Summary
Williams-Beuren syndrome (WBS) is a developmental disorder caused by haploinsufficiency for genes in a 2-cM region of chromosome band 7q11.23. With the exception of vascular stenoses due to deletion of the elastin gene, the various features of WBS have not yet been attributed to specific genes. Although ⩾16 genes have been identified within the WBS deletion, completion of a physical map of the region has been difficult because of the large duplicated regions flanking the deletion. We present a physical map of the WBS deletion and flanking regions, based on assembly of a bacterial artificial chromosome/P1-derived artificial chromosome contig, analysis of high-throughput genome-sequence data, and long-range restriction mapping of genomic and cloned DNA by pulsed-field gel electrophoresis. Our map encompasses 3 Mb, including 1.6 Mb within the deletion. Two large duplicons, flanking the deletion, of ⩾320 kb contain unique sequence elements from the internal border regions of the deletion, such as sequences from GTF2I (telomeric) and FKBP6 (centromeric). A third copy of this duplicon exists in inverted orientation distal to the telomeric flanking one. These duplicons show stronger sequence conservation with regard to each other than to the presumptive ancestral loci within the common deletion region. Sequence elements originating from beyond 7q11.23 are also present in these duplicons. Although the duplicons are not present in mice, the order of the single-copy genes in the conserved syntenic region of mouse chromosome 5 is inverted relative to the human map. A model is presented for a mechanism of WBS-deletion formation, based on the orientation of duplicons' components relative to each other and to the ancestral elements within the deletion region.
Introduction
Williams-Beuren syndrome (WBS [MIM 194050]) is caused by a submicroscopic deletion of band 7q11.23 (Ewart et al. 1993; Francke 1999). Identification of patients with single-gene defects has confirmed that haploinsufficiency for the elastin (ELN) gene is responsible for vascular pathology—and, possibly, for bladder and intestinal diverticuli—but has no clear relation to the other connective-tissue problems of WBS, including inguinal hernias, contractures of the joints, some of the characteristic facial features, and premature aging of the skin (Ewart et al. 1993; Li et al. 1997; Tassabehji et al. 1997). Other phenotypic features of WBS include growth retardation, renal anomalies, transient hypercalcemia, hyperacusis, anxiety disorder, attention-deficit/hyperactivity disorder, and mental retardation (Pober and Dykens 1996; Kaplan et al., in press). To date, the specific genes within the deletion to which these effects are attributable have not been identified. The pattern of cognitive dysfunction in WBS is characterized by pronounced difficulty with processing of visual-spatial information and relative preservation of linguistic ability (Dilts et al. 1990; Wang and Bellugi 1993; Wang et al. 1995; Karmiloff-Smith et al. 1997). The contribution of deletion of LIM-kinase 1 (LIMK1) to the visual-spatial learning difficulty is not yet clear. Evidence for (Frangiskakis et al. 1996) or against (Tassabehji et al. 1999) this hypothesis has rested on the ascertainment of partial-deletion families whose phenotype specifically includes or excludes the “WBS cognitive profile.”
A common WBS-deletion region, estimated at 2 cM, was defined by the genotyping of multiple affected individuals for microsatellite markers from 7q11.23 (Pérez-Jurado et al. 1996; Wu et al. 1998). The physical size of the deletion was predicted to be ∼2 Mb, on the basis of the genetic map and the fact that it is visible on high-resolution chromosomes (Pérez-Jurado et al. 1996; Francke 1999). Osborne et al. (1997a, 1997b, 1999) provided an incomplete physical map based on assembly of a P1-derived artificial chromosome (PAC)/cosmid contig that included 1.1 Mb fully contained within the deletion. Restriction mapping of similar bacterial artificial chromosome (BAC)/PAC clone contigs by two independent groups estimated the common deletion as being 1.5 Mb (Meng et al. 1998a) or 1.4 Mb (Hockenhull et al. 1999).
Genes identified within the common WBS deletion include a human homologue of the Drosophila frizzled receptor (FZD9 [Wang et al. 1997]); syntaxin 1A (STX1A [Osborne et al. 1997b]); CYLN2/CLIP-115, encoding an intracellular linkage protein (De Zeeuw et al. 1997; Hoogenraad et al. 1998) that covers the partial transcripts WSCR3 and WSCR4 (identified by Osborne et al. 1996); EIF4H, a translation-initiation factor (Richter-Cook et al. 1998) covering the partial transcript WSCR1 (also identified by Osborne et al. 1996); GTF2I, encoding the transcription factor TFII-I/SPIN/BAP-135 (Pérez-Jurado et al. 1998); replication factor–complex C subunit 2 (RFC2 [Osborne et al. 1996; Peoples et al. 1996]); FKBP6, an immunophilin FK-506–binding protein–family member (Meng et al. 1998b); BCL7B, a sequence related to a gene identified from a Burkitts lymphoma translocation cell line (Jadayel et al. 1998; Meng et al. 1998b); TBL2/WS-ßTRP, a member of the beta-transducin gene family (Meng et al. 1998b; Pérez-Jurado et al. 1999); a gene, preliminarily named “WS-bHLH,” for the presence of a helix-loop-helix motif (Meng et al. 1998a); WBSCR9/WSTF, a large transcript encoding a putative transcriptional coactivator (Lu et al. 1998; Peoples et al. 1998); CPETR1 and CPETR2, named for their pathological function as Clostridium perfringens–enterotoxin receptors that belong to the claudin family of tight-junction proteins (Paperna et al. 1998); a putative transcription-factor gene with GTF2I-related repeats, GTF2IRD1 (Franke et al. 1999; Osborne et al. 1999); and two incompletely characterized transcripts, designated “WBSCR2” and “WBSCR5” (Osborne et al. 1996).
The WBS-deletion region is flanked by highly conserved duplicated elements within which the common breakpoints cluster. Homologous recombination between these nearly identical regions is believed to account for the high incidence of de novo deletion formation. Genotyping of flanking markers in informative extended families has revealed that deletions arise from interchromosomal recombination events in approximately two-thirds of cases and from intrachromosomal events in approximately one-third of cases (Dutly and Schinzel 1996; Baumer et al. 1998). The flanking duplications were first identified by studies of the microsatellite marker D7S489, primers for which amplify alleles that cluster in three different size ranges, corresponding to three distinct polymorphic loci (D7S489A, -B, and -C [Pérez-Jurado et al. 1996; Robinson et al. 1996]). The upper (D7S489B, 170–178 bp) and lower (D7S489A, 140–144 bp) loci map near the proximal and distal boundaries of the deletion, the former falling within the deletion and the latter outside the deletion. D7S489C-sized alleles (156–158 bp) have been mapped to a poorly defined locus just outside the deletion region, variably placed on the centromeric (Pérez-Jurado et al. 1996; Osborne et al. 1997a) or telomeric (Robinson et al. 1996) side. Subsequently, Pérez-Jurado et al. (1998) identified the GTF2I gene in the telomeric, and the GTF2IP1 pseudogene in the centromeric, breakpoint regions. Whereas the 5′ unique region of GTF2I extends into the deletion, the pseudogene is centromeric to the common breakpoints.
Evidence for a third GTF2I locus outside but very close to the deletion has been emerging. The map published by Osborne et al. (1997a) placed two copies of a GTF2I-like sequence at the centromeric, and one at the telomeric, breakpoint regions, each copy in association with a PMS2-like gene (PMS2L). By using FISH, they showed that the PMS2L genes map to the three sites within 7q11.23 and to another locus at 7q22, as well. Görlach et al. (1997) described the p47-phox gene (NCF1) and pseudogene (NCF1P1) and provided evidence for a second copy of the pseudogene at an unknown locus. The map published by Hockenhull et al. (1999) includes two GTF2I/NCF1 duplications, one each at the centromeric and telomeric breakpoint regions; furthermore, typing of a YAC disclosed the presence of another NCF1 pseudogene somewhere near the telomeric duplication.
Recently, DeSilva et al. (1999) used FISH with duplicated-region clones and found the duplications to be present in nonhuman primates, including chimpanzees, gorillas, orangutans, and gibbons. As in humans, hybridization signals were also detected at 7q22 and 7p22 in chimpanzees and gorillas but not at the homologous sites in gibbons and orangutans. Relative to the human sequence, chromosomes underwent peri- and paracentric inversions between the 7q11.23-q21 region and 7q22 in the gorilla or 7p22 in the orangutan. In mice, only a single site was present, consistent with the finding that Gtf2i is a single-copy gene in the mouse (Wang et al. 1998b).
We have constructed a physical map of the WBS deletion by assembling a BAC/PAC clone contig and by restriction mapping of clones and of genomic DNA from normal and WBS deletion–carrying human chromosomes 7. Our estimate of the common deletion size is 1.5–1.7 Mb. The deletion region is flanked by two highly homologous duplicons of 320–500 kb. These duplicons are a patchwork of at least four definable repeats that are present, in different orientations, within the duplicons. Each repeat is composed of stretches of DNA with high homology in coding and noncoding sequences. We have adopted the term “duplicon” to designate these large genomic regions. This term, first proposed by Eichler et al. (1997), was recently used by Christian et al. (1999) for similar observations in the region flanking the Prader-Willi syndrome/Angelman syndrome–deletion region on chromosome 15. We propose an evolutionary model for the complex organization by serial duplications of ancestral elements, some of which are located within band 7q11.23 and others of which are elsewhere on chromosome 7.
Material and Methods
Samples and DNA Isolation
Human genomic DNA was obtained from subjects with WBS who had the common deletion, from their parents, and from normal controls, under institutional review board–approved protocols. Clinical criteria for inclusion within this study and deletion characterization have been described elsewhere (Pérez-Jurado et al. 1996). DNA was isolated from peripheral blood lymphocytes or Epstein-Barr virus–immortalized lymphoblastoid cell lines (LCLs), either in solution or as intact chromosomes imbedded in agarose blocks, for pulsed-field gel electrophoresis (PFGE; see below). Fusion of fresh leukocytes from a subject with WBS and a Chinese hamster fibroblast line generated two hybrid lines retaining WBS-deletion chromosome 7 (SCH DEL-1 and SCH DEL-2, previously called “53-7” and “53-13” [Peoples et al. 1996]) and two hybrid lines retaining the normal chromosome 7 (SCH NONDEL-1 and SCH NONDEL-2, previously called “53-8” and “53-15”).
BAC-Library Screening
A human genomic BAC library (Kim et al. 1996) was screened by PCR assay of plate pools obtained from Research Genetics (release IV). Forty-seven clones identified were purchased as agar stabs, and DNA was isolated by use of Qiagen Maxiprep reagents and a modified low-copy plasmid–preparation protocol. Three PAC clones (Ioannou et al. 1994) were purchased from Research Genetics. For sequence-tagged site (STS) content mapping, PCR amplifications were carried out in 25-μl reactions with 1.5 mM MgCl2, and either 10–20 ng of clone DNA or 50–100 ng of genomic DNA, for 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s, in an MJ PTC-200 thermocycler.
STS Generation
Primer sequences for STS markers used in this study are given intable 1. Primer sequences for some STS markers were taken from the Human Chromosome 7 Mapping and Sequencing Web site (Bouffard et al. 1997); the Stanford Human Genome Center RH-mapping web site (Stewart et al. 1997); and the STS-Based Map of the Human Genome Web site, release 12, July 1997 (Hudson et al. 1995). D7S489 amplimers were sized by GeneScan analysis, as described elsewhere (Peoples et al. 1998).
Table 1.
