Abstract
Acyl-CoA dehydrogenase (ACAD) defects in isoleucine and valine catabolism have been proposed in clinically diverse patients with an abnormal pattern of metabolites in their urine, but they have not been proved enzymatically or genetically, and it is unknown whether one or two ACADs are involved. We investigated a patient with isolated 2-methylbutyrylglycinuria, suggestive of a defect in isoleucine catabolism. Enzyme assay of the patient's fibroblasts, using 2-methylbutyryl-CoA as substrate, confirmed the defect. Sequence analysis of candidate ACADs revealed heterozygosity for the common short-chain ACAD A625 variant allele and no mutations in ACAD-8 but a 100-bp deletion in short/branched-chain ACAD (SBCAD) cDNA from the patient. Our identification of the SBCAD gene structure (11 exons; >20 kb) enabled analysis of genomic DNA. This showed that the deletion was caused by skipping of exon 10, because of homozygosity for a 1228G→A mutation in the patient. This mutation was not present in 118 control chromosomes. In vitro transcription/translation experiments and overexpression in COS cells confirmed the disease-causing nature of the mutant SBCAD protein and showed that ACAD-8 is an isobutyryl-CoA dehydrogenase and that both wild-type proteins are imported into mitochondria and form tetramers. In conclusion, we report the first mutation in the SBCAD gene, show that it results in an isolated defect in isoleucine catabolism, and indicate that ACAD-8 is a mitochondrial enzyme that functions in valine catabolism.
Introduction
The individual role of the different acyl-CoA dehydrogenases (ACADs) in isoleucine and valine catabolism is at present unclear, and the underlying enzymatic defects causing accumulation of metabolites derived from isobutyryl-CoA and 2-methylbutyryl-CoA are poorly understood. In 1994, a cDNA encoding a human homologue of rat 2-methyl–branched-chain ACAD was cloned and characterized (Rozen et al. 1994). The enzyme was named “short/branched-chain ACAD” (SBCAD), because it differed from its rat homologue in that it exhibited highest activity with butyryl-CoA and 2-methylbutyryl-CoA and low or no activity (Rozen et al. 1994; Binzak et al. 1999) with isobutyryl-CoA, suggesting a primary role in isoleucine catabolism and, perhaps, also in the catabolism of (short-chain) fatty acids, but not in valine catabolism. So far, none of the ACADs have been demonstrated to have significant enzyme activity with isobutyryl-CoA as substrate. However, the recently identified ACAD-8 (Telford et al. 1999), with unknown substrate specificity, exhibits the highest degree of sequence similarity to SBCAD and to short-chain ACAD (SCAD). Therefore, it is a candidate enzyme for branched-chain amino acid catabolism.
In the present study, we characterize the human SBCAD gene structure and describe the identification and characterization of the first mutation in the human SBCAD gene from a patient with 2-methylbutyrylglycinuria. Moreover, we investigate the mitochondrial import, processing, and enzyme activities of overexpressed human SBCAD and ACAD-8.
Patient and Methods
Case History
This 3-year-old boy is the second of three children of a marriage between first cousins from Pakistan. Pregnancy, delivery, and the neonatal period were without complications. At age 3 mo, he was referred to the local hospital, after 2 d of low caloric intake, because of a feverish illness. During his 2d year of life, he became increasingly floppy. He has since had retarded motor development, generalized muscular atrophy, and strabismus. At age 3 years he is able to walk with support. Magnetic-resonance imaging was normal at age 2 years. Urinary organic acid analysis revealed a consistent 2-methylbutyrylglycinuria, with excretion of only trace amounts of isobutyrylglycine. Plasma concentrations of free and total carnitine were slightly below normal ranges. Analysis of the newborn blood spot from the patient showed a normal acylcarnitine profile, as measured by electrospray tandem mass spectrometry (Rashed et al. 1995).
Both parents, a brother (age 12 mo), and a sister (age 4.5 years) of the patient have been without symptoms of disease. However, analysis of the family members showed that, in addition to the index case, the mother also excreted 2-methylbutyrylglycine.
Extraction of Total RNA, Northern Blot Analysis, cDNA Synthesis, PCR Amplification, and Sequence Analysis of SBCAD and ACAD-8 cDNA
Extraction of total RNA from fibroblasts was performed using an RNAzol kit (WAK-Chemie). Fifteen micrograms total RNA was used for northern blot analysis using a SBCAD cDNA (position −5 to 1374) hybridization probe. Blots were visualized on a Molecular Dynamics PhosphorImager. First-strand cDNA synthesis was performed from total RNA using the first-strand cDNA synthesis kit (Clontech). PCR amplification of the entire coding region of SBCAD cDNA (position −5 to 1374) and ACAD-8 cDNA (position −44 to 1489) was performed using standard conditions. The PCR products were subjected to bidirectional cycle sequencing, using a sequencing kit (BigDye terminator; PE Biosystems), and analyzed on ABI 373A and ABI 377 sequencers (PE Biosystems).
