Abstract
Type 2 diabetes mellitus is a complex disorder encompassing multiple metabolic defects. We report results from an autosomal genome scan for type 2 diabetes–related quantitative traits in 580 Finnish families ascertained for an affected sibling pair and analyzed by the variance components-based quantitative-trait locus (QTL) linkage approach. We analyzed diabetic and nondiabetic subjects separately, because of the possible impact of disease on the traits of interest. In diabetic individuals, our strongest results were observed on chromosomes 3 (fasting C-peptide/glucose: maximum LOD score [MLS] = 3.13 at 53.0 cM) and 13 (body-mass index: MLS = 3.28 at 5.0 cM). In nondiabetic individuals, the strongest results were observed on chromosomes 10 (acute insulin response: MLS = 3.11 at 21.0 cM), 13 (2-h insulin: MLS = 2.86 at 65.5 cM), and 17 (fasting insulin/glucose ratio: MLS = 3.20 at 9.0 cM). In several cases, there was evidence for overlapping signals between diabetic and nondiabetic individuals; therefore we performed joint analyses. In these joint analyses, we observed strong signals for chromosomes 3 (body-mass index: MLS = 3.43 at 59.5 cM), 17 (empirical insulin-resistance index: MLS = 3.61 at 0.0 cM), and 19 (empirical insulin-resistance index: MLS = 2.80 at 74.5 cM). Integrating genome-scan results from the companion article by Ghosh et al., we identify several regions that may harbor susceptibility genes for type 2 diabetes in the Finnish population.
Introduction
Type 2 diabetes is a heterogeneous disease characterized by insulin resistance and β-cell dysfunction (DeFronzo 1987; Bergman 1989). Left untreated, the fasting hyperglycemia can lead to a variety of secondary complications, including retinopathy, nephropathy, and neuropathy. Type 2 diabetes is also a risk factor for cardiovascular disease and hypertension (DeFronzo and Ferrannini 1991; Harris 1990) and contributes significantly to disease morbidity and mortality. Although the clinical characteristics of type 2 diabetes are well documented, the specific physiologic or biochemical defects responsible for the pathogenesis of the disease remain unclear. There is good evidence for a genetic component to type 2 diabetes (Rich 1990; Ghosh and Schork 1996). It is also clear that environmental factors play a major role in disease pathogenesis.
Several groups have performed genome scans for type 2 diabetes, in attempts to identify disease-susceptibility loci (Hanis et al. 1996; Mahtani et al. 1996; Hanson et al. 1998; Imperatore et al. 1998; Pratley et al. 1998; Duggirala et al. 1999; Elbein et al. 1999; Ghosh et al. 1999; Ehm et al. 2000). Although evidence has been reported for chromosomal regions that may contain type 2 diabetes–susceptibility genes, until recently only genes for specific subtypes of type 2 diabetes, such as maturity-onset diabetes of the young (MODY), have appeared in the literature (Bell et al. 1991; Froguel et al. 1992; Vaxillaire et al. 1995; Horikawa et al. 1997; Stoffers et al. 1997). These relatively rare subtypes of type 2 diabetes have been determined to be monogenic in nature, and multiple studies suggest that these genes play, at most, a minor role in genetic susceptibility for the more general form(s) of type 2 diabetes (Hanis et al. 1996; Mahtani et al. 1996; Elbein et al. 1999). Recently, the chromosome 2 locus linked to type 2 diabetes in Mexican Americans (Hanis et al. 1996) has been cloned and identified as calpain-10 (Horikawa et al. 2000). This raises the possibility that additional type 2 diabetes–susceptibility genes might be cloned and identified in the near future.
In this article and the companion article (Ghosh et al. 2000 [in this issue]), we describe genomewide results from the Finland–United States Investigation of NIDDM Genetics (FUSION) study, a multicenter effort to positionally clone genes for type 2 diabetes in the Finnish population. As described in the companion article by Ghosh et al. (2000 [in this issue]), we recruited and tested 580 families ascertained for an affected sibling pair. An important component of FUSION was the measurement of a variety of type 2 diabetes–related quantitative traits in subjects with and without disease. As part of our efforts to identify disease-predisposing loci, we performed an autosomal genome scan using these disease-related traits. The rationale behind this approach is that, because the traits examined are closely related to type 2 diabetes, any quantitative-trait locus (QTL) that we identify may also act as a disease-susceptibility or modifier locus. Coupled with the results from our type 2 diabetes genome scan (Ghosh et al. 2000 [in this issue]), we identify along the genome several regions that may harbor type 2 diabetes–susceptibility genes and that warrant further examination.
Subjects and Methods
Subjects and Phenotyping
The FUSION study design, recruitment, and phenotyping have been detailed elsewhere (Valle et al. 1998), and details regarding genotyping can be found in the companion article by Ghosh et al. (2000 [in this issue]). Therefore, in this article we only briefly review recruitment and phenotyping. Index cases were ascertained as those with age at onset of 35–60 years and at least one sibling diagnosed with type 2 diabetes. We successfully recruited and phenotyped 580 families, each with a minimum of one affected sibling pair (ASP) but, whenever possible, also collected additional siblings with type 2 diabetes. Two hundred ten of the families were also extended to include both a nondiabetic spouse and two or more nondiabetic offspring of the index case and/or diabetic siblings.
For all subjects, we collected basic demographic, anthropometric, and extensive medical-history information. Fasting glucose, insulin, lipids, and resting blood pressure were measured in all study subjects. Fasting C-peptide and glutamic acid decarboxylase antibody (GAD Ab) were measured in diabetic individuals, and these values were used in conjunction with insulin-treatment information to identify possible cases of late-onset type 1 diabetes, as described elsewhere (Valle et al. 1998). An oral glucose-tolerance test (OGTT), conforming to World Health Organization (1985) standards, was performed both in all nondiabetic subjects, to determine their glucose-tolerance status, and in diabetic subjects whose affection status could not be readily confirmed, yielding 2-h glucose and insulin measurements in these individuals. Because of the small number of diabetic individuals who underwent the OGTT, 2-h glucose and insulin data were analyzed only for the nondiabetic subjects.
Nondiabetic spouses and offspring of either index cases or diabetic siblings were also invited to undergo a tolbutamide-modified frequently sampled intravenous glucose-tolerance test (FSIGT), with Minimal Model analysis (Bergman et al. 1979; Steil et al. 1993), to derive quantitative estimates of glucose effectiveness (SG) and insulin sensitivity (SI). For these subjects, the acute insulin response to glucose (AIR) was computed as the incremental integrated area under the insulin curve for the first 8 min of the FSIGT and was used as an index of insulin secretion. We also computed the disposition index (DI), the product of SI and AIR, as a resistance-corrected index of β-cell function (Bergman et al. 1981; Kahn et al. 1993).
