di Rocco et al. 1993; Incerti et al. 1993; Candito et al. 1994; Parenti et al. 1995; Santamaria et al. 1996a; Parenti et al. 1998).
The Japanese patient presented with hepatosplenomegaly at age 3 mo. Hyperammonemia and seizures were reported at age 18 mo. The diagnosis of LPI was defined at age 10 years. At the moment, mental development is normal, and no complications have been observed.
Nine families originated from southern Italy, one from Tunisia, and one from Japan. Six of the 14 patients (patients 1A, 1B, 2, 3, 9A, 9B, and 11) were born to consanguineous parents. DNA samples from parents and unaffected sibs, where available, were also investigated to confirm the correct segregation of each mutation.
Structure of the SLC7A7 Gene
The human genomic PAC library RPCI-5 was screened by use of two sets of primers located at the 5′ end (forward, 5′-GGAGATCTCACTGCTTAACGG-3′, and reverse, 5′-AGGCGGCTGGCAGCATAAG-3′) and at the 3′ end (forward, 5′-AAATTGGAGCATTGTGGGC-3′, and reverse, 5′-AGCCTCACTTCCTTTGGAGG-3′) of SLC7A7 cDNA, and two positive clones were identified. Automated sequencing by means of an Applied Biosystem ABI 377 fluorescent sequencer of PAC DNA and PCR-amplified genomic DNA was done by using gene-specific oligonucleotide primers with ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit (PE Biosystems).
Genomic DNA Preparation and Amplification of the SLC7A7 Gene
Genomic DNA was extracted by standard methods from peripheral blood samples of members of all families, cultured skin fibroblasts, and Epstein-Barr virus–transformed lymphocytes (Sambrook et al. 1989). Exons of the SLC7A7 gene were amplified by PCR (Therm Cycler 480; PE Biosystems) with primers designed for exons 1–10. Exon 2 was amplified in two pieces (2a and 2b) by use of two sets of primers.
PCR reactions were done in a total volume of 25 μl, which contained 50 ng of template DNA, 1 × AmpliTaq reaction buffer (PE Biosystems), 250 μM of each nucleotide, 50 ng of each primer, and 0.5 U of AmpliTaq DNA polymerase (PE Biosystems). The reactions were done for 39 cycles, with denaturation at 94°C for 1 min, annealing at 50°C–60°C for 1 min, and extension at 72°C for 1 min. In the first cycle, denaturation was done for 7 min, and, in the final cycle, the extension lasted 7 min.
Southern Hybridization
Genomic DNA from 14 patients with LPI and two controls were digested with EcoRI and PstI restriction enzymes, according to the manufacturer's recommendations. After electrophoresis in a 1% agarose gel, the digested DNAs were transferred onto a Hybond-N membrane (Amersham). The membrane was hybridized at 65°C in a buffer containing 6 × SSC, 1 × Denhardt's solution, 0.5% SDS, 0.2 mg/ml sonicated herring-sperm DNA, and 32PdCTP-labeled full-length SLC7A7 cDNA probe.
SSCP Analysis and Direct Sequencing
For the SSCP analysis, 0.4 μl of 32PdCTP (3,000 Ci/mmol) were added to the PCR-mix reaction. Primers used for PCR-SSCP are listed in table 2, and the conditions are the same as those described for genomic DNA amplifications. SSCP was done on a 6% polyacrylamide nondenaturing gel. Direct sequencing was done on PCR products showing SSCP band shifts. Both strands of the PCR products were sequenced by an automated system (ABI PRISM 377).
Table 2.