Identification of the Human SBCAD Gene Structure and Sequence Analysis of the Patient's Genomic DNA
tBLASTn searches of the GenBank database, with the human SBCAD amino acid sequence, identified the following bacterial artificial chromosomes (BACs): AC012391, AQ306749, and AQ779739. BAC AC012391 has been mapped to chromosome 10q25. BACs were obtained from The Sanger Centre or Research Genetics, and DNA was prepared with a Midi prep kit (Qiagen). Intron sizes were estimated from agarose gel electrophoresis of PCR products amplified with exon-located primers. Regions of the SBCAD gene not covered by the BACs were sequenced from amplified genomic DNA from controls. All sequencing was performed with a combination of primers located in introns and exons of the SBCAD gene. The resulting sequence information was used to design primers (sequences are available on request) for amplification and sequence analysis of all exons and part of the flanking introns. All exons of the SCAD gene were amplified and sequenced using intron-located primers designed on the basis of the gene structure (Corydon et al. 1997). PCRs were carried out under standard conditions in an automated thermal cycler (Thermal cycler 480; PE Biosystems).
Expression of Wild-Type and Mutant SBCAD and ACAD-8 in COS-7 Cells
Human ACAD-8 cDNA (position −44 to position 1489) was cloned between the HindIII and ApaI sites of the expression vector pcDNA3.1+ (InVitrogen), and wild-type and patient SBCAD cDNA (position −5 to position 1374, with or without the 100-bp deletion) was inserted between the HindIII and XhoI sites of pcDNA3.1+; error-free clones were named “pACAD-8-WT,” “pSBCAD-WT,” and “pSBCAD-MUT,” respectively. Transfection of COS-7 cells was performed using FuGene 6 (Boehringer Mannheim), and cells were harvested 48 h posttransfection.
The activity of the overexpressed SBCAD and ACAD-8 proteins was measured in cleared cell lysates, in a standard reaction medium (100 mM Tris, pH 8, and 100 μM flavin adenine dinucleotide) supplemented with 0.2 mM of either (S)-2-methylbutyryl-CoA, isobutyryl-CoA, butyryl-CoA, or isovaleryl-CoA, with ferricenium as the electron acceptor. The formation of tiglyl-CoA and 2-methyl-3-hydroxybutyryl-CoA (from 2-methylbutyryl-CoA), either methacrylyl-CoA and 2-methyl-3-hydroxypropionyl-CoA (from isobutyryl-CoA) or butenoyl-CoA and 3-hydroxy butyryl-CoA (from butyryl-CoA), and 3-methyl-crotonyl-CoA and 3-hydroxy-isovaleryl-CoA (from isovaleryl-CoA) was quantified using high-performance liquid chromatography (HPLC) (C18 reversed phase, 250×10 mm, methanol gradient, detection at 263 nm). Isobutyryl-CoA, butyryl-CoA, and isovaleryl-CoA (Sigma) and (S)-2-methylbutyryl-CoA (synthesized by either the authors or by Jerry Vockley [Mayo Clinic]) were used as substrates. Transfections were performed at least twice, and enzyme activity was measured in duplicate.
In Vitro Mitochondrial Import and Stability of Wild-Type ACAD-8, Wild-Type SCAD, and Wild-Type and Mutant SBCAD
In vitro transcription and translation was carried out using the TnT coupled transcription/translation kit, [35S]-methionine, and plasmids pSBCAD-WT, pSBCAD-MUT, pSCAD-WT, and pACAD-8-WT, according to the manufacturer’s protocol (Promega). The pSCAD-WT plasmid was constructed by subcloning the coding region of human SCAD from pMP6 harboring SCAD cDNA (Corydon et al. 1998) into the polylinker of pcDNA3.1+. Mitochondria were isolated from rat liver, as described elsewhere (Gregersen 1979). The stability and import of the translation products into mitochondria were assayed essentially as described by Volchenboum and Vockley (2000). After SDS-PAGE and Native PAGE (ReadyGels; BioRad), radioactive bands were visualized on a Molecular Dynamics PhosphorImager and were quantified using Imagequant software (Molecular Dynamics).
An ACAD-8 antibody was raised, in rabbits, against a recombinant his-tag fusion protein of human ACAD-8 (amino acids 1–415) expressed in Escherichia coli. In situ immunostaining, localization, and confocal laser scanning–microscopy analysis of ACAD-8 in transfected COS-7 cells was performed as described elsewhere (Corydon et al. 1998).
Results
Identification of Acyl-CoA Dehydrogenase Deficiency
Organic acid analysis of the patient’s urine consistently demonstrated the presence of 2-methylbutyryl-glycine, suggesting an ACAD defect in the isoleucine pathway. To evaluate this possibility, we set up an assay to determine the ACAD activity toward 2-methylbutyryl-CoA in fibroblasts. HPLC was used to separate and quantify the substrate and the reaction products from lysed cells, using ferricenium as an electron acceptor. The enzyme activity in the patient's fibroblasts when 2-methylbutyryl-CoA was used as substrate was shown to be decreased to 10% of controls (control, 0.157 nmol/min/mg protein; patient, 0.016 nmol/min/mg protein [±10%]).
