Abstract
We performed a genome scan at an average resolution of 8 cM in 719 Finnish sib pairs with type 2 diabetes. Our strongest results are for chromosome 20, where we observe a weighted maximum LOD score (MLS) of 2.15 at map position 69.5 cM from pter and secondary weighted LOD-score peaks of 2.04 at 56.5 cM and 1.99 at 17.5 cM. Our next largest MLS is for chromosome 11 (MLS = 1.75 at 84.0 cM), followed by chromosomes 2 (MLS = 0.87 at 5.5 cM), 10 (MLS = 0.77 at 75.0 cM), and 6 (MLS = 0.61 at 112.5 cM), all under an additive model. When we condition on chromosome 2 at 8.5 cM, the MLS for chromosome 20 increases to 5.50 at 69.0 cM (P=.0014). An ordered-subsets analysis based on families with high or low diabetes-related quantitative traits yielded results that support the possible existence of disease-predisposing genes on chromosomes 6 and 10. Genomewide linkage-disequilibrium analysis using microsatellite marker data revealed strong evidence of association for D22S423 (P=.00007). Further analyses are being carried out to confirm and to refine the location of these putative diabetes-predisposing genes.
Introduction
Diabetes is a heterogeneous disorder characterized by a chronic elevation of plasma glucose. Type 2 diabetes (previously known as “non–insulin-dependent diabetes mellitus” [NIDDM]) accounts for ∼90% of cases, whereas type 1 diabetes, an autoimmune disorder, explains most of the remainder. The metabolic hallmarks of type 2 diabetes are insulin resistance, impaired pancreatic beta-cell function, and increased hepatic glucose production. It is thought that both genetic and environmental factors contribute to the pathogenesis of type 2 diabetes, but its mode of inheritance remains unclear (Rich 1990). A rare, monogenic form of type 2 diabetes—maturity-onset diabetes of the young (MODY)—accounts for ∼1%–5% of type 2 diabetes cases, depending on the population (Shuldiner and Silver 1996). MODY has an autosomal dominant mode of inheritance and an earlier age at onset, compared with most other forms of type 2 diabetes. At least six different genes have been isolated that independently cause MODY within single pedigrees (Vionnet et al. 1992; Yamagata et al. 1996a, 1996b; Horikawa et al. 1997; Stoffers et al. 1997; Malecki et al. 1999). MODY genes are thought to play, at most, a minor role in common type 2 diabetes (Shuldiner and Silver 1996).
There have been numerous studies using the candidate-gene approach for type 2 diabetes, but, to date, none have revealed loci that determine disease in a large proportion of diabetics (Ghosh and Schork 1996; Kahn et al. 1996; Elbein 1997). Others have taken a positional cloning approach, and genome scans for type 2 diabetes have now been conducted in several samples from different populations (Hanis et al. 1996; Mahtani et al. 1996; Hanson et al. 1998; Duggirala et al. 1999; Elbein et al. 1999; Ehm et al. 2000). Recently, the chromosome 2 locus linked to type 2 diabetes in Mexican Americans, also known as “NIDDM1” (Hanis et al. 1996), has been identified and cloned (Horikawa et al. 2000).
Type 2 diabetes affects nearly 6% of the U.S. and 4% of the Finnish population 45–64 years of age (Tuomilehto et al. 1991; Bennett et al. 1992; Kenny et al. 1995; Valle et al. 1997). The Finnish population is believed to derive largely from a small group of founders, with little subsequent immigration (Workman et al. 1976), and has been relatively isolated geographically, linguistically, culturally, and, hence, genetically. Finland also has comprehensive population and medical databases that greatly simplify the identification of family material, and extensive church records make it possible to link up families back through several centuries. Finally, Finland boasts a well-educated population strongly supportive of medical research. These features make Finland an excellent choice for conducting genetic studies.
The FUSION (Finland–United States Investigation of NIDDM Genetics) study is an international collaborative effort to map and positionally clone genes predisposing to type 2 diabetes and intermediate quantitative traits in Finnish subjects. We have previously undertaken exclusion mapping for NIDDM1 in our Finnish sample (Hanis et al. 1996; Ghosh et al. 1998). In addition, we have reported positive linkage results for chromosome 20 (Ghosh et al. 1999), giving strong support to similar results from other groups (Bowden et al. 1997; Ji et al. 1997; Zouali et al. 1997). In the present article, we describe extended results for chromosome 20 and the entire autosomal genome scan for type 2 diabetes in a carefully ascertained Finnish sample of affected-sib-pair (ASP) families. In the accompanying article by Watanabe et al. (2000 [in this issue]), we analyze important diabetes-related quantitative traits in affected and unaffected individuals from the same families.