Empirical Metabolic Indices
Because we did not perform FSIGTs in our diabetic individuals, we could not scan for loci linked to the FSIGT-derived measures of insulin resistance and insulin secretion in these individuals. We attempted to circumvent this problem by constructing empirical indices for insulin resistance and insulin secretion, using the fasting-glucose, insulin, and C-peptide measurements from all our diabetic and nondiabetic subjects. For insulin resistance, we used the index described by Sluiter et al. (1976): SI(EST) = 1/(glucose × insulin). This empirical index for insulin resistance, SI(EST), was correlated with SI from the Minimal Model, in nondiabetic spouses and offspring (Spearman correlation: r=.54; P=.0001).
Although fasting insulin or C-peptide might be considered reflections of insulin secretion, these measures do not account for the variability introduced by the underlying stimulatory effect of the fasting-glucose concentration. Thus, we constructed empirical indices for insulin secretion, as follows: IRI = insulin/glucose and IRC = C-peptide/glucose. Since our indices for insulin secretion are derived from fasting measurements, they likely reflect basal insulin secretion, rather than stimulated insulin secretion. Therefore, compared with the correlation between SI and SI(EST), AIR was less strongly correlated with IRI in the nondiabetic spouses and offspring (Spearman correlation: r=.39; P=.0001). Fasting C-peptide was not measured in nondiabetic subjects; therefore, a comparison between AIR and IRC was not possible. However, given the equimolar relationship between insulin and C-peptide (Rubenstein et al. 1969), we would expect the correlation to be similar.
Medication Exclusions
Prior to clinical testing, all subjects, diabetic and nondiabetic, were asked to refrain from taking any of their prescription medications during the day of clinic visits. Data for any subject who, on the morning of blood sampling, took medications likely to affect the outcome of interest were excluded from analyses.
QTL Linkage Analysis
We used a multipoint variance-components approach for our QTL genome scan (Amos 1994; Blangero and Almasy 1997). We modeled the mean level of each quantitative-trait value as a linear function of age and sex. Because obesity is a known risk factor for type 2 diabetes (Lillioja and Bogardus 1988; Ohlson et al. 1988), and because genes for obesity may overlap with those for type 2 diabetes, we also tested models with additional adjustment for either body-mass index (BMI) or waist-to-hip ratio (WHR). Both BMI and WHR were also examined as quantitative traits. For this article, we chose to report only those results from the age- and sex-adjusted and age-, sex-, and BMI-adjusted traits. We report results from either the unadjusted trait or the additional adjustment for WHR only when they provide notably stronger evidence for linkage.
The total variance (σ2) for a quantitative trait was partitioned into components due to a major gene (σ2mg), additive polygenes (σ2pg), and individual-specific environment (σ2e): σ2=σ2mg+σ2pg+σ2e. Similarly, the covariance between the trait values for two noninbred individuals i and j was defined as Cov(Xi,Xj)=σ2mgIBDij+σ2pg2φij+σ2eδij. Here, IBDij is the estimated proportion of genes shared identical by descent (IBD) by i and j at the putative major gene; φij is the kinship coefficient, the probability that alleles drawn at random from individuals i and j, at a given locus, are IBD (Jacquard 1974); and δij=1 if i=j and δij=0 if i≠j.
All data were transformed to approximate univariate normality prior to analysis. Square root (in the case of C-peptide, DI, SI(EST), IRI, and IRC) or natural-logarithm transformations (in the case of fasting and 2-h glucose and insulin, SI) were used for most variables. Exceptions were SG (y0.25) and AIR (y0.185). Also, we used the natural log–transformed BMI and WHR values in the mean model, since their distributions were skewed. Maximum-likelihood parameter estimates were derived by use of the computer program USERFQTL. This software merges the IBD-estimation algorithm from SIBLINK (Hauser and Boehnke 1998) with the variance-components algorithm from the computer program FISHER (Lange et al. 1988). The presence of a major trait gene was assessed at each evaluation point along the genome, by the likelihood-ratio statistic (Λ), in which we compared models with the major-gene variance component set to 0 against those in which it was allowed to be positive. Because of the one-sided nature of this test, Λ is asymptotically distributed as a 50:50 mixture of χ2 on 1 df and a point mass at 0 (Self and Liang 1987), so that the P values are half as large as those in the two-sided case. For presentation purposes, in this article we have converted Λ to maximum LOD scores (MLSs).
Analyses were performed on nuclear families, with diabetic siblings analyzed separately from nondiabetic spouses and offspring, because of differences in trait distributions. In the case of nondiabetic-subject analyses, in which a diabetic index case or sibling served as a parent in the nuclear-family structure, the diabetic individual’s genotype information was included for the purpose of IBD estimation, but his/her quantitative trait values were set to missing, so as not to contribute to the variance-covariance structure. In addition, we performed analyses on the offspring only, to protect against possible violations of model assumptions by inclusion of the nondiabetic spouses. In the majority of cases, the offspring-only analyses gave results similar to those of the nondiabetic spouses and offspring together. Thus, we report only results from the combined nondiabetic spouse and offspring analyses.
We made the a priori decision to analyze diabetic and nondiabetic subjects separately, because of differences in the trait distributions in the two groups. These differences are likely due either to the traits being influenced by the diabetes per se or to the variety and variable efficacy of treatment regimens that the diabetic subjects were undergoing at the time of testing. However, for several traits, we observed overlapping linkage signals between diabetic and nondiabetic subjects. For these situations, we combined the diabetic and nondiabetic trait data and examined whether the trait distributions were approximately univariate normal (data not shown). If the combined trait distribution was approximately univariate normal, we combined the data from diabetics and nondiabetics in a single joint analysis. In these joint analyses, we included disease affection status as an additional covariate in our models. In addition, because USERFQTL limits the analysis to nuclear families, the extended family structure was broken down into individual nuclear families for analysis.
Statistical Significance
Lander and Kruglyak (1995) have proposed guidelines for interpretation of linkage results from genomewide scans. As such, their recommendations are applicable only for interpretation on a genomewide level. Furthermore, their simulations were designed to assess linkage based on affection status and not for QTLs. Therefore, we performed computer simulations to assess pointwise statistical significance more accurately for our most interesting results (MLS ⩾2.50). In each replicate sample, marker genotypes were simulated under the null hypothesis of no linkage, with the observed nuclear-family structures and phenotypes. Simulations of 10,000 or 100,000 replicates were generated, and the resulting LOD-score distribution was used to generate empirical P values. In general, the empirical P values were slightly larger than the nominal P values, suggesting that the nominal values tend to slightly overstate the evidence for linkage (data not shown). Thus, for MLS ⩾2.50 we report the empirical P values from our simulations.