Sequence Analysis
We decided to analyze the cDNA sequence of the ACAD-8 and SBCAD genes from our patient and from two controls, to see whether the excreted 2-methylbutyrylglycine and the deficient enzyme activity with 2-methylbutyryl-CoA as substrate could be caused by a defect in one or both of these enzymes. Moreover, because patients with SCAD deficiency may have small amounts of 2-methylbutyrylglycine in their urine (Tein et al. 1999), we also amplified and sequenced all exons of our patient’s SCAD gene. The patient was found to be heterozygous for the A625 polymorphism (Corydon et al. 1996). No other changes were identified. PCR amplification and sequencing of the ACAD-8 protein–coding region from cDNA revealed no differences from the published sequence (Telford et al. 1999), either in our patient or in the two controls. Interestingly, two of the PCR products, produced with two different SBCAD sense primers and a common SBCAD antisense primer, were ∼100 bp shorter when amplified from the patient's cDNA than when amplified from control cDNA (fig. 1A). Sequence analysis revealed a 100-bp deletion of cDNA positions 1129–1228 in the patient. SBCAD mRNA with this deletion has a shifted reading frame after glutamate 376 and a premature stop codon 12 codons downstream. In addition, the patient and one of the controls were homozygous for the silent mutations 351T→A and 639T→C. The second control was homozygous for 351T→A and heterozygous for 639T→C. A tBLASTn search of the human expressed-sequence-tag database at GenBank indicated that these are normal sequence variations. No other changes in the coding region of SBCAD cDNA were observed in either the patient or the controls.
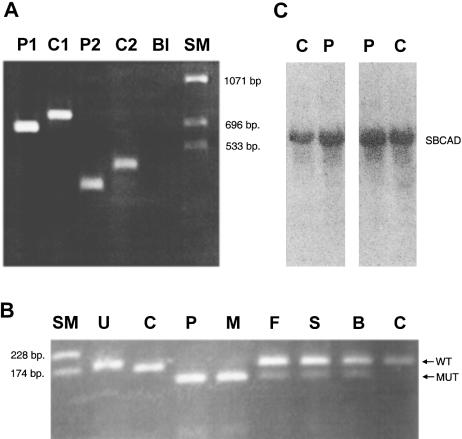
Figure 1 .
A, PCR of SBCAD cDNA from the index patient (lane P1) and a normal control (lane C1), using a sense primer located in exon 6 and an antisense primer located in exon 11. “P2” and “C2” result from identical amplifications as P1 and C1, respectively, except that another sense primer, located in exon 8, was used. A blank amplification is marked by “Bl.” The size marker is indicated by “SM.” B, Agarose gel electrophoresis after restriction digestion of PCR products from genomic DNA from the index patient (lane P), his mother (lane M), his father (lane F), his healthy sister (lane S), his healthy brother (lane B), and a normal control (lane C), using a modified sense primer (5′-ATTACCCTGTGGAGAAATACTGAGATGCAAAGACT-3′) located in exon 10 of the SBCAD gene and an antisense primer (5′-AAATCCAGCCACATAACTAGTGCCACCAAACATATACTG-3′) located in intron 10. Undigested PCR products (U) are 187 bp long. After digestion with the SpeI restriction enzyme, PCR products harboring the 1228G→A mutation are 130 bp long, and PCR products with the wild-type sequence are 167 bp. C, Two different northern blot analyses of 15 μg total fibroblast RNA from the patient (lane P) and a control (lane C), using a SBCAD cDNA probe.
Identification of the SBCAD Gene Structure and Investigation of Genomic DNA from the Patient, His Family Members, and a Control
To investigate the molecular basis for the deletion, we identified and characterized the genomic structure of the human SBCAD gene (accession numbers AF260668–AF260678) (see Patient and Methods section). The SBCAD gene structure (table 1) was confirmed by PCR and by direct sequencing of all exons from genomic DNA from a control.
Table 1.
