Abstract
Mucolipidosis type IV (MLIV) is an autosomal recessive lysosomal storage disorder characterized by severe psychomotor retardation and ophthalmologic abnormalities, including corneal opacity, retinal degeneration, and strabismus. Unlike the situation in other lysosomal disorders, the accumulation of heterogeneous storage material observed in MLIV does not result from a block in the catabolic pathways but is due to an ill-defined transport defect in the late steps of endocytosis. With the aim of cloning the MLIV gene, we searched in the 19p13.2-13.3 region, where the locus previously had been assigned by linkage mapping. In this region, we have identified a novel gene that is mutated in all patients with MLIV who were enrolled in our study. One patient was homozygous for the splice-acceptor mutation, and another was homozygous for a deletion removing the first six exons of the gene. In addition, four compound heterozygotes for these two mutations were identified. Haplotype analysis indicates that we have identified the two major founder mutations, which account for >95% of MLIV chromosomes in Ashkenazi Jewish patients. The gene, ML4, encodes a protein named “mucolipidin,” which localizes on the plasma membrane and, in the carboxy-terminal region, shows homologies to polycystin-2, the product of the polycystic kidney disease 2 gene (PKD2) and to the family of transient receptor potential Ca2+ channels. Mucolipidin is likely to play an important role in endocytosis.
Introduction
Mucolipidosis type IV (MLIV [MIM 252650]) is a progressive neurological disease that usually presents during the 1st year of life, with mental retardation, corneal opacities, and delayed motor milestones (Berman et al. 1974; Amir et al. 1987). Marked heterogeneity in the clinical symptoms has been observed, even among siblings. Patients from >70 families have been described; >90% of them are Ashkenazi Jewish (Raas-Rothschild et al. 1999). The name “mucolipidosis” derives from the presence of cytoplasmic inclusions of sphingolipids, phospholipids, and acid mucopolysaccharides called “storage bodies,” which are seen (by electron microscopy) in almost every cell type of the patients and are similar to those observed in individuals with mucopolysaccharide and lipid-storage disorders (Tellez-Nagel et al. 1976; Bargal and Bach 1988, 1997; Folkerth et al. 1995; Goldin et al. 1995). In contrast to what is seen in most other lipid-storage diseases in which lysosomal hydrolases or activator proteins are defective, extensive studies have shown normal activities of all lysosomal hydrolases participating in the catabolism of the stored macromolecules in MLIV cells, suggesting an alternative mechanism for storage in this cell type (Chen et al. 1998).
Further evidence that the catabolic pathways are not affected in MLIV comes from studies performed with labeled phosphatidylcholine, one of the major lipid classes that accumulate in MLIV (Bargal and Bach 1989), studies showing increased retention in the lysosomes of MLIV fibroblasts but normal degradation (Bargal and Bach 1997). In addition, a substantial alteration of the movement of various endocytic markers along the lysosomal pathway has been demonstrated in MLIV cells (Chen et al. 1998). Taken together, these findings indicate that the cellular phenotype observed in patients with MLIV is likely due to a defect in sorting and/or transport along the late endocytic pathway. This could explain both the absence of massive accumulation of material (an accumulation that is seen in other storage disorders) and the lack of progression of clinical manifestations of the disease during ⩾2–3 decades of life (Chen et al. 1998). The mapping of the MLIV locus to chromosome 19p13.2-13.3 has been defined, by linkage analysis, as a 5.62-cM interval between markers D19S1034 and D19S884 (Slaugenhaupt et al. 1999). Several markers within the linked interval display significant linkage disequilibrium with the disorder. Furthermore, haplotype analysis suggests that two independent gene mutations account for >95% of MLIV chromosomes in Ashkenazi Jewish patients (Slaugenhaupt et al. 1999).
Patients, Material, and Methods
Patients' Cell Lines
Fibroblast cell lines were obtained from the Repository for Mutant Human Cell Strains, Montreal Children’s Hospital (for patients WG0987, WG0988, and WG0909) and from the Coriell Cell Repositories, Camden, NJ (for patients GM02525, GM02526, GM02527, and GM02529). For patient GM02533A, we purchased genomic DNA only.
ML4 cDNA cloning
Expressed Sequence Tags database (Boguski et al. 1993) searches were performed as described elsewhere (Banfi et al. 1998). Selected cDNA clones (IMAGE clones 209402, 21977, and 2517653) were obtained from the UK Human Genome Mapping Project Resource Centre.
DNA Sequencing
Automated sequencing (by an Applied Biosystems ABI 377 fluorescent sequencer) was performed with both vector- and gene-specific oligonucleotide primers. The nucleotide sequence of the human ML4 cDNA has been submitted to GenBank, under accession number AJ293659.
Computer Sequence Analysis
BLAST searches using chromosome 19 markers as queries was performed against both the High Throughput Genomic Sequences and the Non-Redundant Sequences nucleotide databases of the National Center for Biotechnology Information. Human plasmid artificial chromosome (PAC) or human bacterial artificial chromosome (BAC) genomic sequences spanning the MLIV critical region were analyzed by the NIX—Identity Unknown Nucleic Sequence software available from the Human Genome Mapping Project Resource Centre (Borsani et al. 1998).