Primers and Amplimer Sizes of STSs Used in Clone Contig Mapping and for SSN Assays
|
Primers(5′→3′) |
|||||
| STS | Forward | Reverse | Size(bp) | GenBank Accession Number | Reference |
| Clone contig mapping: | |||||
| 5C19L | TACATGAACACAGCACTCCATG | TTGTAGAAATGCGGTCTCACC | 197 | AF166309 | Present study |
| 5C19R | GATGAGCTGACTTTCACAGGC | TGATGTTGAAGATTTGGGCA | 251 | AF166310 | Present study |
| 7H23R | GAGAGGACACAGCCTCTGCT | TGGAGATCCTGGGTGAATG | 120 | AF166315 | Present study |
| 7H23L | GGTTTGATAGTGGCGTCTTAGG | CAAGAAAAGTGGGAGAGAGCA | 120 | AF166314 | Present study |
| 7I15R | CATCAGTGTTTTGGGGGTG | AGCTTCCCTCAACATGAGACA | 193 | AF166317 | Present study |
| 17SP | CCCCAACTTCTCTGTATTTG | AATGTAGCTCCTTGTTCCC | 105 | AF166283 | Present study |
| 30E19L | ACCCAGCAACCAACAATAGC | TGGTACCAAGGGTAACCCG | 159 | AF166294 | Present study |
| 30E19R | CAAGCCCTGAGCCTAATCC | CCTGAGATTAGGAGGGGAGC | 150 | AF166295 | Present study |
| 34N24R | TCATAGGGGAGCAGGTGG | GGTCTCCAGTGAGACCCAGA | 112 | AF166297 | Present study |
| 39H4L | AGCGGCCTCTCTAGTGAGTG | ATTTAAGCAGAGGTTGAGCTGC | 105 | AF166300 | Present study |
| 39H4R | TGCATGGGTGCACATACAC | CCACTGTGTAGCAGCAAAACA | 248 | AF166301 | Present study |
| 51J24R | GATCAAGGGGTCAAGTGCAT | AGCTTAGTCATGGGCCTCAA | 100 | AF166306 | Present study |
| 137N19R | GGATTTCACCATGCTGGC | CCCTTCACCCACCAACTCTA | 305 | AF166278 | Present study |
| 163N16L | GGAGAAGGACACAGCCTCTG | TCCTGCCACTGTCCCAAC | 100 | AF166279 | Present study |
| 163N16R | GCTGGTACTGGGTAAGAAATCA | GACCAGCAGCAAAGTAGATGG | 144 | AF166280 | Present study |
| 171C15L | AGAGGAAGCTTCAGACAAGTGG | TTAAAACCATTGTGCTCTGGC | 155 | AF166281 | Present study |
| 171C15R | ATATAGTTAGTGTGGCAG | CAGCCTTAAAATATACTACC | 76 | G30693 | Bouffard et al. (1997) (sWSS3379) |
| 248G1L | TGACCTTAGGTTAGGTAGGCAA | GTTGCAACAAAAAAGTGTCCTG | 101 | AF166288 | Present study |
| 248G1R | TACAGGCTGAACTAGAACGTGG | ACCGTAGACCACTGCTATCCA | 90 | AF166289 | Present study |
| 269P13L | CTTCCGCAAATGTGGGAC | GCTCACCCTAGCATTGAAGC | 172 | AF166290 | Present study |
| 269P13M | TGCAGGGGGAAAAATAGTTG | GGCTCACAATGTCAAACCCT | 150 | AF166291 | Present study |
| 270D13L | GTATCCTTTAGTTCAATAAACTTATTGTT | AGTCCCAGCTACTTGAGAGGC | 175 | AF166292 | Present study |
| 270D13R | GAGCCTTGGCACCACTCTC | ACTGGCGAAAAGAAGTTAAACC | 107 | AF166293 | Present study |
| 340G9R | GTGTCCTGCGGGTTAATAGTG | ATGGTTGCACACCTCTGTGA | 181 | AF166296 | Present study |
| 350L10L | GCTAAGATGCAGGCACATCA | TGTTACCAGACAAATCCCTGC | 106 | AF166298 | Present study |
| 435J21R | CCATGTTGTCAGCCCAGAC | AGTCTGGGAATCAGGCCC | 178 | AF166304 | Present study |
| 537A20R | TAAATTGGGAAGACATCCGC | GAAGCCCTTCAGACTACCCC | 189 | AF166308 | Present study |
| 763H7L | CAAAAGAGCTGATTCCAATC | ATAGCGAGACCCCATTTC | 310 | AF166311 | Present study |
| 763H7R | AAAGGATCTGGGAAGTATTTG | ATAATCTTTTCCTGGACAAGG | 1,500 | AF166312 | Present study |
| 797L | AGTGCTTGCATGCCTTAG | AAGCACCACCTCTACTCTCA | 156 | AF166313 | Present study |
| 953F13R | ACCGTCTGCTGCTTTGAGAT | ATTGCCCATGCTAAGGACAC | 159 | AF166321 | Present study |
| 965F7L | CGAGACAGAGCTGTGTTGTA | CTTGACCTCCCAAAGTGAT | 278 | AF166322 | Present study |
| AFMb055xe5 | GCTGCACTTTCAGTTTGAATG | CTCAGCAGAGGGACTTCACC | 230 | Z67541 | Hudson et al. (1995), Dib et al. (1996) |
| BCL7B | TGCCTCTTGTCACAAACTGC | ACTCACTGTTGCCCATTTCC | 190 | AJ223979 | Present study |
| CPETR1 | GTACGACTCGCTGCTGG | TCCAGGGAAGAACAAAGC | 500 | AB000712 | Present study |
| CPETR2 | CATCACGTCGCAGAACATCT | CGGATAATGGTGTTGGCC | 315 | AF007189 | Present study |
| CYLN2 IN2 | CCAGCCTGGCAACAGAGT | AGGTAATGTTTACACCCATGGC | 173 | AC004851 | Present study |
| D7S489 | CTGTTGACTTTCCCACACTC | GGCAACTCGAGACGTTAGTT | 140–170 | Z16646 | Hudson et al. (1995), Dib et al. (1996) |
| D7S613 | CAGCCTGGGTAACAAAAGC | CCTCCCTCCCTAATCCATG | 100 | G18333 | Hudson et al. (1995), Dib et al. (1996) |
| D7S788 | CCTCATGGAACTGATTTCCAG | ATTCAACCCTGGCTTTGGTG | 85 | L10529 | Hudson et al. (1995), Dib et al. (1996) |
| D7S789 | ATTGCTTTTTGCCCACCTTC | ACTTAGACTGTAGTCTCTAC | 110 | L10530 | Hudson et al. (1995), Dib et al. (1996) |
| D7S1624 | ATGGAAGAGCTTACACTG | AAGACCCTGAATGTCTTG | 96 | G00136 | Hudson et al. (1995), Dib et al. (1996) |
| D7S1633 | CTATAAGTGTAGAGTTCTGG | GAAACTGTTGAAAGCATAGG | 102 | G00101 | Hudson et al. (1995), Dib et al. (1996) |
| D7S1778 | AGCTTGCCTAGGTTTTGCTG | TGGTCCCTTGAAGATACGTG | 200 | Z67766 | Perez-Jurado et al. (1998) |
| D7S1870 | TTCACTCAGGAAGTGGC | TGGTGATGTGCTTTACTACG | 120 | Z51768 | Gilbert-Dussardier et al. (1995) |
| D7S2024 | ATTACAGGCGTGAACTAC | TACTATGAGAATACAGAGAAGG | 105 | G00215 | Hudson et al. (1995), Dib et al. (1996) |
| D7S2472 | TCTAAAGTCTGCCAGGCTAC | GCAGCGAGACTCCATC | 100 | Z53057 | Hudson et al. (1995), Dib et al. (1996) |
| D7S2476 | GGGCAACATAGCACGATT | CAGGAGTCAGTTAGATAAGGTCAC | 150 | Z53107 | Hudson et al. (1995), Dib et al. (1996) |
| D7S2714 | CTCTGGGTTTCTGCTGAAGTTTG | AGTGACCTTTTTGGGATGAGAATG | 158 | G10931 | Hudson et al. (1995), Dib et al. (1996) |
| ELN 3′UTR | ATCCCATGCCCCTCCGATTC | GGCTTCAGGTGCTTGGGTAC | 400 | U62292 | Present study |
| ELN 5′UTR | CCAGCAGCGAAAGAACAGTC | GGAGGGGACAATTACGAAAG | 180 | U62292 | Present study |
| EST00085 | TGCCAAGCCTGAATCAATGT | GCTCCAAGAGCTTCTCCCTT | 119 | G31686 | Bouffard et al. (1997) (D7S534e) |
| FKBP6-EX7 | TTGAAGGTAATCAAAGGG | TTGTTCTTTACAGCAAGG | 142 | G13134 | Bouffard et al. (1997) (sWSS3352) |
| FZD9 | ATTTCATGTCACTGGTGGTG | ACCTTGACAGATGGGCAGCT | 350 | NM_003508 | Wang et al. (1997) |
| GTF2I 3′UTR | TCACAGAGCCTAGCTTCTTG | CCGGCATTATTTCCTAGTTC | 184 | AF035737 | Perez-Jurado et al. (1998) |
| GTF2I EX10 | GTGGGCCAATGCTAATTCTC | CTTCAGAAACAAGTGAGGACCC | 254 | AF035737 | Present study |
| GTF2IRD1-ex7 | GGATGGCGGGCGGGACTCGAA | AGCTCTCGGATGGCGTGGTTG | 198 | AF156489 | Present study |
| GTF2IRD1-ex22 | ACGGATCGACATCGCCAACAC | CAGGGCTTTCGGAACGGGATT | 135 | AF156489 | Present study |
| HIP1-3′UTR | GCATCCTCTTGAATAGGAAGATCG | CCATCTAGAAGAGGAAAAGTGCTG | 465 | Y09420 | Wedemeyer et al. (1997) |
| HIP1 EX2 | GGGCACCCACCATGAGAAAG | CGTTCGGGTGTCCATCTCG | 230 | Y09420 | Present study |
| HIP1 EX12 | GACCACTTAATTGAGCGACTATAC | CCTTCAGCTGCAGCACAACC | 1,300 | Y09420 | Present study |
| LIMK1 | TTTTATTGTTCTGCGTCTGGG | CAGTGCACTTTGAACCTGGA | 130 | U62293 | Present study |
| POM121-EX5 | TGAGATGCCTCGAGTGGAG | GGGTCTCTGAAGAGAGGCCT | 165 | AC006014 | Present study |
| POM121-EX11 | CCCACGTTGAAGGCAAAC | CTTTTGGAAACTCTGCAGCC | 520 | AC006014 | Present study |
| RFC2 | GCAGAGACTTCACTGACTGAC | TGACCTCAGGTGATCCACCTG | 194 | NM_002914 | Okumura et al. (1995) |
| SHGC4006 | AAGACTTTTAGGGATGTGAGGGG | AGCTCGTGTGCATCAGTTGTTTC | 156 | G17117 | Stewart et al. (1997) |
| SHGC-31781 | ACCAAAAGGCAGAAAATAGACTT | TATCCCCAAGGCTCAGCTG | 150 | G27203 | Stewart et al. (1997) |
| STX1A | CCACTCCACTCCAGGTGG | TACTGAAGGCAAGGAAGCGT | 299 | U87315 | Present study |
| sWSS3308 | CAGAAAACTTGAAACAGG | GTTGAGTTGTATGAGTGG | 60 | G30690 | Bouffard et al. |
| sWSS3369 | GAAGGAAGAGGATCTTAC | ATGCTAAGCCCTTTCTTG | 223 | G13142 | Bouffard et al. (1997) |
| sWSS3501 | CTCACTTTAACTTCACAAC | GAAATAGTCATTTTGGACAG | 108 | G30771 | Bouffard et al. (1997) |
| sWSS3873 | GCAAAAGGAACTTCATGG | CTTTTCATCTCTAACCTAAC | 88 | G30873 | Bouffard et al. (1997) |
| TBL2 | TCCCCAGCTCATATTTATTTGG | AGGTCTGGAAGAAAAGTAGAAAAGA | 265 | AF056184 | Perez-Jurado et al. (1999) (WI-6911) |
| WBSCR9-EX1 | GTGTGCGCGGGAACTCTG | GCGGGAAGGGCTTGCGGC | 242 | AF084479 | Peoples et al. (1998) |
| WBSCR9-EX7 | GAAGTCATTGAGTGGCTCGC | CCGACAGCTTCATTCCCAAT | 1,010 | AF084479 | Present study |
| WSCR5 | TCCCATGAGACAGTCACAACA | CCAGAACAGGGCAGAGTAGG | 104 | G07044 | Osborne et al. (1996) (WI-8920) |
| SSN assays: | |||||
| 23I15L | GGCCAGGTTTCTGTTCAAAC | GAGAGGACGATCAGCCTCAG | 251 | AF169396 | Present study |
| 68E13L | TCTTAATGTCACAAGCAGGAGA | AGCTAGTTTACCTCAGTTCCGC | 202 | AF169398 | Present study |
| 208H19M | AGGGACTTGAAGCCAGCC | CGCTCCCCAAACTCTCATAG | 479 | AF169397 | Present study |
| 269P13R | CTGAAATTGGGGACACCATT | AGTCTGGTGGGAGAGGGATC | 318 | AF169397 | Present study |
| GTF2I EX19-20 | TCTTGGACTCACCGAGGC | TCCAGAAACGACTACAGTGGC | 211 | AF169393 | Present study |
| GTF2I EX28 | ACCTGGAAATCAGCTCCATG | AGCAGCCATGGATAATACGG | 361 | AF169394 | Present study |
| NCF1 EX2 | CTTTCTGCAATCCAGGACAA | ATCACCTGGGCTAAGGTCCT | 305 | M25665 | Gorlach et al. (1997) |
| NCF1 EX3-4 | GGCGATCAATCCAGAGAACA | TGAGCCTTGGTTTCCTCATC | 392 | AF169392 | Present study |
| POM121 EX2-3 | CCCAGTGACTGTGAGGATCG | CTTCTTCTTCTCTTTGAGGGC | 479 | AF169391 | Kipersztok et al. (1995) |
Original STSs were generated from analysis of either clone sequence or GenBank sequence, with either the program PRIMER 0.5 (Whitehead Institute 1991) or OLIGO 5.1 (National Biosciences). To develop new STSs, sequences from BAC and PAC ends were obtained either by direct sequencing (Peoples et al. 1998) or by a modified vectorette-PCR protocol (Riley et al. 1990). For the latter, primer sequences (5′→3′) were R1 (CTCGTATGTTGTGTGGAATGTGAGC), R2 (TTTCACACAGGAAACAGCTATGACCATG), L1 (GGGTTTTCCCAGTCACGACG), and L2 (GTCGACCTGCAGGCATGCAA); enzymes for BAC digestion were BsaAI, BstUI, and a combination of EcoRV, PvuII, StuI, and XmnI. Direct sequencing of amplimers was performed by use of the L2 and R2 primers, as described elsewhere (Peoples et al. 1998). The PAC end sequence 953F13R was derived by direct sequencing by use of the primer (5′→3′) CCGTCGACATTTAGGTGACAC. Markers 763H7L, 763H7R, 965F7L, 797L, and 17SP were derived, by vectorette PCR (Riley et al. 1990), from the left and right arms of CEPH YACs 763H7 and 965F7 (Dausset et al. 1992; Hudson et al. 1995), the left arm of YAC HSC7E797 (Kunz et al. 1994), and the SP arm of P1 clone RMC1317 (Shepherd et al. 1994), identified by screening for the D7S489C locus. GenBank accession numbers for all sequences are listed intable 1.