Results
The mean, median, and SD of trait values for subjects analyzed for this QTL genome scan are shown in table 1. Table 2 provides a breakdown of analyses, by quantitative trait and group. As noted above, neither FSIGT-derived traits for diabetic subjects nor fasting C-peptide and, consequently, IRC, for nondiabetic subjects was available for analysis.
Table 1.
Phenotypes
|
Nondiabetic Subjects |
|||
| Diabetic Index Cases/Siblings (n=1,175) | Spouses (n=194) | Offspring (n=521) | |
| Ratio(Female:Male) |
|||
| Sex | 599:576 | 132:62 | 261:260 |
| Mean ± SD [Median] |
|||
| Age (years) | 64.2 ± 8.3 [64.8] | 61.5 ± 7.7 [61.3] | 34.6 ± 7.4 [34.8] |
| BMI (kg/m2) | 29.8 ± 4.8 [29.3] | 28.4 ± 4.5 [28.1] | 25.9 ± 4.5 [25.3] |
| WHR | .94 ± .08 [.94] | .88 ± .08 [.88] | .86 ± .08 [.85] |
| Fasting glucose (mM) | 10.2 ± 3.5 [9.6] | 5.2 ± .6 [5.2] | 5.1 ± .6 [5.0] |
| 2-h glucose (mM) | … | 6.2 ± 1.8 [6.0] | 5.3 ± 1.5 [5.2] |
| Fasting insulin (pM) | 111 ± 67 [96] | 76 ± 49 [66] | 67 ± 35 [60] |
| 2-h insulin (pM) | … | 427 ± 315 [348] | 317 ± 245 [228] |
| Fasting C-peptide (nM) | 1.59 ± .93 [1.44] | … | … |
| SI(EST) (×100) | .18 ± .31 [.11] | .35 ± .32 [.29] | .40 ± .34 [.34] |
| IRI | 12.0 ± 8.1 [10.3] | 14.4 ± 8.2 [12.9] | 13.2 ± 6.4 [12.3] |
| IRC | .17 ± .11 [.14] | … | … |
| SG (×100 min−1) | … | 1.65 ± .57 [1.59] | 1.77 ± .58 [1.70] |
| SI [×10−5min−1/pM) | … | 5.88 ± 3.41 [5.41] | 7.57 ± 4.48 [6.71] |
| AIR (pM ×8 min) | … | 2,331 ± 1,624 [1,868] | 2,188 ± 1,528 [1,897] |
| DI | … | 12,244 ± 8,726 [9,983] | 14,178 ± 9,163 [12,538] |
Table 2.
Analyses of Diabetic and Nondiabetic Subjects
| Diabetic Subjects | Nondiabetic Subjects | |
| BMI | • | • |
| WHR | • | • |
| Fasting glucose | • | • |
| 2-h glucose | • | |
| Fasting insulin | • | • |
| 2-h insulin | • | |
| Fasting C-peptide | • | |
| SI(EST) | • | • |
| IRI | • | • |
| IRC | • | |
| SG | • | |
| SI | • | |
| AIR | • | |
| DI | • |
Tables 3 and 4 summarize MLS ⩾1.18 (corresponding to P<.01) for diabetic and nondiabetic subjects, respectively. If more than one model yielded a result greater than this cutoff for a given trait, we report only the result with the highest MLS. For 13% of the results reported in tables 3 and 4, one of the alternative models using different covariates yielded an MLS that was both either lower or higher than the value reported in the table and above the 1.18 MLS cutoff value.
Table 3.
MLS ⩾1.18 in Diabetic Subjects
| Position(cM) | Trait | Covariatesa | MLS | Pb |
| Chromosome 1: | ||||
| 98.0 | BMI | 1 | 1.19 | .0097 |
| 164.5 | IRC | 1 | 1.45 | .0049 |
| Chromosome 3: | ||||
| 21.0 | IRI | 2 | 1.39 | .0057 |
| 53.0 | IRC | 2 | 3.13 | .0001 |
| 56.0 | BMI | 1 | 1.19 | .0097 |
| 58.0 | C-peptide | 2 | 2.26 | .0006 |
| Chromosome 4: | ||||
| 28.5 | IRI | 1 | 1.38 | .0058 |
| Chromosome 5: | ||||
| 198.0 | BMI | 1 | 1.24 | .0084 |
| Chromosome 7: | ||||
| 74.5 | C-peptide | 2 | 1.63 | .0031 |
| 74.5 | IRC | 2 | 1.76 | .0022 |
| 101.0 | Insulin | 2 | 1.94 | .0014 |
| Chromosome 8: | ||||
| 103.5 | Glucose | 1 | 1.23 | .0042 |
| Chromosome 9: | ||||
| 0.0 | Insulin | 1 | 1.32 | .0068 |
| 5.0 | C-peptide | 2 | 1.36 | .0061 |
| Chromosome 11: | ||||
| 89.5 | Insulin | 2 | 2.07 | .0010 |
| 91.0 | IRI | 2 | 2.32 | .0005 |
| Chromosome 13: | ||||
| 7.0 | BMI | 1 | 2.95 | .0002 |
| 39.5 | C-peptide | 1 | 1.18 | .0098 |
| 41.0 | Insulin | 1 | 1.22 | .0090 |
| Chromosome 14: | ||||
| 96.5 | SI(EST) | 2 | 1.19 | .0096 |
| 102.0 | Insulin | 1 | 2.67 | .0008 |
| Chromosome 17: | ||||
| 0.0 | SI(EST) | 2 | 1.54 | .0039 |
| Chromosome 19: | ||||
| 8.5 | BMI | 1 | 1.45 | .0049 |
| 73.5 | SI(EST) | 1 | 1.82 | .0019 |
1 = age and sex; 2 = age, sex, and BMI.
For MLS ⩾2.50, empirical P values from simulations are reported. For details, see the Subjects and Methods section.
Table 4.