For sequence assembly and editing, we used both the AutoAssembler version 1.4 (PE Biosystems) and the DNA Strider 1.2 software programs (Marck 1988). Multiple sequence alignment (MSA) was performed by the Clustal W algorithm (Thompson et al. 1994). ML4 nucleotide and amino acid sequences were compared with the Non-Redundant Sequences databases of the National Center for Biotechnology Information, by BLAST version 2.0 (Altschul et al. 1997). Protein sequence analysis was achieved by the PIX—Identity Unknown Protein Sequence tool available from the Human Genome Mapping Project Resource Centre; in particular, the prediction of transmembrane domains was performed by the TMpred—Prediction of Transmembrane Regions and Orientation program. The search against a ProfileScan Server database was performed at ISREC-Bioinformatics.
Immunofluorescence
For immunofluorescence studies, COS7 cells were grown in 8-well slide culture chambers (Nunc) with Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. The cDNA region containing the entire ML4 open reading frame (ORF) was amplified by PCR using cloned Pfu polymerase (Stratagene) and 21977 IMAGE cDNA clone as template. The following sense and antisense primers containing suitable restriction sites were used to amplify the ML4 coding region: 129/BGL (5′-ATAGATCTATGACACCTTCGCAGCCTACAC-3′) and 129/ECO (5′-GGAATTCTCAATTCACCAGCAGCGAATG-3′), for cloning in pMHA vector, and MY129ECO (5′-AGGAATTCATGACACCTTCGCAGCCTACAC-3′) and MY129SAL (5′-AAGTCGACAATTCACCAGCAGCGAATG-3′), for cloning in pMT21 vector. 129-pMHA contains the amplified insert 5′ in-frame with the hemagglutinin (HA) epitope within plasmid pMHA, whereas, in the case of the 129-pMT21 construct, the insert was 3′ in-frame with the Myc epitope tag within the plasmid pMT21. Transfections were performed overnight, with increasing amounts (0.5–1.5 μg) of recombinant plasmid DNA for each well and with LipofectAMINE reagent, according to the manufacturer’s guidelines (Life Technologies). Transfection experiments using the same amount of vector DNA (pMT21 and pMHA) were performed as a control. COS7 cells were grown for an additional 48 h, were rinsed with PBS, and then were fixed immediately with 4% (w/v) PBS-buffered paraformaldehyde. Cells were subsequently permeabilized with 0.2% (v/v) Triton X-100 and were blocked with 10% (v/v) porcine serum and incubated, either with anti-HA monoclonal antibody (4 μg/ml) (Boehringer Mannheim) or with anti- Myc monoclonal antibody, in PBS plus 5% (v/v) porcine serum and 0.1% (v/v) Triton X-100. Staining was performed after incubation with fluorescein 5–isothiocyanated conjugated isotype-specific antibodies (1:150 dilution) (DAKO). Fluorescence microscopy was performed by an Axioplan microscope equipped with an MC 100 Spot Device camera (Zeiss).
Expression Studies
Northern blot of poly(A)+ RNA from various human adult tissues was purchased from Clontech and was hybridized by means of standard protocols (Sambrook et al. 1989). The cDNA insert of the human IMAGE clone 209402 (covering the 1,093–2,245 bp ML4 cDNA region) was used as a probe. Reverse transcriptase–PCR (RT-PCR) experiments were performed according to the manufacturer’s recommended conditions (Gibco BRL).
Mutation Analysis
Oligonucleotide primers for the amplification of all the ML4 exons were designed in the intronic flanking sequences and are available on request. For mutation analysis, the following primers were used: Δ6432 mutation, ZFP2F (5′-AAGCCCTGCATCCATGATG-3′) and 129 6R (5′-CACATGAACCCCACAAACTCC-3′; 486−2 A→G mutation at the genomic level, 1293F (5′-CGGACTCACAGGCCCTCC-3′) and 1293R (5′-CCAACAGTGAAGCCTCGTCC-3′); and 486−2 A→G mutation at the cDNA level, MC21B (5′-GAGCTGCTTTCCCAAAAGG-3′) and MC21E (5′-ACACCTTCGCAGCCTACAC-3′).
Haplotype Analysis
Fluorescence-labeled oligonucleotides for MLIV–critical region polymorphic markers were designed on the basis of sequences publicly available. PCR products were detected by an Applied Biosystems ABI 377 fluorescent sequencer and were analyzed by GENESCAN, version 2.1 (Applied Biosystems).
Results
Identification of the ML4 Gene—and Mutation Analysis in Patients with MLIV
To identify the gene responsible for MLIV, we analyzed the genomic sequences located within the MLIV critical region (Slaugenhaupt et al. 1999). Thirty-two chromosome 19 markers, including sequence-tagged sites and short tandem repeats, were retrieved from The Genetic Location Database and were used as queries in the BLAST sequence-homology search against nucleotide-sequence databases. Using a set of bioinformatic tools (see the Patients, Material, and Methods section), we selected and analyzed 30 of either PAC or BAC sequences, which led us to the identification of a number of putative novel genes.