Hybridization Probes
Probes for PFGE hybridization experiments were either intact cDNA clones without prior separation of insert or gel-purified PCR products (Wizard PCR Preps), except as noted. In the former case, unless otherwise cited, IMAGE-consortium cDNA clones were obtained from Research Genetics, and DNA was isolated by use of a Qiagen miniprep protocol. Specific probe information is given in table 2.
Table 2.
Hybridization Probes for PFGE Blots
| Probe | Clone | Size(bp)a | GenBank Accession Number | Reference | |
| 17SP | P1 clone RMC1317 (Colin Collins, UCSF/LBL), SP vectorette product | 900 | Present study | ||
| CPETR1 | cDNA clone | ? | R48300 | Paperna et al. (1998) | |
| CPETR1 | cDNA clone | ? | W74492 | Paperna et al. (1998) | |
| ELN | cDNA clone containing ELN ORF (Joel Rosenbloom, U. Pennsylvania) | 2,200 | U62292 | Present study | |
| FZD9 | cDNA clone containing FZD9 ORF | 2,200 | NM_003508 | Wang et al. (1997) | |
| GTF2I | cDNA clone 86072 (IB291) (ATCC) | 1,500 | T03439 | Perez-Jurado et al. (1998) | |
| GTF2IRD1 | cDNA clone hbc694 (exon 24) (Graeme Bell, U. Chicago) | ? | T10636 | Franke et al. (1999) | |
| IB2070 | cDNA clone | ? | R20285 | Present study | |
| POM121 | cDNA clone | ? | R87509 | Present study | |
| RFC2 | cosmid RFCp40-#3, T7 vectorette product | 1,900 | AF045555 | Peoples et al. (1996) | |
| SHGC-31781 | cDNA clone | ? | R52511 | Present study | |
| STAG3L | cDNA clone IB1445 (ATCC), 5′ end Alu/vector product | 900 | T03379 | Present study | |
| TBL2 | cDNA clone C-0td07 | ? | Z42768 | Perez-Jurado et al. (1999) | |
| WSCR5 | cDNA clone 52119, HindIII fragment |
1,400 | H23535 | Present study | |
|
Amplimers (5′→3′) |
|||||
| Forward |
Reverse |
||||
| CPETR2 | TATGGAGCCGAGCCGTTAGC | CGGATAATGGTGTTGGCC | 600 | AF007189 | Paperna et al. (1998) |
| CYLN2 (EX3) | CAGAGCCGCTGTCTGAGAG | CCCCACTGCACAAACAGTC | 530 | AJ228871 | Present study |
| ELN | GCTTGCAGATCCACAGGGCAAG | GCGAATCCAGCTTTGAGGCTTCA | 368 | U63721 | Wedemeyer et al. (1997) |
| GTF2I-5′ | ATGTCCACCCTCCCCGTTGA | GGTGGCTTCCTTGAATGTTA | 800 | AF035737 | Perez-Jurado et al. (1998) |
| HIP1 | Same as STS in table 1 | 465 | Y09420 | Wedemeyer et al. (1997) | |
| NCF1 (EX 2) | CACACAGCAAAGCCTCTTTG | TTCTGGGTTCTGCAGTTTCC | 240 | M25665 | Present study |
| POM121-ZP3 | TCACTCTTTAAAGGGTTGAGGG | TGCTATATTTCCCCTACATGCC | 722 | U10099 | Present study |
| STX1A (EX 6) | Same as STS in table 1 | 299 | U87315 | Present study | |
| WBSCR9 | ATGACTTTGTTGGATATGGC | CTTTCCGTTCTTCAGAC | 322 | AF084479 | Peoples et al. (1998) |
| ZP3 | CCCCAGCCTTAGAAACAGC | TGGATGGAGACCACTTTATGC | 802 | X56777 | Present study |
A question mark (?) indicates unknown.
High-Throughput Genome-Sequence (HTGS) Data Analysis
BACs for which HTGS data were available were identified by a BLAST 2.0 alignment search (Altschul et al. 1990, 1997) of the HTGS database maintained by NCBI, with selected STS sequences used as probes (Ouellette and Boguski 1997; Sulston and Waterston 1998). Genomic sequence information was obtained from the Genome Sequencing Center database, Washington University (St. Louis), except for 239C10, which was obtained from GenBank (table 3). Comparison between the sequence of HTGS clone and STS sequences was performed with Sequencher 3.0 alignment software (GeneCodes), at 85% homology. All STS data are available from GenBank (table 1). Table 4 lists sequences from GenBank that are used for comparisons.
Table 3.
BACs Used for HTGS
| Clone | GenBank Accession Number |
| 239C10 | AC004166 |
| hDJ0665P05 | AC004851 |
| hDJ0771P04 | AC004883 |
| hDJ0953A04 | AC006014 |
| hDJ1158B01 | AC004980 |
| hGS166C05 | AC004851 |
| hNH0313P13 | AC005488 |
| hNH0340A14 | AC007078 |
| hNH0396K03 | AC006995 |
| hNH0479C13 | AC005236 |
| hRG023I15 | AC005049 |
| hRG051J22 | AC005056 |
| hRG052H06 | AC005057 |
| hRG208H19 | AC005074 |
| hRG269P13 | AC005080 |
| hRG270D13 | AC005081 |
| hRG315H11 | AC005089 |
| hRG350L10 | AC005098 |
Table 4.
Sequences from HTGS Database That Were Used for Comparisons
| Sequence | Nucleotides from GenBank Sequence | GenBank Accession Number | Reference |
| 23I15R | 1–114 | AF166287 | Present study |
| 51J24L | 1–429 | AF166305 | Present study |
| 93N13L | 1–141 | AF166320 | Present study |
| 194I16L | 1–188 | AF166284 | Present study |
| 194I16R | 1–313 | AF166285 | Present study |
| 350L10R | 1–225 | AF166299 | Present study |
| 426A23R | 1–146 | AF166302 | Present study |
| 435J21L | 1–137 | AF166303 | Present study |
| 537A20L | 1–368 | AF166307 | Present study |
| CYLN2-ex15 | 10,6623–10,6826 | AJ228878 | Hoogenraad et al. (1998) |
| EIF4H | 24,126–24,263, 24,554–24,693 | AF045555 | Osborne et al. (1996) |
| FKBP6-ex1-4 | 1–116, 117–162, 163–334, 335–537 | NM_003602 | Meng et al. 1998b |
| GTF2IP1-C | 1–109 | AF036613 | Perez-Jurado et al. (1998) |
| GTF2I-ex2 | 416–689 | AF035737 | Perez-Jurado et al. (1998) |
| GTF2I-ex11 | 1259–1371 | AF035737 | Perez-Jurado et al. (1998) |
| PMS2L | 46–187, 188–274, 275–377 | U13696 | Osborne et al. (1997a) |
| POM121-ex13 | 89,948–90,240 | AC006014 | Present study |
| STAG3L | 41,849–49,348 | AC006014 | Present study |
| sWSS3380 | 1–150 | G30694 | Bouffard et al. (1997) |
| WS-bHLH | 1,277–1,378, 1,875–2,026 | AF056184 | Meng et al. 1998a |
| ZP3-ex3 | 432–535 | X56777 | Kipersztok et al. (1995) |
| ZP3-ex7 | 924–1,060 | X56777 | Kipersztok et al. (1995) |
Site-Specific Nucleotide (SSN) Assays of BACs Covering Duplicons
PCR primers for STS markers containing SSNs are given in table 1. STS marker GTF2I-ex19–20 PCR contains the codon-changing nucleotide 2217 site difference, and GTF2I-ex28 the silent nucleotide 3130 site difference, for the gene and pseudogene (Pérez-Jurado et al. 1998); the 303/305-bp NCF1–exon 2 containing a GT-dinucleotide deletion of the pseudogene was described by Görlach et al. (1997). Assessment of presence or absence of the PstI and TaqI restriction sites of the NCF1–ex3-4 amplimers was performed by PCR amplification of human, human × Chinese hamster somatic-cell hybrid DNA, hamster control DNA, and BAC clone DNA, as described above, followed by restriction digestion of products, in 50-μl volumes, with each of these enzymes (New England Biolabs), followed by size-fractionation on 2.5% agarose gels. PCR and digestion conditions for assessment of the XcmI and SacII restriction sites on 208H19M amplimers were the same as those for NCF1–ex3-4. For all SSN markers, PCR was carried out as above, in 50-μl volumes, by use of BAC-clone DNA as template. DNA from these experiments was gel purified and directly sequenced as described above. Results of NCF1–ex3-4 and 208H19M digestion were also verified by direct sequencing of PCR products in this manner. SSN positions are given in table 5 and in figure 4 and refer to the nucleotide number of the corresponding GenBank entries.
Table 5.
Restriction-Endonuclease Fragment Sizes, as Determined by PFGE[Note]
|
Fragment Size(s) |
||||||
|
NotI |
PmeI |
AscI |
||||
| Enzyme SCH line | Deleted | Nondeleted | Deleted | Nondeleted | Deleted | Nondeleted |
| Unique loci: | ||||||
| Within deletion: | ||||||
| CPETR1 | 140 kb | 90 kb | 330 kb | |||
| CPETR2 | 140 kb | 220 kb | 330 kb | |||
| CYLN2 | 200 kb | 120 kb | 450 kb | |||
| ELN | 470 kb | 220 kb | 200 kb | |||
| FZD9 | 200 kb | 180 kb | 370 kb | |||
| GTF2IRD1 | 190 kb | 300 kb | 210 kb | |||
| RFC2 | 470 kb | 160 kb | 450 kb | |||
| STX1A | 160 kb | 220 kb | 330 kb | |||
| TBL2 | 160 kb | 220 kb | 330 kb | |||
| WBSCR9 | 200 kb | 180 kb | 370 kb | |||
| WSCR5 | 470 kb | 160 kb | 450 kb | |||
| Outside deletion: | ||||||
| HIP1 | 4 Mb | 1 Mb | 160 kb | 160 kb | 380 kb | 380 kb |
| IB2070 | 4 Mb | 3 Mb | 160 kb | 160 kb | >1.6 Mb | >1.6 Mb |
| SHGC-31781 | 4 Mb | 1 Mb | 280 kb | 280 kb | 320 kb | 320 kb |
| Multicopy loci: | ||||||
| 17SP | 4 Mb | 200 kb, 1 Mb, 3 Mb | 210 kb, 220 kb, 280 kb | 180 kb, 210 kb, 220 kb, 280 kb | 300 kb, 380 kb | 300 kb, 370 kb, 380 kb |
| GTF2I | 4 Mb | 1 Mb, 3 Mb | 140 kb | 140 kb | 300 kb, 320 kb | 300 kb, 320 kb, 370 kb |
| POM121 | 4 Mb | 1 Mb, 3 Mb | 210 kb, 220 kb | 140 kb, 210 kb, 220 kb | 380 kb | 370 kb, 380 kb |
| STAG3L | 4 Mb | 1 Mb, 3 Mb | 210 kb, 220 kb, 280 kb | 210 kb, 220 kb, 280 kb | 300 kb, 380 kb | 300 kb, 380 kb |
Note.— Fragments present in SCH-DEL lines are listed in the columns denoted “Deleted”; fragments present in SCH-NONDEL lines are listed in the columns denoted “Nondeleted.”
Figure 4.
