Abstract
Cytochrome c oxidase (COX) catalyzes both electron transfer from cytochrome c to molecular oxygen and the concomitant vectorial proton pumping across the inner mitochondrial membrane. Studying a large family with multiple cases of neonatal ketoacidotic comas and isolated COX deficiency, we have mapped the disease locus to chromosome 17p13.1, in a region encompassing two candidate genes involved in COX assembly—namely, SCO1 and COX10. Mutation screening revealed compound heterozygosity for SCO1 gene mutations in the patients. The mutated allele, inherited from the father, harbored a 2-bp frameshift deletion (ΔGA; nt 363–364) resulting in both a premature stop codon and a highly unstable mRNA. The maternally inherited mutation (C520T) changed a highly conserved proline into a leucine in the protein (P174L). This proline, adjacent to the CxxxC copper-binding domain of SCO1, is likely to play a crucial role in the tridimentional structure of the domain. Interestingly, the clinical presentation of SCO1-deficient patients markedly differs from that of patients harboring mutations in other COX assembly and/or maturation genes.
Introduction
Mitochondrial respiratory-chain deficiency is a clinically and genetically heterogeneous condition that accounts for an ever-increasing spectrum of clinical presentations in humans (DiMauro et al. 1999; Wallace 2000). This broad clinical heterogeneity stems from both the dual genetic origin of respiratory-chain components (mtDNA and nuclear DNA) and the number of nuclear genes involved in the assembly and maintenance of the respiratory chain (Leonard and Schapira 2000b). Mutations have been reported in both the mtDNA and the nuclear genes encoding mitochondrial proteins (Leonard and Schapira 2000a), but the molecular bases of respiratory-chain deficiency remain unknown in most patients, a feature that makes genetic counseling in respiratory-chain deficiency particularly difficult (von Kleist-Retzow et al. 1999).
The mitochondrial cytochrome c oxidase (COX) is the terminal complex of the respiratory chain and catalyzes electron transfer from reduced cytochrome c to oxygen. The isolated deficiency of this enzyme is the most frequent and clinically heterogeneous cause of respiratory-chain deficiency in our series (accounting for ∼30% of those [von Kleist-Retzow et al. 1998]). This clinical variability is probably due to the number of nuclear genes involved in the expression, maturation, and assembly of the 13 COX subunits (Taanman 1997). So far, COX deficiency has been ascribed to mutations of either mtDNA-encoded COX subunits (Keightley et al. 1996; Comi et al. 1998; Hanna et al. 1998; Bruno et al. 1999; Clark et al. 1999; Rahman et al. 1999; Hoffbuhr et al. 2000) or nuclear genes involved in assembly of functional complexes—namely, SURF1, SCO2, and COX10. Indeed, mutations in the nuclear SURF1 gene (MIM 185620 and MIM 256000) usually cause Leigh subacute necrotizing encephalomyopathy (Zhu et al. 1998; Tiranti et al. 1999; Poyau et al. 2000) and have occasionally been associated with other clinical presentations (von Kleist-Retzow et al., in press). On the other hand, patients harboring SCO2 mutations (MIM 604272 and MIM 604377) have presented with encephalocardiomyopathy (Papadopoulou et al. 1999; Jaksch et al. 2000), whereas COX10 mutations (MIM 602125) have accounted for tubulopathy and leukodystrophy in one family (Valnot et al. 2000).
By studying a family with early-onset hepatic failure and neurological involvement, we have mapped the disease locus to chromosome 17p13.1 and have ascribed this condition to mutations of SCO1, a protein that plays an important role for proper assembly of the COX.
Patients and Methods
Patients
A boy (patient II.1) born to unrelated healthy parents after a term pregnancy (birth weight 2,700 g; length 45 cm; head circumference 34 cm) was found to be hypotonic and lethargic and required immediate assistance for respiratory distress. He had a severe metabolic acidosis (pH 7.19; plasma bicarbonate 7.7 mM; lactate 12.7 mM normal; <2.5), with abnormally high urinary lactate, fumarate, and succinate; hepatocellular dysfunction was noted at day 4 (factor V 25%; fibrinogen 1.2 g/liter), and he recovered thereafter. However, severe axial hypotonia, hypoglycemia, and hyperlactatemia (3.5 mM) persisted, and liver enlargement was observed. Then he presented with recurrent episodes of apnea and bradycardy and died at age 2 mo. For investigation of respiratory-chain activities, immediate postmortem liver and skeletal muscle biopsies were performed, and blood lymphocytes were isolated. Histopathological examination of the liver showed swollen hepatocytes, with microvesicular lipid vacuoles and panlobular steatosis. Furthermore, histological study of a muscle-biopsy sample revealed an accumulation of lipid droplets. One year later, a second boy was born at week 34 of pregnancy (patient II.2; birth weight 1,750 g). He presented with a severe neurological distress and metabolic acidosis and died at age 5 d.