One of the genes that we identified shares significant sequence identity with polycystin-2, an integral membrane protein encoded by the polycystic kidney disease 2 (PKD2) gene, which is mutated in one form of autosomal dominant polycystic kidney disease (Cai et al. 1999) (see below). On the basis of the data presented in this article, we have named the gene “ML4.” ML4 maps to PAC RP11-492L14 (GenBank accession number AC021153) and BAC CTD-2207023 (GenBank accession number AC008878). These genomic clones contain the sequences of the “core markers” D19S873, D19S905, and D19S901, which are included within the 1.2-cM region defined by linkage disequilibrium (Slaugenhaupt et al. 1999). ML4 is flanked by the gene for FLJ10390 (GenBank accession number AK001252, which is similar to the human zinc-finger protein 135 gene), and by the gene for neuropathy target esterase (NTE [GenBank accession number AJ004832]).
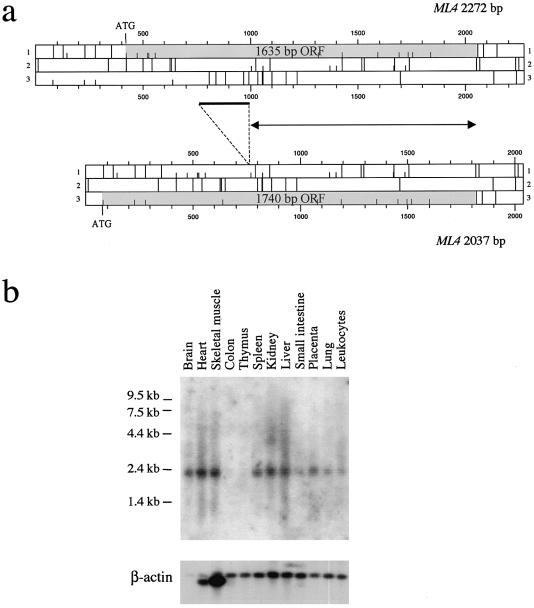
A cDNA contig of 2,273 bp (GenBank accession number AJ293659) was assembled by full-insert sequence analysis of IMAGE clones 209402, 21977, and 2517653, corresponding to expressed-sequence tags (ESTs) of the transcript. This transcript contains an ORF of 1,635 bp encoding a protein of 545 amino acids (fig. 1a). The codon for the translation-initiator methionine (Met-1), which does not fulfill the Kozak consensus sequence (Kozak 1984), is preceded by several in-frame stop codons. Sequence comparison of clones 21977 and 2517653, both from libraries of brain cDNA, indicates that clone 2517653 represents a shorter (2,037 bp), alternatively spliced transcript missing a portion (235 bp) of exon 5 (for ML4 gene structure, see below) (fig. 1a). A corresponding alternatively spliced form has been observed in ESTs from other species (data not shown). Since the size of the spliced region is not an exact multiple of 3, this alternative ML4 transcript presents a longer ORF (fig. 1a), with the first Met-1 (which only marginally fulfills the Kozak consensus sequence) located in exon 1 on a different frame.
Figure 1.
a, ORF map of ML4 cDNA. Short vertical bars denote possible translation start sites; long bars denote stop codons. The ML4 coding region is gray shaded. The 2,272-bp ML4 transcript contains an ORF of 1,635 bp encoding a protein of 545 amino acids. A 2,037-bp alternative spliced form, missing a 235-bp region that is denoted by the thick bar, also is shown. Since te size of the spliced region is not an exact multiple of 3, this alternative ML4 transcript presents a longer ORF (1,740 bp, encoding a 580-amino-acid protein), with the first Met located at nucleotide position 81. The proteins encoded by the two forms differ in their NH2 portions, whereas they are identical for the last 345 residues, because of the usage of the same reading frame in the two transcripts (the region denoted by the double-arrowheaded line) downstream of the splicing event. b, Northern blot analysis of the ML4 gene. A cDNA probe spanning the entire coding sequence of ML4 was hybridized to commercially available northern blot containing 2 μg of poly(A)+ RNA from various human adult tissues. A band of ∼2.2 kb, consistent with the size of the cDNA contig, is detected in all tissues tested, with the exception of colon and thymus. Hybridization to a human β-actin cDNA revealed almost equal amounts of poly(A)+ RNA in each lane.
The predicted protein product is 580 amino acids long (GenBank accession number AJ293970) and, because of the usage of the same reading frame in the two forms downstream of the splicing event, is identical to the 545-amino-acid form in the last 353 residues (fig. 1a). RT-PCR analysis on RNA from several human tissues suggests that the longer transcript, corresponding to clone 21977, is clearly the most abundant one (data not shown).
Comparison of the ML4 cDNA sequence with the sequences of genomic clones RP11-492L14 and CTD-2207O23 revealed that the gene is organized into 13 exons spanning 11.3 kb. All the ML4 exon-intron junctions agree with the GT-AG consensus sequence.