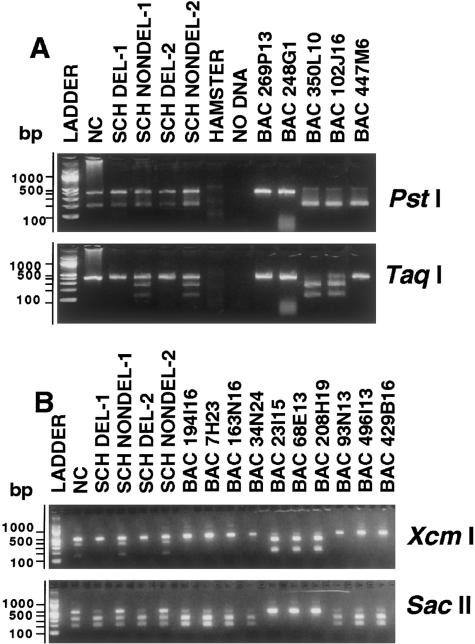
A, Identification of the third GTF2I/NCF1 locus by restriction digestion of the 392-bp amplimer of NCF1 exon 3–4 from human cells, SCH, and BACs. These PCR products contain two SSN differences that change restriction-enzyme sites: a PstI site at position 213 of the 350L10 amplimer (A at 213) is absent on 269P13 (G at 213), and a TaqI site at position 262 of 350L10 (C at 263) is also absent on 269P13 (T at 263). Intact and digested PstI fragments were generated from both the deleted and nondeleted SCH lines. Amplimers of BACs 269P13 and 248G1 (GTF2IP1/NCF1P1) are not cut, whereas those of BACs 350L10 and 102J16 (GTF2I/NCF1) and 447M6 (GTF2IP2/NCF1P2) are. In contrast, TaqI-cleaved bands were apparent only in the SCH NONDEL-1 and -2 lanes, consistent with deletion of the NCF1 locus residing on BACs 350L10 and 102J16(GTF2I/NCF1), which also show TaqI digestion. Amplimers from BACs 269P13 and 248G1(GTF2IP1/NCF1P1) and BAC 447M6 (GTF2IP2/NCF1P2) are not cut, consistent with their mapping to loci outside the deletion. The combined TaqI and PstI data are consistent with a model placing BAC 447M6 at a third NCF1 locus, outside the deletion. Incomplete TaqI digestion of the BAC 102J16 amplimer does not affect the conclusions, because the presence of a single locus with the TaqI site has been confirmed by direct sequencing of the amplimer. B, Assignment of BACs to repeat clusters by restriction digestion of human, SCH, and BAC amplimers of the 479-bp STS 208H19M (in REP B1) containing two SSN differences. Sequence comparisons revealed the presence of an XcmI site at position 362 for the 208H19 amplimer (CT at 366–367) and its absence on that for 313P13 (2-bp deletion, relative to 208H19); in contrast, a SacII site was predicted at position 324 for 313P13 (T at 326) but not for 208H19 (G at 326). XcmI restriction digestion generated, for BACs 23I15, 68E13, and 208H19, predicted fragments of 330 and 150 bp that were absent in all other BACs tested. Bands of the same size were also generated from the SCH NONDEL lines but not from the SCH DEL lines. In contrast, SacII yielded predicted bands of 300 and 180 bp only for BACs 194I16, 7H23, 163N16, and 34N24. In the case of SacII, uncut fragments were of significantly lower intensity when they were from the SCH DEL lines. These results provide strong evidence for the presence of a locus on BACs 23I15, 68E13, and 208H19 within the WBS deletion, a locus that is highly homologous to extradeletion sequences on BACs 194I16, 7H23, 34N24, 93N13, 496I13, 429B16, and 163N16.
PFGE
Agarose blocks were prepared from LCLs from the donor of the SCH lines, 12 other sporadic WBS patients, two sets of unaffected parents, 17 unrelated normal controls, and human × Chinese hamster somatic-cell hybrid lines immobilized in low-melting-temperature agarose, at a concentration of ∼107 cells/ml. High-molecular-weight DNA was prepared by incubation of agarose blocks with sodium sarkosyl and proteinase K. After having been washed in buffer, blocks were incubated with the following restriction endonucleases (New England Biolabs): NotI, PmeI, AscI, PacI, SfiI, BssHII, AvrII, NheI, and SpeI. Digest products were size-fractionated in either 1% (100-kb–1.6-Mb resolution) or 0.7% (1–5-Mb resolution) agarose gel, by PFGE with use of a CHEF gel apparatus (Biorad) under the following conditions: 100–800-kb resolution at 10–50-s pulse times, 200 V, 24 h; 200-kb–1.5-Mb resolution at 40–150-s pulse times, 200 V, 24 h; 2–4-Mb resolution at 1-h pulse time, 50 V, 120 h, followed by 90-s pulse time, 50 V, 24 h. Undigested Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Hansenula wingeii chromosomes were used as size markers. Gels were imaged with ethidium bromide and were transferred to nylon filters by use of the Southern blot technique. BAC and PAC DNA in solution (see above) was digested with NotI, NotI/PmeI, and NotI/AscI, in 30-μl volumes. Electrophoresis was performed for 2–20-s pulse times at 200 V for 24 h in 1% agarose gels.
Probe labeling and hybridization techniques have been described elsewhere (Peoples et al. 1998). Each probe listed in table 2 was hybridized to a panel of blots containing at least four WBS and four normal control samples, as well as the four hybrid cell lines described above.
Results
A 3-Mb Clone Contig and PFGE Map of the WBS Deletion and Flanking Regions
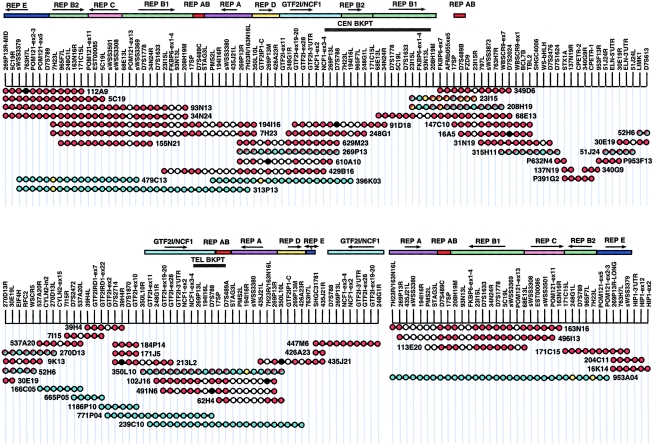
To assemble a clone contig covering the ∼2-cM region of the WBS deletion, we started with a YAC contig assigned to the flanking duplicated regions, as reported elsewhere (Pérez-Jurado et al. 1996). Because the YACs within the deletion appeared to be unstable and/or rearranged, we initiated a BAC library–screening effort. A total of 58 markers, derived from gene sequences, public databases, and YAC end sequences, were used in the initial round of BAC-library screening (table 1). Ordering of BACs, deepening of coverage, and closure of gaps were accomplished through generation of new STSs derived from BAC end sequences (table 1). An ∼500-kb region between TBL2 and ELN was difficult to map and appears to be underrepresented in the BAC library that we used. This gap was closed by the serendipitous mapping of two new intradeletion genes within this region (Paperna et al. 1998) and by incorporation of PAC 953F13 (Osborne et al. 1997a, 1997b) and PACs 632N4 and 391G2 (Meng et al. 1998a) into our contig. The STS-content map of our clone contig, shown in figure 1, covers the WBS deletion and both flanking regions. HTGS data analyzed by sequence alignment were also used in the construction of this map (table 3).
Figure 1.
STS-content map of BAC/PAC contig of WBS deletion and flanking regions. Red circles denote STS present by PCR; black circles denote STS absent by PCR; aqua circles denote STS present by sequence alignment with HTGS data; yellow circles denote STS absent by sequence alignment; white circles denote STS not assessed. Multicopy sequence clusters (REPs) are presented as colored bars above the contig. Arrows indicate orientation (arbitrarily defined) of these REPs relative to each other. The centromeric and telomeric breakpoint limits (black bars) were determined by SSN assays of hybrid cell lines derived from one individual with WBS.
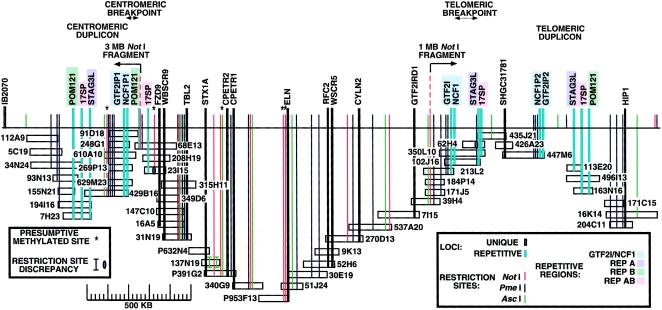
Long-range restriction mapping of contig clones and genomic DNA provided a size estimate of 3 Mb for our map of the deletion and flanking regions (fig. 2). The deletion is covered by unique clones that are unambiguously ordered and that span a distance of 1.6 Mb (figs. 1 and 2). A gap remains in the telomeric flanking region within the telomeric duplicon between clone 447M6 and clones 163N16, 113E20, and 496I13 (figs. 1 and 2).
Figure 2.
Long-range restriction map of the WBS deletion and flanking repeat–containing regions, as derived from PFGE studies of BACs, human × hamster somatic-cell hybrids, and human lymphoblasts. The map covers 3 contiguous Mb, including 1.5–1.7 Mb within the deletion. The centromeric and telomeric breakpoint limits were determined by PFGE blot hybridization data. Restriction sites of methylation-sensitive enzymes identified in BACs but not in genomic DNA are indicated by asterisks (*). The few unexplained discrepancies between genomic and clone restriction sites are also indicated.
Comparison of PFGE data (table 5), obtained by use of genomic versus clone DNA cleaved with three different enzymes and analyzed under a variety of electrophoretic conditions, resulted in excellent agreement. In a few instances, the sizes of fragments generated by the methylation-sensitive enzymes NotI and AscI were consistent with CpG methylation of genomic DNA sites that were sensitive to digestion in the BACs and PACs. Presumptive methylation sites and the few restriction-site discrepancies are indicated in figure 2.
Long-Range Restriction Mapping of Duplicons in the Flanking Regions
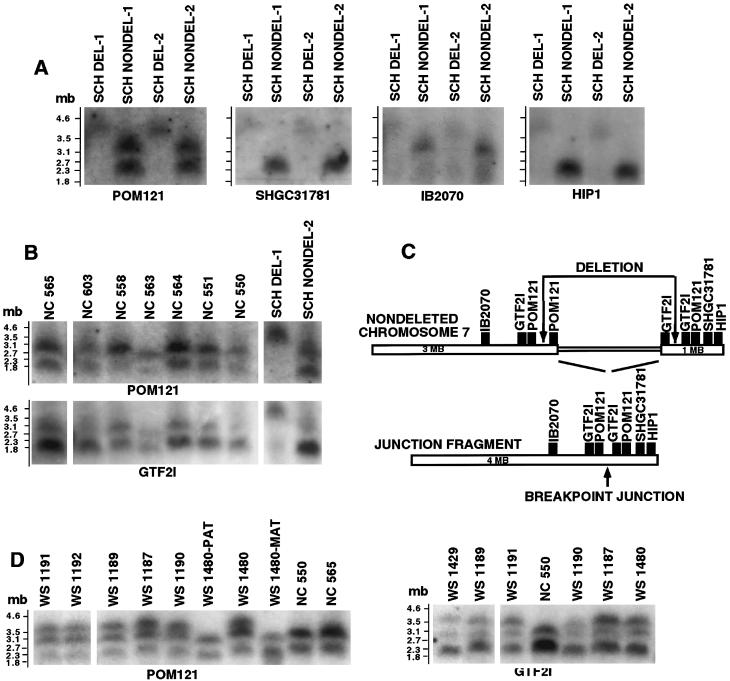
In search of deletion junction fragments, we hybridized, with the GTF2I probe IB291, PFGE blots with DNA from individuals with WBS, normal controls, and WBS-deletion and nondeleted chromosome 7 somatic-cell hybrids digested with SfiI, BssHII, AvrII, NheI, or SpeI. For all enzymes and all samples, single restriction fragments of ⩽150 kb were seen. Conservation of restriction sites resulted in junction fragments identical in size to the two fragments contributing to them. When NotI fragments were separated under a range of conditions, allowing effective resolution of fragments of 100 kb–5 Mb, a 4-Mb junction fragment was detected in somatic-cell–hybrid lines containing the chromosome 7 with the WBS deletion (fig. 3A). Both NotI fragments (1 and 3 Mb) derived from the normal chromosome 7 contribute to formation of the 4-Mb junction fragment (fig. 3B). This conclusion was confirmed when NotI blots were hybridized with single-copy probes mapping outside the deletion, on opposite sides (fig. 3A). SHGC-31781 from the telomeric side recognizes the 1-Mb fragment, whereas IB2070 from the centromeric side recognizes the 3-Mb fragment. The telomeric 5′-GTF2I–specific probe from within the deletion (Pérez-Jurado et al. 1998) also recognizes the 1-Mb fragment (data not shown). The SHGC-31781 locus maps to the telomeric YAC HSC7E640, whereas IB2070 maps to the centromeric CEPH YAC 855H10 as mapped by Pérez-Jurado et al. (1996). The size of the smaller fragment was determined to be ∼1 Mb, on gels resolving fragments of 200 kb–1.6 Mb; this fragment appears to run higher on the 2–4-Mb-resolution gels shown here. The orientation of the NotI fragments is shown in figure 3C. Hybridization with a probe for the single-locus gene HIP1 shows that it also maps to the 1-Mb fragment (fig. 3A) and is therefore on the telomeric side.