Exon-Intron Organization of the Human SBCAD Gene[Note]
| Intron 3′ Splice Site | Exon | 5′ Exon Sequence | ExonSize(bp) | Exon StartPosition | 3′ Exon Sequence | Intron 5′Splice Site | Intron Size |
| .......... | 1a | CAGCAGGCTG | gtgagtgcgt | ??? | |||
| ttccctacag | 2 | CTAAGAAGAA | 160 | 43 | AAGAGTTCAG | gtaagtaaat | ≈2.7 kb |
| ctgcccacag | 3 | TTAAAAAATT | 101 | 203 | TCAACAAGGG | gtacagttca | ≈2.3 kb |
| ttttctttag | 4 | TTGATGGGTA | 207 | 304 | TACAGAAAAA | gtgagttgag | ≈0.4 kb |
| ccatatgtag | 5 | GTAGGAAGTT | 171 | 511 | CCCTACCATT | gtaagtttga | ≈1.4 kb |
| ttctttttag | 6 | GGATATAAGG | 126 | 682 | AAATGTCAAG | gtgggtatcg | ≈1.0 kb |
| gtattttcag | 7 | GTTCCAGAAG | 93 | 808 | TGCTGCACAG | gtaagtcaga | ≈2.8 kb |
| gttttttcag | 8 | ATGCTGGGAC | 90 | 901 | TGATTTTCAG | gtatgtaatt | ≈6.5 kb |
| actgttacag | 9 | GGCCTCCAAC | 138 | 991 | TGCATCAGAG | gtaaaaaaaa | 1,874 bp |
| cttttggtag | 10 | ATTGCAGGAC | 100 | 1129 | GCAAAGATTG | gtaaatagat | 534 bp |
| tttgcttaagb | 11 | GTACGATATA | ??? | 1229 | |||
| Consensus (yyyyyyynag) | G......... | .......AAG | gtragt.... |
Note.— Start position of each exon is indicated relative to the corresponding position in the cDNA sequence; intron and exon sizes are indicated, as well as 10 bp of the exon sequence and 10 bp of the intron sequence at each junction. The position of the 1228G→A mutation observed in the patient is indicated by underlining. The sequence data have been submitted to the GenBank database (accession numbers AF260668–AF260678).
Transcription initiation site not determined.
Poly A addition sites at cDNA positions 1388–92, 1916–20, and 2646–51.
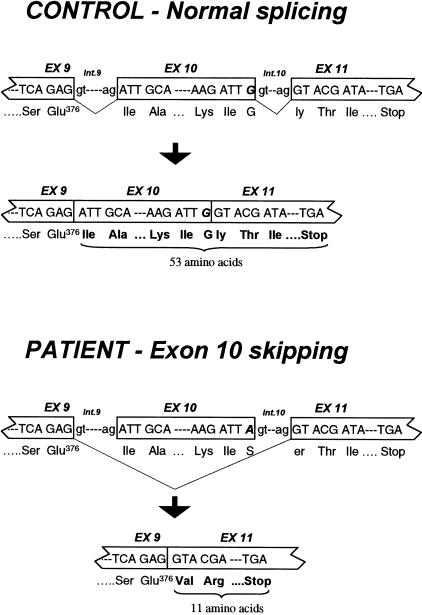
PCR amplification and sequence analysis of genomic DNA from a control and from the patient showed that the 100-bp sequence deleted in the patient's cDNA was present in his genomic DNA, excluding a genomic deletion. The deleted sequence corresponds to exon 10 (table 1), indicating that aberrant splicing of SBCAD mRNA causes the deletion. Sequence analysis of amplified genomic DNA showed that the patient was apparently homozygous for a single G→A transition (1228G→A) in exon 10. Amplification of genomic DNA with primers that introduced a SpeI restriction site only in mutant DNA, followed by digestion with SpeI (fig. 1B), showed homozygosity for the 1228G→A mutation in the patient and heterozygosity in the father and in both healthy siblings (fig. 1B). Interestingly, the mother was also found to be homozygous for the mutation. This was confirmed by sequence analysis of a PCR product from the mother that harbored exon 10. Analysis of the family members showed that, just like the patient, the mother excretes 2-methylbutyrylglycine in her urine. The 1228G→A mutation changes the codon for glycine 410 to that for serine and is located at the last nucleotide in the skipped exon 10 (table 1 and fig. 2). Sequence analysis of both introns flanking exon 10, in both a control and the patient, revealed no other changes in the splice consensus sequences. Therefore, we believe that the 1228G→A mutation causes the observed skipping of exon 10 in the patient's SBCAD mRNA. Analysis by PCR amplification of two different cDNAs from the patient, using different sense and antisense primers located in different exons flanking the skipped exon, indicated that no normal-sized SBCAD mRNA is detectable (fig. 1A and not shown) and thus that the 1228G→A mutation affects splicing severely. Northern blot analysis of two different preparations of total RNA, from the patient's and from control fibroblasts, showed that the steady-state amounts of SBCAD mRNA are comparable (fig. 1C). Sequence analysis of exon 10 amplified from genomic DNA from 59 Danish controls (118 alleles) showed that the 1228G→A mutation was not present.
Figure 2 .
Position, in the SBCAD gene, of the 1228G→A mutation, and the resulting aberrant splicing in the patient, compared with the normal splicing pattern in a control person. The position of the 1228G→A mutation observed in the patient is indicated by boldface italic type.