Methods
Enzyme assays.—COX (E.C.1.9.3.1), succinate quinone DCPIP reductase, and succinate cytochrome c reductase activities were measured spectrophotometrically on tissue homogenates, as described elsewhere (Rustin et al. 1994).
DNA analyses.—DNA was extracted from leukocytes, muscle, or chorionic villus biopsies. For genotyping, microsatellite markers from the Généthon database were used (Dib et al. 1996). For chromosome X markers, the PCR products were loaded onto a polyacrylamide gel and were transferred to a positive nylon membrane (Nytran plus; Schleicher and Schuell). Membranes were hybridized with a (CA)12 probe. The labeling of the probe and the revelation of the blots were performed using the ECL direct nucleic acid–labeling and –detection system (Amersham Pharmacia Biotech). The blots were autoradiographed with Kodak-X-OMAT films for 2–30 min.
For the genomewide search, 382 pairs of fluorescent oligonucleotides from the GENESCAN linkage mapping set, version 2 (PE Biosystems), were used under conditions recommended by the manufacturer. Amplified fragments were electrophoresed and analyzed with an automatic sequencer (ABI 377). The polymorphic markers had an average spacing of 10 cM throughout the genome. Linkage analysis was performed using M-LINK and LINKMAP, version 5.1.
Sequence analysis.—The six exons of the SCO1 gene (MIM 603644) were amplified by PCR using the following SCO1-specific intronic primers (forward/reverse, 5′→3′)—exon 1, TACCGGAAATCGCGGGGA and AGAAGGGTTCCAGGTGTGC; exon 2, TTTATGTTTGAAATCCCTGCC and TACAGGGCTGAGCAGATGAT; exon 3, TGTCGATATGTTTTTGTCTCCT and CTTTGTTTAGTTAGTGATGGCT; exon 4, TTAGGGTGTGAATACGGAC and AGGCACTGTAAGGTTCAAAT; exon 5, TAGAGATGGTTGTTTTACTGG and TGGTCCATGGGTTAAAACTG; and exon 6, TTGGTAATCTTTGTCACACTC and TTAGCAAGAGAATACTGCATC. Amplification products were electrophoresed through a 2% low-melting-point agarose gel, were purified, and were directly sequenced using the PRISM Ready Reaction Sequencing Kit (PE Biosystems) on an automatic sequencer (ABI 373; PE Biosystems). SSCP analysis was performed using the GeneGel Excel 12.5/24 kit from Pharmacia. Exon 3 primers were used to amplify a 253-bp fragment surrounding the C520T mutation that creates a XbaI restriction site. The XbaI restriction fragments were separated on a 3% agarose gel.
Results
Enzymological Studies
A severe isolated COX deficiency with markedly altered activity ratios was found in a postmortem liver-biopsy sample from patient II.1 (table 1). The same enzyme deficiency was found in skeletal muscle and circulating lymphocytes of both the patient (not shown) and his younger brother (patient II.2; not shown). Subsequently, two prenatal diagnoses were performed, and the two male fetuses showed COX deficiency, in both chorionic villi and amniotic cell fluid, at weeks 11 and 16 of pregnancy, respectively.
Table 1.
Respiratory-Chain Enzyme Activities in Liver and Muscle Homogenate of Patient II.1 and in Controls[Note]
| Patient II.1 | Control | |
| Activities (nmol/min/mg protein): | ||
| Liver (n = 26): | ||
| CI | 28 | 11–31 |
| CII | 79 | 76–194 |
| CIII | 150 | 95–246 |
| CIV | 17 | 84–245 |
| Muscle (n = 37): | ||
| CII+III | 30 | 19–37 |
| CIV | .5 | 94–196 |
| Activity ratios: | ||
| Liver (n = 26): | ||
| CIV/CI | .6 | 8.2 ± 2.3 |
| CIV/CII | .2 | 1.6 ± .2 |
| CIV/CIII | .1 | 2.4 ± .2 |
| CIII/CII | 1.9 | 2.4 ± .2 |
| Muscle (n = 37): | ||
| CIV/CII+III | .02 | 4.7 ± 1.3 |
Note.— Both absolute activities and ratios are indicated. The absence of a normal distribution of absolute control values precluded the use of SDs. Because control activity ratios follow a Gaussian distribution (Chretien et al. 1998), these values are presented as mean ± 1 SD. Experimental conditions are as described in the Patients and Methods section. Abnormal values are underlined.