Figure 3.
NotI fragment analysis of the flanking and deletion-junction regions. A, 2–4-Mb resolution Southern blot of the four WBS SCH lines, hybridized successively with flanking region probes. The probe for POM121, which is part of REP B, hybridized to both normal (3 and 1 Mb) NotI fragments and the 4-Mb junction fragment present only in the SCH DEL lanes. A probe for the telomeric locus SHGC-31781 hybridized to the lower fragment and to the junction fragment and thus localized the 1-Mb fragment to the telomeric side. The IB2070 probe from the centromeric flanking region recognized only the upper fragment and the junction fragment, consistent with the 3-Mb NotI fragment's being derived from the centromeric side. The HIP1 locus, previously localized outside the WBS deletion (Wedemeyer et al. 1997), was localized to the telomeric side, by hybridization to the 1-Mb fragment in the SCH NONDEL lanes and to the junction fragment in the SCH DEL lanes. B, Southern blot of seven human normal control samples (NC), an SCH DEL line, and an SCH NONDEL line, hybridized successively with probes for POM121 and GTF2I (IB291). Both probes recognized the 1- and 3-Mb bands flanking the deletion, as well as the 4-Mb deletion-junction fragment. For POM121, differential hybridization intensities suggest a 2:1 copy-number ratio for the 3-Mb fragment, relative to the 1-Mb fragment, whereas for GTF2I the relative gene dosage is reversed. C, Map of the NotI fragments flanking the WBS deletion and of the deletion-junction fragment. IB2070, SHGC-31781, and HIP1 are single-copy loci mapped to either the centromeric (IB2070) or the telomeric (SHGC-31781 and HIP1) flanking region. GTF2I and POM121 exist in three copies. Both probes hybridize to the 4-Mb fragment with greater intensity than do the single copy probes suggesting that two copies of these loci are flanking the deletion and that one is within the deletion. D, PFGE Southern blots containing samples from affected (WS) individuals, parental controls (PAT and MAT), and unrelated normal controls (NC), hybridized with either the POM121 or the GTF2I probes, documenting the consistency of the 4-Mb junction fragments in a larger number of individuals with WBS. As in B, both probes recognize the 1- and 3-Mb NotI fragments in all samples. The 4-Mb fragment is seen with either probe in all seven unrelated WS samples. The de novo occurrence of this deletion-junction fragment in case WS 1480 is illustrated by its absence in both parents.
Inspection of the GTF2I/NotI PFGE hybridization data suggested a twofold dosage of the 1-Mb fragment, relative to the 3-Mb fragment (fig. 3B, bottom). Comparison with results obtained for the same blots hybridized with the POM121 probe (fig. 3B, top) confirm that this is not an effect of less-efficient transfer of DNA for the larger fragment relative to the smaller. Hybridizations, with the NCF1 probe, to filters of DNA cleaved with NotI, PmeI, and AscI yielded the same results as did hybridizations with the GTF2I probe. These results suggested the presence of a third GTF2I/NCF1 locus mapping outside the deletion to the same telomeric 1-Mb NotI fragment involved in junction-fragment formation as the original GTF2I/NCF1. The mechanism of deletion formation appears to be consistent in WBS. As shown in figure 3D, the same 4-Mb junction fragment is present in seven unrelated individuals with WBS and not in unaffected parents.
Identification of a Third GTF2I/NCF1 Locus
We began mapping the duplicons of the WBS deletion and flanking regions by looking for single-nucleotide differences that distinguish the GTF2I and NCF1 genes from their pseudogenes. BACs 350L10 and 269P13 were assigned to the telomeric GTF2I/NCF1 locus and the centromeric GTF2IP1/NCF1P1 locus, respectively, on the basis of the three locus-specific single-nucleotide differences in the GTF2I/GTF2IP1 3′ common region and the 5′ GTF2I-specific region (Pérez-Jurado et al. 1998) and the GT tandem repeat in exon 2 (Görlach et al. 1997), which distinguishes NCF1 from the NCF1P1 pseudogene. BAC 239C10, for which HTGS data are available, also contains the GTF2I and NCF1 genes, on the basis of the presence of the same nucleotide variants used to define 350L10. In turn, HTGS data for BAC 396K03 confirm that it contains the GTF2IP1 locus. Surprisingly, the 136-bp GTF2IP1-specific 5′ exon sequence identified by Pérez-Jurado et al. (1998) was also present in BAC 239C10, establishing that this sequence is not unique to the centromeric locus but, rather, is a member of another duplicated element.
On the basis of sequence information from BACs 350L10 and 269P13, a 392-bp amplimer from intron 2 to intron 4 of the NCF1/NCF1P1 locus (NCF1–ex3-4) was designed, incorporating four SSN differences, two of which were discernible by restriction digestion. Results of restriction digestion of amplimers from the WBS-deletion and nondeletion chromosome 7 SCHs and selected clones from the duplicons are shown in figure 4A. The presence of a PstI-sensitive amplimer in the SCHs carrying the deleted chromosome 7, as well as the absence of TaqI sensitivity in these same amplimers, suggests the presence of a third locus outside the deletion. The digestion pattern of the amplimer from BAC 447M6 indicates that it maps to this site. Amplimers of all BACs were sequenced and the results of the nucleotide differences defined three distinct sets, all entirely in agreement with the results of the restriction digestions (table 6). Comparison of the other GTF2I/GTF2IP1 and NCF1/NCF1P1 amplimer sequences of these clones disclosed that, for all other defined SSN differences, BAC 447M6 shares homology with the centromeric clones. The clones from the duplicons were thus mapped to three loci that are distinguishable by the SSNs present in the NCF1–ex3-4 amplimer. The loci represented by clone 447M6 are henceforth referred to as “GTF2IP2” and “NCF1P2.”
Table 6.
Assignment of Duplicon-Region BACs to Discrete Groups, by Locus-Specific Sequence Assays and HTGS Comparisons[Note]
| BAC | GTF2I R634H(151) | GTF2I nt3130(312) | NCF1 exon 2(144) | NCF1 ex3-4(112) | NCF1 ex3-4(187) | NCF1 ex3-4(213) | NCF1 ex3-4(263) | Locus | |
| GTF2I/NCF1 loci: | |||||||||
| 1 Group 1 BACsa | A | G | ( ) | A | A | G | T | Cen duplicon | |
| 2 Group 2 BACsb | G | C | GT | C | G | A | C | GTF2I/NCF1 | |
| 3 447M6 | A | G | ( ) | A | A | A | T | Tel duplicon | |
| 4 hRG269P13 sequence | A | G | ( ) | A | A | G | T | Cen duplicon | |
| 5 hRG350L10 sequence | G |
C |
GT |
C | G | A | C | GTF2I/NCF1 | |
| 269P13R-74 |
269P13R-159 |
269P13R-241 |
|||||||
| 269P13R: | |||||||||
| 1 Group 1 BACsc | G | T | A | GTF2I/NCF1 | |||||
| 2 Group 2 BACsd | A | C | G | Cen duplicon | |||||
| 3 Group 3 BAC: 194I16 | A | C | A | Cen duplicon | |||||
| 4 Group 3 BAC: 496I13 | A | C | A | Tel duplicon | |||||
| 5 BAC 435J21 | G | T | A | GTF2I/NCF1 | |||||
| 6 BAC 429B16 | A | C | A | Cen duplicon | |||||
| 7 hRG269P13 sequence | A | C | G | Cen duplicon | |||||
| 8 hNH0396K0 sequence | A | C | A | Cen duplicon | |||||
| 9 hRG350L10 sequence | G | T | A | GTF2I/NCF1 | |||||
| 10 239C10 sequence | G | T | A | GTF2I/NCF1 | |||||
| 11 hDJ0953A04 sequence | A | C | A | Tel duplicon | |||||
| 12 hNH0313P13 sequence | A | C | A | Cen duplicon | |||||
| 13 hNH0340A1 sequence | A | T | A | ZP3 | |||||
| 14 hDJ1158B01 sequence | A |
T |
A |
POM121-ZP3 | |||||
| POM121-72 |
POM121-99 |
POM121-114 |
POM121-160 |
POM121-170 |
POM121-199-209 |
POM121-401 |
POM121-480 |
||
| POM121-ex2-3: | |||||||||
| 1 Group 1 BACse | T | G | G | A | T | AGCACAGACTT | T | C | Cen duplicon |
| 2 Group 2 BACsf | A | A | G | G | A | ( ) | T | C | POM121/ZP3 |
| 3 BAC 171C15 | A | A | C | A | C | ( ) | C | T | Tel duplicon |
| 4 hNH0479C1 sequence | T | G | G | A | T | AGCACAGACTT | T | C | Cen duplicon |
| 5 hNH0313P13 sequence | T | G | G | A | T | AGCACAGACTT | T | C | Cen duplicon |
| 7 hDJ1158B01 sequence | A | A | G | G | A | ( ) | T | C | POM121-ZP3 |
| 6 hDJ0953A04 sequence | A |
A |
C |
A |
C |
( ) |
C |
T |
Tel duplicon |
| 23I15L-109 |
23I15L-110 |
23I15L-133 |
23I15L-176 |
23I15L-180 |
23I15L-190 |
23I15L-214 |
23I15L-229 |
||
| 23I15L: | |||||||||
| 1 Group 1 BACsg | C | A | A | C | G | A | A | T | Cen duplicon |
| 2 Group 2 BACsh | C | A | A | C | G | A | A | T | Tel duplicon |
| 3 BAC 23I15 | T | G | G | A | C | G | G | A | Ancestral REP B |
| 4 BAC 68E13 | T | G | G | A | C | G | G | A | Ancestral REP B |
| 5 FKBP6 EXON 4 | T | G | G | Ancestral REP B | |||||
| 6 hRG023I15 sequence | T | G | G | A | C | G | G | A | Ancestral REP B |
| 7 hDJ0953A04 sequence | C | A | A | C | G | A | A | T | Tel duplicon |
| 8 hNH0313P13 sequence | C |
A |
A |
C |
G | A | A | T | Cen duplicon |
| 68E13L-73 |
68E13L-147 |
68E13L-172 |
68E13L-173 |
||||||
| 68E13L: | |||||||||
| 1 Group 1 BACsg | A | C | T | T | Cen duplicon | ||||
| 2 Group 2 BACsh | A | T | C | T | Tel duplicon | ||||
| 3 BAC 68E13 | G | C | C | A | Ancestral REP B | ||||
| 4 hDJ0953A04 sequence | A | T | C | T | Tel duplicon | ||||
| 5 hNH0313P13 sequence | A | C | T | T | Cen duplicon |
Note.— ( ) = sequence absent.
269P13, 248G1, 629M23, and 429B16.
350L10, 102J16, and 62H4.
350L10 and 62H4.
610A10, 269P13, and 629M23.
34N24, 5C19, 112A9, and 93N13.
23E9 and 83O6.
5C19, 7H23, 34N24, 93N13, 155N21, 373H3, and 194I16.
163N16, 496I13, and 113E20.
Presence of the unique-site STS SHGC-31781 (UniGene Hs.5291) unequivocally established contiguity of BACs 447M6 and 435J21 (fig. 2). Another SSN assay confirmed overlap of clone 435J21 with the GTF2I/NCF1-containing clones 350L10, 102J16, 62H4, and 491N6 and with 239C10, by use of HTGS data. Results of direct sequencing of amplimers of 269P13R (overlapping sWSS3499), including three SSNs predicted by sequence comparison of HTGS data from BACs 313P13, 350L10, and 269P13 are shown intable 6. Because the three duplicons represent paralogues, we defined the term “paralotype” as the set of locus-specific nucleotides identified by a given SSN assay. Presence of the paralotype GTA for amplimers, and/or HTGS data from each of these clones, is evidence that these clones contain a common 269P13R locus. Contiguity was thus provided for the two GTF2I/NCF1 paralogues mapping to the telomeric deletion–flanking region, in agreement with the PFGE data.