Expression of Wild-Type and Mutant SBCAD, ACAD-8, and SCAD in COS-7 Cells
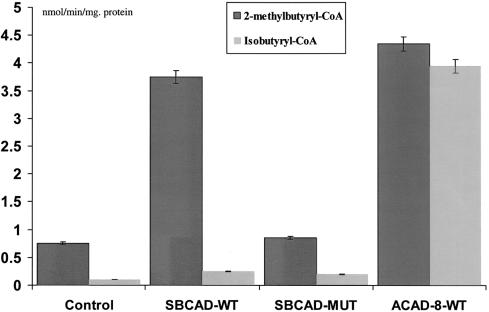
To demonstrate the disease-causing nature of the 1228G→A mutation, we analyzed both the mutant SBCAD protein encoded by the mRNA resulting from exon skipping and the wild-type SBCAD protein resulting from overexpression. In addition, we overexpressed wild-type human ACAD-8, with the purpose of comparing the substrate specificity to that of SBCAD. Our results showed that mutant SBCAD protein has no enzyme activity and that the substrate for SBCAD wild-type protein is 2-methylbutyryl-CoA (fig. 3). Furthermore, our results showed that the main substrate for ACAD-8 is isobutyryl-CoA but that it also has activity with 2-methylbutyryl-CoA. None of the enzymes had significant activity toward either butyryl-CoA or isovaleryl-CoA.
Figure 3 .
Enzyme activities after expression of wild-type and mutant SBCAD and ACAD-8 in COS-7 cells. The results from measuring ACAD activity in COS-7 cells transfected with pSBCAD-WT, pSBCAD-MUT, or pACAD-8-WT expression vectors are shown. The control is COS-7 cells harboring the expression vector pcDNA3.1+ with no cDNA insert (Control). Activity measurements are given in nmol/min/mg protein. Blackened bars show the enzyme activity measured with 2-methylbutyryl-CoA as substrate, and gray bars show the enzyme activity measured with isobutyryl-CoA as substrate. Error bars indicate the range of the values measured. None of the transfected cells showed activity above background when either butyryl-CoA or isovaleryl-CoA was used as substrate (not shown).
In Vitro Mitochondrial Import and Stability of Wild-Type and Mutant SBCAD, ACAD-8, and SCAD
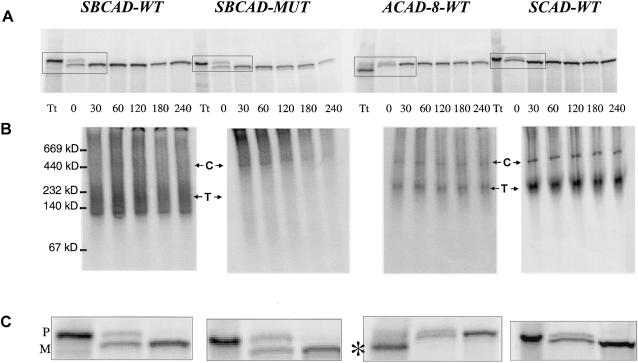
To further study the molecular defect of the mutant SBCAD protein and to investigate whether precursor SBCAD, SCAD, and ACAD-8 proteins are imported into mitochondria and are correctly processed, we used a coupled in vitro transcription/translation system to synthesize radioactively labeled precursor proteins and incubated them with isolated rat mitochondria. Our analysis by SDS-PAGE showed that both mutant and wild-type precursor SBCAD protein, precursor wild-type ACAD-8, and precursor wild-type SCAD proteins were imported into mitochondria and were processed to mature size (fig. 4A and C). The stability of the imported proteins was investigated by testing samples withdrawn at different time points, after removal of all unimported precursor proteins by trypsin digestion and washing. This indicated that mutant SBCAD protein decayed slightly faster than wild-type SBCAD (fig. 4A). Investigation of the imported proteins by native PAGE showed that wild-type SBCAD, SCAD, and ACAD-8 proteins formed larger complexes inside the mitochondria, corresponding in size to tetramers, and that mutant SBCAD protein failed to do so (fig. 4B). Furthermore, a high-molecular-weight band shown, by Western blot analysis (authors' unpublished results), to comigrate with mHsp60, was much more prominent in extracts from cells overexpressing mutant SBCAD protein than in extract from cells overexpressing wild-type SBCAD.
Figure 4 .
In vitro mitochondrial import and stability of wild-type and mutant SBCAD, wild-type ACAD-8, and wild-type SCAD. Wild-type and mutant SBCAD, wild-type ACAD-8, and SCAD wild-type precursor proteins were synthesized by coupled in vitro transcription/translation and were incubated with isolated rat mitochondria for 30 min. After removal, by washing, of unimported precursor proteins, the fate of the imported proteins was followed for different time intervals. Twenty-microliter samples of the mitochondria were collected at the time points indicated, were washed two times, and were incubated with trypsin, followed by addition of trypsin inhibitor. Then samples were lysed and subjected to centrifugation. Finally, the supernatant fraction was subjected to either SDS-PAGE (A and C) or native PAGE (B). A 1-μl sample of the total translatate (Tt) withdrawn prior to incubation with mitochondria was also subjected to electrophoresis. The positions and molecular masses of coelectrophoresed marker proteins are indicated on the left margin. Positions of the precursor (P) and the mature (M) form of the respective proteins are shown on the right margin, and the position corresponding to the tetrameric proteins (T) and mHsp60 chaperone complexes (C) are indicated. The positions of the chaperone complexes were demonstrated by western blot analysis using an anti-mHsp60 antibody (authors' unpublished results). In the total translatate produced from the pACAD-8-WT plasmid, a lower-molecular-weight band (denoted by the asterisk [*]) was present, but the corresponding polypeptides were not imported into mitochondria. We presume that this band represents ACAD-8 proteins in which an internal in-frame ATG codon has been used as the translation start signal. Such internal ATGs, which are present in a more optimal sequence context for translation initiation (Kozak 1989) than are the correct ATG, are located at positions 115–117 and 178–180 in the ACAD-8 cDNA. Initiation of translation at either of these positions will result in production of proteins without a mitochondrial targeting sequence and with a size corresponding to the lower-molecular-weight band.
