Abstract
The authors had previously mapped a new locus—DFNA17, for nonsyndromic hereditary hearing impairment—to chromosome 22q12.2-q13.3. DFNA17 spans a 17- to 23-cM region, and MYH9, a nonmuscle–myosin heavy-chain gene, is located within the linked region. Because of the importance of myosins in hearing, MYH9 was tested as a candidate gene for DFNA17. Expression of MYH9 in the rat cochlea was confirmed using reverse transcriptase–PCR and immunohistochemistry. MYH9 was immunolocalized in the organ of Corti, the subcentral region of the spiral ligament, and the Reissner membrane. Sequence analysis of MYH9 in a family with DFNA17 identified, at nucleotide 2114, a G→A transposition that cosegregated with the inherited autosomal dominant hearing impairment. This missense mutation changes codon 705 from an invariant arginine (R) to histidine (H), R705H, within a highly conserved SH1 linker region. Previous studies have shown that modification of amino acid residues within the SH1 helix causes dysfunction of the ATPase activity of the motor domain in myosin II. Both the precise role of MYH9 in the cochlea and the mechanism by which the R705H mutation leads to the DFNA17 phenotype (progressive hearing impairment and cochleosaccular degeneration) remain to be elucidated.
Introduction
We had previously mapped a new locus for nonsyndromic hereditary hearing impairment (HHI), DFNA17 (MIM 603622), to chromosome 22q12.2-q13.3. DFNA17 maps to a relatively large genetic region of 17–23 cM, typical for the size of the family studied (Lalwani et al. 1999). The low resolution of the linked region precluded the use of the positional-cloning approach to identify the disease gene. The candidate-gene approach, the default alternative to positional cloning, was therefore pursued (Collins 1995). Analysis of the chromosome 22 gene map identified 163 candidate genes within the region spanning the DFNA17 locus (Dunham et al. 1999). One of the candidate genes encoding the nonmuscle–myosin heavy-chain A (MYH9) was selected for further analysis. The selection of MYH9 (which encodes a member of the class II, or conventional, myosins) was based on identification of mutations in several other myosin heavy-chain genes that are pathogenically linked to HHI (Friedman et al. 1999).
The class II myosins are broadly expressed in skeletal, cardiac, and smooth muscles, as well as in nonmuscle tissues, and they consist of a pair of heavy chains, a pair of light chains, and a pair of regulatory light chains (Sellers 2000). The N-terminal motor domain is the most highly conserved region of the myosin heavy chain and contains the ATP- and actin-binding sites. The myosin, also known as the “sarcomeric myosin,” that mediates skeletal muscle contraction represents the most well-characterized representative of the class II myosin family. Cardiac muscle cells and smooth muscle cells also possess isoforms of class II myosin that are distinct from the sarcomeric myosin and that mediate contraction in these muscle cells.
Characterization of myosins in the nonmuscle cells has identified two distinct isoforms of class II myosin: nonmuscle myosin–IIA (MYH9) (Saez et al. 1990; Simons et al. 1991) and nonmuscle myosin–IIB (MYH10) (Simons et al. 1991). MYH9 and MYH10 have been mapped to 22q11.2 and 17q13, respectively, and exhibit 85% identity in their motor domains. Several skeletal muscle heavy-chain genes have also been localized to the region containing MYH10. Most cells express relatively equal amounts of each of these myosins, with a few exceptions. For example, platelets express MYH9 only (Maupin et al. 1994; Phillips et al. 1995), whereas neuronal tissues predominantly express MYH10. MYH9 and MYH10 demonstrate overlapping but distinct intracellular locations when coexpressed within the same cell type (Kolega 1998a, 1998b). In vitro motility studies of these two isoforms have shown that MYH9 is severalfold faster than MYH10, both in its rate of ATP hydrolysis and in its rate of movement of actin filaments (Kelley et al. 1996). The differences in their localization and in their in vitro characteristics suggest differing in vivo functions.
Our understanding of the biological role of nonmuscle myosin is derived in part from studies in invertebrate animals and by analogy with skeletal and smooth muscle myosin II. Use of invertebrate model systems has shown that nonmuscle myosin II is directly involved in diverse cellular functions, including the mediation of cytokinesis in Dictyostelium species (De Lozanne and Spudich 1987), the morphogenesis during the development of Drosophila species (Edwards and Kiehart 1996), and the establishment of cellular polarity in Caenorhabditis elegans embryos (Guo and Kemphues 1996). In vertebrate animals, the biological role of MYH10 has been investigated through the generation of knockout mice (Tullio et al. 1997). The MYH10−/− fetuses die in utero or perinatally because of congestive heart failure, demonstrating that MYH10 is required for normal cardiac development. The biological role of MYH9 has not yet been investigated or described in vertebrates.
