Introduction
In 1995, much to the delight of mitochondrial Eve (Cann et al. 1987), the search for a suitable partner resulted in the discovery of Y-chromosomal Adam (Dorit et al. 1995; Pääbo 1995). Like Eve, he was traced back to sub-Saharan Africa, although with a date of 270,000 years ago, he seems a bit older than she. On the basis of the absence of sequence variation in a part of the ZFY gene in 38 globally dispersed male subjects, this date was not undisputed (Donnelly et al. 1996; Fu and Li 1996; Rogers et al. 1996; Weiss and von Haeseler 1996). Later in the same year, two additional studies did reveal a limited number of sequence variants on other parts of the Y chromosome. Because of these variants, estimates were obtained of the times back to our most recent common (male) ancestor of 37,000–49,000 years ago (Whitfield et al. 1995) and of 51,000–411,000 years ago (Hammer 1994).
Now, just 5 years later, with simple PCR strategies, ⩾250 polymorphic loci scattered over the entire nonrecombining part of the human Y chromosome can be identified. Among these polymorphisms are (1) biallelic markers with a low mutation rate representing unique (or near-unique) mutation events (UMEs) in human evolution, such as single base-pair substitutions (Underhill et al. 1997), an ALU insertion/deletion polymorphism (Hammer 1994), or a LINE insertion (Santos et al. 2000); (2) moderately fast-evolving microsatellites or simple-tandem repeats (STRs), with an average mutation frequency of ∼.2% per generation (Heyer et al. 1997; Jobling et al. 1999; Kayser et al. 2000); and (3) fast-evolving loci, such as the minisatellite locus MSY1 (Jobling et al. 1998) with a mutation frequency of 6%–11% per generation.
With the exception of the two pseudoautosomal regions, the almost 60–Mb nonrecombining part of the Y chromosome is transmitted strictly from father to son without recombination (Jobling and Tyler-Smith 1995). This renders the Y chromosome probably the most versatile haplotypic genotyping system of the human genome. It is thus not surprising that chromosome-Y polymorphisms have been used to follow the migration patterns of our male ancestors from the recent past (Heyer et al. 1997; Foster et al. 1998) through historical times (Skorecki et al. 1997; Hammer et al. 2000), to the origins of modern humans (Hammer et al. 1998).
Recently, two excellent review articles featuring the Y chromosome were published. The first (Bertranpetit 2000) addresses the difficulties of reliably tracing back human origins solely on the basis of Y-chromosomal UMEs. The second (Jobling and Tyler-Smith 2000) gives a detailed discussion of many genetic aspects of the Y chromosome in the context of disease and selection. This editorial will be restricted to the combined use of UMEs and STRs on the Y chromosome to reconstruct our genetic history. This application has received considerable attention in recent articles published in this journal and elsewhere (see, e.g., Zerjal et al. 1997; Bianchi et al. 1998; Hurles et al. 1998, 1999; Kittles et al. 1998; Bosch et al. 1999; Karafet et al. 1999; Lahermo et al. 1999; Ruiz-Linares et al. 1999; Helgason et al. 2000b; Hill et al. 2000; Santos et al. 2000). That the combined use of Y-chromosomal UMEs and STRs is not without any caveats will be explained below.
The Y Chromosome: A Collection of Bottlenecks
For a better understanding of the power and pitfalls of the combined use of Y-chromosomal UMEs and STRs, it is essential to explain its theoretical basis in some detail. Figure 1 illustrates the combined genotyping of Y-chromosomal UMEs and STRs in a hypothetical population sample. We assume that (1) no strange Y lineages were introduced into this population by means of migration, (2) each UME occurred for the first time within our population, (3) each UME occurred only once, and (4) we have detected all UMEs present in the male subjects studied. In our example, genotypic information from three UMEs and a number of STRs was available. Distinct Y chromosomes, defined solely on the basis of UME character states, are designated as “haplogroups.” Distinct Y chromosomes identified by STRs are designated as “haplotypes,” and Y chromosomes that are defined by the combination of UMEs and STRs are called “lineages.”
Figure 1.