PCR Primer Sets for Amplifying Exons of SLC7A7[Note]
|
Primer |
|||
| Exon | Forward | Reverse | Product Size (bp) |
| 1 | 5′-ACTGCTGGCTGGGAAGGAG-3′ | 5′-gagggttagcaaggtaagtgg-3′ | 158 |
| 2a | 5′-gccaaccacctaatcgtatag-3′ | 5′-CAGATTTCTTAATGGTGGTGCCC-3′ | 384 |
| 2b | 5′-GCTGTCGGGGGCCTCTTCTCCG-3′ | 5′-tagcctcctaccccagccc-3′ | 295 |
| 3 | 5′-gagtctaaggccctagggtgg-3′ | 5′-ctgcaggctgtttcctcatcc-3′ | 343 |
| 4 | 5′-tgtcagtcctttacttgaccatc-3′ | 5′-tatggaaagttggtggtgcag-3′ | 252 |
| 5 | 5′-gagatagggatatgactgtggg-3′ | 5′-cagtaccccacaagacaccc-3′ | 227 |
| 6 | 5′-agatcctgctaatgactcttcc-3′ | 5′-gttctgtccactgcatagccc-3′ | 200 |
| 7 | 5′-ccaattttctcagcttctcc-3′ | 5′-cgcaatctggctttcagtctc-3′ | 173 |
| 8 | 5′-aaggccctgagaagagctg-3′ | 5′-ttcaggtggagcagaggtagg-3′ | 225 |
| 9 | 5′-gaatcctgagttgtccactcc-3′ | 5′-agctctgcaatagctgagcgg-3′ | 270 |
| 10 | 5′-tgagttccattcagtgacacctg-3′ | 5′-AAATTGGAGCATTGTGGGC-3′ | 500 |
Note.—Intron sequences are shown in lowercase letters. Exon sequences are shown in uppercase letters.
Restriction Enzyme Digestion of PCR Products
For those mutations (242A→C, 967G→A, 1399C→A) introducing or abolishing the recognition sequence of a restriction enzyme, the specific PCR products were digested with the requested restriction enzyme (NcoI, BsmAI, HinfI) following the manufacturer's instructions (New England Biolabs). Digests were analyzed on a 2% Separide Gel Matrix (Gibco BRL).
Results
Structure of the SLC7A7 Gene
The RPCI-5 PAC library was screened by use of two sets of primers located at the 5′ and 3′ ends of SLC7A7 cDNA, and two clones were identified. One of them, PAC clone 830D3, contains the entire gene. The location and sequence of all exon-intron boundaries (table 1) were determined by direct sequencing of either the PAC clone or the PCR products obtained from amplification of genomic DNA with cDNA-derived oligonucleotide primers.
Table 1.
Exon-Intron Boundaries and Sizes of Exons of the SLC7A7 Gene
| Exon | 3′ Splice Sitea | 5′ Splice Sitea | Exon sizeb |
| 1 | … | CAAGGAgtgagtgtgc | ? |
| 2 | ttccttaaagGGCAAT | GCATCTgtaagcgggg | 540 |
| 3 | tttctttcagGTCTCT | GCCAGGgttaagaaat | 125 |
| 4 | tgggttccagGAGCCT | TGAGAGgtaggtactg | 144 |
| 5 | ttggctgtagGAACCT | GCTGTGgtaaaggatt | 123 |
| 6 | tttcccttagACTTTT | TTCTAGgtaagagtgg | 103 |
| 7 | cctaacatagGCTTTT | TTCAATgtaagtgaac | 97 |
| 8 | acgcttacagGGTATC | CTCAAGgtaaggcaac | 183 |
| 9 | gtgcccttagCTCAGC | TCGTGGgtaagccgtg | 99 |
| 10 | ctctttgtagGGTCTG | … | ? |
Intron sequences are shown in lowercase letters. Exon sequences are shown in uppercase letters.
? = Exon size unknown.
The SLC7A7 gene was shown to be organized in 10 exons with sizes in the range of 97–540 bp. All of the exon-intron junctions agree with the gt–ag consensus sequence. Primers for the amplification of all SLC7A7 exons were designed in the intronic flanking sequences and are reported in table 2. The codon for the translation-initiator methionine (Met1) is located in exon 2 at position 242, whereas the termination codon, TAA, is located at position 1775 in exon 10.
Analysis of the SLC7A7 Gene in Patients with LPI
Southern blot analysis of genomic DNA revealed no detectable differences in the patients with LPI vis-à-vis the controls, excluding major rearrangements (data not shown). The search for mutations was then done by SSCP analysis of each of the 10 exons, including the exon-intron boundaries, in 14 patients with LPI and two controls. Abnormal SSCP conformers were detected for exons 2, 3, 4, 6, 8, and 9. By direct sequencing, we found eight novel mutations and one previously reported mutation (table 3). All patients were homozygous for a specific mutation, except the Japanese patient.