Genetic Studies
Before the birth of individual II.6, the observation of four affected males (two neonates and two fetuses) and a healthy girl in a nonconsanguineous family was consistent with either a maternal or an X-linked mode of inheritance. Both screening of the mtDNA, for large rearrangements and common point mutations known to cause COX deficiency (MERRF and MELAS mutations), and sequencing of mitochondrial COX genes and surrounding tRNA genes failed to detect any disease-causing mtDNA mutation. Extensive genotyping using 40 microsatellite DNA markers of chromosome X excluded an X-linked mode of inheritance in this family (not shown).
A genomewide linkage search revealed that the four affected individuals carried an identical genotype at locus D17S1852 on chromosome 17, whereas the two healthy sibs carried different haplotypes at this locus (fig. 1). No other region of haploidentity was identified.
Figure 1 .
Pedigree and haplotype analyses. Haplotypes are given for loci D17S938, D17S1852, D17S799, and D17S921 (top to bottom).
Screening for Mutations in the SCO1 Gene
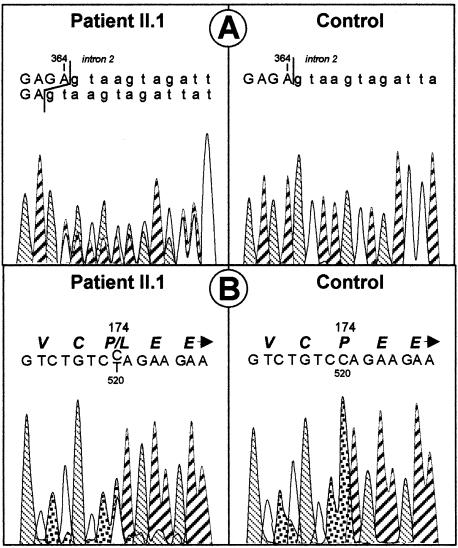
Two genes were regarded as excellent candidates, by virtue of both position and function. The COX10 gene maps to human chromosome 17p13 and encodes a heme A:farnesyltransferase involved in the assembly of the COX I prosthetic group. This gene has recently been shown to account for COX deficiency in a family with tubulopathy and encephalomyopathy (Valnot et al. 2000). On the other hand, SCO1 also maps to chromosome 17p13 and codes for a protein involved in mitochondrial copper import and/or insertion into COX. In yeast, mutations in either genes are known to result in the absence of functional heme in COX. Because an informative intragenic polymorphism allowed us to exclude COX10 (not shown), a search for mutations in the SCO1 gene was undertaken by directly sequencing the six exons of the gene. All affected individuals were compound heterozygotes for SCO1 mutations. A 2-bp deletion (ΔGA; nt 363–364), inherited from the father, resulted in both a frameshift and a premature stop codon in exon 2 (figs. 2A and 3B). The mutated mRNA was predicted to encode a truncated protein missing the functionally conserved putative core region of the enzyme and was highly unstable, since it could not be detected by reverse transcription–PCR amplification of the patient’s liver RNA (fig. 3A). A second allelic mutation, inherited from the mother, was found in exon 3 of the SCO1 gene (figs. 2B and 3C). This mutation, a C→T transition at nt 520, changed a highly conserved proline into a leucine in the protein (P174L; fig. 3D). This proline is also conserved in human and yeast SCO2, a mitochondrial protein that is highly homologous to SCO1 and that is also involved in copper insertion within COX. The P174L transition created a XbaI restriction site and was absent from 110 controls of the same ethnic origin. No SCO1, SCO2, or COX10 mutations were found in 18 patients with isolated COX deficiency and similar clinical presentation.
Figure 2 .
Sequence analysis of SCO1. A, Paternally inherited frameshift mutation in exon 2. B, Maternally inherited mutation in exon 3.
Figure 3 .