In the present study, we have identified an MYH9 mutation that is linked to nonsyndromic deafness and is known as “DFNA17.” The mutation, R705H, the consequence of a G→A transposition in the nucleotide sequence, is considered critical for the ATPase activity of the myosin motor domain. The cosegregation of the mutant MYH9 with nonsyndromic hearing impairment demonstrates a biologically significant role for MYH9 in hearing and an organ-specific pathology associated with the mutant allele.
Family, Material, and Methods
Family Data
The family has been described in detail elsewhere (Lalwani et al. 1997, 1999). In brief, affected family members exhibit nonsyndromic HHI with an autosomal dominant mode of transmission. The hearing impairment begins at age 10 years and initially involves only the high frequencies; by the third decade of life, affected family members have moderate-to-severe deafness. Histological examination of the proband’s temporal bone revealed classical cochleosaccular dysplasia (CSD) with degeneration of the organ of Corti, the saccular epithelium, and the stria vascularis. In addition, the proband had asymptomatic loss of neurons, and gliosis was present in the inferior olivary nucleus. The deafness in the family was mapped, by linkage analysis, to 22q12.2-22q13.3, a region spanning 17–23 cM.
Reverse Transcriptase–PCR Assay of MYH9 Expression in the Rat Cochlea
One microgram of total RNA from rat kidney, lung, and cochlea was extracted and then reverse transcribed into cDNA by use of MMLV (Moloney murine leukemia virus) reverse transcriptase (RT) (Life Technologies) and random hexamers. The reaction was heat inactivated, and 1 μl of a total volume of 20 μl was used in the PCR to assay, by rat MYH9–specific primers, for the presence of MYH9 expression. The primers were designed using Primer3 software and the rat MYH9 sequence (GenBank accession number GI 6981235). PCR was carried out in a PTC-200 thermocycler (MJ Research) in a total volume of 20 μl with 1.5 mM MgCl2, 200 mM dNTP, 5 pmol each of forward and reverse primers, and a unit of Taq DNA polymerase. An initial denaturing period of 3 min at 94°C was followed by 35 cycles of 15 s at 94°C, 20 s at 55°C, and 30 s at 72°C and by a final extension period of 5 min at 72°C. The PCR products from two sets of primer pairs—MYH9(2163)A (ggagttccggcagaggtatgag) and MYH9(2994)B (cttggccagtttgcagttttga) and MYH9(2504)A (ggctcttcaccaaggtcaag) and MYH9(3201)B (gctgaggtctgtggagtctc)—were sequenced to assess their identity. The MYH9 primer pairs delimit a region of >1 kb midway across the MYH9 cDNA sequence. To confirm the integrity of cDNA, the tissue cDNA was also characterized for expression of a housekeeping gene, cytochrome p450 reductase (CYPR), by CYPR-specific primers CYPR(1141)A (ccgcacggccctcacctacta) and CYPR(1511)B (tggccacccccttgttcactc).
Immunodetection of MYH9 in the Rat Cochlea
The rat cochlea was harvested, decalcified, and embedded, as described elsewhere (Mhatre et al. 1999). The paraffin-embedded tissues were sectioned (6 μm) and mounted on Superfrost-pretreated glass slides (Fisher Scientific). Cochlear sections were deparaffinized, rehydrated, and then incubated with a blocking reagent, I-PBT (0.2% I-block [Tropix] and 0.05% Tween-20 in PBS). The blocked sections were incubated in the presence or absence of varying dilutions (1:25–1:200) of a polyclonal anti-MYH9 antibody (Biomedical Technologies). The anti-MYH9 antibody is prepared in rabbits, from human platelet myosin, and is not cross-reactive with any other protein, including myosins of skeletal, smooth, and cardiac muscle). The tissue sections were washed with PBS and incubated with a biotin-conjugated anti–rabbit IgG secondary antibody (Vector Laboratories) at a dilution of 1:250 in I-PBT for 1 h at room temperature. The bound label (biotin) was amplified with alkaline phosphatase–conjugated streptavidin diluted to 1:500 in I-PBT for 1 h at room temperature. The bound label was detected using NBT-BCIP, an alkaline phosphatase color substrate, and the slides were coverslipped for microscopy.