MLS ⩾1.18 in Nondiabetic Subjects[Note]
| Position(cM) | Trait | Covariates | MLS | P |
| Chromosome 1: | ||||
| 131.5 | SG | 1 | 1.41 | .0054 |
| 165.5 | Glucose | 1 | 1.67 | .0028 |
| 265.0 | Insulin | 2 | 1.37 | .0060 |
| 267.0 | 2-h insulin | 2 | 2.63 | .0003 |
| Chromosome 2: | ||||
| 97.0 | SG | 1 | 1.22 | .0090 |
| 145.0 | SI | 2 | 1.43 | .0051 |
| Chromosome 3: | ||||
| 38.0 | DI | 1 | 1.31 | .0071 |
| 40.0 | Glucose | 2 | 2.22 | .0008 |
| 65.0 | BMI | 1 | 2.16 | .0008 |
| Chromosome 6: | ||||
| 41.0 | Glucose | 2 | 1.92 | .0015 |
| 69.0 | BMI | 1 | 1.18 | .0100 |
| 119.5 | Insulin | 1 | 1.85 | .0019 |
| 119.5 | IRI | 1 | 2.02 | .0012 |
| Chromosome 7: | ||||
| 0.0 | Glucose | 2 | 1.24 | .0085 |
| 82.0 | BMI | 1 | 2.21 | .0007 |
| Chromosome 8: | ||||
| 13.0 | DI | 1 | 1.32 | .0069 |
| 135.5 | BMI | 1 | 2.29 | .0006 |
| Chromosome 9: | ||||
| 51.0 | 2-h insulin | 2 | 1.19 | .0097 |
| 114.5 | AIR | 2 | 1.97 | .0013 |
| Chromosome 10: | ||||
| 12.5 | Glucose | 1 | 1.23 | .0087 |
| 21.0 | AIR | 2 | 3.11 | .0001 |
| 102.0 | SG | 1 | 1.63 | .0031 |
| Chromosome 12: | ||||
| 129.5 | BMI | 1 | 1.36 | .0061 |
| Chromosome 13: | ||||
| 62.0 | IRI | 2 | 1.37 | .0059 |
| 65.5 | 2-h insulin | 1 | 2.86 | .0002 |
| Chromosome 14: | ||||
| 0.0 | DI | 1 | 1.19 | .0096 |
| 1.0 | SI | 2 | 1.46 | .0047 |
| 5.5 | 2-h insulin | 2 | 1.51 | .0042 |
| 25.5 | Glucose | 1 | 2.01 | .0012 |
| Chromosome 15: | ||||
| 74.0 | SI(EST) | 1 | 1.18 | .0100 |
| 74.0 | Insulin | 2 | 1.53 | .0040 |
| 75.0 | IRI | 2 | 1.48 | .0045 |
| Chromosome 16: | ||||
| 42.0 | SI(EST) | 1 | 1.47 | .0046 |
| 44.5 | IRI | 2 | 1.89 | .0016 |
| 45.0 | Insulin | 2 | 1.71 | .0025 |
| 65.0 | 2-h insulin | 1 | 1.28 | .0077 |
| 78.0 | SG | 1 | 1.33 | .0067 |
| Chromosome 17: | ||||
| 0.0 | 2-h glucose | 2 | 1.44 | .0051 |
| 9.0 | Insulin | 1 | 2.97 | .0001 |
| 9.0 | 2-h insulin | 2 | 2.71 | .0006 |
| 9.0 | SI(EST) | 1 | 2.25 | .0006 |
| 9.0 | IRI | 1 | 3.20 | <.0001 |
| 46.5 | SG | 1 | 1.27 | .0077 |
| Chromosome 18: | ||||
| 36.0 | WHR | 1 | 2.61 | .0003 |
| Chromosome 19: | ||||
| 72.5 | 2-h insulin | 1 | 1.39 | .0057 |
| 74.0 | IRI | 1 | 1.52 | .0040 |
| 75.5 | Insulin | 1 | 1.38 | .0058 |
| 76.5 | SI(EST) | 1 | 1.94 | .0014 |
| 79.5 | 2-h glucose | 1 | 1.40 | .0065 |
| 94.5 | Glucose | 1 | 2.05 | .0011 |
| Chromosome 20: | ||||
| 9.5 | 2-h glucose | 2 | 1.76 | .0022 |
| 19.5 | IRI | 1 | 1.19 | .0095 |
| Chromosome 22: | ||||
| 0.0 | 2-h insulin | 1 | 2.08 | .0010 |
Note.— Data are as defined in the footnotes to table 3.
Diabetic Subjects
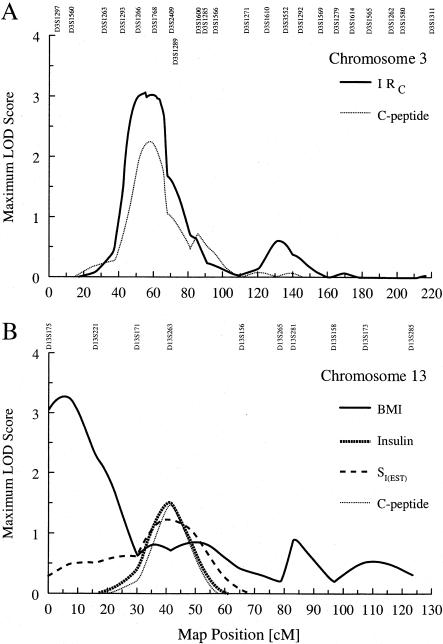
In the analyses of our diabetic subjects, the strongest evidence for QTL linkage was observed on chromosomes 3 and 13, for IRC and BMI, respectively (fig. 1). On chromosome 3 (fig. 1A), the MLS for age-, sex-, and BMI-adjusted IRC was 3.13 at 53.0 cM, between markers D3S1266 and D3S1768, and had a 1-LOD support interval of 44.0–67.0 cM. As might be expected, because it is a component of IRC, age-, sex-, and BMI-adjusted fasting C-peptide also showed evidence for linkage (MLS = 2.26) in the same region of chromosome 3, and it yielded a maximum at 58.0 cM near marker D3S1768.
Figure 1.
A, MLS curves for age-, sex-, and BMI-adjusted IRC and for age-, sex-, and BMI-adjusted fasting C-peptide, for chromosome 3 in diabetic subjects. B, MLS curves for results for chromosome 13 in diabetic subjects. Unadjusted BMI showed the strongest evidence for linkage. However, a cluster of other traits—fasting insulin; SI(EST); and age-, sex-, and WHR-adjusted fasting C-peptide—showed weaker evidence for linkage at a different locus. Marker names and positions are given at the top of each graph.
On chromosome 13 (fig. 1B), the MLS for BMI was 3.28 at 5.0 cM, between markers D13S175 and D13S221, and had a 1-LOD support interval of 0.0–16.0 cM. With adjustment for age and sex, this signal dropped to an MLS of 2.95 at 7.0 cM. There were no other linkage peaks in this region of the chromosome, for any of the other traits. A second region along chromosome 13 (fig. 1B) showed more-modest evidence for linkage to fasting insulin (MLS = 1.52 at 41.0 cM), SI(EST) (MLS = 1.47 at 41.5 cM), and age-, sex-, and WHR-adjusted fasting C-peptide (MLS = 1.23 at 40 cM). All three signals maximized near marker D13S263.