Table 3.
Mutations of the SLC7A7 Gene in Italian, Tunisian, and Japanese Patients with LPI
|
Mutation |
||||
| Patienta | Ethnicity | Allele 1 | Allele 2 | Source of Data |
| 1A | Italian (Puglia) | M1L | M1L | Present study |
| 1B | Italian (Puglia) | M1L | M1L | Present study |
| 2 | Italian (Calabria) | S386R | S386R | Present study |
| 3 | Italian (Calabria) | S386R | S386R | Present study |
| 4 | Italian (Calabria) | W242X | W242X | Present study |
| 5 | Italian (Lazio) | W242X | W242X | Present study |
| 6A | Italian (Sicilia) | 786insT | 786insT | Present study |
| 6B | Italian (Sicilia) | 786insT | 786insT | Present study |
| 7 | Italian (Molise) | 455delCTCT | 455delCTCT | Present study |
| 8 | Italian (Campania) | 1425delTTCT | 1425delTTCT | Present study |
| 9A | Tunisian | 1425delTTCT | 1425delTTCT | Present study |
| 9B | Tunisian | 1425delTTCT | 1425delTTCT | Present study |
| 10 | Japanese | IVS3 +1G→A | IVS6 +1G→T | Present study |
| 11 | Italian (Campania) | 1625insATCA | 1625insATCA | Present study |
| 12 | Italian (Campania) | 1625insATCA | 1625insATCA | III-2, F2 in Borsani et al. (1999) |
| 13 | Italian (Campania) | 1625insATCA | 1625insATCA | III-3, F3 in Borsani et al. (1999) |
| 14C | Italian (Campania) | 1625insATCA | 1625insATCA | III-6, F4 in Borsani et al. (1999) |
| 15C | Italian (Campania) | 1625insATCA | 1625insATCA | IV-2, F4 in Borsani et al. (1999) |
| 16 | Italian (Campania) | 197-740delb | 197-740delb | IV-2, F1 in Borsani et al. (1999) |
“A” and “B” indicate that the patients are siblings; “C” indicates that the patients are consanguineous.
Previously reported as 197Δ543 mutation by Borsani et al. (1999)
A missense mutation at codon 1 (M1L)
On both alleles of two affected brothers, patients 1A and 1B born to consanguineous parents, we identified a 242A→C transversion that caused a methionine-to-leucine change at codon 1 in exon 2 (M1L). An NcoI site is destroyed by this mutation. By NcoI digestion of exon 2, we confirmed that these patients are homozygous for this mutation and that the parents and the unaffected sister are heterozygous.
A missense mutation at codon 386 (S386R)
In two independent patients (2 and 3), both born to first-cousin parents, we identified a 1399C→A transversion that changed a serine to arginine at codon 386 (S386R) in exon 8. Both patients are homozygous for this mutation, which introduces a new HinfI site in exon 8. In both patients, sequencing of the remainder of the coding region and the exon-intron boundaries of the gene did not detect other mutations. By HinfI digestion of exon 8, we confirmed that patients 2 and 3 are homozygous for the S386R mutation and that all of their parents are heterozygous. Two unaffected sisters of patient 2 were heterozygous and homozygous for the wild-type allele, respectively. This mutation was not detected in HinfI digests of amplified exon 8 from 54 unrelated control DNAs (data not shown).
A nonsense mutation at codon 242 (W242X)
In two independent patients (4 and 5), a 967G→A transition creates a termination codon TGA (W242X) in exon 4. Both patients are homozygous for this mutation, which introduces a new BsmAI site in exon 4. By BsmAI digestion of exon 4, we confirmed that patients 4 and 5 are homozygous for the 967G→A mutation and all of their parents are heterozygous. An unaffected brother of patient 4 was heterozygous.