Molecular analysis of SCO1 mutations. A, Sequence analysis of cDNA of patient II.1. B, SSCP screening for the paternally inherited frameshift mutation in the family. C, Screening of the maternally inherited mutation (C520T) in exon 3 by XbaI restriction analysis. D, Sequence alignment of the SCO1 protein from patient II.1, from controls, and from nonhuman sources, as well as of human and yeast SCO2 proteins. F = father; M = mother; C = control; ND = nondigested control; MW = molecular weight.
Discussion
Combining conventional linkage analysis and the candidate-gene approach in a nonconsanguineous family with multiple cases of neonatal-onset hepatic failure and ketoacidotic comas, we have shown here that mutations in the COX-assembly gene SCO1 can cause severe isolated COX deficiency.
The SCO1 gene product is believed to transfer copper from Cox17p, a copper-binding protein of the cytosol and mitochondrial intermembrane space, to the mitochondrial COX subunit II. Mutational analyses of the yeast SCO1 gene have shown that the CxxxC copper-binding motif is essential for the protein function, since mutant proteins failed to restore the respiratory competence of the sco1 null mutant (Rentzsch et al. 1999). The human cDNA counterpart has been identified by sequence homology with the yeast SCO1 gene (Petruzella et al. 1998). The human and yeast proteins share a 40% identity, suggesting a similar function of the protein in the two organisms. The patients reported here were compound heterozygotes for a frameshift mutation resulting in both a premature stop codon and a proline-to-leucine missense mutation immediately adjacent to the CxxxC copper-binding motif, in a highly conserved domain of the SCO1 protein. Because prolines are known to bend proteins, this substitution in a highly conserved region may interfere with the function of SCO1, modifying the tertiary structure of the copper-binding domain. These data suggest that the missense mutation combined with the loss of function of the second SCO1 allele is responsible for the isolated COX deficiency in this family.
Interestingly, another SCO-like protein, SCO2, has been identified in both humans and yeast. SCO1 and SCO2 are highly homologous, since the proteins share a 40% identity, especially in the core region of the protein. Although the respective roles of the two proteins remain to be fully established, several lines of evidence suggest that both proteins are involved in mitochondrial copper trafficking from the intermembrane space to the inner mitochondrial membrane, copper being initially directed to the mitochondria by Cox17p (Glerum et al. 1996). The two SCO proteins are similarly associated with the inner mitochondrial membrane (Buchwald et al. 1991; Glerum et al. 1996; Paret et al. 1999), and overexpression of either SCO1 or SCO2 partially rescues a Cox17 null–mutant yeast strain, suggesting a partial functional redundancy of these two genes (Glerum et al. 1996). Finally, the SCO1 and SCO2 genes present a largely similar pattern of expression in human tissues (Papadopoulou et al. 1999).
Recently, mutations in the SCO2 gene have been reported in COX-deficient patients with cardioencephalomyopathy (Papadopoulou et al. 1999; Jaksch et al. 2000). Moreover, a recurrent mutation converted a glutamic acid into a lysine (E140K) in the region surrounding the CxxxC copper-binding domain of the SCO2 protein and was believed to displace copper from the copper-binding domain. This glutamic acid corresponds to amino acid residue 176 in the human SCO1 protein and is very close to the modified proline identified in our patients (residue 174).
The clinical consequences of the SCO1 mutations are markedly different from those caused by mutations in the functionally related SCO2 gene. Indeed, SCO1 mutations caused neonatal hepatic failure and ketoacidotic comas as the onset symptoms, whereas SCO2 mutations developed hypertrophic cardiomyopathy and hypotonia during the first weeks of life. Whether the clinical discrepancies between closely related gene mutations are significant, fortuitous, or related to the small number of reported cases is still debatable.
To date, mutations in four nuclear genes involved in maturation or assembly of COX have been identified in humans—namely, SURF1, COX10, SCO1, and SCO2. All four gene mutations caused severe isolated COX deficiency, with an abnormal pattern of COX subunits. Again, no correlation between mutant genotypes and clinical phenotypes could be made. Indeed, patients with SURF1 mutations presented with Leigh syndrome (Zhu et al. 1998; Tiranti et al. 1999), SCO2 mutations caused cardioencephalomyopathy (Papadopoulou et al. 1999; Jaksch et al. 2000), and COX10 mutations caused tubulopathy and encephalopathy, whereas SCO1 mutations caused neonatal hepatic failure and ketoacidotic comas. The onset of the disease was consistently very early, with no symptom-free period, suggesting an antenatal expression of the disease. Expression studies have shown that SURF1 (Yao and Shoubridge 1999), SCO1, and SCO2 (Papadopoulou et al. 1999) are ubiquitously expressed in human adult tissues, the higher expression being observed in mitochondria-rich tissues—namely, heart, skeletal muscle, liver, and kidney. However, brain shows a lower expression, compared with the highly aerobic tissues mentioned above. On the contrary, COX10 is highly expressed in skeletal muscle, heart, and testis but is less highly expressed in liver and kidney, whereas patients with COX10 presented with tubulopathy and encephalopathy (Murakami et al. 1997). The variations, in the steady-state levels of the four COX assembly–gene mRNAs, among adult tissues cannot be related to the clinical expression of the SURF1-, SCO1-, SCO2-, and COX10-deficient patients. Thus, one can hypothesize that the different clinical presentations are related to different patterns of expression of the genes during embryonic or fetal life. Studying the temporospatial expression pattern of these four genes, as well as other COX-assembly genes in human embryos, will help in the elucidation of both their respective requirement during development and the variable clinical presentations of the patients.