The bottleneck model. It is assumed that no strange Y lineages were introduced into our population by means of migration and that each UME occurred only once. In our example, we have genotypic information from three UMEs and from a number of STRs. A, From sequence information obtained from nonhuman primates, we know the ancestral status of each of these UMEs, here designated as “0.” Subsequently, the derived state of each UME is designated as “1.” From the four different haplogroups, A (0–0–0) is the most ancestral one. Nonrecurrent mutation events at T1, T2, and T3 result in the derived haplogroups B (0–1–0), C (0–1–1), and D (1–1–1). B, Within each haplogroup, a large number of different STR-defined haplotypes can be distinguished. The STR haplotype of the haplogroup A Y chromosome, from which at time T1 the first haplogroup B arose, defines the ancestral STR status of all subsequent B Y lineages. Because of a number of successive ultimate genetic bottleneck events (from a single chromosome, a whole new group of chromosomes grows), two additional Y haplogroups are born at times T2 and T3.
In this idealized example, because of three independent mutation events at times T1, T2, and T3, four distinct Y haplogroups can be identified (fig. 1a). If one would type only UMEs, this would be all the Y variability there is to be detected. In reality, however, each haplogroup consists of a variable number of Y chromosomes that share the same UME character state but vary in Y-STR haplotype. These haplotypes connect to a network, instead of to a simple haplotype tree, because of recurrent and/or parallel mutation events of Y STRs (fig. 1B). Subsequently, from this collection of chromosomes, because of a unique, nonrecurrent mutation event, a single chromosome with a specific STR haplotype mutates to the first Y chromosome with a new UME character state. Over time, because of the relatively high mutation frequency of STRs, a new STR network arises from this single Y chromosome. This process defines the ultimate genetic bottleneck. In our example, the first haplogroup A, B, C, and D Y chromosomes arose at different times during evolution. If a constant Y-STR mutation rate were assumed, the accumulated genetic STR variance within each different haplogroup can be used as an indication of the age of these haplogroups; the older the haplogroup, the more variable the STR-haplotypes network will be. Also note that one haplogroup (C) is absent from the sample of present-day Y chromosomes. It either went extinct or was not picked up by our sampling scheme in this hypothetical population.
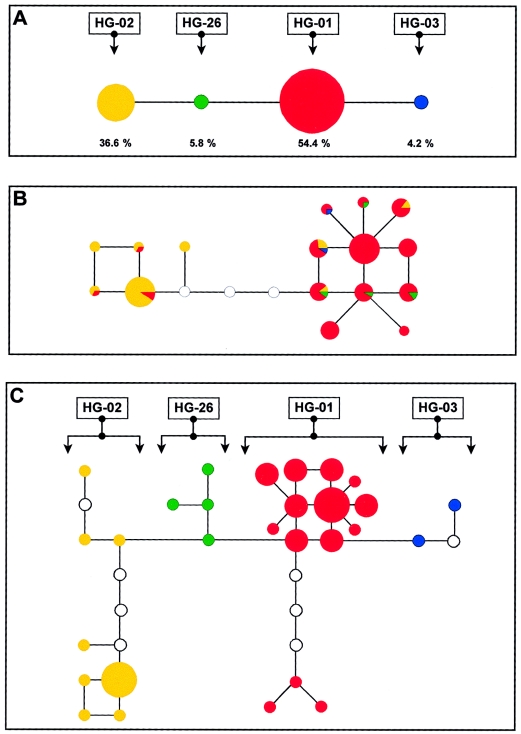
For most human populations, the simplistic assumptions from the above model population will be incorrect. Many population samples will contain immigrants. Consequently, these population samples will harbor UMEs that arose for the first time somewhere else. Also, we will never be able to identify all UMEs in a number of male subjects. Nevertheless, we can still make important observations. To illustrate these, the Y-chromosomal genotyping results from 261 Dutch male subjects (Kraayenbrink et al., unpublished results) are summarized in figure 2 (for full details, see the legend to that figure).
Figure 2.
Genotyping results of three Y UMEs and 6 Y-STR loci among Dutch male subjects. A, A maximum parsimony tree connecting four Y haplogroups observed in 261 Dutch male subjects. Haplogroup frequencies are indicated below each pie. Nomenclature of these UME-defined haplogroups is according to Jobling and Tyler-Smith (2000). B, A median-joining network (Bandelt et al. 1999) connecting Y-STR haplotypes. In the 261 male subjects, only the 16 most frequently observed haplotypes (i.e., those shared between three or more male subjects) are illustrated. These 16 haplotypes occurred in a total of 126 (48%) male subjects. The diameter of each circle corresponds to a categorical absolute frequency (n=3–5, n=6–15, or n=16–25). Multiple colored pies indicate haplotypes that are found in different haplogroups. Within each pie, again, the absolute frequency is indicated. Unblackened circles indicate missing haplotypes. Circles are connected by single STR-repeat mutation steps. C, A median joining network connecting Y lineages. In the same 126 male subjects, 28 distinct Y lineages could be identified. The diameter of each circle corresponds to a categorical absolute frequency (n=3–5, n=6–15, or n=16–25). Unblackened circles indicate missing haplotypes. Circles are connected by single STR mutation steps or by single UME mutation steps.