A single-base insertion in exon 3 (786insT)
In two affected brothers, patients 6A and 6B, a T insertion in exon 3, 786insT, causes a frameshift and introduces a termination codon (TAA) at position 812. Both patients are homozygous for this mutation. Direct sequencing of exon 3 revealed a carrier status of the parents for this mutation.
A 4-bp deletion in exon 2 (455delCTCT)
Patient 7 was homozygous for a 4-bp deletion in exon 2, 455delCTCT, which causes a frameshift and introduces a termination codon (TAA) at position 743. Direct sequencing of exon 2 revealed that the parents and an unaffected brother are heterozygous for this mutation.
A 4-bp deletion in exon 8 (1425delTTCT)
A 4-bp deletion in exon 8, 1425delTTCT, was found in homozygosity in patient 8 and in two affected Tunisian brothers (9A and 9B). This mutation causes a frameshift that abolishes the termination codon at position 1775, introducing a new TGA at position 1790. By SSCP analysis of exon 8, we found all parents were heterozygous for this mutation as was an unaffected sister of patient 8.
Two splice mutations at IVS3 +1G→A and IVS6 +1G→T
In patient 10, from Japan, two splice mutations were found in heterozygosis. The first one, inherited from the mother, is a G→A transition at the donor site of intron 3. The second one, inherited from the father, is a G→T transversion at the donor site of intron 6. The sequences of the remainder of the coding region and the exon-intron boundaries of the gene were identical to the wild type. Two unaffected sibs were heterozygous for IVS3 +1G→A and IVS6 +1G→T, respectively.
1625insATCA mutation
In patient 11, born to first-cousin parents, we found homozygosity for 1625insATCA, a mutation previously reported (Borsani et al. 1999). This mutation results in a frameshift that disrupts the reading frame at codon 462 and introduces a translation termination codon (TGA) 13 bp downstream. Direct sequencing of exon 9 from genomic DNA of the parents revealed their carrier status for this mutation.
Discussion
The present study provides the complete structure of the LPI gene, SLC7A7, including sequences and PCR conditions, to enable mutational analysis of all 10 exons of the gene. Southern blot analysis of patients' genomic DNA did not reveal gross gene rearrangements as previously observed in patient 16. We then searched for mutations of the SLC7A7 gene by using SSCP analysis in 1 Japanese, 2 Tunisian, and 11 Italian patients, representing 18 affected independent alleles. This study led to the identification of eight novel mutations and an already-known mutation. All the affected alleles were identified, and each patient was homozygous for a specific mutation, except the Japanese patient (table 3). The segregation of each mutation in the pedigree was coherent with the recessive mode of inheritance of the disease.
Characteristics of the Mutations
The missense M1L changes the first methionine (Met1) to leucine. Mutations of the translation-initiator ATG (Met1) have rarely been observed in inherited diseases. However, such a mutation was recently found in the RHAG gene in patients affected by Rhmod syndrome (Huang et al. 1999), in which use of alternative ATGs in the RHAG mRNA was allowed by some in-frame ATGs sharing significant homology with Kozak's consensus sequence (Kozak 1987). In our case, the sequence context of the subsequently available ATG (Met50, GGGAACATGA) does not conform to Kozak's consensus (CCA[G/A]CCATGG). If we used this ATG, the mutated protein would lack the first 49 amino acid residues corresponding to the intracellular N-terminus and part of the first TM domain. In addition, the N-terminus truncation would affect the insertion of the protein into the lipid bilayer (Singer 1990). A better Kozak consensus is found for the following ATG (Met143), but more dramatic alteration of the protein should be expected.
The missense mutation S386R was found in two independent patients originating from the same geographic area (Calabria). We do not have formal proof that S386R is a disease-causing mutation. However, its causative effect is strongly supported by the following: (1) absence of other mutations within the coding region and the exon-intron boundaries of the SLC7A7 gene in both patients; (2) absence of this mutation in 108 control alleles, which excludes it as a polymorphic change; (3) the mutation coherently segregates with the phenotype; and (4) the serine residue at position 386 is conserved in the mouse ortholog (my+LAT1) and in homologous proteins, such as hy+LAT2, hLAT1, and IU12 (Torrents et al. 1998; Pfeiffer et al. 1999). Linkage analysis with markers of the LPI locus showed that the two patients carry different haplotypes (Lauteala et al. 1998). This suggests that the same mutation arose independently in the two pedigrees. Alternatively, multiple recombinational events on a common ancestral allele would have been required.