Northern blot analysis of poly(A)+ RNA, using a ML4 cDNA probe from several human adult tissues, identified a single 2.2-kb band present in all tissues tested, with the exception of colon tissue and thymus tissue (fig. 1b). The ML4 transcript can also be detected in fibroblast cell lines, by RT-PCR (see below). These data support an almost ubiquitous expression of the gene.
ML4 mutation analysis was performed in six patients with MLIV who are from five different families. Fibroblast cell lines and genomic DNAs were obtained from cell repositories (see the Patients, Material, and Methods section). Biological samples from parents were available for one patient only (WG909; see below).
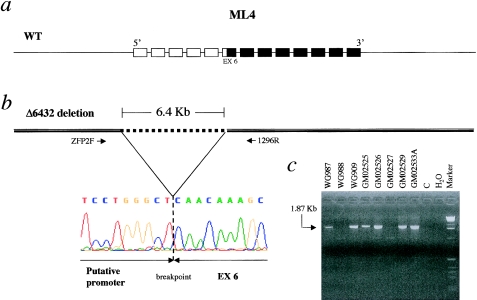
Patient GM02525 is homozygous for a deletion removing 6,432 bp (Δ6432), spanning a region from the 5′ end of the gene to exon 6 (fig. 2). The absence of ML4 transcript from this patient’s fibroblasts was detected by RT-PCR using primers designed from the sequence of exons 7–13 (data not shown). This result suggests that the deletion encompassing 938 bp upstream of the cDNA sequence removes ML4 transcription regulatory elements.
Figure 2.
Mutation Δ6432 in patients with MLIV. a, ML4 gene, which is organized into 13 exons. The unblackened boxes denote the ML4 exons removed by the Δ6432 deletion; the blackened boxes denote the exons that are retained. b, Δ6432 Interstitial deletion removing 6,432 bp of genomic DNA, denoted by the dotted line. The primer pair (i.e., ZFP2F oligonucleotide, designed on the basis of the sequence 2.5 kb upstream of the first base of the ML4 cDNA, and 1296R, designed on the basis of intron 7; see the Patients, Material, and Methods section) used to amplify the breakpoint is also shown. The electropherogram at the bottom left corresponds to the nucleotide sequence across the breakpoint as determined in patient GM02525. c, Segregation of the 1.87-kb junction fragment in patients WG909, GM02525, GM02526, GM02529, and GM02533A (see text) and in the carrier father, WG987. The wild-type allele could not be amplified by PCR, because of its large size (8.3 kb), as shown in control genomic DNA (lane C).
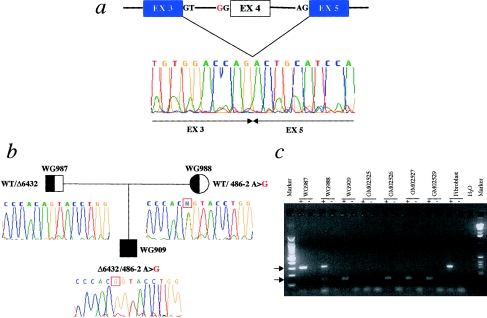
Patient GM02527 is either homozygous or hemizygous for an A→G transition at position −2 of the AG acceptor site of the intron flanking exon 4 (mutation 486−2 A→G), which disrupts the GT-AG rule of splicing. Consequently, the resulting transcript lacks 165 bp, because of the skipping of exon 4 (fig. 3a). This causes a frameshift leading to a premature translation termination 374 bp downstream. The predicted truncated protein retains only the first 21 amino acids of the wild-type protein. All the other patients, including WG909 GM02526, GM02529, and GM02533A (who is affected brother of GM02529), were found to be compound heterozygotes for the aforementioned Δ6432 and 486−2 A→G mutations (figs. 2c and 3c). As shown in figure 3b, the 486−2 A→G mutation segregates with the disease in the family of patient WG909.
Figure 3.
Mutation 486-2A→G in patients with MLIV. a, Lack of 165 bp, corresponding to exon 4 skipping, observed, at the cDNA level, by RT-PCR. The electropherogram shows the cDNA sequence across the deletion. The mutated allele encodes a truncated protein that retains only the first 21 amino acids of the wild-type allele. The region of homology to the PKD2 gene family is deleted in this mutant. b, 486-2A→G mutation, which segregates with the disease in the family of patient WG909. The electropherograms indicate the genomic sequences surrounding the 486-2A→G mutation. The mother is a carrier of the mutation, showing heterozygosity at the 486-2 position, whereas the father, who is a carrier of the Δ6432 allele, is hemizygous for a portion of the ML4 gene (see fig. 2b and text). The WG909 proband is a compound heterozygote for the 486-2A→G and Δ6432 mutations. c, RT-PCR of RNA from fibroblast cell lines, performed with primers located on exons 3 (MC21E) and 5 (MC21B) (see the Patients, Material, and Methods section). An altered size fragment, of 142 bp (lower arrow), is amplified in patients WG909, GM02526, GM02527, and GM02529 and in the carrier mother, WG988. The wild-type fragment, of 307 bp (upper arrow), is present only in the unaffected parents—WG987 and WG988—and in the fibroblast control cDNA sample. A plus sign (+) and a minus sign (−) denote, respectively, the presence and absence of RT in the RT-PCR. H2O = the negative control. No amplification product is observed in patient GM02525, who is homozygous for the Δ6432 deletion, which removes this portion of the gene. The cDNA of patient GM02533A was not available.