First, an STR-haplotype network, connecting seemingly unrelated male subjects from a particular population, will almost always underestimate the true genetic variation present in these male subjects (compare the networks in figs. 2b and 2c). More single-mutation steps are necessary to connect Y-chromosome lineages (UMEs and STRs combined; fig. 2c) than are STR-haplotype networks (fig. 2b). Because of this, Y-STR mutation rates will always be underestimated if based on STR-only networks (Cooper et al. 1996; Forster et al. 2000). Similarly, reliable inferences about Y-STR mutation models cannot be obtained from STR networks (Cooper et al. 1999). Such inferences should be based only on direct observations (Kayser et al. 2000).
Second, the bottleneck model predicts that within each UME-defined haplogroup, the STR allele frequency distribution entirely depends on the original ancestral STR haplotype. Thus, in many cases, the allele frequency distribution of Y-STR loci will differ between haplogroups. The Y-STR allele frequency distribution of any random male population sample will be the product of the relative frequencies of haplogroups and the STR allele frequency distribution within these haplogroups. This effect is illustrated, in figure 3, for two chromosome Y STR loci—DYS390 and DYS392—in 261 Dutch male subjects. The allele frequency distribution of these two loci in the combined population sample reflects the sum of the allele frequency distributions among haplogroup 2 and haplogroup 1 chromosomes. Because of locus-specific differences in the shape of the distribution between the two haplogroups, the resulting combined distribution is unimodal for DYS390 and bimodal for DYS392.
Figure 3.
Allele frequency distribution of the Y STRs DYS390 and DYS392 in 261 Dutch male subjects. The frequencies in the total sample (n=261), in haplogroup 1 male subjects (see fig. 2; n=142), and in haplogroup 2 male subjects (n=93) are shown. Colors correspond to those in figure 2. The blue/green hatched box of the total sample represents the relative contribution of haplogroup 3 and 26 male subjects.
The net effect of these first two points is to cause a breakdown of “linkage” between the various STR loci on the Y chromosome, resulting in marked differences in genetic variance per locus, per haplogroup, and per population sample. Therefore, general statistical properties of Y STRs cannot be reliably measured in a global population sample without considering these confounding aspects (Goldstein et al. 1996).
Third, two neighboring Y haplogroups will initially share a single completely identical Y chromosome (apart from the single mutation event separating them). However, over time, because of parallel STR mutations, different haplogroups can share more STR-defined haplotypes. The pattern of haplotype similarities between haplogroups will be shaped primarily by differences in the mutation rate of Y STRs (Kayser et al. 2000). In figure 2b, this is illustrated for the Dutch male subjects, where the multicolored pies indicate STR haplotypes that are found to be shared between different haplogroups. This again underlines that one underestimates the true genetic variance by means of a Y-STR–only network.
Fourth, it will take considerable time before sufficient STR variability will arise within each haplogroup. Thus, time is a major driving force, shaping the genetic variance pattern of Y haplogroups. If a haplogroup is very old, too many parallel STR mutation events will result in an underestimate of the age of the haplogroup if it is estimated on the basis of accumulated STR variance. On the other hand, the age of a very recent haplogroup can be overestimated, because of drift effects of individual Y lineages. Over time, haplogroups will also be distributed from one population to another. This, in addition to population growth and decline (population bottlenecks), will strongly influence the reliability of coalescence estimates. Thus, there will be only a limited number of cases in which the application of such coalescent approaches is possible and reliable.
Examples of Calculating the Age of Y Haplogroups
On the basis of accumulated genetic (STR) variance, a number of different methods have been used to date the age of a Y haplogroup or a specific migration event (Bertranpetit and Calafell 1996; Goldstein et al. 1996; Thomas et al. 1998; Wilson and Balding 1998). Such estimates can be made only on the basis of genetic information from the population in which these haplogroups (probably) arose. These methods invariably assume a single mutation rate that is constant over time for all Y STRs considered, and they do not allow population growth. There is now sufficient evidence of a significant difference in mutation rate among Y STRs (Heyer et al. 1997; Jobling et al. 1999; Kayser et al. 2000). From our bottleneck model, it will be apparent that accumulation of STR variability without population growth is a very unlikely model (which was recently confirmed in the study by Pritchard et al. 1999). To illustrate the difficulties associated with dating haplogroups and migration events, two recent studies that calculated age estimates from Y-UME and Y-STR data are contrasted below.