Subjects and Methods
Subjects
The design of the FUSION study has been described elsewhere (Valle et al. 1998). In brief, we identified 580 families with an ASP. Index cases were ascertained primarily on the basis of the National Hospital Discharge Registry, and diabetes was diagnosed according to World Health Organization (1985) criteria. We selected families for study using the following rules: (1) 35–60 years as age at diagnosis of type 2 diabetes in the index case; (2) at least one living affected sibling; and (3) at least one parent reported to be unaffected. Our age-at-diagnosis criterion was chosen to be late enough to exclude most cases of type 1 diabetes and MODY but early enough to increase familiality of the disease (Mitchell et al. 1994). In addition, nondiabetic spouses and at least two unaffected offspring were ascertained in 210 families. Finally, 231 elderly controls, with two normal oral glucose-tolerance tests documented at ages 65 and 70 years, were collected. All diabetic subjects had C-peptide and glutamic acid decarboxylase (GAD) antibody measurements performed, in addition to fasting-insulin and fasting-glucose levels. We also developed criteria based on fasting C-peptide and GAD levels (Valle et al. 1998) in addition to insulin-treatment history, to identify affected siblings with possible late-onset type 1 diabetes. On the basis of these criteria, 41 families with probable type 1 diabetes were excluded from the present analysis. Family studies were approved by Institutional Review Boards at the National Institutes of Health (assurance number SPA S-5737-05) and at the National Public Health Institute in Helsinki.
Genotyping
DNA samples were isolated from whole blood by a salting-out procedure (GENTRA DNA isolation kit). Microsatellite markers with heterozygosities generally >.7 were chosen from published maps. Genotypes were determined as described elsewhere (Magnuson et al. 1996; Ghosh et al. 1997). Most of the fluorescently labeled microsatellite repeat markers genotyped in this study were modified from the ABI PRISM Linkage Mapping Set (PE Biosystems); in addition, some in-house–designed primer pairs were used (Magnuson et al. 1996). Single-plex or multiplex PCRs were performed under cycling conditions of either 3-step + 10-min extension at 72°C or a touchdown protocol (Magnuson et al. 1996; Ghosh et al. 1997, 1998).
Statistical Analysis
Map construction and linkage analysis
We used RELPAIR (Boehnke and Cox 1997) to detect possible pedigree errors. We removed cases of non-Mendelian inheritance, which had been identified by MENDEL (Lange et al. 1988) through calculation of a zero likelihood. Marker order and sex-averaged intermarker distances were estimated, on the basis of the 210 extended FUSION families and the CEPH (Centre d'Etude Polymorphisme Humaine) reference pedigrees, by maximum likelihood calculated using CRI-MAP and MultiMap (Lander and Green 1987; Matise et al. 1994).
For ASP linkage analysis, we employed the multipoint maximum-likelihood method initially suggested by Risch (1990), as programmed in the computer program SIBLINK (Hauser et al. 1996; Hauser and Boehnke 1998). This method is parameterized in terms of the probabilities—z0, z1, and z2—that the ASP shares, respectively, 0, 1, or 2 genes identical by descent (IBD) at a putative disease-predisposing locus. We estimated IBD status every 0.5 cM along each chromosome, using all available marker information for the chromosome and marker-allele frequencies estimated on the basis of our family data. We maximized the likelihood at each point under either additivity (z1= 1/2) or the “possible triangle constraints” (Holmans 1993). We performed these analyses twice, first by treating all pairs as independent and, second, by weighting the contribution of each ASP by 2/s, where s is the number of affected sibs in a family (Suarez and Hodge 1979).
After genotyping had been completed, 61 families were excluded from the original 580, in addition to the 41 type 1 diabetic families. Of these 61 families, 1 was removed because of multiple unresolvable pedigree errors. The remaining 60 families were removed because, after identifiable errors were removed, the families had no genotyped ASP. From our final analyzed sample of 478 families, we constructed a total of 719 ASPs, approximately equivalent to 586 (s-1) independent sib pairs (Suarez and Hodge 1979). This comprised 385 families with two, 84 with three, 6 with four, and 1 each with five, six, and seven affected sibs. In addition, 452 affected sibships had no living parents, 23 had one, and 3 had two.
We present weighted ASP results, recognizing that this approach is, in general, somewhat conservative (Meunier et al. 1997). Furthermore, only results from the additive model are described, since the possible-triangle results are very similar.
Ordered-subsets analysis
Because of the phenotypic and likely genotypic heterogeneity underlying type 2 diabetes, we performed ordered-subsets analysis (Hauser et al. 1998). In this analysis, we sought to identify more-homogeneous groups of families, on the basis of their mean levels for a diabetes-associated quantitative trait. For this purpose, we ranked families according to their mean sibship value for the quantitative trait of interest—for example, body-mass index (BMI). Starting with the family with the lowest BMI, family-specific LOD scores were added in, one family at a time, in rank order, until all families were included. After each family was added, we determined the MLS for the current subset of families. The overall MLS obtained for any subset of the families was finally reported. We then repeated the analysis, starting with the family with the highest mean BMI.
We performed this ordered-subsets analysis on the basis of mean values for fasting glucose, fasting C-peptide, fasting insulin, SI(EST) (1/[fasting glucose × fasting insulin]), IRI (fasting insulin/fasting glucose), IRC (fasting C-peptide/fasting glucose), age at diagnosis, and BMI. The empirical values—SI(EST) for insulin resistance and IRI and IRC for insulin secretion—were derived to allow approximate measures of these indices in affected individuals, since frequently sampled intravenous glucose-tolerance test analyses were performed only in unaffected relatives (see the accompanying paper by Watanabe et al. [2000]).