Mutation Analysis of MYH9 in the Family with DFNA17
cDNA analysis
The MYH9 transcript of affected (III-4) and unaffected (V-2) members of the family with DFNA17 was detected and characterized using total RNA harvested from Epstein-Barr virus–transformed lymphocytes. One microgram of the total RNA was reverse transcribed into cDNA, by MMLV RT (Life Technologies) and random hexamers. The cDNA was amplified with human MYH9–specific primer sets that yielded overlapping fragments spanning the entire length of the MYH9 coding sequence (5,886 bp). The amplified fragments were purified and then sequenced bidirectionally, by an ABI automated sequencer (Applied Biosystems) with appropriate primers. Sequencher software (Gene Codes) was used for sequence analysis.
Genomic analysis
The 40 exons, including the intron-exon junctions, of MYH9 were directly assessed using DNA harvested from the peripheral blood leukocytes of individuals III-4 and V-2. Approximately 50 ng of genomic DNA was amplified with MYH9-specific primer pairs for each of the 40 MYH9 exons (Dunham et al. 1999), and the amplified products were sequenced using an ABI automated sequencer. Sequencher software was used for sequence analysis. After the presence of a mutation in exon 16 was established, this exon was amplified from all members of the family with DFNA17 and was sequenced using primer pairs MYH9-16F1 (tgttctaggcacggaagtgag) and MYH9-16R1 (ttctacatggatggaggacg) or MYH9-16F2 (tcttgccctgtcaggttcat) and MYH9-16R2 (agcccacctctgggactc). The MYH9-16F1/R1 and MYH9-16F2/R2 primer pairs yielded a 295- and a 284-bp fragment, respectively.
Restriction analysis
Mutation detection by sequence analysis was corroborated through restriction-enzyme analysis. MYH9 exon 16 was PCR-amplified from genomic DNA of the members of the family with DFNA17 and from 50 control individuals with normal hearing. The amplified product was digested with the restriction enzyme Fnu4HI at 37°C for 1 h and was then resolved on a 10% acrylamide gel. The mutant sequence alteration eliminates an Fnu4HI restriction site in MYH9 exon 16 of the affected individuals, thereby yielding a restriction-digestion pattern different from that of unaffected individuals.
Results
Expression Analysis of MYH9 in the Rat Cochlea
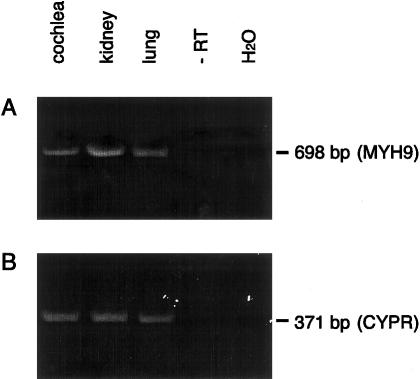
Tissue samples of rat kidney, lung, and cochlea were screened for expression of MYH9 transcript. The primer pair MYH9(2504)A and MYH9(3201)B (see Family, Material, and Methods section) was used to amplify a 698-bp DNA from kidney, lung, and cochlear cDNA (fig. 1A). A similar tissue-expression pattern was also found, by primers MYH9(2163)A and MYH9(3201)B to amplify adjacent MYH9 coding sequence (data not shown). cDNA from all three tissue types was positive for CYPR expression, confirming the integrity of the cDNA (fig. 1B). Fragments from the cochlea that were PCR-amplified using MYH9-specific primers were sequenced and were found to be nearly identical with the rat MYH9 sequence published elsewhere (GenBank accession number GI 6981235). The nucleotide sequence of the amplified PCR fragments, spanning >1 kb of coding region, contained four single-base-pair differences in four separate codons from the published sequence of rat MYH9. Three of these substitutions are silent changes. The fourth, a G→A transposition at nucleotide 3019, which represents the first base in codon 1007, changes the amino acid from aspartate (D) to leucine (N); N is also present at codon 1007 in human MYH9. The D3019N alteration is considered to be a polymorphism.
Figure 1 .