In one study (Hurles et al. 1999), the authors presented estimates of the date of an Iberian-specific UME (SRY-2627 C→T). They compared three classical “constant rate & size” models (Bertranpetit and Calafell 1996; Goldstein et al. 1996; Thomas et al. 1998) with a new method that does allow individual Y-STR mutation rates and population growth (BATWING [Bayesian Analysis of Trees with Internal Node Generation; accessible at the BUGS Analyses Web site], an extension of the model described by Wilson and Balding 1998). Roughly the same recent origin of this specific mutation was found in all four models (1,650–3,452 years with 95% confidence interval [CI] 1,044–16,001 years), with the growth model predicting the most recent origin. It was also found that, despite its recent origin, haplogroup 22 shares seven STR haplotypes with its ancestral haplogroup 1, which perfectly illustrates the rapid parallel evolution of Y STRs (see also Jobling and Tyler-Smith 2000).
A second example of the combined use of Y UMEs and Y STRs is the study of Helgason et al. (2000b). In that article, the authors speculate on the origin of the male founders of the Icelandic population. On the basis of classical markers, it has been shown that different parts of Iceland have been settled by a variable number of people from Gaelic (Scottish and Irish) and/or Viking (Scandinavian) origin between 850 and 1200 A.D. (Williams 1993). Helgason et al. (2000a, 2000b) now show that most of the present-day Y chromosomes are of Scandinavian origin, whereas most of mtDNA lineages were of Gaelic origin.
On the basis of accumulated Y-STR variance among haplogroups 1, 2, and 3 in the combined group of Icelandic, Scandinavian, and Gaelic male subjects, the authors also provide estimates of the coalescence dates for these haplogroups. However, in contrast with the example above (the Iberian haplogroup 22), these three haplogroups did not arise in any of these three populations. These haplogroups have a much wider distribution across the world, suggesting a more ancient and non–western European origin (Jobling and Tyler-Smith 2000). Because the present-day haplogroup 1, 2, and 3 Y lineages have such a complex history before they arrived in western Europe, we are confronted with a kind of black-box scenario. We know only that these haplogroups arose outside western Europe and that men with such lineages arrived at the western fringe of Europe at some date. However, we have no clue about the migration history of these lineages. This, effectively, prevents us from making reliable coalescent estimates using western European populations, so the estimates derived from these data should be interpreted with caution.
Helgason et al. (2000b) also estimate the time-depth of divergence of Icelandic STR haplotypes from the two source populations. From the accumulated STR variance of Iceland-specific Y-STR haplotypes, if a generation time of 35 years and an average Y-STR mutation rate of .21% were assumed, and using a coalescent model that does not allow for population growth, the authors derive an estimate of divergence of the Icelanders from the founding populations of 2,717 years (95% CI 1,164–9,508), which clearly predates the settling of Iceland. The authors hypothesize that this discrepancy could be due to the fact that many of the Iceland-specific haplotypes have yet to be sampled from the ancestral populations. This would reduce the number of Iceland-specific haplotypes, reduce the accumulated variance, and result in a more recent estimate. Another, perhaps more likely, hypothesis might be that the present-day Icelandic chromosome Y gene pool has been shaped by the peculiar population demographic history of the Icelandic population, combined with the sensitivity for genetic drift of the Y chromosome. This, together with the use of an analytical model that does not allow for population growth and variable Y-STR mutation rates, could have easily influenced the divergence estimates from this fascinating study.
Conclusions
The increasing amount of polymorphic markers on the Y chromosome, combined with its unique haplotypic inheritance pattern, renders this chromosome a tantalizing simple genetic tool to infer human migration events. Because of the lack of recombination, the Y chromosome seems the perfect model to test coalescence processes before they are used for autosomal inferences. The isolated use of STRs or UMEs gives useful and comparable information (Forster et al. 2000). Evidently, by using them both, a more complete and accurate picture can be obtained. It is thus not surprising that the combined use of these two different types of polymorphisms has become increasingly popular. However, this appealing simplicity is also misleading. Only with care and in the appropriate populations can accumulated Y-STR variance be used to make coalescent estimates of haplogroup ages, as has been done, for example, by Hurles et al. (1999).