The nonsense mutation W242X in exon 4 was found in two independent patients originating from different geographic areas (Calabria and Lazio). The linkage analysis with markers of the LPI locus showed different haplotypes in the two patients, thus suggesting independent mutational events. The predicted mutated protein would lack 7 of the 12 TM domains and the intracellular C-terminus, which should yield a nonfunctional product.
The splice mutations found in the Japanese patient, IVS3 +1G→A and IVS6 +1G→T, disrupt the GT rule of the donor splice sites of introns 3 and 6, respectively. Unfortunately, none of the patient's cell lines were available for mRNA studies. Nevertheless, the kind of mutations, in addition to the segregation data and the absence of changes in the coding region and in the other exon-intron boundaries, all appear to be conclusive for a disease-causing role.
The 786insT mutation causes a frameshift and a premature termination of translation at position 812. The predicted mutated protein would be severely altered because of the lack of the last 9 TM domains and the intracellular C-terminus.
Even more dramatic is the effect of the 455delCTCT mutation. The frameshift introduces a premature termination codon (TAA) in the reading frame at position 743. The predicted mutated protein lacks all of the TM domains and the intracellular C-terminus.
The 1425delTTCT mutation causes a frameshift that abolishes the wild-type termination codon, introducing a new one (TGA) at position 1790, 15 bp downstream of the former. The resulting protein would differ from the wild-type sequence in the last 116 amino acid residues, with a gain of 5 residues at the intracellular C-terminus stretch. The hydrophobicity plot of the mutated protein predicts a dramatic change of the last four TM domains of the wild-type protein, with loss of an entire domain and shortening of the C-terminus stretch.
Interestingly, this mutation was found in both Italian and Tunisian patients. Haplotype analysis might help us to understand if migrations within the Mediterranean basin contributed to the spread of this mutation, as has been observed for several other inherited diseases.
The 1625insATCA mutation was found in patient 11, who originates from the same geographic area (Campania) as four other patients already reported (12, 13, 14C, and 15C in table 3). These four patients are homozygous for the same mutation, and the affected allele might descend from a common ancestor, as suggested by pedigree analysis and haplotypes linked to the LPI locus (Borsani et al. 1999). Surprisingly, only a single allele of patient 11 shares the common haplotype carried in homozygosity by the four other patients (Lauteala et al. 1998). This diversity might be explained by an independent mutational event on the chromosome carrying the different haplotype. However, the consanguinity of the parents of patient 11 and the geographic origin in common with the other four patients might favor the hypothesis of multiple recombinational events between the LPI locus and the markers. Similarly, the Finnish founder mutation was also found in a single patient without the common 6-4 haplotype (Torrents et al. 1999).
The present results indicate that, at least in Italy, LPI is caused by several independent mutational events, a fact that seems unusual for a rare disease. In particular, we found three different mutations (967G→A, 1625insATCA, 197-740del) in a small geographic area (places of patients' origin are <50 km apart). This recalls the so-called “reunion paradox” and another similar phenomenon observed in Galilee (Beckmann 1996; Zlotogora et al. 1996). Two different genetic mechanisms were postulated to explain the occurrence of different mutations in a genetic isolate. The first one relies on a digenic model, the second on both a high mutation rate and a selective advantage for the carriers. In our case, we do not have data supporting either of these hypotheses. Only a mini-founder effect can be considered for 1625insATCA in Campania. Indeed, “private” or rare mutations were frequently detected in Italy for other inherited metabolic disorders, such as phenylketonuria (Guzzetta et al. 1997), aldolase B deficiency (Santamaria et al. 1996b), and homocystinuria caused by cystahionine β-synthase deficiency (Sebastio et al. 1995). In contrast, a single mutation accounts for most of the Finnish LPI alleles, in agreement with the concept of “the Finnish disease heritage” (Norio et al. 1973).