RT-PCR analysis of MYH9 expression in rat cochlear cDNA. cDNA from several tissues, including the cochlea, was assayed for MYH9 expression via PCR using MYH9-specific primers. A, cDNA from the cochlea, kidney, and lung. All samples yielded the expected 698-bp fragment amplified using rat MYH9-specific primers. Cochlear RNA, in the absence of RT, and water (H2O) served as negative controls. B, cDNA from all tissues tested for MYH9 expression. All samples are positive for expression of CYPR, a housekeeping gene, using CYPR-specific primers that yield the expected 371-bp fragment.
MYH9 was immunolocalized in cochlear sections with a polyclonal anti-MYH9 antibody prepared against purified platelet myosin (Biomedical Technologies). The MYH9 immunoreactivity was detected in various cochlear tissues (fig. 2A); these included outer hair cells in the organ of Corti, the subcentral region of the spiral ligament, and the Reissner membrane. MYH9 was not observed in the auditory neurons or within the stria vascularis, a vascular tissue that forms the lateral wall of the cochlear duct. Despite the presence of MYH9 in the auditory neuroepithelia, MYH9 immunoreactivity was absent in the cupular vestibular neuroepithelia.
Figure 2 .
Immunolocalization of MYH9 in the rat cochlea. Radial sections of the rat cochlea were hybridized with anti-MYH9 antibody, were developed with a biotinylated, alkaline phosphatase–linked amplification system and were stained with NBT-BCIP. A, Photomicrograph of rat cochlear duct, illustrating the presence of anti-MYH9 immunoreactivity in several regions of the cochlea, including outer hair cells, cells of the organ of Corti (OC), the subcentral region of the spiral ligament (SL), and the Reissner membrane (RM). SGN = spiral ganglion neurons; SV = stria vascularis. B, The cochlear sections incubated without the anti-MYH9 antibody. The sections are free of immunoreactivity.
Analysis of MYH9 in the Family with DFNA17
Sequence analysis of MYH9 in the family with DFNA17 was undertaken after demonstration that its transcript and the translated product were present in the inner ear of the rat cochlea. The MYH9 was initially screened by sequence analysis of its cDNA, prepared from reverse-transcribed RNA harvested from peripheral blood lymphocytes of the affected and unaffected members of the DFNA17 pedigree shown in figure 3A. The sequence from an affected member (III-4) contained a single-base-pair heterozygosity at two locations (nucleotides 2114 and 4902) within the coding sequence, a heterozygosity that was absent in the MYH9 cDNA sequence of an unaffected member (V-2). The G→A transposition at nucleotide 4902, at the third position of codon 1634, is a silent base-pair alteration, because it does not change the amino acid at this site. The second of these two differences resulted in an altered MYH9 polypeptide. This sequence alteration was observed at nucleotide 2114, representing the second nucleotide in codon 705. Although the unaffected individuals displayed G at nucleotide 2114, the affected individuals displayed both G and A. The presence of A at nucleotide 2114 alters codon 705, causing substitution of arginine by histidine. The amino acid substitution caused by this single-base-pair transposition is located within exon 16 of the MYH9 gene.
Figure 3.
A, Pedigree with DFNA17. The affected members of five generations of the family with DFNA17 are denoted by blackened symbols. B, Sequence analysis of MYH9 in the family with DFNA17. MYH9 was sequenced in genomic DNA from the affected and unaffected members of the family with DFNA17. Although a single peak (G) is observed at nucleotide 2114 in the MYH9 sequence from the unaffected member V-2 (upper panel), a doublet (A/G) is observed at nucleotide 2114 (N) in the affected member III-4 (lower panel). The G→A transposition alters the codon from arginine (R) to histidine (H). C, Restriction analysis of MYH9 exon 16 2114G→A mutation eliminates the Fnu4HI restriction site, and this is reflected in the restriction digest pattern of MYH9 exon 16 from affected (III-4) and unaffected (V-2) individuals from the DFNA17 pedigree. The 284-bp PCR-amplified fragment contains three recognition sites for Fnu4HI—at 130, 135 and 168 bp from the 5′ end, of which the last site is absent in the mutant allele. Numbers at left are sizes of DNA ladder bands.