Other noteworthy results include those for age- and sex-adjusted fasting insulin on chromosome 14 (MLS = 2.67 at 102.0 cM, near marker D14S95), as well as both IRI (MLS = 2.32 at 91.0 cM, near D11S1311) and age-, sex-, and BMI-adjusted fasting insulin, on chromosome 11. The MLS curve for fasting insulin maximized at 90.0 cM (MLS = 2.26 near D11S1311), and there was a secondary peak at 53.5 cM (MLS = 1.52). When SI(EST) was examined, this secondary peak showed evidence for linkage (MLS = 1.68). Given that SI(EST) is derived on the basis of fasting insulin, this suggests that on this chromosome there may be a QTL involved in some complex interaction between insulin and glucose.
Nondiabetic Subjects
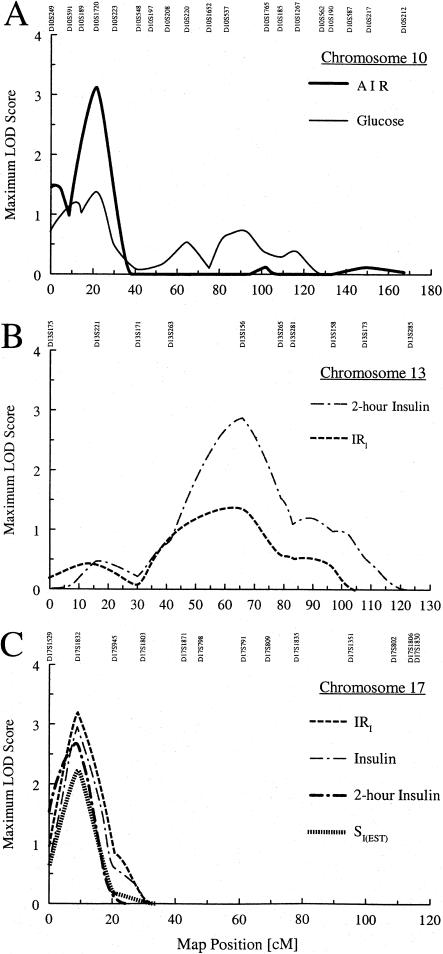
In the analysis of the nondiabetic spouses and offspring, the strongest evidence for linkage was observed on chromosomes 10, 13, and 17 (fig. 2). On chromosome 10 (fig. 2A), the MLS for age-, sex-, and BMI-adjusted AIR was 3.11 at 21.0 cM, near marker D10S1720, with a 1-LOD support interval of 13.0–27.5 cM. Fasting glucose also showed a weaker linkage peak in the same region (MLS = 1.38 at 21.0 cM). Unadjusted fasting insulin and the empirical indices IRI, IRC, and SI(EST) all showed very modest linkage peaks (MLS = 0.58–0.89) in the same region (data not shown).
Figure 2.
A, MLS curves for age-, sex-, and BMI-adjusted AIR and for fasting glucose, for chromosome 10 in nondiabetic subjects. B, MLS curves for age- and sex-adjusted 2-h insulin and for age-, sex-, and BMI-adjusted IRI, for chromosome 13 in nondiabetic subjects. C, MLS curves for age- and sex-adjusted IRI, age- and sex-adjusted fasting insulin, for age-, sex-, and BMI-adjusted 2-h insulin, and for age- and sex-adjusted SI(EST), for chromosome 17 in nondiabetic subjects.
On chromosome 13, the MLS for age- and sex-adjusted 2-h insulin was 2.86 at marker D13S156 at 65.5 cM (fig. 2B); however, the 1-LOD support interval was very wide (50.0–76.5 cM). Age-, sex-, and BMI-adjusted IRI also showed evidence for linkage in the same region (MLS = 1.37 at 62.0 cM). On chromosome 17 (fig. 2C), age- and sex-adjusted IRI showed the strongest evidence for linkage, with MLS = 3.20 at 9.0 cM (1-LOD support interval 4.0–15.0 cM), near marker D17S1832. Not surprisingly, age- and sex-adjusted fasting insulin (MLS = 2.97) and SI(EST) (MLS = 2.25) both showed evidence for linkage in this region. However, age-, sex-, and BMI-adjusted 2-h insulin also showed evidence for linkage at the same location (MLS = 2.71), with a 1-LOD support interval of 0.5–13.5 cM (fig. 2C). Other noteworthy results in our nondiabetic subjects include 2-h insulin, on chromosome 1 (MLS = 2.70 at 270.0 cM); BMI, on chromosome 3 (MLS = 2.52 at 64.0 cM); age- and sex-adjusted WHR, on chromosome 18 (MLS = 2.61 at 36.0 cM); and age-, sex-, and WHR-adjusted 2-h insulin, on chromosome 22 (MLS = 2.70 at 0.0 cM).
Overlap of Results for Diabetic and Nondiabetic Subjects
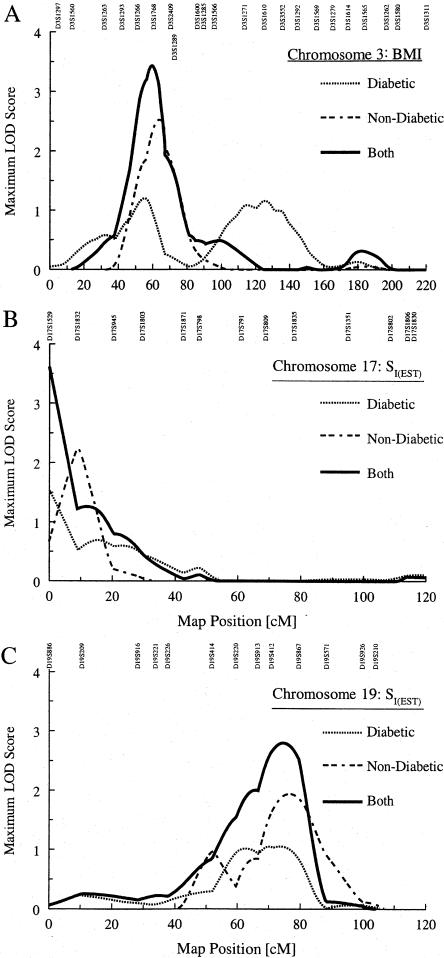
We observed some regions where evidence for linkage to a specific quantitative trait overlapped between diabetic and nondiabetic subjects. Specifically, on chromosome 3 we observed a stronger signal (MLS = 2.52 at 64.0 cM) for BMI in our nondiabetic subjects and a weaker signal in the same proximity in our diabetic subjects (MLS = 1.19 at 56.0 cM), after adjustment for age and sex. We also observed a similar situation with SI(EST) on chromosomes 17 and 19. For these chromosomes, we repeated QTL linkage analyses for these traits, combining data from both groups in a joint analysis.