These results may indicate that the mutant SBCAD protein is unable to form tetramers and, instead, remains in complex with chaperones, such as mHsp60, a tendency previously observed with mutant medium-chain ACAD (MCAD) (Saijo et al. 1994; Andresen et al. 1997; Bross et al. 1998).
Finally, we analyzed intact COS-7 cells overexpressing ACAD-8, using in situ immunostaining (see Patient and Methods section). Using our anti–ACAD-8 antibody together with a monoclonal anti-mitochondrial Hsp60 antibody, followed by confocal laser scanning–microscopy analysis, we could demonstrate that ACAD-8 and endogenous mHsp60 colocalize (results not shown).
Discussion
The number and the individual role of the candidate enzymes for the ACAD step in the catabolism of isoleucine and valine are still not elucidated, and neither genetic nor enzymatic evidence for inherited defects has so far been reported. In the present study, we show, using 2-methylbutyryl-CoA as enzyme substrate, severely decreased enzyme activity in the fibroblasts from a patient with 2-methylbutyrylgycinuria. This demonstrates, for the first time, the existence of an ACAD defect in isoleucine metabolism. Consequently, we initiated a molecular genetic characterization of the candidate genes, SBCAD, ACAD-8, and SCAD, from our patient. We found that he had no changes in the ACAD-8 gene and was only heterozygous for the SCAD A625 variant. Instead, the enzymatic defect could be explained by our finding that he is homozygous for a 1228G→A mutation that changes the codon for glycine 410 to serine in the SBCAD gene. It is, however, obvious that the deleterious effect of the mutation does not result from this amino acid change, since the mutation results in complete skipping of the constitutive exon (fig. 2). Mutations (such as 1228G→A) of the consensus G at the −1 position of the 5′ splice site usually result in exon skipping, and G→A is observed in two-thirds of all such mutations (Nakai and Sakamoto 1994). Our analysis of the patient's cDNA shows that no normal splicing occurs (fig. 1A). It is not obvious why this −1G→A change has such a drastic effect. At first glance, this 5′ splice site of the human SBCAD gene (table 1) conforms well with human splice consensus sequences (Zhang 1998). However, the sequence of the last three nucleotides of SBCAD exon 10 is TTG, which is much less frequently observed than the consensus, AAG (Zhang 1998). This could perhaps be of importance, since it has been demonstrated that, with smaller-than-average-size exons (such as SBCAD exon 10), the requirement for a consensus splice sequence is more stringent (Zhang 1998).
SBCAD mRNA with the 100-bp deletion encodes a protein in which the reading frame is shifted after glutamate 376, ending with a premature translation termination codon (PTC) 12 codons downstream (fig. 2). This PTC is located in the last exon of the SBCAD gene, which explains why we observe normal amounts of mutant SBCAD mRNA, despite the fact that introduction of a PTC will usually result in low steady-state amounts of mRNA (Maquat 1995). Evidence from several genes shows that a PTC has to be located >50–55 nucleotides upstream of the most 3′ exon-exon junction to have an effect (Nagy and Maquat 1998).
Because the mutant mRNA is stable, it is likely that the encoded mutant protein is produced. It is, however, very unlikely that this mutant protein has any enzyme activity, since several functionally important amino acids, such as the active-site glutamate 414 (Binzak et al. 1998), are missing. Moreover, overexpression of wild-type and mutant SBCAD protein in COS-7 cells and in vitro transcription/translation experiments have shown that the mutant protein has no detectable enzyme activity (fig. 3) and that it has a slightly reduced stability and a lack of ability to form tetramers (fig. 4). This is consistent with previous observations from characterization of two disease-causing PTC mutations (R324X and E359X) in MCAD (Andresen et al. 1994) and by recent experiments, with mutant isovaleryl-CoA dehydrogenase proteins (Volchenboum and Vockley 2000), that show that the carboxy-terminal part of an ACAD is necessary for subunit interaction and tetramer stability.