The Flanking-Region Duplicons and Junction-Fragment Formation
Extension of our contig by STS-content mapping of clones contiguous with the GTF2I/NCF1 pseudogene loci was hampered by the very high degree of sequence identity among them. PFGE data allowed mapping of the duplicated elements prior to development of assays that could discriminate among duplicons. Fragment sizes for the nonunique probes POM121, STAG3L, and 17SP are summarized in table 5. These probes were identified as mapping to the flanking regions by STS-content mapping and HTGS-data analysis. POM121 sequences are predicted to code for a 121-kD integral-membrane protein containing a nucleoporin-like region (Hallberg et al. 1993). STAG3L sequences show strong homology to the Stromalin antigen 3 cDNA sequence (L. Pérez-Jurado, personal communication), and 17SP is an anonymous probe derived from P1 clone RMC1317. All three probes mapped to two nearly identical sites flanking the deletion contained on two 380-kb AscI fragments and on 210- and 220-kb PmeI fragments. POM121 and 17SP also mapped, within the deletion, to a 370-kb AscI fragment in common with the intradeletion loci FZD9 and WBSCR9 and with the centromeric GTF2IP1/NCF1P1 locus. STAG3L and 17SP also mapped to a third extradeletion locus, on the same 280-kb PmeI fragment as SHGC-31781 mapped.
In the SCH-DEL lines, junction fragments for both PmeI and AscI are recognized by the GTF2I probe. Whereas the three GTF2I loci demonstrate near-complete site conservation for PmeI, AscI sites differentiate the loci. For PmeI, identically sized, 140-kb donor fragments contribute to the 140-kb junction fragment. AscI sites are found ∼100 kb centromeric of both the GTF2IP1/NCF1/P1 locus and the GTF2I/NCF1 locus. Therefore, recombination between the 370-kb centromeric GTF2IP1/NCF1/P1-containing AscI fragment and the 300-kb telomeric GTF2I/NCF1 fragment results in a 300-kb AscI junction fragment nearly identical to the GTF2I/NCF1-locus donor fragment. Deletion breakpoints must occur at nearly homologous sites within or near the centromeric GTF2IP1/NCF1P1 and telomeric GTF2I/NCF1 duplications. Therefore, the common recombination event occurs between the centromeric duplicon GTF2IP1/NCF1P1 locus and the ancestral GTF2I/NCF1 locus, which are in the same orientation.
Repeat (REP) Elements A–E
The PFGE data suggested a model in which two highly homologous duplicons, each including one GTF2I/NCF1 pseudogene locus, mapped on either side of the deletion, in inverted orientation with respect to each other (fig. 2). STS-content mapping of BAC clones contiguous with both the GTF2I/NCF1 gene and pseudogene loci disclosed the presence of discrete clusters of sequence elements that occurred within and flanking the WBS deletion. These elements are defined as REPs A, AB, B1, B2, and C–E (fig. 1). The relative orientation of these repeat clusters, where discernible, is indicated.
SSN assays of POM121 exons 2 and 3 in REP C and of 23I15L and 68E13L in REP B1 allowed for the discrimination of centromeric from telomeric duplicon clones (table 6). Sequencing of amplimers unequivocally placed BACs 112A9, 5C19, 93N13, 34N24, 155N21, 194I16, and 7H23 in one duplicon and BACs 171C15, 113E20, 163N16, and 496I13 in the other. HTGS analysis localized BACs 479C13 and 313P13 with the former group and 953A04 with the latter.
Sequence analysis of the 318-bp 269P13R (REP A) amplimers, followed by comparison with HTGS data, failed to discriminate between the duplicons (table 6). The ACA and ACG paralotypes were both associated with the centromeric duplicon and likely represent a polymorphism. Larger products were amplified from several clones because of variably present 20- and 50-bp repeats within the amplimer. These were called “269P13R-MID,” for the 340–380-bp products, and “269P13R-LONG,” for the 460-bp products (fig. 1). These longer 269P13R sequences were associated with REP E, not with REP A. BAC 171C15 was established as contiguous with the telomeric gene locus HIP1, by the presence of 269P13R-LONG in common with HIP1 clones 204C11 and 16K14. Together, these data established the 171C15/953A04 duplicon as telomeric and the 194I16/313P13 duplicon as centromeric.
The Telomeric Duplicon: Incomplete Clone Coverage
The inverted telomeric duplicon contains a gap in clone coverage, between the GTF2IP2/NCF1P2-carrying BAC 447M6 and BACs 163N16, 113E20, and 496I13 (fig. 1). Genomic PFGE data predict the gap to be ⩾80 kb. The absence of NotI sites on the telomeric duplicon and contiguous HIP1-containing clones, as well as the requirement that GTF2I/NCF1, GTF2IP2/NCF1P2, the telomeric duplicon REPs A–E, and the HIP1 locus all reside on the same 1-Mb fragment, suggests that the 140-kb PmeI and 320-kb AscI fragments occupied by GTF2IP2/NCF1P2 are contiguous with the 210- or 220-kb PmeI and 380-kb AscI fragments carrying the telomeric duplicon probes STAG3L, 17SP, and POM121. Therefore, the genomic DNA–derived PFGE map is essentially intact. Comparison with HTGS data from centromeric flanking BACs 269P13 and 396K03 shows that the region of the centromeric duplicon that corresponds to the gap in the telomeric duplicon contig covers ⩾60 kb; however, HTGS data in this region are discontiguous for both clones.
Estimation of Size and Degree of Homology of the Duplicons
The largest restriction fragments showing restriction-site conservation between the centromeric and telomeric duplicons are the 380-kb (REPS A and B) and 300- or 320-kb (GTF2I/NCF1) AscI fragments, suggesting that the duplication could be as large as 680 kb. Since the former fragment contains at least one unique sequence (i.e., HIP1) on the telomeric side, the duplicons must be smaller than this estimated maximum. When the proximal PmeI site of the centromeric POM121/17SP/STAG3L duplicon fragment and the distal limit of the NCF1P1 locus are used as boundaries, the duplicon size is estimated at ∼320 kb. Analysis of HTGS data from BACs 269P13, 396K03, and 313P13 predicts a size of ⩾280 kb, although the sequences are discontiguous, with several gaps.
For comparison, sequences of the overlapping 170 kb from HTGS data for BACs 313P13 (centromeric duplicon) and 953A04 (telomeric duplicon) were assembled around selected markers from each REP area and were analyzed for their degree of identity. At REPs A and AB, sequence differences were found at a frequency of 1–2/1,000 bp. REP B1 sequence differences ranged from ∼5/1,000, around FKBP6 ex1-4, to ∼12/1,000, at 68E13L; POM121 ex11 in REP C demonstrated differences at 17/1,000, and the REP B2 loci 171C15L and 965F7L differed at a frequency of ∼40/1,000. HTGS data from the 120 kb of overlapping sequence of BACs 269P13/396K03 (GTF2IP1/NCF1P1) and 350L10/239C10 (GTF2I/NCF1) also revealed a high level of identity, with nucleotide differences of only 1–2/1,000. Results of SSN assays in this region suggested the same degree of identity between the GTF2IP1/NCF1P1 locus and the GTF2IP2/NCF1P2 locus. Therefore, sequence conservation is significantly higher near the GTF2I/NCF1 repeats and falls off toward REP B2. Comparison of sequence information for the centromeric duplicon BAC 313P13 versus that for the intradeletion REP B BAC 208H19 disclosed that homology between these regions is substantially lower, with differences occurring at frequencies from ∼20/1,000 bp, around 17SP (REP AB), to 60/1,000, around 93N13L (REP B1).
Origin and Distribution of the REP Elements
The REP regions were first identified on the basis of the presence of multiple loci, mapping within and immediately flanking the deletion, that are recognized by primers for D7S489. Our experimental data confirm that the D7S489A locus maps just distal of GTF2I/NCF1, whereas D7S489B maps proximally between FKBP6 and FZD9. D7S489C-sized alleles of 156 and 158 bp are amplified from clones in both the centromeric and telomeric duplicons. Thus, D7S489C is duplicated and cannot serve to discriminate between the two duplicons. D7S489ABC, along with a new, neighboring anonymous marker, 17SP, form an unusual element, referred to here as “REP AB.” Both sequences are found, by STS-content mapping and by sequence analysis of HTGS clones, close to the ancestral REP B1 element (D7S489B; BAC 68E13, etc.), between REPs A and B of the centromeric and telomeric duplicon clones (D7S489C; BACs 194I16, 7H23, 313P13, etc.), and with REP A sequences only (D7S489A; BAC 350L10, etc.). We assume that the D7S489B/BAC 68E13 REP B1 locus is “ancestral” because it contains the complete FKBP6 gene, including the 3′ exons 5–8 not otherwise associated with the duplicon REP B1 sequences, which contain only FKBP6 exons 1–4. Also, FKBP6 exon 4 overlaps the 23I15L SSN-assay locus, matching the paralotype associated with BACs 23I15 and 68E13 and distinguishing it from the duplicon sequences (table 6). Interestingly, both probe STAG3L and probe 17SP also hybridize weakly to 200-kb NotI fragments in the SCHs DEL-1, DEL-2, and NONDEL-2 hybrid lines but not in the SCH NONDEL-1. This result is consistent with the presence of a STAG3L/17SP locus on 7q22, since the single chromosome 7 in the SCH NONDEL-1 has undergone a terminal deletion distal to 7q11.23 (data not shown).
In figure 1, the REP B locus has been divided into REPs B1 and B2 because of an intervening duplicated element, called “REP C,” within the duplicon. This element has not been assayed by hybridization, because we were unable to generate an effective probe from this region. We have not ruled out the possibility that, in this region, sequence homology between the ancestral REP B locus and the duplicons is too low for reliable PCR amplification. REP C is interesting, in that, with REP B1, it contains sequences from the POM121 gene, which is involved in another recombination event with the chromosome 7 ZP3 gene (Kipersztok et al. 1995). HTGS data available for BACs 340A14 and 1158B10 show that they contain REP E sequences (sWSS3379; 763H7L and 1158B01 only), REP A sequences (sWSS3380 and 435J21L), and the common unique paralotype ATA at 269P13R (table 6). These clones contain the ZP3 locus and the POM121-ZP3 locus, respectively. Both genes have previously been mapped to 7q11.23 (Kipersztok et al. 1995). PFGE results with the ZP3-specific and POM121-ZP3–specific probes revealed that these loci are beyond the immediate WBS flanking region (data not shown).
The Telomeric and Centromeric Deletion Breakpoints
In our WBS-deletion SCHs, the telomeric breakpoint is defined by the presence of D7S489A and by the absence of GTF2I (fig. 1). These boundaries are consistent with those in the majority of individuals with WBS, as determined by assessment of a GTF2I/GTF2IP1 dosage-sensitive, site-specific PCR-RFLP assay (Pérez-Jurado et al. 1998) and by simple sequence tandem repeat (SSTR) typing at D7S489A of informative families (Pérez-Jurado et al. 1996; Wu et al. 1998). The telomeric breakpoint as determined by PFGE mapping falls between GTF2I/NCF1 (deleted) and STAG3L/17SP (retained) (fig. 2).
At the centromeric breakpoint, the internal limit for the common WBS deletion has previously been defined by the absence of both the FZD9 gene (Wang et al. 1997) and the SSTR locus AFMb055xe5 (Peoples et al. 1998). These data have placed the centromeric breakpoint onto or proximal to BAC 68E13. PFGE mapping with PmeI has placed the centromeric breakpoint between the centromeric GTF2IP1/NCF1P1 locus (retained) and POM121 on BAC 68E13 (deleted) (fig. 2 and table 5).
Meng et al. (1998a) placed the centromeric breakpoint on BAC 68E13 by use of FISH studies of WBS chromosomes. Recognizing that BAC 68E13 contains nonunique sequences, making FISH-based determination of deletion/nondeletion status unreliable, we looked for a way to discriminate the 68E13 REP B sequence from that of the extradeletion copies. By comparing HTGS data from BACs 313P13 and 208H19, we designed an SSN assay (208H19-M) incorporating several predicted restriction-site differences for the two loci. Results of SSN assays of BACs and SCHs indicate that the locus defined by BACs 68E13, 23I15, and 208H19 is within the deletion (fig. 4B).
By comparison with HTGS data from the homologous REP B region of BAC 313P13, representing the centromeric duplicon, BAC 68E13 is predicted to contain ∼42 kb of REP B sequence; 208H19-M maps 37 kb from the centromeric limit, at 68E13L. However, PFGE data show that the 140-kb PmeI fragment containing the ancestral REP B1 locus of BAC 68E13, recognized by the probe POM121, is not present in the SCHs retaining the deleted chromosome 7. The PmeI junction fragment is also absent, consistent with the complete deletion of this locus on the WBS chromosome in the SCH lines. Sequence information obtained from the 5′ and 3′ ends of cDNA 166272 containing POM121-ex13, compared with BAC 313P13 HTGS data, shows that this cDNA is predicted to match the homologous regions of BAC 68E13, at both ends. Furthermore, the 3′ sequence of this cDNA is contiguous with the 68E13L sequence, confirming that it is homologous to the centromeric limit of BAC 68E13. Therefore, PFGE hybridization results are consistent with the centromeric breakpoint's lying proximal of BAC 68E13 but distal of the PmeI-site cluster on the GTF2IP1/NCF1P1 BACs 91D18, 269P13, etc.