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www3.ncbi.nlm.nih.gov/Omim/ (for SURF1 mutations [MIM 185620 and MIM 256000], COX10 mutations [MIM 602125], SCO2 mutations [MIM 604272 and MIM 604377], SCO1 [MIM 603644]
References
- Bruno C, Martinuzzi A, Tang Y, Andreu AL, Pallotti F, Bonilla E, Shanske S, Fu J, Sue CM, Angelini C, DiMauro S, Manfredi G (1999) A stop-codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. Am J Hum Genet 65:611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald P, Krummeck G, Rodel G (1991) Immunological identification of yeast SCO1 protein as a component of the inner mitochondrial membrane. Mol Gen Genet 229:413–420 [DOI] [PubMed] [Google Scholar]
- Chretien D, Gallego J, Barrientos A, Casademont J, Cardellach F, Munnich A, Rötig A, Rustin P (1998) Biochemical parameters for the diagnosis of mitochondrial respiratory chain deficiency in humans, and their lack of age-related changes. Biochem J 329:249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KM, Taylor RW, Johnson MA, Chinnery PF, Chrzanowska-Lightowlers ZM, Andrews RM, Nelson IP, Wood NW, Lamont PJ, Hanna MG, Lightowlers RN, Turnbull DM (1999) An mtDNA mutation in the initiation codon of the cytochrome C oxidase subunit II gene results in lower levels of the protein and a mitochondrial encephalomyopathy. Am J Hum Genet 64:1330–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi GP, Bordoni A, Salani S, Franceschina L, Sciacco M, Prelle A, Fortunato F, Zeviani M, Napoli L, Bresolin N, Moggio M, Ausenda CD, Taanman JW, Scarlato G (1998) Cytochrome c oxidase subunit I microdeletion in a patient with motor neuron disease. Ann Neurol 43:110–116 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- DiMauro S, Bonilla E, De Vivo DC (1999) Does the patient have a mitochondrial encephalomyopathy? J Child Neurol Suppl 1:S23–S35 [DOI] [PubMed]
- Glerum DM, Shtanko A, Tzagoloff A (1996) SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J Biol Chem 271:20531–20535 [DOI] [PubMed] [Google Scholar]
- Hanna MG, Nelson IP, Rahman S, Lane RJ, Land J, Heales S, Cooper MJ, Schapira AH, Morgan-Hughes JA, Wood NW (1998) Cytochrome c oxidase deficiency associated with the first stop-codon point mutation in human mtDNA. Am J Hum Genet 63:29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffbuhr KC, Davidson E, Filiano BA, Davidson M, Kennaway NG, King MP (2000) A pathogenic 15-base pair deletion in mitochondrial DNA-encoded cytochrome c oxidase subunit III results in the absence of functional cytochrome c oxidase. J Biol Chem 275:13994–14003 [DOI] [PubMed] [Google Scholar]
- Jaksch M, Ogilvie I, Yao J, Kortenhaus G, Bresser HG, Gerbitz KD, Shoubridge EA (2000) Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum Mol Genet 9:795–801 [DOI] [PubMed] [Google Scholar]
- Keightley JA, Hoffbuhr KC, Burton MD, Salas VM, Johnston WS, Penn AM, Buist NR, Kennaway NG (1996) A microdeletion in cytochrome c oxidase (COX) subunit III associated with COX deficiency and recurrent myoglobinuria. Nat Genet 12:410–416 [DOI] [PubMed] [Google Scholar]
- Leonard JV, Schapira AH (2000a) Mitochondrial respiratory chain disorders. I. Mitochondrial DNA defects. Lancet 355:299–304 [DOI] [PubMed] [Google Scholar]
- ——— (2000b) Mitochondrial respiratory chain disorders. II. Neurodegenerative disorders and nuclear gene defects. Lancet 355:389–394 [DOI] [PubMed] [Google Scholar]
- Murakami T, Reiter LT, Lupski JR (1997) Genomic structure and expression of the human heme A:farnesyltransferase (COX10) gene. Genomics 42:161–164 [DOI] [PubMed] [Google Scholar]