Together with our finding that the mutation was not present in 118 control chromosomes, these results clearly demonstrate that the 1228G→A mutation is deleterious. We therefore conclude that the enzyme defect in our patient is caused by homozygosity for the SBCAD 1228G→A mutation.
The fact that isolated accumulation of 2-methylbutyrylglycine with only trace amounts of isobutyrylglycine was observed in our patient, together with the fact that human SBCAD has no detectable activity with isobutyryl-CoA indicates that, in humans, SBCAD deficiency affects isoleucine catabolism and not valine catabolism. It is therefore obvious that another ACAD serves the valine pathway. A likely candidate for this is ACAD-8, since we have shown in the present study that this enzyme has very high activity toward isobutyryl-CoA.
Recently, another patient, who showed a metabolite pattern (2-methylbutyrylcarnitine and 2-methylbutyrylglycine in body fluids) similar to that of our patient was reported (Gibson et al. 1999). The initial clinical presentation of this patient (presentation, at day 3 of life, with hypoglycemia, lethargy, and apnea) was different from that observed in our patient, although both have developed neurological symptoms. This indicates that patients with SBCAD deficiency may present differently. Such clinical heterogeneity is common in ACAD defects (Andresen et al. 1997, 1999). Likewise, the finding that the clinically asymptomatic mother has a genotype identical to that of the index patient and that she excretes 2-methylbutyrylglycine in her urine is not so surprising; it may illustrate the emerging fact that the mere presence of the genetic defect is not always enough to cause clinical disease in inherited metabolic disorders. We have previously observed the same phenomenon in families suffering from other ACAD defects, such as MCAD and glutaryl-CoA dehydrogenase deficiency, in which asymptomatic genetically affected siblings and parents of a clinically affected patients have been identified (Amir et al. 1989; Andresen et al. 1997). The finding that the clinical manifestation of SBCAD deficiency in our patient was preceded by episodes of infectious disease further corroborates our notion (Andresen et al. 1997) that environmental factors, such as metabolic stress and fever, may be crucial determinants of the clinical phenotype in genetically affected individuals. An explanation for this could be that overlapping enzyme activity from ACAD-8 and SCAD is sufficient to avoid harmful accumulation of metabolites from isoleucine metabolism in patients with SBCAD deficiency when they are not subjected to metabolic stress.
Acknowledgments
We are grateful to Hans Eiberg for generously providing DNA from normal controls. We thank Jerry Vockley (Mayo Clinic, Rochester, MN) for providing a sample of 2-methylbutyryl-CoA. Clones RP11-162A23 and bA564D11 were generously provided by Amy Taylor (Chromosome 10 Mapping Group, The Sanger Centre, Hinxton, United Kingdom). Thanks to Silje, Emil, and Olav for their kind support. This work was supported by Danish Medical Research Council grants 9702280 (to N.G.) and 9802627 (to B.S.A.) and by the Karen Elise Jensen Foundation.
Note added in proof.—
After submission of this manuscript, a paper describing heterozygosity for a mutation SBCAD cDNA in a patient with 2-methylbutyrylglycinuria was reported by Gibson et al. (2000).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Genbank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for human SBCAD gene [accession numbers AF260668–AF260678])
References
- Amir N, Elpeleg ON, Shalev RS, Christensen E (1989) Glutaric aciduria type I: enzymatic and neuroradiologic investigations of two kindreds. J Pediatr 114:98–99 [DOI] [PubMed]
- Andresen BS, Bross P, Udvari S, Kirk J, Gray RGF, Kmock S, Chamoles N, Knudsen I, Winter V, Wilcken B, Yokota I, Hart K, Packman S, Harpey JP, Saudubray JM, Hale DE, Bolund L, Kolvraa S, Gregersen N (1997) The molecular basis of medium-chain acyl-CoA dehydrogenase (MCAD) deficiency in compound heterozygous patients: is there a correlation between genotype and phenotype? Hum Mol Genet 6:695–708 [DOI] [PubMed] [Google Scholar]
- Andresen BS, Jensen TG, Bross P, Knudsen I, Winter V, Kølvraa S, Bolund L, Ding JH, Chen YT, VanHove JLK, Curtis D, Yokota I, Tanaka K, Kim JJP, Gregersen N (1994) Disease-causing mutations in exon 11 of the medium-chain acyl-CoA dehydrogenase (MCAD) gene. Am J Hum Genet 54: 975–988 [PMC free article] [PubMed] [Google Scholar]