To assess the relationship of pairs of loci in the predisposition to type 2 diabetes, and to identify possible interactions, we also carried out ordered-subsets analysis at one locus (the analysis locus), on the basis of evidence for linkage at another locus (the conditioning locus). In this case, the covariate for addition of families was the average, over all ASPs within the family, of the estimated IBD values at the conditioning locus. Furthermore, we varied the conditioning point ±5 cM around the peak of the MLS curve on the conditioning chromosome. An analogous approach to assessment of interaction has recently been described by Cox et al. (1999).
We estimated P values for the ordered-subsets MLS by using a permutation-test framework (Hauser et al. 1998). Under this permutation framework, the observed ordered-subsets MLS was compared with the ordered-subsets MLS obtained at any point along the chromosome when families were added in a random order. The empirical P value was the proportion of 5,000 random orderings of the families that gave an MLS greater than the observed ordered-subsets MLS.
Association analysis
In addition to linkage analysis, we also carried out linkage-disequilibrium analysis of our genome-scan data. Although the prior likelihood that linkage disequilibrium would be identified by use of a 10-cM map of markers was low, we felt that the time and expense of gathering the data justified the effort. In our analysis, we compared the 524 index cases with a control group comprising the 231 elderly controls and 162 nondiabetic spouses of either the index cases or their affected siblings. We pooled the latter two groups after first showing that their allele-frequency distributions were similar across the markers tested.
Three tests were undertaken for each marker. First, we calculated a standard Pearson χ2 test of association, using a 2×N table, where N is the number of marker alleles. Second, in the best-allele test, each marker allele was compared, in turn, against the pool of all other marker alleles, and the maximum of these N 2×2 Pearson χ2 test statistics was taken. This test is tailored to the situation of a single disease-associated marker allele. Third, the marker alleles that were more common in affected individuals were pooled, as were those more common in unaffected individuals. This high-versus-low test is aimed at the situation of two or more disease-associated marker alleles. Each test was performed initially with all alleles included and then was repeated, but with the exclusion of alleles present in <10 copies. We undertook this exclusion to safeguard against the disproportionate impact of rare alleles on the test statistics. Significance levels for all three statistics were assessed by permutation, because of the sparse data and the data manipulation for the best-allele and high-versus-low tests; we used 100,000 permuted data sets for this purpose.
Results
Our affected subjects had a nearly equal sex distribution, were mostly 50–70 years of age (see table 1), and had been diagnosed with diabetes for a mean period of 12 years at the time of study. A total of 408 microsatellite markers were typed in this sample, producing an autosomal map of estimated length 3,323 cM, with an average resolution of ∼8 cM. There were two gaps >20 cM in estimated length: 21.0 cM on chromosome 18, between markers D18S63 and D18S843, and 24.4 cM on chromosome 13, between D13S263 and D13S156.
Table 1.
Phenotype Descriptions of ASPs with Type 2 Diabetes
| Diabetic Index Cases/Siblings (n=1,175) | |
| Female:Male Ratio |
|
| Sex | 599:576 |
| Mean ± SD [Median] |
|
| Age (years) | 64.2 ± 8.3 [64.8] |
| BMI (kg/m2) | 29.8 ± 4.8 [29.3] |
| WHR | .94 ± .08 [.94] |
| Fasting glucose (mM) | 10.2 ± 3.5 [9.6] |
| Fasting insulin (pM) | 111 ± 67 [96] |
| Fasting C-peptide (nM) | 1.59 ± .93 [1.44] |
| SI(EST) (×100) | .18 ± .31 [.11] |
| IRI | 12.0 ± 8.1 [10.3] |
| IRC | .17 ± .11 [.14] |
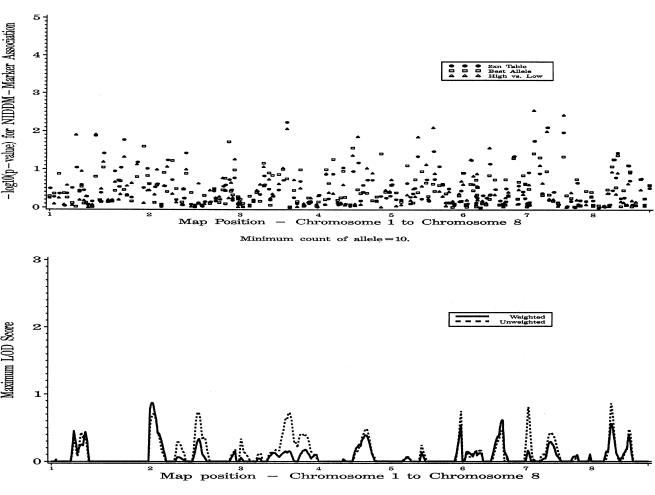
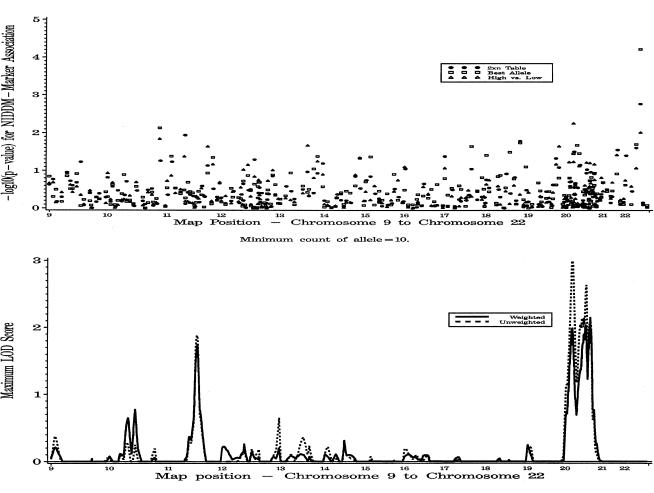
Results for the complete autosomal genome scan, including both weighted and unweighted ASP linkage analysis under the additive model and association analysis for all three tests of association, are presented in figures 1 and 2. In what follows, we present results chosen from among those with ASP MLSs >0.5 and ordered-subsets analysis MLSs with P⩽.05.