Sequence analysis of all 40 coding exons of the MYH9 gene from the genomic DNA harvested from representative members of the family with DFNA17 confirmed the presence of the G→A transposition in exon 16 of the affected members but not in the unaffected members (fig. 3B). The G→A transposition in the second nucleotide of codon 705 causes the elimination of Fnu4HI restriction site. Therefore, the Fnu4HI digest of exon 16 amplified from affected individuals is expected to yield an additional restriction fragment, of 149 bp, that is not seen in unaffected individuals. This marker was used to corroborate the presence of the sequence alteration within exon 16 of MYH9 in the affected members—as well as its absence in the unaffected members—of the family with DFNA17 (fig. 3C). Individuals V-1 and V-3, whose affected status was unknown because of their young age at the time of linkage analysis, were found to be unaffected and affected, respectively, by sequence and restriction analysis. The G→A transposition in nucleotide 2114 was absent in the genomic DNA from 50 randomly selected healthy individuals. Sequence alignment of the MYH9 polypeptide spanning the SH1-SH2 linker region of muscle and nonmuscle class II myosins from seven species illustrates both the strict conservation of this linker region and the invariance of arginine (R705) in this sequence motif (fig. 4).
Figure 4 .
Alignment of the SH1 linker region in class II myosins. Alignment of peptide sequence spanning the SH1 linker region of muscle and nonmuscle class II myosins from seven species illustrates both the strict conservation of this linker region and the invariance of arginine (R705) in this sequence motif.
Discussion
Through the use of the candidate-gene approach, we have identified MYH9 as the gene at the DFNA17 locus that is responsible for autosomal dominant HHI. MYH9 represents the first conventional myosin heavy-chain gene linked to hearing impairment, and it joins the growing list of unconventional (non–class II) myosin heavy-chain genes (myosins VI [Avraham et al. 1995], VIIA [Gibson et al. 1995, Weil et al. 1995], and XV [Probst et al. 1998]) associated with HHI. Similar to MYH9, expression of these unconventional myosins is not limited to the cells and tissues of the inner ear, yet the predominant clinical manifestation of their dysfunction is hearing impairment.
The identified mutation, R705H, occurs in a highly conserved and functionally critical region within the carboxy-terminal half of the myosin heavy-chain motor domain that forms the globular head in the hexameric myosin molecule. Within the motor domain, R705 resides in a highly conserved linker region that spans 16 amino acids and that contains two free thiol groups, C704 (SH1) and C694 (SH2). Crystal structure of the globular head of skeletal muscle myosin has shown that the two thiols are part of two short α-helices joined through a kink at the conserved G696 residue (Rayment 1996; Gulick and Rayment 1997). The SH1 and SH2 helices are believed to play a key role in the conformational changes that occur in the myosin head during force generation coupled to ATP hydrolysis. X-ray crystallographic studies suggest that during the power stroke the light-chain–binding domain swings relative to the catalytic/ATP-binding domain. The pivot point of this swinging motion is considered to be in the vicinity of the SH1-SH2 helix (Houdusse et al. 1999). Not surprisingly, in vitro and in vivo modification of the SH1 helix, including cross-linking of SH1-SH2 groups or alteration/substitution of SH1/SH2, has been shown to disrupt the mechanical function of the myosin motor domain (Patterson et al. 1997; Suzuki et al. 1997). These studies have demonstrated that the functional integrity of myosin is critically dependent on its flexibility at the SH1-SH2 helix. The strict conservation of this linker region among myosin II subtypes within and between species further underscores its functional importance. The R705H mutation in the family with DFNA17 may cause an altered conformation of the SH1 helix that affects its flexibility and movement, thereby disrupting the mechanical function of the motor domain.
We have not yet determined the mechanism by which MYH9 dysfunction leads to hearing loss and the observed inner-ear histopathology, i.e., CSD, in the proband with DFNA17. Localization of MYH9 expression in the mammalian cochlea has particular importance for understanding and interpreting the histopathology associated with its dysfunction. Within the rat cochlea, MYH9 expression was localized in three distinct tissue types: the organ of Corti, the subcentral region of the spiral ligament, and the Reissner membrane. Within the organ of Corti, the MYH9 expression was identified throughout the outer hair cells, including those of the cuticular plate. Both myosin VI and myosin VIIA have also been localized within the mechanosensory hair cells. Myosin VI is localized within the actin-rich cuticular plate and in the rootlet actin filaments that descend from stereocilia into the cuticular plate, suggesting a role in stabilizing the basal attachment of stereocilia (Avraham et al. 1997, Hasson et al. 1997). Myosin VIIA is localized in the stereocilia and cell body of hair cells (Hasson et al. 1997). Postulated roles for myosin VIIA include the maintenance of stereocilia integrity and membrane trafficking in the inner hair cells. The role of MYH9 in hair cells remains to be deciphered.