There is clearly a demand for better analytical tools that are capable of using Y-STR data with all its complexity. Currently available software is inadequately calibrated and tested. It is hoped that routines such as BATWING (BUGS Analyses) will eventually be fully developed. For the time being, any published Y-STR–based estimate should be treated with utmost care for one simple reason: its message came through many bottlenecks.
Electronic-Database Information
The URL for data in this article is as follows:
- BUGS Analyses, http://www.maths.abdn.ac.uk/~ijw/ (for BATWING)
References
- Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bertranpetit J (2000) Genome, diversity, and origins: the Y chromosome as a storyteller. Proc Natl Acad Sci USA 97:6927–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertranpetit J, Calafell F (1996) Genetic and geographic variation in cystic fibrosis: evolutionary considerations. In: Chadwick D, Cardew G (eds) Variation in the human genome. John Wiley & Sons, New York, pp 97–118 [Google Scholar]
- Bianchi NO, Catanesi CI, Bailliet G, Martinez-Marignac VL, Bravi CM, Vidal-Rioja LB, Herrera RJ, López-Camelo JS (1998) Characterization of ancestral and derived Y-chromosome haplotypes of New World native populations. Am J Hum Genet 63:1862–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Santos FR, Pérez-Lezaun A, Comas D, Benchemsi N, Tyler-Smith C, Bertranpetit J (1999) Variation in short tandem repeats is deeply structured by genetic background on the human Y chromosome. Am J Hum Genet 65:1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC (1987) Mitochondrial DNA and human evolution. Nature 325:31–36 [DOI] [PubMed] [Google Scholar]
- Cooper G, Amos W, Hoffman D, Rubinsztein DC (1996) Network analysis of human Y microsatellite haplotypes. Hum Mol Genet 5:1759–1766 [DOI] [PubMed] [Google Scholar]
- Cooper G, Burroughs NJ, Rand DA, Rubinsztein DC, Amos W (1999) Markov chain Monte Carlo analysis of human Y-chromosome microsatellites provides evidence of biased mutation. Proc Natl Acad Sci USA 96:11916–11921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P, Tavaré S, Balding DJ, Griffiths RC (1996) Estimating the age of the common ancestor of men from the ZFY intron. Science 272:1357–1359 [DOI] [PubMed]
- Dorit RL, Akashi H, Gilbert W (1995) Absence of polymorphism at the ZFY locus on the human Y chromosome. Science 268:1183–1185 [DOI] [PubMed] [Google Scholar]
- Forster P, Röhl A, Lünnemann P, Brinkmann C, Zerjal T, Tyler-Smith C, Brinkmann B (2000) A short tandem repeat-based phylogeny for the human Y chromosome. Am J Hum Genet 67:182–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster EA, Jobling MA, Taylor PG, Donnelly P, de Knijff P, Mieremet R, Zerjal T, Tyler-Smith C (1998) Jefferson fathered slave’s last child. Nature 396:27–28 [DOI] [PubMed] [Google Scholar]
- Fu Y-X, Li W-H (1996) Estimating the age of the common ancestor of men from the ZFY intron. Science 272:1356–1357 [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Zhivotovsky LA, Nayar K, Ruiz-Linares A, Cavalli-Sforza LL, Feldman MW (1996) Statistical properties of the variation at linked microsatellite loci: implications for the history of human Y chromosomes. Mol Biol Evol 13:1213–1218 [DOI] [PubMed] [Google Scholar]
- Hammer MF (1994) A recent insertion of an ALU element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol 11:749–761 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL (1998) Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol 15:427–441 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonné-Tamir B (2000) Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA 97:6769–6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir S, Gulcher JR, Ward R, Stefánsson K (2000a) mtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet 66:999–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir S, Nicholson J, Sykes B, Hill EW, Bradley DG, Bosnes V, Gulcher JR, Ward R, Stefánsson K (2000b) Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet 67:697–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Puymirat J, Dieltjes P, Bakker E, de Knijff P (1997) Estimating Y-chromosome-specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet 6:799–803 [DOI] [PubMed] [Google Scholar]