Figure 1.
Genomewide single-point association analysis and genomewide weighted and unweighted multipoint linkage analysis for type 2 diabetes, on chromosomes 1–8. In the linkage analysis, results are for the additive analysis. For association, the 2×N, best-allele, and high-versus-low analyses are shown for each marker.
Figure 2.
Genomewide single-point association analysis and genomewide weighted and unweighted multipoint linkage analysis for type 2 diabetes, on chromosomes 9–22.
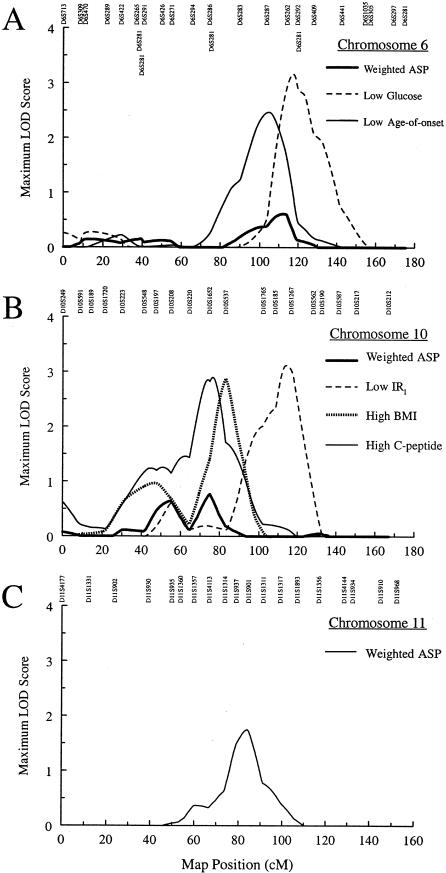
Chromosome 6 (Fig. 3A)
Figure 3.
ASP linkage analysis and ordered-subsets–linkage results for type 2 diabetes. A, ASP linkage results for chromosome 6, for all families, for 94 families with the lowest fasting plasma-glucose levels, and for 74 families with the lowest age at onset. B, ASP linkage results for chromosome 10, for all families, for 40 families with the lowest IRI, for 75 families with the highest BMI, and for 28 families with the highest fasting C-peptide. C, ASP linkage results for chromosome 11.
The MLS was 0.61 at 112.5 cM between markers D6S287 and D6S262. Ordered-subsets analysis identified 94 families with the lowest mean fasting plasma glucose, on the basis of an MLS of 3.17 (P=.013) at 117.0 cM. The range for mean plasma glucose in these sibships was 4.43–8.16 mM. Ordered-subsets analysis also identified 74 families with lowest mean age at diagnosis, on the basis of an MLS of 2.48 (P=.041) at 104.5 cM. The range for the mean age at diagnosis was 29–44 years. Eleven families were common to both ordered subsets. In this analysis, no evidence for linkage with the HLA region was detected, suggesting that our procedure to exclude families with individuals with type 1 diabetes was successful.
Chromosome 10 (Fig. 3B)
MLSs of 0.64 at 54.5 cM (between markers D10S197 and D10S208) and of 0.77 at 75.0 cM (between markers D10S1652 and D10S537) were obtained. Ordered-subsets analysis identified (a) 40 families with the lowest IRI (range of mean IRI 0.81–5.27 pmol/mmol), on the basis of an MLS of 3.12 (P=.015) at 114.5 cM; (b) 75 families with the highest BMI (range of mean BMI 33.7–41.4 kg/m2), on the basis of an MLS of 2.89 (P=.031) at 83.0 cM; and (c) 28 families with the highest fasting C-peptide (range of mean 3.06–4.46 nM), on the basis of an MLS of 2.90 (P=.025) at 76.5 cM. There were no families that appeared in all three ordered subsets. The marker D10S217 at 150.4 cM showed some evidence of association with diabetes (best allele P=.0074).
Chromosome 11 (Fig. 3C)
The MLS was 1.75 at 84.0 cM, close to D11S901. The nearby marker D11S1314 at 73.8 cM was weakly associated with diabetes (best allele P=.042).
Chromosome 2 (Fig. 4A)
Figure 4.
Interaction-analysis plots for chromosome 20, conditional on chromosome 2. A, ASP linkage results for all families, for chromosome 2. B, ASP linkage results for chromosome 20. There is an increase in LOD score for chromosome 20, to 5.50 at 69.0 cM, when we condition on chromosome 2 at 8.5 cM.
The MLS was 0.87 at 5.5 cM between markers D2S319 and D2S162.