To test whether these two mutations represent the “major” and “minor” founder mutations predicted by haplotype studies of families with MLIV (Slaugenhaupt et al. 1999), we performed haplotype analysis on our patients, with four MLIV-CR polymorphic markers (table 1). Of these markers, D19S873 and D19S905 represent two of the core markers described by Slaugenhaupt et al. (1999). The allele phases could be unequivocally established only in patient WG909 (because of the availability of the parents) and in GM02525 (because of the homozygosity for all the markers). In all other patients who are compound heterozygotes, allele phases were arbitrarily inferred to fit with the two most common haplotypes. The major mutation in Ashkenazi Jewish patients with MLIV clearly appears to be 486−2 A→G, since it is associated with the founder allele marked by the 283-120-216-230 haplotype corresponding to markers D19S869, D19S873, D19S905, and D19S884 (Slaugenhaupt et al. 1999) (table 1). The Δ6432 mutation probably represents the minor founder allele, since it is associated with a less frequent haplotype (275-126-234-230) observed in the same population (table 1). Therefore, the two mutations that we identified are likely to account for 96% of the MLIV chromosomes in the Ashkenazi Jewish population (Slaugenhaupt et al. 1999). In patients WG909, GM02526, and GM02527, alleles for the distal markers (indicated by footnote “c” in table 1) differ from those of the major and minor haplotypes. Slaugenhaupt et al. (1999) have reported similar discrepancies, which are probably due to ancestral recombination events, in patients with MLIV.
Table 1.
Mutation and Haplotype Analysis in Patients with MLIV
|
Haplotypeb(Core Markers) |
|||||
| Genotype | D19S869 | D19S873 | D19S905 | D19S884 | |
| Individual (status):a | |||||
| WG987 (carrier) | WT | 275 | 116 | 216 | 238 |
| Δ6432 | 275 | 126 | 234 | 222c | |
| WG988 (carrier) | WT | 275 | 120 | 216 | 232 |
| 486−2 A→G | 283 | 120 | 216 | 230 | |
| WG909 (affected) | Δ6432 | 275 | 126 | 234 | 222c |
| 486−2 A→G | 283 | 120 | 216 | 230 | |
| GM02525 (affected) | Δ6432 | 275 | 126 | 234 | 230 |
| Δ6432 | 275 | 126 | 234 | 230 | |
| GM02526 (affected) | Δ6432 | 275 | 126 | 234 | 230 |
| 486−2 A→G | 285c | 120 | 216 | 230 | |
| GM02527 (affected) | 486−2 A→G | 275c–283 | 120 | 216 | 230–236c |
| 486−2 A→G | 120 | 216 | |||
| GM02529 (affected) | Δ6432 | 275 | 126 | 234 | 230 |
| 486−2 A→G | 283 | 120 | 216 | 230 | |
| GM02533A (affected) | Δ6432 | 275 | 126 | 234 | 230 |
| 486−2 A→G | 283 | 120 | 216 | 230 | |
| Haplotype: | |||||
| Major | 486−2 A→G | 283 | 120 | 216 | 230 |
| Minor | Δ6432 | 275 | 126 | 234 | 230 |
WG987 and WG988 are the unaffected parents of patient WG909; GM02529 and GM02533A are brothers.
The major and minor haplotypes are as reported by Slaugenhaupt (1999).
Allele(s) for distal markers that differ from the major and minor haplotypes.
Sequence Analysis and Cellular Localization of the ML4 Gene Product
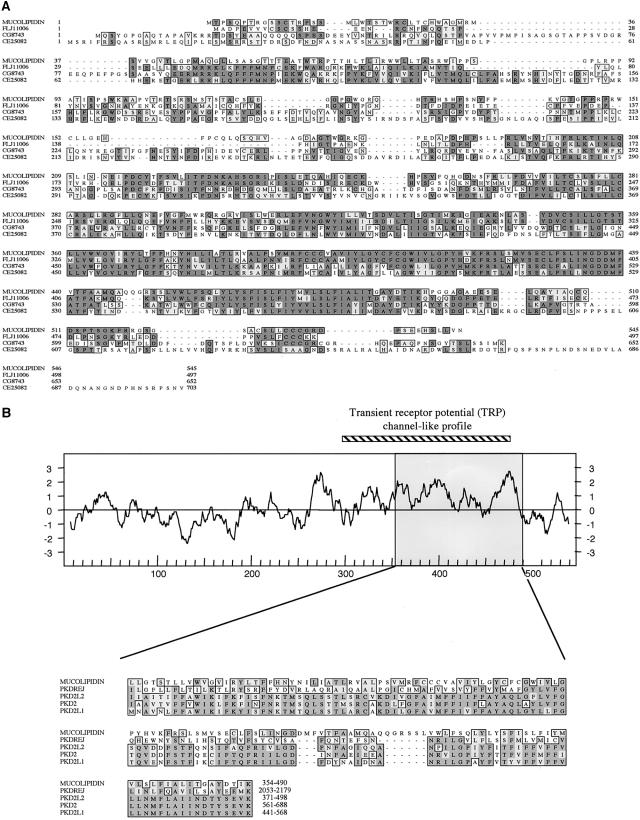
To understand the biochemical function of the ML4 gene product, we performed a sequence database search, using the predicted protein sequence as a query. Significant homologies with three uncharacterized proteins were identified, which were found to spread along most of the length of the ML4 gene. The highest homology was detected with the human protein FLJ11006 (61% identity; 77% similarity), followed by the CG8743 protein from Drosophila melanogaster (44% identity; 64% similarity) and the CE25082 Caenorhabditis elegans polypeptide (40% identity; 63% similarity) (fig. 4a).