- Papadopoulou LC, Sue CM, Davidson MM, Tanji K, Nishino I, Sadlock JE, Krishna S, Walker W, Selby J, Glerum DM, Coster RV, Lyon G, Scalais E, Lebel R, Kaplan P, Shanske S, De Vivo DC, Bonilla E, Hirano M, DiMauro S, Schon EA (1999) Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet 23:333–337 [DOI] [PubMed] [Google Scholar]
- Paret C, Ostermann K, Krause-Buchholz U, Rentzsch A, Rodel G (1999) Human members of the SCO1 gene family: complementation analysis in yeast and intracellular localization. FEBS Lett 447:65–70 [DOI] [PubMed] [Google Scholar]
- Petruzzella V, Tiranti V, Fernandez P, Ianna P, Carrozzo R, Zeviani M (1998) Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitrochondrial respiratory chain. Genomics 54:494–504 [DOI] [PubMed]
- Poyau A, Buchet K, Bouzidi MF, Zabot MT, Echenne B, Yao J, Shoubridge EA (2000) Missense mutations in SURF1 associated with deficient cytochrome c oxidase assembly in Leigh syndrome patients. Hum Genet 106:194–205 [DOI] [PubMed] [Google Scholar]
- Rahman S, Taanman JW, Cooper JM, Nelson I, Hargreaves I, Meunier B, Hanna MG (1999) A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am J Hum Genet 65:1030–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch A, Krummeck-Weiss G, Hofer A, Bartuschka A, Ostermann K, Rodel G (1999) Mitochondrial copper metabolism in yeast: mutational analysis of Sco1p involved in the biogenesis of cytochrome c oxidase. Curr Genet 35:103–108 [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Gerard B, Bourgeron T, Rötig A, Saudubray JM, Munnich A (1994) Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta 228:35–51 [DOI] [PubMed] [Google Scholar]
- Taanman JW (1997) Human cytochrome c oxidase: structure, function, and deficiency. J Bioenerg Biomembr 29:151–163 [DOI] [PubMed] [Google Scholar]
- Tiranti V, Jaksch M, Hofmann S, Galimberti C, Hoertnagel K, Lulli L, Freisinger P, Bindoff L, Gerbitz KD, Comi GP, Uziel G, Zeviani M, Meitinger T (1999) Loss-of-function mutations of SURF-1 are specifically associated with Leigh syndrome with cytochrome c oxidase deficiency. Ann Neurol 46:161–166 [DOI] [PubMed] [Google Scholar]
- Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rötig A (2000) A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum Mol Genet 9:1245–1249 [DOI] [PubMed] [Google Scholar]
- von Kleist-Retzow JC, Cormier-Daire V, de Lonlay P, Parfait B, Chretien D, Rustin P, Feingold J, Rötig A, Munnich A (1998) A high rate (20%–30%) of parental consanguinity in cytochrome-oxidase deficiency. Am J Hum Genet 63:428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kleist-Retzow JC, Vial E, Chantrel-Groussard K, Rötig A, Munnich A, Rustin P, Taanman JW (1999) Biochemical, genetic and immunoblot analyses of 17 patients with an isolated cytochrome c oxidase deficiency. Biochim Biophys Acta 1455:35–44 [DOI] [PubMed] [Google Scholar]
- von Kleist-Retzow JC, Yao J, Taanman JW, Chantrel-Groussard K, Chretien D, Rötig A. Mutations in SURF1 are not specifically associated with Leigh syndrome. J Med Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC (2000) Mitochondrial defects in cardiomyopathy and neuromuscular disease. Am Heart J 39:S70–S85 [DOI] [PubMed]
- Yao J, Shoubridge EA (1999) Expression and functional analysis of SURF1 in Leigh syndrome patients with cytochrome c oxidase deficiency. Hum Mol Genet 8:2541–2549 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 20:337–343 [DOI] [PubMed] [Google Scholar]