Chromosome 20 (Fig. 4B)
In a previous article, we reported MLSs of 1.92 (at 18.5 cM), 2.06 (at 57.0 cM), and 2.00 (at 69.5 cM) for chromosome 20, on the basis of analysis of 38 markers (Ghosh et al. 1999). With the addition of five more markers, the up-to-date results are MLS peaks of 1.99 at 17.5 cM, 2.04 at 56.5 cM, and 2.15 at 69.5 cM. The marker D20S892 at 21.0 cM is associated with diabetes (high-versus-low P=.0055) and is ∼3.5 cM centromeric from the LOD-score peak on the p arm.
Chromosome 20/Chromosome 2 Interaction (Fig. 4B)
In an attempt to increase mapping resolution for chromosome 20, we performed two-locus analysis by carrying out ordered-subsets analysis on IBD sharing in regions suggestive of linkage on chromosomes 2, 7, 10, and 11. When we placed the conditioning point at 5.5 cM on chromosome 2, the MLS for chromosome 20 increased to 5.06 (P=.007) at 69.0 cM. To find the highest MLS for chromosome 20, we also explored moving the conditioning point away from 5.5 cM on chromosome 2. When the conditioning point was placed at 8.5 cM on chromosome 2, the MLS increased to 5.50 (P=.0014) at 69.0 cM on chromosome 20, with a 1-LOD support interval of width 7.5 cM, as opposed to 40 cM for unconditional analysis. Data on 273 families with mean IBD >.48 at position 8.5 cM on chromosome 2 contributed to this interaction MLS. This was the largest conditional MLS for any of our pairs of linkage peaks; it also was the largest conditional MLS for any pair of points along the entire genome.
Chromosome 22
In this genome scan, a marker on chromosome 22 showed the strongest association with diabetes (fig. 2). A single allele for the marker D22S423 (43.6 cM) was present at a frequency of 17.7% in affected individuals and 10.5% in controls and spouses (P=.00007). However, there was no evidence for linkage in this region.
Discussion
Genome scans have been completed for type 2 diabetes and related traits in several populations (Hanis et al. 1996; Mahtani et al. 1996; Stern et al. 1996; Hanson et al. 1998; Imperatore et al. 1998; Pratley et al. 1998; Duggirala et al. 1999; Elbein et al. 1999; Ehm et al. 2000). The genome scan reported here for type 2 diabetes–susceptibility genes in a Finnish sample represents one of the largest genetic studies thus far of this complex metabolic disorder (Valle et al. 1998). We have performed multipoint ASP linkage, ordered-subsets linkage, and association analyses for type 2 diabetes. The ASP analysis uses all sibships, whereas the ordered-subsets analysis is based on selected sibships who have either the largest or smallest mean values for a diabetes-related quantitative trait. Ordered-subsets analysis yields a more homogenous group.
The association analysis comparing probands with unaffected spouses and elderly controls was used as a screening tool for possible linkage disequilibrium. We fully recognize the low prior probability of detecting disequilibrium for a complex disease in a 10-cM genome scan, even in a relatively isolated population such as that of Finland. Our interesting association result with D22S423 must be interpreted with caution. In the accompanying paper by Watanabe et al. (2000 [in this issue]), we describe QTL analysis of metabolic and anthropometric traits, and we report results for both affected and unaffected predisposed family members. We believe that this sort of multipronged attack is a useful approach in the dissection of type 2 diabetes or any complex genetic disease (Ghosh and Schork 1996).
Our LOD scores for the ASP analyses do not reach genomewide significance according to accepted criteria (Lander and Kruglyak 1995). It is clear that some of the results that we report here may well be false positives. However, our multipronged approach, along with reports from other groups, gives support to our results.
Our three largest ASP LOD scores are for chromosome 20—one on the p arm, one near the centromere, and one on the q arm. The interaction result with chromosome 2 increases both the evidence for linkage and the resolution for the locus at 69.5 cM on chromosome 20. In addition, the diabetes-associated marker D20S892 at 21 cM is close to the MLS peak at 17.5 cM. Three other groups have also reported evidence for linkage on 20q (Bowden et al. 1997; Ji et al. 1997; Zouali et al. 1997).
Our next largest MLS is for chromosome 11 at 84.0 cM. Pratley et al. (1998) reported a multipoint MLS of 1.31 for 2-h insulin levels, very near the marker D11S2371, in 388 unaffected Pima Indian sib pairs. Microsatellite mapping data from the Center for Medical Genetics, Marshfield Medical Research Foundation (Broman et al. 1998), suggest that this peak occurs at ∼76 cM on our map. The candidate genes in this region include uncoupling protein 2 (MIM 601693) (∼76 cM on our map), pyruvate carboxylase (MIM 266150) (∼71 cM), and muscle glycogen phosphorylase (MIM 232600) (∼69 cM). The nearby region between D11S2000 and D11S912 (∼101-137 cM on our map) has also demonstrated evidence for linkage, in Pima Indians, to diabetes, BMI (Hanson et al. 1998), percentage of body fat (Norman et al. 1997, 1998), and 2-h insulin levels (Pratley et al. 1998).