Discussion
Completion of the WBS deletion–region map has been hampered by the presence of highly homologous regions, flanking the deletion, within which the deletion breakpoints cluster. Both accurate assessment of the deletion size and definition of precise breakpoint sequences require the establishment of contiguous-clone coverage across these regions. We have identified several site-specific sequence changes that have allowed us to precisely map BACs to specific duplicons. Furthermore, by integration of long-range restriction-mapping data, we have defined the presence of a second GTF2I/NCF1 pseudogene cluster and have localized it within 1 Mb telomeric of the WBS deletion. Mapping reports confounded by these three extremely similar duplicons have led to confusion in the literature. The large-scale chromosome-mapping efforts of the National Human Genome Research Institute (Bouffard et al. 1997; Touchman et al. 1997) and the Whitehead Institute Human Physical Mapping Project (release 12; July, 1997) understandably produced contigs that bounced back and forth across the deletion, from one duplicon to the other.
In this report, we have presented detailed dissections of the centromeric and telomeric duplicons flanking the WBS deletion. Each duplicon is composed of one copy each of a GTF2I/NCF1 pseudogene locus contiguous with newly defined low-copy repeated regions, which we have designated REPs “A,” “AB,” “B1,” “B2,” and “C”–“E.” These duplicons extend over >320 kb; are inverted in orientation, relative to each other; and demonstrate a high level of sequence identity, with only 1–2/1,000 nucleotide differences at the GTF2I/NCF1, REP A, and REP AB loci and with more sequence divergence toward the distal elements of the duplicon. Hockenhull et al. (1999) recognized the presence of a centromeric duplicon with content similar to that of the duplicon described here. However, they incorrectly placed the telomeric BACs 113E20 and 163N16 in this duplicon. They further reported their duplicon to be in opposite orientation to ours, suggesting that they failed to discriminate between the BACs containing the D7S489B and C loci.
Some of the duplicon repeat elements are found in association with the telomeric GTF2I/NCF1 (REPs A, AB, and D) site, which contains the functional loci and is considered to be the ancestral locus of the GTF2IP1/NCF1P1 and GTF2IP2/NCF1P2 pseudogene-containing duplicons. In turn, REPs B1 and B2—associated with FKBP6 on BACs 68E13, 208H19, and 23I15 within the common deletion—represent the ancestral locus for these repeat regions within the duplicons. The origin of the AB elements found in proximity to each ancestral locus is unclear; however, PFGE data provide additional evidence of homology to a non-7q11.23 site. Previous reports suggested that REP A sequences PMS2L and STAG3L may not have originated as part of the WBS region at 7q11.23 but that they may be part of a duplication derived from the 7q22 region, which, in humans, has some homology with the WBS-region repeats (Osborne et al. 1997a; DeSilva et al. 1999; L. Pérez-Jurado, personal communication). It is possible that REP AB is derived from an ancient inversion event that brought 7q11.23 and 7q22 sequences together, the remnants of which are found today as paralogous sequences at both loci. Such inversions were shown, by DeSilva et al. (1999) for gorilla and orangutan. In addition, we present evidence for a smaller inversion event, comprising the WBS-deletion region only, relative to the conserved syntenic region on mouse chromosome 5. We have localized HIP1 to the telomeric side of the WBS-deletion region, whereas, in the mouse, Hip1 is found near Fzd9 and Wbscr9 (Wang et al. 1999; L. Pérez-Jurado, personal communication).
Within the flanking duplicons, the POM121 gene was identified and the low-copy REP elements A and E were defined. Each of these elements is found in BACs containing the POM121-ZP3 fusion gene, whereas the A and B elements are found with the (presumably ancestral) ZP3 locus. Whereas the ZP3 and POM121-ZP3 loci have not been precisely mapped relative to the deletion, both have been localized to 7q11.23. These results provide further evidence for both the mobile nature of these sequence elements on chromosome 7—that is, they can be present beyond the WBS-deletion region—and the complexity of their distribution.
The duplicated regions were first defined by the SSTR D7S489, whose primers generate amplimers from at least three loci designated as “A”–“C” (Pérez-Jurado et al. 1996). BACs from which D7S489C-sized alleles can be amplified have, by some groups, been mapped centromeric to the deletion (Pérez-Jurado et al. 1996; Osborne et al. 1997a), whereas others (Robinson et al. 1996) have placed this locus on the telomeric side. We found D7S489C in both duplicons. One question that remains unanswered is why the D7S489C loci amplify so weakly from genomic DNA, compared with the D7S489A and B loci. On the basis of HTGS data, the A- and C-locus primer-site sequences are identical to those of the primers used for PCR, whereas the B locus has a single mismatch in the reverse primer yet is amplified much more strongly than either of the C loci. Our previous report on GTF2I incorrectly identified D7S489B as part of the centromeric repeat (Pérez-Jurado et al. 1998). We now understand that D7S489B is part of the ancestral REP AB locus and not a part of the complete duplication that is contiguous with GTF2I/NCF1 sequences. This finding is in disagreement with the map presented by Osborne et al. (1997a), which identifies two complete copies of the repeat on the centromeric side of the deletion that are in association with D7S489B.
Our map places each of the centromeric GTF2IP1/NCF1P1, REP A, and REP D loci in the same orientation as the telomeric GTF2I/NCF1 and contiguous REP loci. Since the telomeric breakpoints cluster between the GTF2I/NCF1 locus and the telomeric REP AB sequence D7S489A, the common WBS deletion results from nonhomologous recombination between the GTF2I/NCF1 locus and the GTF2IP1/NCF1P1 locus. Our inability to detect novel junction fragments with GTF2I and NCF1 probes reflects the strong conservation of restriction sites over the 150–200-kb GTF2I/NCF1 loci. Breakpoint heterogeneity on the telomeric side is suggested by rare WBS cases reported to be hemizygous at D7S489A (Wang et al. 1998a). In these cases, an intrachromosomal exchange may have occurred between the centromeric GTF2IP1/NCF1 duplicon and the telomeric GTF2IP2/NCF1P2 duplicon.
Görlach et al. (1997) identified the common NCF1 mutation leading to autosomal chronic granulomatous disease (CGD [MIM 233700]) as being a potential gene conversion between the NCF1 gene and a pseudogene. Carriers of this mutation should be investigated for possible recombination between the NCF1 locus and the telomeric pseudogene NCF1P2 locus. On our map, these loci are in opposite orientation to each other, requiring the consideration of models other than simple nonhomologous crossover events. Patients with WBS with D7S489A deletions do not have more-severe phenotypic findings, consistent with the absence, in individuals hetero- and homozygous for the NCF1 del GT mutation resulting from such a recombination, of pathology beyond CGD. The substantially stronger degree of homology demonstrated among the loci GTF2I/NCF1, REP A, and REP D—as opposed that among to the REP B, C, and E loci—may reflect frequent conversion events preserving homology among the former, which do not occur for the latter.
Our model placing the common centromeric breakpoint at the centromeric GTF2IP1/NCF1P1 locus disagrees with the report by Meng et al. (1998a), who, by use of FISH studies of patients with WBS, mapped the proximal breakpoint on BAC 68E13. We determined, by use of PFGE blot hybridization with the POM121 probe, that the centromeric breakpoint is proximal to BAC 68E13. This BAC contains ∼50 kb of REP element B1 that is also present in both flanking regions outside the deletion. Therefore, FISH studies with this BAC as the probe should be inconclusive.
Duplicon-mediated microscopic and submicroscopic deletions are a relatively common accident of human reproduction. In velo-cardio-facial syndrome (VCFS [MIM 192430]), a 3-Mb deletion in 22q11 is bounded by ∼200-kb duplicons within which the common breakpoints cluster (Edelmann et al. 1999a, 1999b). As described here for the WBS region, the VCFS repeats are also nearly identical with conservation of restriction sites. The Smith-Magenis syndrome (SMS [MIM 182290]) deletion at 17p11.2 is also mediated by nonhomologous recombination at duplicons of ∼200 kb (Chen et al. 1997). The SMS region shares with WBS the presence of a third copy of the repeat, which does not participate in the typical recombination errors. The Prader-Willi syndrome (PWS [MIM 176270])/Angelman syndrome (AS [MIM 105830]) region at 15q11-13 contains at least three to five copies of a 50–200-kb transcriptionally active repeat unit, which serve as foci for the nonhomologous recombination events leading to the common PWS/AS deletions (Amos-Landgraf et al. 1999). Other examples of duplicons associated with chromosomal deletions and/or duplications include the Charcot-Marie-Tooth 1A/hereditary neuropathy with liability to pressure palsies (HNPP [MIM 162500]) region, also at 17p11.2-13, and the spinal muscular atrophy types 1-3 (SMAI, II, and III [MIM 253300, MIM 253550, and MIM 253400]) locus at 5q11-13 (Eichler 1998; Lupski 1998; Mazzarella and Schlessinger 1998).
Although the map presented here of the WBS-deletion region and the flanking duplicons is not entirely complete, it is, to date, the most comprehensive. The BAC contig covering the unique deleted region is complete and should enable the identification of all functional genes that are lost by the deletion formation. Further work should compare the sequence of the telomeric GTF2IP2/NCF1P2 locus with those of the gene and the centromeric pseudogene loci. Placement of BACs into accurate contigs should accelerate the assembly of contiguous sequence generated by the high-throughput genome-sequencing project.