MYH9 expression was also identified in the Reissner membrane, a membranous barrier consisting of two layers of cells that forms the upper boundary of the cochlear duct. The CSD histopathology of the proband affected with DFNA17 demonstrated a collapsed Reissner membrane (Lalwani et al. 1997). MYH9 dysfunction in the Reissner membrane may contribute to its compromised cytoarchitecture, thus disturbing cochlear-fluid homeostasis and effecting cochlear function. Of the various cochlear tissues, the one with the most conspicuous absence of MYH9 expression is the stria vascularis, which forms the lateral wall of the cochlear duct and whose dysfunction is generally considered to be the cause of CSD. The absence of MYH9 expression in the stria vascularis suggests that this tissue type is not directly responsible for the underlying pathophysiology of CSD.
MYH9 is also expressed within the spiral ligament that consists of connective tissue comprising fibrocytes. These fibrocytes of the spiral ligament are classified according to the distinct isoforms of the Na+ and K+ ATPase that they express, as well as by their cytoskeletal components. The subcentric region of the spiral ligament, which is immnoreactive to the MYH9 antibody, is characterized by the presence of type III and type IV fibrocytes. The type III fibrocytes are rich in actin and other contractile and contraction-associated proteins (Henson et al. 1984, 1985). The postulated role for these cells includes providing anchorage for surrounding cells and/or developing or reacting to tension generated in the basilar membrane–spiral ligament complex (Henson et al. 1985). Maintenance of this tension is probably more crucial in the basal turn of the cochlea, where sensitivity to high frequencies resides and where the basilar membrane is wider than at the apex. This may be the simple explanation for the observed high-frequency hearing impairment in DFNA17. The contribution of MYH9 dysfunction to HHI and presbycusis (age-associated hearing impairment similar to the DFNA17 phenotype) remains to be determined. Both the precise role of MYH9 in the cochlea and the mechanism by which R705H mutation leads to the DFNA17 phenotype of progressive hearing impairment and CSD remain to be elucidated.
Acknowledgments
We thank Ryan E. Stern for his assistance. This work was supported in part by National Institute on Deafness and Other Communication Disorders grant K23 DC 00112 (to A.K.L.), the Deafness Research Foundation, the National Organization for Hearing Research, the American Hearing Research Foundation, Hearing Research, Inc., and the University of California at San Francisco REAC Jacobsen Fund.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank (for rat myh9 sequence [accession number GI 6981235])
- Genome Database, http://www.gdb.org
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim (for DFNA17 [MIM 603622])
- Primer3 software, http://www.genome.wi.mit.edu
References
- Avraham KB, Hasson T, Sobe T, Balsara B, Testa JR, Skvorak AB, Morton CC, Copeland NG, Jenkins NA (1997) Characterization of unconventional MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice. Hum Mol Genet 6:1225–1231 [DOI] [PubMed] [Google Scholar]
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA (1995) The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet 11:369–375 [DOI] [PubMed] [Google Scholar]
- Collins FS (1995) Positional cloning moves from perditional to traditional. Nat Genet 9:347–350 [DOI] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA (1987) Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236:1086–1091 [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Edwards KA, Kiehart DP (1996) Drosophila nonmuscle myosin II has multiple essential roles in imaginal disc and egg chamber morphogenesis. Development 122:1499–1511 [DOI] [PubMed] [Google Scholar]
- Friedman TB, Sellers JR, Avraham KB (1999) Unconventional myosins and the genetics of hearing loss. Am J Med Genet 89:147–157 [DOI] [PubMed] [Google Scholar]
- Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD (1995) A type VII myosin encoded by the mouse deafness gene shaker-1. Nature 374:62–64 [DOI] [PubMed] [Google Scholar]
- Gulick AM, Rayment I (1997) Structural studies on myosin II: communication between distant protein domains. Bioessays 19:561–569 [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ (1996) A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature 382:455–458 [DOI] [PubMed] [Google Scholar]