In our data, there is some evidence for a diabetes gene on chromosome 6q that affects age at diagnosis (ordered-subsets MLS = 2.48 at 104.5 cM). Hanson et al. (1998) also reported a single-point MLS of 1.84 for age-adjusted diabetes, at D6S1009 (120.1 cM on our map) in 264 Pima Indian nuclear families containing 966 siblings. In addition, IDDM15 is located at ∼90 cM on our map (Delépine et al. 1997). Since we have removed sibs who either have or are likely to have type 1 diabetes, this could imply that the same susceptibility gene influences both forms of diabetes. Stern et al. (1996) studied 444 individuals from 32 Mexican American families and found an MLS >1 for both fasting and 2-h glucose levels, near D6S292 at 119.5 cM on our map. Hanis et al. (1996) obtained a single-point MLS of 0.77 at D6S262 (114.2 cM on our map) in a separate Mexican American sample.
Our results for chromosome 10 at 75.0 cM suggest a locus that may be involved in both obesity and diabetes. Interestingly, in the accompanying paper (Watanabe et al. (2000 [in this issue]), we report an MLS of 1.67 (P=.0028) for waist-to-hip ratio (WHR) at 75.0 cM in QTL analysis of unaffected individuals. Also, Hager et al. (1998) have reported an MLS of 4.85 for BMI, at the marker D10S197 (47.3 cM on our map) in a collection of French families. This result coincides with a more proximal peak of 0.64 at 54.5 cM on chromosome 10.
Pratley et al. (1998) reported a multipoint MLS of 2.99 for fasting glucose near the marker D22S270 on chromosome 22, which was their best result for the trait. Also, Hanis et al. (1996) found an MLS of 0.61 for diabetes, at D22S270. This marker is ∼2 cM telomeric to D22S423 (on the basis of the Whitehead Institute for Biomedical Research/MIT Center for Genome Research map), the marker that gave our strongest association result.
At present, we are undertaking a number of different strategies to positionally clone genes in areas of interest. Our focus is on chromosome 22, near the marker D22S423, and on chromosome 20, at the 69.5 cM peak. We are typing additional markers in both these regions. We see microsatellite marker-marker disequilibrium on chromosome 20, at distances of <1 cM (authors' unpublished data). Further interaction analyses are being conducted by use of both GENEHUNTER PLUS (Cox et al. 1999) and SIBLINK (Hauser et al. 1996). We will carry out multivariate analyses in regions with overlapping results and have generated phenotype comparisons between apparently linked and unlinked families (authors' unpublished data). Finally, we and others have embarked on a collaboration to map type 2 diabetes–susceptibility genes by combining data sets and integrating maps (Boehnke et al. 1998).
Acknowledgments
The authors thank the thousands of Finnish citizens who volunteered to participate in the FUSION study. We also gratefully acknowledge the hard work of Paula Nyholm, Juoko Sundvall, Tuula Tenkula, and Sanelma Vilkkilä. This project was made possible by intramural funds from National Human Genome Research Institute project OH95-C-N030. The work in Finland was partially supported by Finnish Academy grant 38387. We want to thank Darryl Leja for help with the figures. M.B. is supported by National Institutes of Health (NIH) grant HG00376. R.M.W. was previously supported by individual National Research Service Award DK09525 from the NIH and is now supported by a Career Development Award from the Americans Diabetes Association. R.N.B. is supported by NIH grants DK27619 and DK29867. C.D.L., J.A.D., W.L.D., M.P.E., E.ML., and H.M.S. were previously supported, and T.E.F and R.C.M. are currently supported, by NIH training grant HG00040. We also wish to thank two anonymous reviewers for their helpful comments.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics
- Online Mendelian Inheritance in Man (OMIM), http://www3.ncbi.nlm.nih.gov/Omim/ (for uncoupling protein 2 [MIM 601693], pyruvate carboxylase [MIM 266150], and muscle glycogen phosphorylase [MIM 232600])
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www-genome.wi.mit.edu/
References
- Bennett PH, Bogardus C, Tuomilehto J, Zimmett P (1992) Epidemiology and natural history of NIDDM: non-obese and obese. In: Alberti KGMM, DeFronzo RA, Keen H, Zimmett P (eds) International textbook of diabetes. John Wiley & Sons, New York, pp 147–176 [Google Scholar]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke M, The International Type 2 Diabetes Linkage Analysis Consortium (1998) Lessons learned in a combined linkage analysis (24 datasets, >2000 families) of type 2 diabetes on chromosome 20. Am J Hum Genet Suppl 63:A282 [Google Scholar]