- Hill EW, Jobling MA, Bradley DG (2000) Y-chromosome variation and Irish origins: a pre-neolithic gene gradation starts in the near east and culminates in western Ireland. Nature 404:351–352 [DOI] [PubMed] [Google Scholar]
- Hurles ME, Irven C, Nicholson J, Taylor PG, Santos FR, Loughlin J, Jobling MA, Sykes BC (1998) European Y-chromosomal lineages in Polynesians: a contrast to the population structure revealed by mtDNA. Am J Hum Genet 63:1793–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Veitia R, Arroyo E, Armenteros M, Bertranpetit J, Pérez-Lezaun A, Bosch E, Shlumukova M, Cambon-Thomsen A, McElreavey K, López De Munain A, Röhl A, Wilson IJ, Singh L, Pandya A, Santos FR, Tyler-Smith C, Jobling MA (1999) Recent male-mediated gene flow over a linguistic barrier in Iberia, suggested by analysis of a Y-chromosomal DNA polymorphism. Am J Hum Genet 65:1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Bouzekri N, Taylor PG (1998) Hypervariable digital DNA codes for human paternal lineages: MVR-PCR at the Y-specific minisatellite, MSY1 (DYF155S1) Hum Mol Genet 7:643–653 [DOI] [PubMed]
- Jobling MA, Heyer E, Dieltjes P, de Knijff P (1999) Y-chromosome-specific microsatellite mutation rates re-examined using a minisatellite, MSY1. Hum Mol Genet 8:2117–2120 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C (1995) Fathers and sons: the Y chromosome and human evolution. Trends Genet 11:449–456 [DOI] [PubMed] [Google Scholar]
- ——— (2000) New uses for new haplotypes: the human Y chromosome, disease and selection. Trends Genet 16:356–362 [DOI] [PubMed] [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Krüger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittles RA, Perola M, Peltonen L, Bergen AW, Aragon RA, Virkkunen M, Linnoila M, Goldman D, Long JC (1998) Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 62:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahermo P, Savontaus M-L, Sistonen P, Béres J, de Knijff P, Aula P, Sajantila A (1999) Y chromosomal polymorphisms reveal founding lineages in the Finns and the Saami. Eur J Hum Genet 7:447–458 [DOI] [PubMed] [Google Scholar]
- Pääbo S (1995) The Y chromosome and the origin of all of us. Science 268:1141–1142 [DOI] [PubMed]
- Pritchard JK, Seielstad MT, Pérez-Lezaun A, Feldman MW (1999) Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol Biol Evol 16:1791–1798 [DOI] [PubMed] [Google Scholar]
- Rogers J, Samollow PB, Comuzzie AG (1996) Estimating the age of the common ancestor of men from the ZFY intron. Science 272:1360–1361 [DOI] [PubMed]
- Ruiz-Linares A, Ortíz-Barrientos D, Figueroa M, Mesa N, Múnera JG, Bedoya G, Vélez ID, García LF, Pérez-Lezaun A, Bertranpetit J, Feldman MW, Goldstein DB (1999) Microsatellites provide evidence for Y chromosome diversity among the founders of the New World. Proc Natl Acad Sci USA 96:6312–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Pandya A, Kayser M, Mitchell RJ, Liu A, Sing L, Destro-Bisol G, Noveletto A, Qamar R, Mehdi SQ, Adhikari R, de Knijff P, Tyler-Smith C (2000) A polymorphic L1 retroposon insertion in the centromere of the human Y chromosome. Hum Mol Genet 9:421–430 [DOI] [PubMed] [Google Scholar]
- Skorecki K, Selig S, Blazer S, Bradman R, Bradman N, Waburton PJ, Ismajlowicz M, Hammer MF (1997) Y chromosomes of Jewish priests. Nature 385:32 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein DB (1998) Origins of Old Testament priests. Nature 394:138–140 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G, von Haeseler A (1996) Estimating the age of the common ancestor of men from the ZFY intron. Science 272:1359–1360 [DOI] [PubMed] [Google Scholar]
- Whitfield LS, Sulston JE, Goodfellow PN (1995) Sequence variation of the human Y chromosome. Nature 378:379–380 [DOI] [PubMed] [Google Scholar]
- Williams JT (1993) Origin and population structure of the Icelanders. Hum Biol 65:167–191 [PubMed] [Google Scholar]
- Wilson IJ, Balding DJ (1998) Genealogical inference from microsatellite data. Genetics 150:499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhövel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183 [PMC free article] [PubMed] [Google Scholar]