Figure 4.
MSA using the Clustal W algorithm and hydropathy plot of the mucolipidin protein. a, MSA between mucolipidin and related amino acid sequences found in protein databases. Mucolipidin = human mucolipidin polypeptide (545-amino-acid form); FLJ11006 = hypothetical protein FLJ11006 (GenBank accession number BAA91951) predicted from a cDNA clone identified by a large-scale cDNA sequencing approach; CG8743 (GenBank accession number AE003516) and CE25082 (GenBank accession number not available) = predicted proteins from genomic sequences of Drosophila melanogaster and Caenorhabditis elegans, respectively. Identical residues (shaded) and conservatively substituted residues (unshaded) are boxed. The gaps inserted by the Clustal W program are denoted by broken lines. b, Top, Hydropathy plot of the mucolipidin protein (545-amino-acid form). The amino acid position is plotted as a function of the average hydrophobicity of an 11-amino-acid peptide (Kyte and Doolittle 1982). The region corresponding to the TRPC-like profile is shown above the plot, as a thick diagonally striped line. Bottom, MSA between a 136-amino-acid sequence of mucolipidin (amino acids 354–490, also indicated as a shaded box in the hydropathy plot) and a region of human proteins belonging to the polycystic kidney disease 2 family. PKDREJ = polycystic kidney disease and receptor for egg jelly (sea urchin homologue)–like protein (GenBank accession number NP_006062); PKD2L2 = polycystic kidney disease 2–like 2 (GenBank accession number AAF65622); PKD2 = polycystic kidney disease 2 protein (GenBank accession number NP_032887); PKD2L1 = polycystic kidney disease 2–like 1 protein (GenBank accession number NP_057196). The first and last amino acids of each sequence, numbered according to their database entries, are at the end of each sequence.
In addition to the homology with these proteins of unknown function, the region spanning amino acids 354–490 of the ML4-encoded protein shows a lower but significant identity with proteins belonging to PKD2 (fig. 4b [fig. 4b). Interestingly, a search against a ProfileScan Server database revealed a significant match for a transient receptor potential channel (TRPC)–like pattern seen in a family of putative Ca2+ channels (Zhu et al. 1996; Harteneck et al. 2000). The match is restricted to the region spanning amino acids 296–486, which is highly significant in channel function, since it includes the region believed to constitute the ion pore (Zhu et al. 1996).
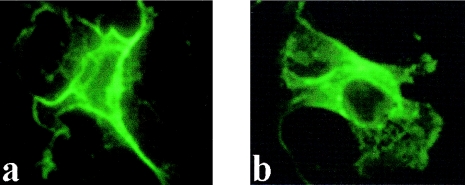
The TMpred membrane-spanning–prediction program (TMpred—Prediction of Transmembrane Regions and Orientation) revealed the presence of six putative transmembrane domains in the C-terminal portion of the ML4-encoded protein (fig. 4b fig. 4b). Of these, the fifth domain, showing a lower score, is thought not to span the membrane completely in TRPC proteins (Birnbaumer et al. 1996). These data suggest that ML4 encodes an integral membrane protein. To provide experimental evidence for this prediction, we performed immunohistochemical analysis of COS7 cells transfected with recombinant ML4 cDNA (encoding for the 545-amino-acid form) containing two different protein tags (HA and Myc) adjacent to the ML4 sequence. The results show a typical plasma-membrane localization (fig. 5), whereas cells transfected with only the vector DNAs do not show any staining (data not shown).
Figure 5.
Immunofluorescence of ML4 encoded protein. COS7 cells were transfected with either 129-pMHA (a) or 129-pMT21 (b) constructs and were grown 48 h prior to fixation and immunofluorescence staining. Cells were fixed with paraformaldehyde and were permeabilized with Triton X-100 and then were treated with either anti-HA (A) or anti-Myc (B) monoclonal antibodies. Staining was performed with fluorescein 5–isothiocyanated isotype-specific antibodies.