- Bowden DW, Sale M, Howard TD, Qadri A, Spray BJ, Rothschild CB, Akots G, Rich SS, Freedman BI (1997) Linkage of genetic markers on human chromosomes 20 and 12 to NIDDM in Caucasian sib pairs with a history of diabetic nephropathy. Diabetes 46:882–886 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosome 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- Delépine M, Pociot F, Habita C, Hashimoto L, Froguel P, Rotter J, Cambon-Thomsen A, Deschamps I, Djoulah S, Weissenbach J, Nerup J, Lathrop M, Julier C (1997) Evidence of a non-MHC susceptibility locus in type I diabetes linked to HLA on chromosome 6. Am J Hum Genet 60:174–187 [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP (1999) Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehm MG, Karnoub MC, Sakul H, Gottschalk K, Holt DC, Weber JL, Vaske D, Briley D, Briley L, Kopf J, McMillen P, Nguyen Q, Reisman M, Lai EH, Joslyn G, Shepherd NS, Bell C, Wagner MJ, Burns DK, American Diabetes Association GENNID Study Group (2000) Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet 66:1871–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC (1997) The genetics of human noninsulin-dependent (type 2) diabetes mellitus. J Nutr 127:1891S–1896S [DOI] [PubMed] [Google Scholar]
- Elbein SC, Hoffman MD, Teng K, Leppert MF, Hasstedt SJ (1999) A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes 48:1175–1182 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hauser ER, Magnuson VL, Valle T, Ally DS, Karanjawala ZE, Rayman JB, et al (1998) A large sample of Finnish diabetic sib-pairs reveals no evidence for a non-insulin-dependent diabetes mellitus susceptibility locus at 2qter. J Clin Invest 102:704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karanjawala ZE, Hauser ER, Ally D, Knapp JI, Rayman JB, Musick A, Tannenbaum J, Te C, Shapiro S, Eldridge W, Musick T, Martin C, Smith JR, Carpten JD, Brownstein MJ, Powell JI, Whiten R, Chines P, Nylund SJ, Magnuson VL, Boehnke M, Collins FS, FUSION (Finland-US Investigation of NIDDM Genetics) Study Group (1997) Methods for precise sizing, automated binning of alleles, and reduction of error rates in large-scale genotyping using fluorescently-labeled dinucleotide markers. Genome Res 7:165–178 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Schork NJ (1996) Genetic analysis of NIDDM: the study of quantitative traits. Diabetes 45:1–14 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Hauser ER, Valle T, Magnuson VL, Erdos MR, Langefeld CD, et al (1999) Type 2 diabetes: evidence for linkage on chromosome 20 in 716 Finnish affected sib pairs. Proc Natl Acad Sci USA 96:2198–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P (1998) A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet 20:304–308 [DOI] [PubMed] [Google Scholar]
- Hanis CL, Boerwinkle E, Chakraborty R, Ellsworth D, Concannon P, Stirling B, Morrison VA, et al (1996) A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a susceptibility locus on chromosome 2. Nat Genet 13:161–166 [DOI] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Faroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M (1998) Genetic linkage analysis of complex genetic traits by using affected sibling pairs. Biometrics 54:1238–1246 [PubMed] [Google Scholar]
- Hauser ER, Boehnke M, Guo S-W, Risch N (1996) Affected-sib-pair interval mapping and exclusion for complex genetic traits: sampling considerations. Genet Epidemiol 13:117–137 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Boehnke M (1998) Stratified linkage analysis of complex genetic traits using related covariates. Am J Hum Genet Suppl 63:A45 [Google Scholar]
- Holmans P (1993) Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet 52:362–374 [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, Lindner T, Yamagata K, Ogata M, Tomonaga O, Kuroki H, Kasahara T, Iwamoto Y, Bell GI (1997) Mutation in hepatocyte nuclear factor-1β gene (TCF2) associated with MODY. Nat Genet 17:384–385 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Imperatore G, Hanson RL, Pettitt DJ, Kobes S, Bennett PH, Knowler WC, Pima Diabetes Genes Group (1998) Sib-pair linkage analysis for susceptibility genes for microvascular complications among Pima Indians with type 2 diabetes. Diabetes 47:821–830 [DOI] [PubMed] [Google Scholar]
- Ji L, Malecki M, Warram JH, Yang Y, Rich SS, Krolewski AS (1997) New susceptibility locus for NIDDM is localized to human chromosome 20q. Diabetes 46:876–881 [DOI] [PubMed] [Google Scholar]
- Kahn CR, Vicent D, Doria A (1996) Genetics of non-insulin dependent (type II) diabetes mellitus. Annu Rev Med 47:509–531 [DOI] [PubMed] [Google Scholar]
- Kenny SJ, Aubert RE, Geiss LS (1995) Prevalence and incidence of non-insulin-dependent diabetes. In: Harris MI, Cowie CC, Stern MP, Boyko EJ, Reiber GE, Bennett PH (eds) Diabetes in America, 2d ed. NIH publ 95-1468. National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, pp 47–67 [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M (1988) Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 5:471–472 [DOI] [PubMed] [Google Scholar]
- Magnuson VL, Ally DS, Nylund SJ, Karanjawala ZE, Rayman JB, Knapp JI, Lowe AL, Ghosh S, Collins FS (1996) Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. Biotechniques 21:700–709 [DOI] [PubMed] [Google Scholar]
- Mahtani MM, Widen E, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, Chan G, Daly M, Forsblom C, Kanninen T, Kirby A, Kruglyak L, Munnelly K, Parkkonen M, Reeve-Daly MP, Weaver A, Brettin T, Duyk G, Lander ES, Groop LC (1996) Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet 14:90–94 [DOI] [PubMed] [Google Scholar]