- Hasson T, Walsh J, Cable J, Mooseker MS, Brown SD, Steel KP (1997) Effects of shaker-1 mutations on myosin-VIIa protein and mRNA expression. Cell Motil Cytoskeleton 37:127–138 [DOI] [PubMed] [Google Scholar]
- Henson MM, Burridge K, Fitzpatrick D, Jenkins DB, Pillsbury HC, Henson OW Jr (1985) Immunocytochemical localization of contractile and contraction associated proteins in the spiral ligament of the cochlea. Hear Res 20:207–214 [DOI] [PubMed] [Google Scholar]
- Henson MM, Henson OW Jr, Jenkins DB (1984) The attachment of the spiral ligament to the cochlear wall: anchoring cells and the creation of tension. Hear Res 16:231–242 [DOI] [PubMed] [Google Scholar]
- Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C (1999) Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell 97:459–470 [DOI] [PubMed] [Google Scholar]
- Kelley CA, Sellers JR, Gard DL, Bui D, Adelstein RS, Baines IC (1996) Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities []. J Cell Biol 134:675–687 (erratum: J Cell Biol 138:215 [1997]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J (1998a) Cytoplasmic dynamics of myosin IIA and IIB: spatial “sorting” of isoforms in locomoting cells. J Cell Sci 111:2085–2095 [DOI] [PubMed] [Google Scholar]
- ——— (1998b) Fluorescent analogues of myosin II for tracking the behavior of different myosin isoforms in living cells. J Cell Biochem 68:389–401 [DOI] [PubMed] [Google Scholar]
- Lalwani AK, Linthicum FH, Wilcox ER, Moore JK, Walters FC, San Agustin TB, Mislinski J, Miller MR, Sinninger Y, Attaie A, Luxford WM (1997) A five-generation family with late-onset progressive hereditary hearing impairment due to cochleosaccular degeneration. Audiol Neurootol 2:139–154 [DOI] [PubMed]
- Lalwani AK, Luxford WM, Mhatre AN, Attaie A, Wilcox ER, Castelein CM (1999) A new locus for nonsyndromic hereditary hearing impairment, DFNA17, maps to chromosome 22 and represents a gene for cochleosaccular degeneration. Am J Hum Genet 64:318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P, Phillips CL, Adelstein RS, Pollard TD (1994) Differential localization of myosin-II isozymes in human cultured cells and blood cells. J Cell Sci 107:3077–3090 [DOI] [PubMed] [Google Scholar]
- Mhatre AN, Steinbach S, Hribar K, Hoque AT, Lalwani AK (1999) Identification of aquaporin 5 (AQP5) within the cochlea: cDNA cloning and in situ localization. Biochem Biophys Res Commun 264:157–162 [DOI] [PubMed] [Google Scholar]
- Patterson B, Ruppel KM, Wu Y, Spudich JA (1997) Cold-sensitive mutants G680V and G691C of Dictyostelium myosin II confer dramatically different biochemical defects. J Biol Chem 272:27612–27617 [DOI] [PubMed] [Google Scholar]
- Phillips CL, Yamakawa K, Adelstein RS (1995) Cloning of the cDNA encoding human nonmuscle myosin heavy chain-B and analysis of human tissues with isoform-specific antibodies. J Muscle Res Cell Motil 16:379–389 [DOI] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, Friedman TB, Camper SA (1998) Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science 280:1444–1447 [DOI] [PubMed] [Google Scholar]
- Rayment I (1996) The structural basis of the myosin ATPase activity. J Biol Chem 271:15850–15853 [DOI] [PubMed] [Google Scholar]
- Saez CG, Myers JC, Shows TB, Leinwand LA (1990) Human nonmuscle myosin heavy chain mRNA: generation of diversity through alternative polyadenylylation. Proc Natl Acad Sci USA 87:1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR (2000) Myosins: a diverse superfamily. Biochim Biophys Acta 1496:3–22 [DOI] [PubMed] [Google Scholar]
- Simons M, Wang M, McBride OW, Kawamoto S, Yamakawa K, Gdula D, Adelstein RS, Weir L (1991) Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res 69:530–539 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ohkura R, Sugiura S, Yasuda R, Kinoshita K Jr, Tanokura M, Sutoh K (1997) Modulation of actin filament sliding by mutations of the SH2 cysteine in Dictyostelium myosin II. Biochem Biophys Res Commun 234:701–706 [DOI] [PubMed] [Google Scholar]
- Tullio AN, Accili D, Ferrans VJ, Yu ZX, Takeda K, Grinberg A, Westphal H, Preston YA, Adelstein RS (1997) Nonmuscle myosin II-B is required for normal development of the mouse heart. Proc Natl Acad Sci USA 94:12407–12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, Kelley PM, Kimberling WJ, Wagenaar M, Levi-Acobas F, Larget-Piet D, Munnich A, Steel KP, Brown SDM, Petit C (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374:60–61 [DOI] [PubMed] [Google Scholar]