Discussion
The severity of the mutations found, as well as their concordance with both disease status and haplotype data, clearly indicate that we have identified the MLIV gene and the two major mutations causing this disorder. Therefore, MLIV is caused by a loss of function of the ML4 gene product, which we therefore have named “mucolipidin.” We have demonstrated that mucolipidin is an integral membrane protein ubiquitously found in almost all tissues tested. Hypotheses about the biological role of mucolipidin may emerge not only from studies of the cellular phenotype observed in MLIV but also from the similarities detected between mucolipidin and other known proteins. Studies performed in MLIV cells strongly suggest that mucolipidin plays a role in transport and/or sorting in the late endocytic pathway. In this context, the presence, within mucolipidin, of a protein profile described in molecules involved in Ca2+ transport (i.e., TRPC [Putney and McKay 1999]) is intriguing. Intracellular Ca2+ levels are of crucial importance in a variety of biological processes, some of which appear to be altered in MLIV; for example, intraorganellar Ca2+ is required for the fusion of late endosomes and lysosomes, the reformation of lysosomes, and the dynamic equilibrium of organelles in the late endocytic pathway (Pryor et al. 2000). In addition, localized Ca2+ levels determine the spatial and temporal targeting of protein kinase C alpha (PKC alpha) inside the cell (Maasch et al. 2000). Interestingly, phorbol myristate acetate–mediated translocation of PKC from the cytosolic to the membranous compartment is perturbed in MLIV fibroblasts (Turgeman and Boneh 1996).
Mucolipidin also shares significant similarities with polycystin-2, the product of the PKD2 gene involved in autosomal dominant polycystic kidney disease (ADPKD [MIM 173910]). Polycystin-2 is also predicted to be an integral membrane protein, with six membrane-spanning domains and intracellular NH2 and COOH termini. Similar to what is seen in mucolipidin, a TRPC domain is present also in polycystin-2. An analysis of membrane transport revealed that basolateral trafficking of proteins and lipids is impaired in polarized ADPKD cells (Charron et al. 2000).
Together, the presence of a TRPC-like domain, the homology with PKD2-like proteins, and the localization to the plasma membrane suggest that mucolipidin mediates membrane trafficking of proteins and lipids. The possibility that mucolipidin participates in intracellular calcium-level regulation, by acting as an ion channel, needs to be further explored by detailed studies. With the cloning of the ML4 gene encoding mucolipidin, functional studies performed in both wild-type and MLIV cells are now feasible. These will not only advance our knowledge of the pathogenesis of this severe disease but also will help to elucidate a fundamental process in cell biology, such as endocytosis.
Acknowledgments
We thank R. Tonlorenzi, L. Perone, and M. Riboni for technical assistance; M. De Fusco for haplotype analyses; G. Meroni and V. Marigo for helpful suggestions; and H. Roth for preparation of the manuscript. This work was supported by EURO-IMAGE grant BIOMED2 CT97-BMH4-2284 EC to A.B.
Note added in proof.—
After the manuscript of this article was submitted for publication, an article (Bargal et al. 2000) was published describing both the cloning of a gene identical to ML4 and the identification of mutations in MLIV.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST
- Expressed Sequence Tags database, http://www.ncbi.nlm.nih.gov/dbEST/index.html
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for BAC CTD-2207O23 [accession number AC008878], C. elegans polypeptide CE25082 [accession number not available], CG8743 [accession number AE003516], FLJ10390 [accession number AK001252], human FLJ11006 protein [accession number BAA91951], human ML4 cDNA nucleotide sequence [accession number AJ293659], NTE [accession number AJ004832], PAC RP11-492L14 [accession number AC021153], PKD2 [accession number NP_032887], PKD2L1 [accession number NP_057196], PKD2L2 [accession number AAF65622], PKDREJ [accession number NP_006062], and 2,273-bp DNA contig [accession numbers AJ293659 and AJ293970])
- Genetic Location Database, The, http://cedar.genetics.soton.ac.uk/public_html/ldb.html
- NIX—Identity Unknown Nucleic Sequence, http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MLIV [MIM 252650] and ADPKD [MIM 173910])
- PIX—Identity Unknown Protein Sequence, http://www.hgmp.mrc.ac.uk/Registered/Webapp/pix/
- ProfileScan Server, http://www.isrec.isb-sib.ch/software/PFSCAN_form.html
- TMpred—Prediction of Transmembrane Regions and Orientation, http://www.ch.embnet.org/software/TMPRED_form.html
- UK Human Genome Mapping Project Resource Centre, http://www.hgmp.mrc.ac.uk (for cDNA clones [IMAGE clones 209402, 21977, and 2517653])
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir N, Zlotogora J, Bach G (1987) Mucolipidosis type IV: clinical spectrum and natural history. Pediatrics 79:953–959 [PubMed] [Google Scholar]