- Malecki MT, Jhala US, Antonellis A, Fields L, Doria A, Orban T, Saad M, Warram JH, Montminy M, Krolewski AS (1999) Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet 23:323–328 [DOI] [PubMed] [Google Scholar]
- Matise TC, Perlin M, Chakravarti A (1994) Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet 6:384–390 [DOI] [PubMed] [Google Scholar]
- Meunier F, Phillipi A, Martinez M, Demenais F (1997) Affected sib-pair tests for linkage: type 1 errors with dependent sib-pairs. Genet Epidemiol 14:1107–1111 [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Reinhart LJ, Stern MP (1994) NIDDM in Mexican American families: heterogeneity by age of onset. Diabetes Care 17:567–573 [DOI] [PubMed] [Google Scholar]
- Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, Garvey WT, Burns D, Knowler WC, Bennett PH, Bogardus C, Ravussin E (1998) Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet 62:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RA, Thompson DB, Faroud T, Garvey WT, Bennett PH, Bogardus C, Ravussin E (1997) Genomewide search for genes influencing percent body fat in Pima Indians: suggestive linkage at chromosome 11q21-q22. Am J Hum Genet 60:166–173 [PMC free article] [PubMed] [Google Scholar]
- Pratley RE, Thompson DB, Prochazka M, Baier L, Mott D, Ravussin E, Sakul H, Ehm MG, Burns DK, Foroud T, Garvey WT, Hanson RL, Knowler WC, Bennett PH, Bogardus C (1998) An autosomal genomic scan for loci linked to prediabetic phenotypes in Pima Indians. J Clin Invest 101:1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich SS (1990) Mapping genes in diabetes. Diabetes 39:1315–1319 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- Shuldiner AR, Silver KD (1996) Candidate genes for non-insulin-dependent diabetes mellitus. In: LeRoith D, Taylor S, Olefsky JM (eds) Diabetes mellitus. Lippincott-Raven, Philadelphia, pp 565–574 [Google Scholar]
- Stern MP, Duggirala R, Mitchell BD, Reinhart LJ, Shivakumar S, Shipman PA, Uresandi OC, Benavides E, Blangero J, O'Connell P (1996) Evidence for linkage of regions on chromsomes 6 and 11 to plasma glucose concentrations in Mexican Americans. Genome Res 6:724–734 [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Ferrer J, Clarke WL, Habener JF (1997) Early-onset type-II diabetes mellitus (MODY 4) linked to IPF1. Nat Genet 17:138–139 [DOI] [PubMed] [Google Scholar]
- Suarez BK, Hodge SE (1979) A simple method to detect linkage for rare recessive diseases: an application to juvenile diabetes. Clin Genet 15:126–136 [DOI] [PubMed] [Google Scholar]
- Tuomilehto J, Korhonen HJ, Kartovaara L, Salomaa V, Stengard JH, Pitkanen M, Aro A, Javela K, Uusitupa M, Pitkaniemi J (1991) Prevalence of diabetes mellitus and impaired glucose tolerance in the middle-aged population of three areas in Finland. Int J Epidemiol 20:1010–1017 [DOI] [PubMed] [Google Scholar]
- Valle T, Tuomilehto J, Bergman RN, Ghosh S, Hauser ER, Eriksson J, Nylund SJ, Kohtamäki K, Toivanen L, Vidgren G, Tuomilehto-Wolf E, Ehnholm C, Blaschak J, Langefeld CD, Watanabe RM, Magnuson V, Ally DS, Hagopian WA, Ross E, Buchanan TA, Collins F, Boehnke M (1998) Mapping genes for NIDDM: design of the Finland-United States Investigation of NIDDM Genetics (FUSION) study. Diabetes Care 21:949–958 [DOI] [PubMed] [Google Scholar]
- Valle T, Tuomilehto J, Eriksson J (1997) Epidemiology of NIDDM in Europids. In: Alberti KGMM, Zimmett P, DeFronzo RA, Keen H (eds) International textbook of diabetes mellitus. John Wiley & Sons, New York, pp 125–142 [Google Scholar]
- Vionnet N, Stoffel M, Takeda J, Yasuda K, Bell GI, Zouali H, Lesage S, Velho G, Iris F, Passa P, Froguel P, Cohen D (1992) Nonsense mutation in the glucokinase gene causes early-onset non-insulin-dependent diabetes mellitus. Nature 356:721–722 [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Ghosh S, Langefeld CD, Valle TT, Hauser ER, Magnuson VL, Mohlke KL, et al (2000) The Finland–United States Investigation of Non–Insulin-Dependent Diabetes Mellitus Genetics (FUSION) Study. II. An autosomal genome scan for diabetes-related quantitative-trait loci. Am J Hum Genet 67:1186–1200 (in this issue) [PMC free article] [PubMed] [Google Scholar]
- Workman PL, Mielke JH, Nevanlinna HR (1976) The genetic structure of Finland. Am J Phys Anthropol 44:341–367 [DOI] [PubMed] [Google Scholar]
- World Health Organization (1985) Report of the expert committee on diabetes. WHO tech rep 727. World Health Organization, Geneva [Google Scholar]
- Yamagata K, Furuta H, Oda N, Kaisaski P, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI (1996a) Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1). Nature 384:458–460 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, Southam L, et al (1996b) Mutations in the hepatocyte nuclear factor-1α gene in maturity-onset diabetes of the young (MODY 3). Nature 384:455–458 [DOI] [PubMed] [Google Scholar]
- Zouali H, Hani EH, Phillipi A, Vionnet N, Beckmann JS, Demenais F, Froguel P (1997) A susceptibility locus for early-onset non-insulin dependent (type 2) diabetes mellitus maps to chromosome 20q, proximal to the phosphoenolpyruvate carboxylase gene. Hum Mol Genet 6:1401–1408 [DOI] [PubMed] [Google Scholar]