Relationship between the Mutations and Clinical Features
We failed to establish a genotype/phenotype correlation. In fact, both intrafamilial and interfamilial phenotypic variability were observed in homozygotes for the same mutation. Both the age at onset and the compliance to the treatment do not seem to explain the differences in the clinical presentation. Patients affected by the 1625insATCA mutation present variable phenotypes: very mild in patient 12; very mild in patient 13, in contrast to the affected brother who died from a sudden hyperammonemic coma; severe in patient 14C with additional findings such as short stature, renal insufficiency, and recurrent pancreatitis; severe in patient 15C, a double first-cousin once removed from patient 14C, affected by early onset of pulmonary insufficiency caused by alveolar proteinosis; and very mild in patient 11. Moreover, in patient 16, a 28-year-old man, homozygosity for the 197-740del mutation, which removes 168 codons at the protein level, allows the mildest phenotype we have ever observed. In contrast, his affected brother died from a severe clinical course with a fatal hyperammonemic coma, at age 6 years.
All patients, except the Japanese patient, were shown to be homozygous for a specific mutation. This indicates that consanguinity, observed in 5 of the 11 families, is often associated with LPI and may explain the rare occurrence of the disease.
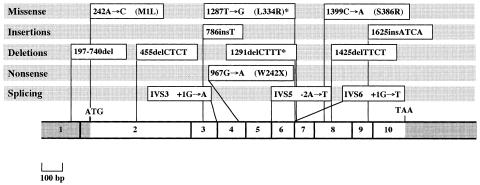
Until now, molecular analysis of patients with LPI has only identified mutations in the SLC7A7 gene (table 4 and fig. 1). Mutations of 4F2hc, the heavy chain of the heterodimer 4F2hc/SLC7A7, were never found, at least in Italian patients (data unpublished). This is in agreement with previous linkage studies that assigned the LPI locus to chromosome 14q11.2 for Finnish, Italian, Swedish, Latvian, Moroccan, Saudi-Arabian, and Turkish patients (Lauteala et al. 1998). Another transcript, SLC7A8, maps to the same chromosomal region and is located in the same YAC 806b10 as SLC7A7 (Bassi et al. [in press]). SLC7A8 shares a high level of homology with SLC7A7, its pattern of expression is compatible with the LPI pathophysiology, and the predicted protein displays 12 TM domains. By direct sequencing of SLC7A8, we did not find disease-causing mutations in Italian and Finnish patients (Borsani et al. 1999). Therefore, genetic homogeneity of LPI is definitely established.
Table 4.
Mutations and Polymorphism of the SLC7A7 Gene
| Mutationa | Sequence Alteration | Effect on Coding Sequence |
| Missense: | ||
| M1L | A→C at 242 in exon 2 | Met1Leu substitution |
| L334Rb | T→G at 1287 in exon 7 | Leu334Arg substitution |
| S386R | C→A at 1399 in exon 8 | Ser386Arg substitution |
| Nonsense: | ||
| W242X | G→A at 967 in exon 4 | Termination at codon 77 |
| Insertion or deletion: | ||
| 786insT | Insertion of a T between 786 and 787 in exon 3 | Frameshift after Leu182, termination at codon 190 |
| 1625insATCA | Insertion of ATCA between 1625 and 1626 in exon 9 | Frameshift after Ile461, termination at codon 467 |
| 197-740delc | Deletion of intron 1 and exon 2 | Deletion of first 168 codons |
| 455delCTCT | Deletion of CTCT at 455–458 in exon 2 | Frameshift after Leu71, termination at codon 167 |
| 1291delCTTTb | Deletion of CTTT at 1291–1294 in exon 7 | Frameshift after Leu334, termination at codon 348 |
| 1425delTTCT | Deletion of TTCT at 1425–1428 in exon 8 | Frameshift after Leu395, termination at codon 516 |
| Splicing: | ||
| IVS3 +1G→A | GT→AT in donor of intron 3 | No mRNA studies |
| IVS5-2A→Td | AG→TG in acceptor of intron 5 | Frameshift after Val298, termination at codon 307 |
| IVS6 +1G→T | GT→TT in donor of intron 6 | No mRNA studies |
| Polymorphism: | ||
| S53S | A→G at 400 in exon 2 | No effect |
Figure 1.