In the joint analysis of all subjects for BMI without adjustment for covariates on chromosome 3, we observed an MLS of 3.43 at 59.5 cM (fig. 3A); when we adjusted for age and sex, the MLS dropped to 2.83. Similarly, for SI(EST) on chromosome 17, when we combined our diabetic and nondiabetic subjects in a joint analysis, we observed an MLS of 3.61 at 0.0 cM, after adjusting for age, sex, and BMI (fig. 3B). The strong telomeric signal suggests the need for additional markers in this region, to refine our linkage signal. For chromosome 19, the joint analysis yielded an MLS of 2.80 at 74.5 cM after adjustment for age, sex, and WHR (fig. 3C).
Figure 3.
A, MLS curves for age- and sex-adjusted BMI in diabetic subjects and for BMI in nondiabetic subjects, for chromosome 3. When data from both groups are analyzed jointly, BMI shows a strong signal at 59.5 cM, with an MLS of 3.43. B, MLS curves for age-, sex-, and BMI-adjusted SI(EST) in diabetic subjects and for age- and sex-adjusted SI(EST) in nondiabetic subjects, for chromosome 17. Joint analysis of age-, sex-, and BMI-adjusted SI(EST) shows a stronger signal (MLS = 3.61) at the p-terminus. C, MLS curves for age- and sex-adjusted SI(EST), in both diabetic and nondiabetic subjects, for chromosome 19. Joint analysis of age-, sex-, and WHR-adjusted SI(EST) shows a stronger signal (MLS = 2.80) at 74.5 cM.
There were other examples of overlapping signals across traits. On chromosome 1, the signal for age- and sex-adjusted IRC in the diabetic subjects (MLS = 1.61) and fasting glucose for the same adjustment in the nondiabetics (MLS = 1.66) both maximized in the same region (∼165 cM). Similarly, on chromosome 3, age-, sex-, and BMI-adjusted fasting C-peptide in the diabetics (MLS = 2.26) and age- and sex-adjusted BMI in the nondiabetics (MLS = 2.52) maximized in close proximity, at 58.0 and 64.0 cM, respectively. Chromosome 19 showed a particularly interesting set of four outcomes: the evidence for linkage to age- and sex-adjusted SI(EST) in diabetic subjects (MLS = 1.82 at 73.5 cM) overlapped with those for 2-h insulin (MLS = 1.59 at 71.0 cM) in the nondiabetic subjects, age- and sex-adjusted SI(EST) (MLS = 1.94 at 76.5 cM) in the nondiabetic subjects, and age- and sex-adjusted IRI (MLS = 1.52 at 74.0 cM) in the nondiabetic subjects.
Discussion
Type 2 diabetes has all the characteristics of a complex disorder: convincing evidence for a genetic component, ambiguous mode of inheritance, strong environmental component, and likely multiple genes of varying effect that work independently and/or in concert (Rich 1990; Ghosh and Schork 1996). The difficult task of identifying the susceptibility genes is complicated by the fact that the interaction of these factors results in a variety of disease etiologies. Thus, although, because of clearly defined diagnostic criteria (National Diabetes Data Group 1979; World Health Organization 1985), individuals can be identified as having type 2 diabetes, not all individuals who eventually develop type 2 diabetes follow the same path toward disease, nor will they follow the same progression toward disease complications. These complex features may account for why, despite the large number of groups performing genomic scans for type 2 diabetes (Hanis et al. 1996; Mahtani et al. 1996; Hanson et al. 1998; Imperatore et al. 1998; Pratley et al. 1998; Duggirala et al. 1999; Elbein et al. 1999; Ghosh et al. 1999; Ehm et al. 2000), there have been, until recently, no published reports of a successful cloning and identification of a major susceptibility gene. However, recently the chromosome 2 locus linked to type 2 diabetes in Mexican Americans, also known as “NIDDM1” (Hanis et al. 1996), has been cloned and identified as calpain-10, a calpain-like cysteine protease (Horikawa et al. 2000).
In this article and the companion article (Ghosh et al. 2000 [in this issue]), we have reported the initial results of a genomewide scan for linkage to diabetes and diabetes-related quantitative traits in the FUSION study. In the companion article (Ghosh et al. 2000 [in this issue]), we have reported results from our genomewide scan of affected sibling pairs sampled from the population of Finland. Given the expected difficulties in doing a genome scan for a complex disease, we augmented our ASP linkage results by doing disease-by-marker association analysis, ordered-subsets linkage analysis of the disease status data, and QTL linkage analysis of our quantitative-trait data. In this discussion, we focus on the results from our QTL genome scan but also draw on results from our other analyses, where our evidence for a possible diabetes-susceptibility gene may be the strongest.
The goal of QTL linkage analysis is to identify loci that influence specific quantitative traits; in our study, the traits that we examined are also intimately associated with type 2 diabetes. Through this approach, we hoped to identify QTLs that might also act as diabetes-susceptibility loci. We identified along the genome several regions that may contain QTLs. In our analytical strategy, we chose to analyze data from diabetic and nondiabetic subjects separately. There were two reasons for taking this approach. First, the characteristics of the trait distribution between diabetic and nondiabetic subjects tended to be different. This likely reflects the possibility that the trait may behave differently in diabetic individuals, because of disease-induced alterations in physiology and/or biochemistry. In addition, >90% of our diabetic volunteers were being treated for their disease through pharmacological intervention, and a large proportion of the remainder were treated via lifestyle modification—that is, changes in diet and/or physical activity. Thus, trait information in these subjects is likely to be confounded by these treatment effects.
Despite these potential differences, we did observe overlapping signals for certain traits across the two groups. We further investigated these traits by combining data from diabetic and nondiabetic subjects after examining the trait distribution of the joint data. In the joint analyses, we observed substantial increases in some of our overall MLSs (fig. 3).
A secondary consideration is the issue of overlapping linkage signals across different traits. We found these overlaps of particular interest, since many of the traits that we examined in FUSION, although clinically or physiologically important, still reflect the net integrated effect of multiple downstream biochemical events. Furthermore, because many of these traits are part of the endocrine hormone–feedback regulatory system, it would not be unusual to see a single locus having pleiotropic effects on two or more traits. Thus, we believe that overlapping signals across traits should be carefully evaluated, taking into account whether such clustering of traits tells a coherent story.
A large proportion of our linkage results involved traits associated with β-cell function: fasting insulin, fasting C-peptide, 2-h insulin, and AIR. This suggests that genes associated with β-cell function may be exerting the strongest genetic effects in the population that we studied, and it is consistent with our observation that, in our sample, heritability for secretion-related traits was stronger than that for insulin resistance–related traits (Watanabe et al. 1999). This pattern was also found in a twin study in the same population (Lehtovirta et al. 2000).