- Andresen BS, Olpin S, Poorthuis B, Scholte HR, Vianey-Saban C, Wanders RJA, Ijlst L, Morris A, Pourfarzam M, Bartlett K, Baumgartner ER, deKlerk JB, Schroeder LD, Corydon TJ, Lund H, Winter V, Bross P, Bolund L, Gregersen N (1999) It is possible to correlate genotype with disease phenotype in very-long-chain acyl-CoA dehydrogenase (VLCAD) deficiency. Am J Hum Genet 64:479–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzak B, Willard J, Vockley J (1998) Identification of the catalytic residue of human short/branched chain acyl-CoA dehydrogenase by in vitro mutagenesis. Biochim Biophys Acta 1382:137–142 [DOI] [PubMed]
- Bross P, Andresen BS, Gregersen N (1998) Impaired folding and subunit assembly as disease mechanism: the example of medium-chain acyl-CoA dehydrogenase deficiency. Prog Nucleic Acid Res Mol Biol 58:301–337 [DOI] [PubMed] [Google Scholar]
- Corydon MJ, Andresen BS, Bross P, Kjeldsen M, Andreasen PH, Eiberg H, Kolvraa S, Gregersen N (1997) Structural organization of the human short-chain acyl-CoA dehydrogenase gene. Mamm Genome 8:922–926 [DOI] [PubMed] [Google Scholar]
- Corydon TJ, Bross P, Jensen TG, Corydon MJ, Lund TB, Jensen UB, Kim JJ, Gregersen N, Bolund L (1998) Rapid degradation of short-chain acyl-CoA dehydrogenase variants with temperature-sensitive folding defects occurs after import into mitochondria. J Biol Chem 273:13065–13071 [DOI] [PubMed] [Google Scholar]
- Corydon MJ, Gregersen N, Lehnert W, Ribes A, Rinaldo P, Kmoch S, Christensen E, Kristensen TJ, Andresen BS, Bross P, Winter V, Martinez G, Neve S, Jensen TG, Bolund L, Kolvraa S (1996) Ethylmalonic aciduria is associated with an amino acid variant of short chain acyl-coenzyme A dehydrogenase. Pediatr Res 39:1059–1066 [DOI] [PubMed] [Google Scholar]
- Gibson KM, Burlingame TG, Hogema B, Jakobs C, Schutgens RB, Millington D, Roe CR, Roe DS, Sweetman L, Steiner RD, Linck L, Pohowalla P, Sacks M, Kiss D, Rinaldo P, Vockley J (2000) 2-Methylbutyryl-coenzyme A dehydrogenase deficiency: a new inborn error of L-isoleucine metabolism. Pediatr Res 47:830–833 [DOI] [PubMed]
- Gibson KM, Sacks M, Kiss D, Pohowalla P, Linck L, Steiner RD, Burlingame T (1999) 2-Methylbutyrylglycinuria in a neonate with CNS dysfunction: evidence for isolated 2-methylbutyryl-CoA dehydrogenase deficiency, an inborn error of L-isoleucine metabolism. J Inherit Metab Dis 22(Suppl)1:O31 [Google Scholar]
- Gregersen N (1979) Studies on the effects of saturated and unsaturated short-chain monocarboxylic acids on the energy metabolism of rat liver mitochondria. Pediatr Res 13:1227–1230 [DOI] [PubMed] [Google Scholar]
- Kozak M (1989) The scanning model for translation: an update. J Cell Biol 108: 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1:453–465 [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23:198–199 [DOI] [PubMed] [Google Scholar]
- Nakai K, Sakamoto H (1994) Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene 141:171–177 [DOI] [PubMed] [Google Scholar]
- Rashed MS, Ozand PT, Bucknall MP, Little D (1995) Diagnosis of inborn errors of metabolism from blood spots by acylcarnitines and amino acids profiling using automated electrospray tandem mass spectrometry. Pediatr Res 38: 324–331 [DOI] [PubMed] [Google Scholar]
- Rozen R, Vockley J, Zhou L, Milos R, Willard J, Fu K, Vicanek C, Low-Nang L, Torban E, Fournier B (1994) Isolation and expression of a cDNA encoding the precursor for a novel member (ACADSB) of the acyl-CoA dehydrogenase gene family. Genomics 24:280–287 [DOI] [PubMed] [Google Scholar]
- Saijo T, Welch WJ, Tanaka K (1994) Intramitochondrial folding and assembly of medium-chain acyl-CoA dehydrogenase (MCAD). J Biol Chem 269:4401–4408 [PubMed] [Google Scholar]
- Tein I, Haslam RH, Rhead WJ, Bennett MJ, Becker LE, Vockley J (1999) Short-chain acyl-CoA dehydrogenase deficiency: a cause of ophthalmoplegia and multicore myopathy. Neurology 52:366–372 [DOI] [PubMed] [Google Scholar]
- Telford EA, Moynihan LM, Markham AF, Lench NJ (1999) Isolation and characterisation of a cDNA encoding the precursor for a novel member of the acyl-CoA dehydrogenase gene family. Biochim Biophys Acta 1446:371–376 [DOI] [PubMed] [Google Scholar]
- Volchenboum SL, Vockley J (2000) Mitochondrial import and processing of wild type and type III mutant isovaleryl-CoA dehydrogenase. J Biol Chem 275:7958–7963 [DOI] [PubMed] [Google Scholar]
- Zhang MQ (1998) Statistical features of human exons and their flanking regions. Hum Mol Genet 7:919–932 [DOI] [PubMed] [Google Scholar]