Localization of all mutations of the SLC7A7 gene identified to date. The asterisk (*) indicates that the nomenclature of these mutations is as reported by Torrents et al. (1999).
The DNA-based tests are now easily accessible for molecular diagnosis, genetic counseling, and prenatal diagnosis of LPI (Sperandeo et al. 1999). Finally, expression studies will help address questions about the effects of some mutations, for example, S386R, and will shed light on the functioning of the 4F2hc/SLC7A7 complex.
Acknowledgments
We thank the families, for contributing to this project, and A. Pepe and M. R. Tuzzi, for technical assistance. We also thank the YAC Screening Center at San Raffaele Biomedical Science Park. Telethon Grant E.652 is gratefully acknowledged. This work was supported by MURST Cofin 1998 (to G.S.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genbank, http://www.ncbi.nlm.gov/Genbank/index.html (for SLC7A7 cDNA, [Y18474])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/OMIM (for LPI [MIM 222700]) [PubMed]
References
- Antonarakis SE (1998) Recommendations for a nomenclature system for human gene mutations: Nomenclature Working Group. Hum Mutat 11:1–3 [DOI] [PubMed]
- Bassi MT, Sperandeo MP, Incerti B, Bulfone A, Pepe A, Surace EM, Gattuso C, et al. SLC7A8, a gene mapping within the lysinuric protein intolerance critical region, encodes a new member of the glycoprotein associated amino acid transporter family. Genomics (in press) [DOI] [PubMed] [Google Scholar]
- Beckmann JS (1996) The reunion paradox and the digenic model. Am J Hum Genet 59:1400–1402 [PMC free article] [PubMed]
- Borsani G, Bassi MT, Sperandeo MP, De Grandi A, Buoninconti A, Riboni M, Manzoni M, et al (1999) SLC7A7, encoding a putative permease-related protein, is mutated in patients with lysinuric protein intolerance. Nat Genet 21:297–301 [DOI] [PubMed]
- Candito M, Vianey-Saban C, Ferraci JP, Bebin B, Chazalette JP, Sebag F, Mathieu M, et al (1994) Lysinuric protein intolerance: urinary amino acid excretion at 2 and 9 days of age. J Inherit Metab Dis 17:252–253 [DOI] [PubMed]
- Di Rocco M, Garibotto G, Rossi GA, Caruso U, Taccone A, Picco P, Borrone C (1993) Role of haematological, pulmonary and renal complications in the long-term prognosis of patients with lysinuric protein intolerance. Eur J Pediatr 152:437–440 [DOI] [PubMed]
- Guzzetta V, Bonapace G, Dianzani I, Parenti G, Lecora M, Giannattasio S, Concolino D, et al (1997) Phenylketonuria in Italy: distinct distribution pattern of three mutations of the phenylalanine hydroxylase gene. J Inherit Metab Dis 20:619–624 [DOI] [PubMed]
- Huang C, Cheng GJ, Reid ME, Chen Y (1999) Rhmod syndrome: a family study of the translation-initiator mutation in the Rh50 glycoprotein gene. Am J Hum Genet 64:108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incerti B, Andria G, Parenti G, Sebastio G, Ghezzi M, Strisciuglio P, Sperlì D, et al (1993) Lysinuric protein intolerance: studies on 17 Italian patients. Am J Hum Genet Suppl 53:908 [Google Scholar]
- Kozak M (1987) At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 196:947–950 [DOI] [PubMed]
- Lauteala T, Mykkanen J, Sperandeo MP, Gasparini P, Savontaus ML, Simell O, Andria G, et al (1998) Genetic homogeneity of lysinuric protein intolerance. Eur J Hum Genet 6:612–615 [DOI] [PubMed]
- Norio R, Nevanlinna HR, Perheentupa J (1973) Hereditary diseases in Finland: rare flora in rare soul. Ann Clin Res 5:109–141 [PubMed]