Both the National Institutes of Health (NIH) group in Phoenix that is studying the Pima Indians (Pratley et al. 1998) and the group studying the Old Order Amish (St. Jean et al. 1999) have completed QTL genome scans for type 2 diabetes–related quantitative traits, and the Phoenix group has completed linkage analyses for diabetic nephropathy and retinopathy (Imperatore et al. 1998). Also, several groups have reported QTL linkage results for specific chromosomes (Thompson et al. 1995; Duggirala et al. 1996, 1999; Stern et al. 1996; Comuzzie et al. 1997). In what follows, we note regions along the genome where evidence for linkage in our QTL genome scan appears to overlap with evidence reported by these different studies and where results from our own ASP linkage analyses provide additional evidence for regions potentially harboring type 2 diabetes–susceptibility genes.
On chromosome 3 in Pima Indians, two peaks for fasting insulin (MLS = 1.30 at 35 cM and MLS = 1.25 at 45 cM) that flank our linkage peak (MLS = 2.22) for fasting glucose at 40 cM in nondiabetic subjects have been reported (Pratley et al. 1998). Also, the GENNID study recently reported evidence for linkage (MLS = 3.91) to type 2 diabetes in Mexican Americans, at marker D3S2432 (∼50 cM) (Ehm et al. 2000). We also have observed weaker linkage signals for BMI (MLS = 1.27 at 55.0 cM) in our diabetic subjects and for both AIR (MLS = 1.12 at 47.0 cM) and DI (MLS = 1.31 at 38.0 cM) in our nondiabetic subjects. This clustering of insulin secretion–related traits in our sample suggests that this QTL may be involved in the regulation of β-cell function.
The gene for peroxisome-proliferator-activator receptor-γ2 (PPARG [MIM 601487]) is near this region of chromosome 3. PPARG plays an important role in adipogenesis and insulin sensitization (Auwerx 1999). Recent reports, including our own study (J. A. Douglas, M. R. Erdos, R. M. Watanabe, A. Braun, C. L. Johnston, P. Oeth, K. L. Mohlke, T. T. Valle, C. Ehnholm, T. A. Buchanan, R. N. Bergman, F. S. Collins, M. Boehnke, and J. Tuomilehto, unpublished data), have shown an association between the presence of the Pro12Ala polymorphism in PPARG and both type 2 diabetes (Deeb et al. 1998) and obesity (Beamer et al. 1998).
We also observed linkage signals proximal to the PPARG region. These include fasting C-peptide (MLS = 2.26 at 58.0 cM) and IRC (MLS = 3.13 at 53.0 cM) in our diabetic subjects and BMI (MLS = 2.52 at 64.0 cM) in our nondiabetic subjects. In Pima Indians, peaks for fasting insulin (MLS = 1.22 at 188 cM [Pratley et al. 1998]) and for diabetic nephropathy (MLS = 2.03 at 181.1 cM [Imperatore et al. 1998]) have been reported near the signal that we found for SI(EST) (MLS = 1.59) at 196 cM. This region includes the gene for somatostatin (SST [MIM 182450]).
The Phoenix group also has reported, on chromosome 6, possible linkage to fasting glucose (MLS = 1.79 at 41 cM [Pratley et al. 1998]), which coincides with our evidence for linkage (MLS = 1.92) for the same trait and chromosomal location (table 4) in our nondiabetic subjects. Stern et al. (1996) have reported linkage to fasting glucose, on chromosome 6 at D6S300, ∼50 cM away from the location of our linkage peak, toward the q-terminus. This 40–50-cM region of chromosome 6 includes a variety of plausible candidate genes, including those for the major histocompatibility complex, tumor necrosis factor-α (TNF [MIM 191160]), and glucagon-like peptide-1 receptor (GLP1R [MIM 138032]).
In our nondiabetic subjects, we also had, for both fasting insulin (MLS = 1.85 at 119.5 cM) and IRI (MLS = 2.02 at 119.5 cM), interesting QTL results that overlapped with those of our ASP-based linkage analysis (MLS = 0.61-0.78 near 113 cM) (Ghosh et al. 2000 [in this issue]). Ordered-subsets analysis revealed two subsets with strong evidence for linkage to this region: low age at diagnosis (74 families; MLS = 2.48 at 104.5 cM) and low fasting glucose (94 families; MLS = 3.17 at 117.0 cM). In addition, in our nondiabetic subjects, we observed weaker QTL linkage results for 2-h insulin (MLS = 1.16 at 128.0 cM) and SI(EST) (MLS = 1.17 at 125.0 cM). Stern et al. (1996) have reported, near this region of chromosome 6 in their sample of Mexican Americans, evidence for linkage to fasting glucose (MLS = 2.17). Also, linkage to age-adjusted type 2 diabetes has been reported in this region (Z-score = 1.39 at 128 cM) in the Pima Indians (Hanson et al. 1998).
The Phoenix group has reported evidence for linkage, on chromosome 9, to both 2-h insulin (MLS = 2.16 at 98 cM [Pratley et al. 1998]) and diabetic nephropathy (two-point MLS = 1.28 at 114.6 cM [Imperatore et al. 1998]), near our peak (MLS = 1.97 at 114.5 cM) for AIR. This region includes the genes for muscle and hepatic fructose-1,6 bisphosphatase (FBP2 [MIM 603027] and FBP1 [MIM 229700]), a key rate-limiting enzyme for gluconeogenesis.
In the region on chromosome 19 where we observed evidence for linkage to multiple traits, Pratley et al. (1998) have reported linkage to 2-h glucose (MLS = 1.14 at 75 cM). We also observed a weak association between marker D19S867 at 79.5 cM and affection status (P=.05; Ghosh et al. 2000 [in this issue]). This region includes genes for the muscle form of creatine kinase (CKM [MIM 123310]), gastric inhibitory peptide receptor (GIPR [MIM 137241]), hepatocyte nuclear factor 3γ (HNF3G [MIM 602295]), and muscle glycogen synthase I (GYS1 [MIM 138570]). GYS1 is the rate-limiting step for glycogen formation in muscle and has been examined as a candidate gene, by both Groop et al. (1993) and the Phoenix group (Majer et al. 1996). Groop et al. (1993) noted a specific allele that identified a subgroup of their diabetic subjects with both a strong family history of disease and a higher prevalence of hypertension and insulin resistance. In contrast, Majer et al. (1996) found no mutations in the GYS1 gene or alterations in mRNA expression or enzymatic activity.