Acknowledgments
We are grateful to Dr. Paige Kaplan for clinical samples; to Vida Meyers, Jai Saxena, Erika Valero, Christiane Versbach, Skye Mayo, Jac Luna, and Xianyu Zhang for technical assistance; and to Kathy Redman for administrative assistance. We thank Rachel Wevrick, Joe Giacalone, and Xu Li for helpful discussion. This work was supported by NIH research grants HG00298 and HD33505 (to U.F.) and by the Howard Hughes Medical Institute, of which U.F. is an investigator and Y.-K.W. an associate. R.P. was supported by Institutional Postdoctoral NRSA GM08404 and Clinical Investigator Award HD01181, Y.F. by a fellowship from the Deutsche Forschungsgemeinschaft, and T.P. by a Lynn Marie Chandler Research Fellowship and by the Evelyn L. Neizer Fund.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Web/Genbank/Genbank/Overview.html
- Genome Sequencing Center, Washington University, St. Louis, http://genome.wustl.edu/gsc/
- Human Chromosome 7 Mapping and Sequencing, http://genome.nhgri.nih.gov/chr7/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for WBS [MIM 194050], CGD [MIM 233700], VCFS [MIM 192430], SMS [MIM 182290], PWS [MIM 176270], AS [MIM 105830], HNPP [MIM 162500], SMAI, II, and III [MIM 253300, MIM 253550, and MIM 253400])
- Stanford Human Genome Center, http://www-shgc.stanford.edu/Mapping/index.html
- STS-Based Map of the Human Genome, http://carbon.wi.mit.edu:8000/cgi-bin/contig/phys_map
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 [DOI] [PMC free article] [PubMed]
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, et al (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer A, Dutly F, Balmer D, Riegel M, Tukel T, Krajewska-Walasek M, Schinzel AA (1998) High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum Mol Genet 7:887–894 [DOI] [PubMed]
- Bouffard GG, Idol JR, Braden VV, Iyer LM, Cunningham AF, Weintraub LA, Touchman JW, et al (1997) A physical map of human chromosome 7: an integrated YAC contig map with average STS spacing of 79 kb. Genome Res 7:673–692 [DOI] [PubMed]
- Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, et al (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH (1999) Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 8:1025–1037 [DOI] [PubMed]
- Dausset J, Ougen P, Abderrahim H, Billault A, Sambucy JL, Cohen D, Le Paslier D (1992) The CEPH YAC library. Behring Inst Mitt 91:13–20 [PubMed]
- DeSilva U, Massa H, Trask B, Green E (1999) Comparative mapping of the region of human chromosome 7 deleted in Williams syndrome. Genome Res 9:428–436 [PMC free article] [PubMed]
- De Zeeuw CI, Hoogenraad CC, Goedknegt E, Hertzberg E, Neubauer A, Grosveld F, Galjart N (1997) CLIP-115, a novel brain-specific cytoplasmic linker protein, mediates the localization of dendritic lamellar bodies. Neuron 19:1187–1199 [DOI] [PubMed]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed]
- Dilts CV, Morris CA, Leonard CO (1990) Hypothesis for development of a behavioral phenotype in Williams syndrome. Am J Med Genet Suppl 6:126–131 [DOI] [PubMed]
- Dutly F, Schinzel A (1996): Unequal interchromosomal rearrangements may result in elastin gene deletions causing the Williams-Beuren syndrome. Hum Mol Genet 5:1893–1898 [DOI] [PubMed]
- Edelmann L, Pandita RK, Morrow BE (1999a) Low-copy repeats mediate the common 3-mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet 64:1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RSK, et al (1999b) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 [DOI] [PubMed]
- Eichler EE (1998) Masquerading repeats: paralogous pitfalls of the human genome. Genome Res 8:758–762 [DOI] [PubMed]
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, et al (1997) Interchromosomal duplications of the adrenoleukodystrophy locus: a phenomenon of pericentromeric plasticity. Hum Mol Genet 6:991–1002 [DOI] [PubMed]
- Ewart A, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, et al (1993) Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet 5:11–16 [DOI] [PubMed]
- Francke U (1999) Williams-Beuren syndrome: genes and mechanisms. Hum Mol Genet 8:1947–1954 [DOI] [PubMed]
- Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, et al (1996) LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell 86:1–20 [DOI] [PubMed]
- Franke Y, Peoples RJ, Francke U (1999) Identification of GTF2IRD1, a putative transcription factor within the Williams-Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet 86:296–304 [DOI] [PubMed]
- Gilbert-Dussardier B, Bonneau D, Gigarel N, Le Merrer M, Bonnet D, Philip N, Serville F, et al (1995) A novel microsatellite DNA marker at locus D7S1870 detects hemizygosity in 75% of patients with Williams syndrome. Am J Hum Genet 56:542–544 [PMC free article] [PubMed]
- Görlach A, Lee PL, Roesler J, Hopkins PJ, Christensen B, Green ED, Chanock SJ, et al (1997) A p47-phox pseudogene carries the most common mutation causing p47-phox-deficient chronic granulomatous disease. J Clin Invest 100:1907–1918 [DOI] [PMC free article] [PubMed]
- Hallberg E, Wozniak RW, Blobel G (1993) An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol 122:513–521 [DOI] [PMC free article] [PubMed]
- Hockenhull EL, Carette MJ, Metcalfe K, Donnai D, Read AP, Tassabehji M (1999) A complete physical contig and partial transcript map of the Williams syndrome critical region. Genomics 58:138–145 [DOI] [PubMed]
- Hoogenraad CC, Eussen BHJ, Langeveld A, van Haperen R, Winterberg S, Wouters CH, Grosveld F, et al (1998) The murine CYLN2 gene: genomic organization, chromosome localization, and comparison to the human gene that is located within the 7q11.23 Williams syndrome critical region. Genomics 53:348–358 [DOI] [PubMed]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, et al (1995) An STS-based map of the human genome. Science 270:1945–1954 [DOI] [PubMed]
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, et al (1994) A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet 6:84–89 [DOI] [PubMed]
- Jadayel DM, Osborne LR, Coignet LJA, Zani VJ, Tsui LC, Scherer SW, Dyer MJS (1998) The BCL7 gene family: deletion of BCL7B in Williams syndrome. Gene 224:35–44 [DOI] [PubMed]
- Kaplan P, Wang P, Francke U. Williams (Williams-Beuren) syndrome: a distinct neurobehavioral disorder. J Child Neurol (in press) [DOI] [PubMed] [Google Scholar]
- Karmiloff-Smith A, Grant J, Berthoud I, Davies M, Howlin P, Udwin O (1997) Language and Williams syndrome: how intact is “intact”? Child Dev 68:246–262 [PubMed]
- Kim UJ, Birren BW, Slepak T, Mancino V, Boysen C, Kang HL, Simon MI, et al (1996) Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213–218 [DOI] [PubMed]
- Kipersztok S, Osawa GA, Liang L-F, Modi WS, Dean J (1995) POM-ZP3, a bipartite transcript derived from human ZP3 and a POM121 homologue. Genomics 25:354–359 [DOI] [PubMed]
- Kunz J, Scherer SW, Klawitz I, Soder S, Du YZ, Speich N, Kalff-Suske, et al (1994) Regional localization of 725 human chromosome 7-specific yeast artificial chromosome clones. Genomics 22:439–448 [DOI] [PubMed]
- Li DY, Toland AE, Boak BB, Atkinson DL, Ensing GJ, Morris CA, Keating MT (1997) Elastin point mutations cause an obstructive vascular disease, supravalvular aortic stenosis. Hum Mol Genet 6:1021–1028 [DOI] [PubMed]
- Lu X, Meng X, Morris CA, Keating MT (1998) A novel human gene, WSTF, is deleted in Williams syndrome. Genomics 54:241–249 [DOI] [PubMed]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease. Trends Genet 14:417–422 [DOI] [PubMed]
- Mazzarella R, Schlessinger D (1998) Pathological consequences of sequence duplications in the human genome. Genome Res 8:1007–1021 [DOI] [PubMed]
- Meng X, Lu X, Li Z, Green ED, Massa H, Trask BJ, Morris CA, et al (1998a) Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes. Hum Genet 103:590–599 [DOI] [PubMed]
- Meng X, Lu X, Morris CA, Keating MT (1998b) A novel human gene FKBP6 is deleted in Williams syndrome. Genomics 52:130–137 [DOI] [PubMed]
- Okumura K, Nogami M, Taguchi H, Dean FB, Chen M, Pan ZQ, Hurwitz J, et al (1995) Assignment of the 36.5 kDa (RFC5), 37 kDa (RFC4), 38 kDA (RFC3), and 40 kDa (RFC2) subunit genes of replication factor C to chromosome bands 12q24.2-q24.3, 3q27, 13q12.3-q13, and 7q11.23. Genomics 25:274–278 [DOI] [PubMed]
- Osborne LR, Campbell T, Daradich A, Scherer SW, Tsui LC (1999) Identification of a putative transcription factor gene (WBSCR11) that is commonly deleted in Williams-Beuren syndrome. Genomics 57:279–284 [DOI] [PubMed]
- Osborne LR, Herbrick JA, Greavette T, Heng HHQ, Tsui LC, Scherer SW (1997a) PMS2-related genes flank the rearrangement breakpoints associated with Williams syndrome and other diseases on human chromosome 7. Genomics 45:402–406 [DOI] [PubMed]
- Osborne LR, Martindale D, Scherer SW, Shi XM, Huizenga J, Heng HHQ, Costa T, et al (1996) Identification of genes from a 500-kb region at 7q11.23 that is commonly deleted in Williams syndrome patients. Genomics 36:328–336 [DOI] [PubMed]
- Osborne LR, Soder S, Shi XM, Pober B, Costa T, Scherer SW, Tsui LC (1997b) Hemizygous deletion of the syntaxin 1A gene in individuals with Williams syndrome. Am J Hum Genet 61:449–452 [DOI] [PMC free article] [PubMed]
- Ouellette B, Boguski M (1997) Database divisions and homology search files: a guide for the perplexed. Genome Res 7:952–955 [DOI] [PubMed]
- Paperna T, Peoples R, Wang Y-K, Kaplan P, Francke U (1998) Genes for the CPE-receptor (CPETR1) and the human homolog of RVP1 (CPETR2) are localized within the Williams-Beuren syndrome deletion. Genomics 54:453–459 [DOI] [PubMed]
- Peoples RJ, Cisco MJ, Kaplan P, Francke U (1998) Identification of the WBSCR9 gene, encoding a novel transcriptional regulator, in the Williams-Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet 82:238–246 [DOI] [PubMed]
- Peoples R, Pérez-Jurado L, Wang Y-K, Kaplan P, Francke U (1996) The gene for replication factor C subunit 2 (RFC2) is within the 7q11.23 Williams syndrome deletion. Am J Hum Genet 58:1370–1373 [PMC free article] [PubMed]
- Pérez-Jurado LA, Peoples R, Kaplan P, Hamel BCJ, Francke U (1996) Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am J Hum Genet 59:781–792 [PMC free article] [PubMed]
- Pérez-Jurado LA, Wang Y-K, Francke U, Cruces J. (1999) TBL2, a novel transducin family member in the WBS: characterization of the complete sequence, genomic structure, transcriptional variants and the mouse ortholog. Cytogenet Cell Genet 86:277–284 [DOI] [PubMed]
- Pérez-Jurado LA, Wang Y-K, Peoples R, Coloma A, Cruces J, Francke U (1998) A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Hum Mol Genet 7:325–334 [DOI] [PubMed]
- Pober BR, Dykens EM (1996) Williams syndrome: an overview of medical, cognitive and behavioral features. Child Adolesc Psychiatr Clin North Am 5:929–943 [Google Scholar]
- Richter-Cook NJ, Dever TE, Hensold JO, Merrick WC (1998) Purification and characterization of a new eukaryotic protein translation factor: eukaryotic initiation factor 4H. J Biol Chem 273:7579–7587 [DOI] [PubMed]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, et al (1990) A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res 18:2887–2890 [DOI] [PMC free article] [PubMed]
- Robinson WP, Waslynka J, Bernasconi F, Wang M, Clark S, Kotzot D, Schinzel A (1996) Delineation of the 7q11.2 deletions associated with Williams-Beuren syndrome and mapping of a repetitive sequence to within and to either side of the common deletion. Genomics 34:17–23 [DOI] [PubMed]
- Shepherd NS, Pfrogner BD, Coulby JN, Ackerman SL, Vaidyanathan G, Sauer RH, Balkenhol TC, et al (1994) Preparation and screening of an arrayed human genomic library generated with the P1 cloning system. Proc Natl Acad Sci USA 91:2629–2633 [DOI] [PMC free article] [PubMed]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, et al (1997) An STS-based radiation hybrid map of the human genome. Genome Res 7:422–433 [DOI] [PubMed]
- Sulston JE, Waterston R (1998) Toward a complete human genome sequence. Genome Res 8:1097–1108 [DOI] [PubMed]
- Tassabehji M, Metcalfe K, Donnai D, Hurst J, Reardon W, Burch M, Read AP (1997) Elastin: genomic structure and point mutations in patients with supravalvular aortic stenosis. Hum Mol Genet 6:1029–1036 [DOI] [PubMed]
- Tassabehji M, Metcalfe K, Karmiloff-Smith A, Carette MJ, Grant J, Dennis N, Reardon W, et al (1999) Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet 64:118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchman JW, Bouffard GG, Weintraub LA, Idol JR, Wang L, Robbins CM, Nussbaum JC, et al (1997) 2006 expressed-sequence tags derived from human chromosome 7-enriched cDNA libraries. Genome Res 7:281–292 [DOI] [PubMed]
- Wang MS, Schinzel A, Kotzot D, Casey R, Chodirker BN, Petersen MB, Gyftdimou J, et al (1998a) Clinical correlations in Williams-Beuren syndrome (WBS): no evidence of a parent of origin effect or influence of elastin (ELN) polymorphism. Am J Hum Genet Suppl 63:A690 [Google Scholar]
- Wang PP, Bellugi U (1993) Williams syndrome, Down syndrome and cognitive neuroscience. Am J Dis Child 147:1246–1251 [DOI] [PubMed]
- Wang PP, Doherty S, Rourke SB, Bellugi U (1995) Unique profile of visuo-perceptual skills in a genetic syndrome. Brain Cogn 29:54–65 [DOI] [PubMed]
- Wang Y-K, Harryman Samos C, Peoples R, Pérez-Jurado LA, Nusse R, Francke U (1997) A novel human homologue of the Drosophila frizzled wnt receptor gene binds wingless protein and is in the Williams syndrome deletion at 7q11.23. Hum Mol Genet 6:465–472 [DOI] [PubMed]
- Wang Y-K, Pérez-Jurado LA, Francke U (1998b) A mouse single-copy gene, Gtf2i, the homolog of human GTF2I, that is duplicated in the Williams-Beuren syndrome deletion region. Genomics 48:163–170 [DOI] [PubMed]
- Wang Y-K, Spörle R, Paperna T, Schughart K, Francke U (1999) Characterization and expression pattern of the frizzled gene Fzd9, the mouse homolog of FZD9 which is deleted in Williams-Beuren syndrome. Genomics 57:235–248 [DOI] [PubMed]
- Wedemeyer N, Peoples R, Himmelbauer H, Lehrach H, Francke U, Wanker E (1997) Localization of the human HIP1 gene close to the elastin (ELN) locus on 7q11.23. Genomics 46:313–315 [DOI] [PubMed]
- Wu YQ, Sutton R, Nickerson E, Lupski JR, Potocki L, Korenberg JR, Greenberg F, et al (1998) Delineation of the common critical region in Williams syndrome and clinical correlation of growth, heart defects, ethnicity and parental origin. Am J Med Genet 78:82–89 [DOI] [PubMed]