- Banfi S, Guffanti A, Borsani G (1998) How to get the best of dbEST. Trends Genet 14:80–81 [DOI] [PubMed] [Google Scholar]
- Bargal R, Avidan N, Ben-Asher E, Olender Z, Zeigler M, Frumkin A, Raas-Rothschild, Glusman G, Lancet D, Bach G (2000) Identification of the gene causing mucolipidosis type IV. Nat Genet 26:118–123 [DOI] [PubMed] [Google Scholar]
- Bargal R, Bach G (1988) Phospholipids accumulation in mucolipidosis IV cultured fibroblasts. J Inherit Metab Dis 11:144–150 [DOI] [PubMed] [Google Scholar]
- ——— Bargal R, Bach G (1989) Phosphatidylcholine storage in mucolipidosis IV. Clin Chim Acta 181:167–174 [DOI] [PubMed] [Google Scholar]
- ——— (1997) Mucolipidosis type IV: abnormal transport of lipids to lysosomes. J Inherit Metab Dis 20:625–632 [DOI] [PubMed] [Google Scholar]
- Berman ER, Livni N, Shapira E, Merin S, Levij IS (1974) Congenital corneal clouding with abnormal systemic storage bodies: a new variant of mucolipidosis. J Pediatr 84:519–526 [DOI] [PubMed] [Google Scholar]
- Birnbaumer L, Zhu X, Jiang M, Boulay G, Peyton M, Vannier B, Brown D, Platano D, Sadeghi H, Stefani E, Birnbaumer M (1996) On the molecular basis and regulation of cellular capacitative calcium entry: roles for Trp proteins. Proc Natl Acad Sci USA 93:15195–15202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Lowe TMJ, Tolstoshev CM (1993) dbEST—database for “expressed sequence tags”. Nat Genet 4:332–333 [DOI] [PubMed] [Google Scholar]
- Borsani G, Ballabio A, Banfi S (1998) A practical guide to orient yourself in the labyrinth of genome databases. Hum Mol Genet 7:1641–1648 [DOI] [PubMed] [Google Scholar]
- Cai Y, Maeda Y, Cedzich A, Torres VE, Wu G, Hayashi T, Mochizuki T, Park JH, Witzgal R, Somlo S (1999) Identification and characterization of polycystin-2, the PKD2 gene product. J Biol Chem 274:28557–28565 [DOI] [PubMed] [Google Scholar]
- Charron AJ, Nakamura S, Bacallao R, Wandinger-Ness A (2000) Compromised cytoarchitecture and polarized trafficking in autosomal dominant polycystic kidney disease cells. J Cell Biol 149:111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Bach G, Pagano RE (1998) Abnormal transport along the lysosomal pathway in mucolipidosis, type IV disease. Proc Natl Acad Sci USA 95:6373–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkerth RD, Alroy J, Lomakina I, Skutelsky E, Raghavan SS, Kolodny EH (1995) Mucolipidosis IV: morphology and histochemistry of an autopsy case. J Neuropathol Exp Neurol 54:154–164 [PubMed] [Google Scholar]
- Goldin E, Blanchette-Mackie EJ, Dwyer NK, Pentchev PG, Brady RO (1995) Cultured skin fibroblasts derived from patients with mucolipidosis 4 are auto-fluorescent. Pediatr Res 37:687–692 [DOI] [PubMed] [Google Scholar]
- Harteneck C, Plant TD, Schultz G (2000) From worm to man: three subfamilies of TRP channels. Trends Neurosci 23:159–166 [DOI] [PubMed] [Google Scholar]
- Kozak M (1984) Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res 12:857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte JP, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132 [DOI] [PubMed] [Google Scholar]
- Maasch C, Wagner S, Lindschau C, Alexander G, Buchner K, Gollasch M, Luft FC, Haller H (2000) Protein kinase calpha targeting is regulated by temporal and spatial changes in intracellular free calcium concentration [Ca(2+)](i). FASEB J 14:1653–1663 [DOI] [PubMed] [Google Scholar]
- Marck C (1988) ‘DNA Strider': a ‘C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res 16:1829–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor PR, Mullock BM, Bright NA, Gray SR, Luzio JP (2000) The role of intraorganellar Ca(2+) in late endosome-lysosome heterotypic fusion and in the reformation of lysosomes from hybrid organelles. J Cell Biol 149:1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney JW Jr, McKay RR (1999) Capacitative calcium entry channels. Bioessays 21:38–46 [DOI] [PubMed] [Google Scholar]
- Raas-Rothschild A, Bargal R, DellaPergola S, Zeigler M, Bach G (1999) Mucolipidosis type IV: the origin of the disease in the Ashkenazi Jewish population. Eur J Hum Genet 7:496–498 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Slaugenhaupt SA, Acierno JS Jr, Helbling LA, Bove C, Goldin E, Bach G, Schiffmann R, Gusella JF (1999) Mapping of the mucolipidosis type IV gene to chromosome 19p and definition of founder haplotypes. Am J Hum Genet 65:773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez-Nagel I, Rapin I, Iwamoto T, Johnson AB, Norton WT, Nitowsky H (1976) Mucolipidosis IV: clinical, ultrastructural, histochemical, and chemical studies of a case, including a brain biopsy. Arch Neurol 33:828–835 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeman D, Boneh A (1996) Protein kinase C activation and phosphate uptake are altered in intact mucolipidosis type-4 skin fibroblasts. Biochem Mol Med 59:33–37 [DOI] [PubMed] [Google Scholar]
- Zhu X, Jiang M, Peyton M, Boulay G, Hurst R, Stefani E, Birnbaumer L (1996) trp, a novel mammalian gene family essential for agonist-activated capacitative Ca2+ entry. Cell 85:661–671 [DOI] [PubMed] [Google Scholar]