- Palacin M, Estevez R, Bertran J, Zorzano A (1998) Molecular biology of mammalian plasma membrane amino acid transporters. Physiol Rev 78:969–1054 [DOI] [PubMed]
- Parenti G, Larocca MR, Sperandeo MP, Buoninconti A, Passariello A, Sebastio G, Andria G (1998) Clinical variability of lysinuric protein intolerance in a large pedigree. Am J Hum Genet Suppl 63:1567 [Google Scholar]
- Parenti G, Sebastio G, Strisciuglio P, Incerti B, Pecoraro C, Terracciano L, Andria G (1995) Lysinuric protein intolerance characterized by bone marrow abnormalities and severe clinical course. J Pediatr 126:246–251 [DOI] [PubMed]
- Parini R, Vegni M, Pontiggia M, Melotti D, Corbetta C, Rossi A, Piceni Sereni L (1991) A difficult diagnosis of lysinuric protein intolerance: association with glucose-6-phosphate dehydrogenase deficiency. J Inherit Metab Dis 14:833–834 [DOI] [PubMed]
- Pfeiffer R, Rossier G, Spindler B, Meier C, Kuhn L, Verrey F (1999) Amino acid transport of y+L-type by heterodimers of 4F2hc/CD98 and members of the glycoprotein-associated amino acid transporter family. EMBO J 18:49–57 [DOI] [PMC free article] [PubMed]
- Sambrook J, Fritsh EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Santamaria F, Parenti G, Guidi G, Rotondo A, Grillo G, La Rocca MR, Celentano L, et al (1996a) Early detection of lung involvement in lysinuric protein intolerance: role of high resolution computed tomography and radioisotopic methods. Am J Respir Crit Care Med 153:731–735 [DOI] [PubMed]
- Santamaria R, Tamasi S, Del Piano G, Sebastio G, Andria G, Borrone C, Faldella G, et al (1996b) Molecular basis of hereditary fructose intolerance in Italy: identification of two novel mutations in the aldolase B gene. J Med Genet 33:786–788 [DOI] [PMC free article] [PubMed]
- Sebastio G, Sperandeo MP, Panico M, de Franchis R, Kraus JP, Andria G (1995) The molecular basis of homocystinuria due to cystathionine beta-synthase deficiency in Italian families, and report of four novel mutations. Am J Hum Genet 56:1324–1333 [PMC free article] [PubMed]
- Simell O (1995) Lysinuric protein intolerance and other cationic aminoacidurias. In: Scriver CR, Beaudet AL, Sly WS, Valle DT (eds) The metabolic and molecular bases of inherited disease. Vol 3. McGraw-Hill, New York, pp 3603–3627 [Google Scholar]
- Singer SJ (1990) The structure and insertion of integral proteins in membranes. Annu Rev Cell Biol 6:247–296 [DOI] [PubMed]
- Sperandeo MP, Buoninconti A, Passariello A, Scala I, Adami A, Lauteala T, Mykkanen J, et al (1999) Feasibility of prenatal diagnosis of lysinuric protein intolerance: a case report. Prenat Diagnosis 19:771–773 [PubMed] [Google Scholar]
- Torrents D, Estevez R, Pineda M, Fernandez E, Lloberas J, Shi YB, Zorzano A, et al (1998) Identification and characterization of a membrane protein (y+L amino acid transporter-1) that associates with 4F2hc to encode the amino acid transport activity y+L: a candidate gene for lysinuric protein intolerance. J Biol Chem 273:32437–32445 [DOI] [PubMed]
- Torrents D, Mykkanen J, Pineda M, Feliubadalo L, Estevez R, de Cid R, Sanjurjo P, et al (1999) Identification of SLC7A7, encoding y+LAT-1, as the lysinuric protein intolerance gene. Nat Genet 21:293–296 [DOI] [PubMed]
- Zlotogora J, Gieselmann V, Bach G (1996) Multiple mutations in a specific gene in a small geographic area: a common phenomenon? Am J Hum Genet 58:241–243 [PMC free article] [PubMed]