Elsewhere, we have reported, for chromosome 20, modest QTL results (Ghosh et al. 1999) that overlapped with the ASP linkage peak toward the p-terminus of the chromosome. We have since genotyped an additional 5 markers, bringing the total to 43 markers genotyped on chromosome 20. The information provided by these markers modestly increased our QTL linkage signals for 2-h insulin (from MLS = 1.06 at 19.0 cM to MLS = 1.09 at 18.5 cM), IRI (from MLS = 1.12 at 19.5 cM to MLS = 1.19 at 19.5 cM), and 2-h glucose (from MLS = 1.59 at 9.5 cM to MLS = 1.70 at 9.0 cM).
Finally, on the p-terminus of chromosome 22, the Phoenix group reports evidence for linkage to high-dose M-value from the glucose clamp (MLS = 2.12), corresponding to our result for 2-h insulin (MLS = 2.70). We are not aware of any obvious candidate genes in this region.
We identified other candidate genes that, on the basis of our QTL genome scan, fall into regions of interest. Many, if not all, have been examined as candidate genes in a variety of type 2 diabetes populations. On chromosome 1, the gene for pyruvate kinase (PK1 [MIM 266200]), a key glycolytic enzyme, and the gene for the G-protein coupled potassium channel (KCNJ9 [MIM 600932], also known as “GIRK3” or “Kir3.3”) fall within the region in which we observed evidence for linkage both to IRC in our diabetic subjects (MLS = 1.61) and to fasting glucose in our nondiabetic subjects (MLS = 1.66). Genes for insulin-like growth factor–binding proteins 1 and 3 (IGFBP1 [MIM 146730] and IGFBP3 [MIM 146732]) are near the chromosome 7 region in which we observed results, in diabetic subjects, for fasting C-peptide (MLS = 1.63) and IRC (MLS = 1.76). IGF-binding proteins regulate IGF activity and, under some conditions, may play a role in development of diabetic complications (Bach and Rechler 1995). Glucokinase (GCK [MIM 138079]), the key rate-limiting enzyme for glycolysis, is found just proximal to this region, and polymorphisms in this gene are responsible for MODY-2 (Froguel et al. 1992; Vionnet et al. 1992) St. Jean et al. (1999) have noted, on chromosome 14, evidence for linkage to fasting glucose (MLS = 2.00 at 29 cM) and to 2-h glucose (MLS = 2.53 at 22 cM), in large pedigrees from the Old Order Amish. These results coincide with our fasting-glucose result (MLS = 2.28 at 24.5 cM) in nondiabetic subjects. The genes for phosphoenolpyruvate carboxykinase 2 (PCK2 [MIM 261650]) and hepatocyte nuclear factor 3α (HNF3A [MIM 602294]) flank this region of chromosome 14. Finally, on chromosome 17, genes for tyrosine kinase nonreceptor 1, glucose transporter 4 (SLC2A4 [MIM 138190]), mitogen-activated protein kinase kinase 4 (MAP2K4 [MIM 601335]), and adenosine A2b receptor (ADORA2B [MIM 600446]) fall into the region of our linkage peaks for fasting insulin (MLS = 2.97), 2-h insulin (MLS = 2.71), SI(EST) (MLS = 2.25), and IRI (MLS = 3.20), at 9.0 cM in our nondiabetic subjects.
In summary, we report the results from both a type 2 diabetes genome scan (Ghosh et al. 2000 [in this issue]) and an autosomal genome scan for type 2 diabetes–related quantitative traits. Our QTL linkage analyses reveal along the genome several regions that may include genes related to both adiposity and insulin secretion in our Finnish cohort. Furthermore, there is overlap between our QTL linkage results and results from our disease status–based linkage results. When we integrated the results from all of our analyses, we identified, on chromosomes 1–3, 6, 7, 10, 11, 17, 19, and 20, regions that have evidence for possible diabetes-susceptibility loci. We have now targeted these regions for further statistical analyses and/or additional genotyping, to further refine our evidence for linkage.
Acknowledgments
The authors thank the thousands of Finnish citizens who volunteered to participate in the FUSION study. We also gratefully acknowledge the hard work of the numerous field workers in Finland, as well as the work of Paula Nyholm, Jouko Sundvall, Tuula Tenkula, and Sanelma Vilkkilä. We would also like to thank Darryl Leja at the National Human Genome Research Institute (NHGRI) for his assistance in preparing the figures for this article. This project was made possible by intramural funds from NHGRI project OH95-C-N030 and by NIH grant HG00376 (to M.B.); the project was also partly supported by the Finnish Academy. Family studies were approved by institutional review boards at the NIH (assurance number SPA S-5737-05) and at the National Public Health Institute in Helsinki. R.M.W. was previously supported by individual NIH National Research Service Award DK09525 and is now supported by a Career Development Award from the American Diabetes Association. R.N.B. is supported by NIH grants DK27619 and DK29867. C.D.L., J.A.D., W.L.D., M.P.E., E.M.L., and H.M.S. were previously supported, and T.E.F. and R.C.M. are currently supported, by NIH training grant HG00040.
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www3.ncbi.nlm.nih.gov/Omim (for PPARG [MIM 601487], SST [MIM 182450], TNF [MIM 191160], GLP1R [MIM 138032], FBP1 [MIM 229700], FBP2 [MIM 603027], CKM [MIM 123310], GIPR [MIM 137241], HNF3G [MIM 602295], GYS1 [MIM 138570], PK1 [MIM 266200], KCNJ9 [MIM 600932], IGFBP1 [MIM 146730], IGFBP3 [MIM 146732], GCK [MIM 138079], PCK2 [MIM 261650], HNF3A [MIM 602294], SLC2A4 [MIM 138190], MAP2K4 [MIM 601335], and ADORA2B [MIM 600446])
References
- Amos CI (1994) Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Auwerx J (1999) PPARgamma, the ultimate thrifty gene. Diabetologia 42:1033–1049 [DOI] [PubMed] [Google Scholar]
- Bach LA, Rechler MM (1992) Insulin-like growth factors and diabetes. Diabetes Metab Rev 8:229–257 [DOI] [PubMed] [Google Scholar]
- Beamer BA, Yen C-J, Andersen RE, Muller D, Elahi D, Cheskin LJ, Andres R, Roth J, Shuldiner AR (1998) Association of the Pro12Ala variant in the peroxisome proliferator-activated receptor-γ2 gene with obesity in two Caucasian populations. Diabetes 47:1806–1808 [DOI] [PubMed] [Google Scholar]
- Bell GI, Xiang KS, Newman MV, Wu SH, Wright LG, Fajans SS, Spielman RS, Cox NJ (1991) Gene for non-insulin-dependent diabetes mellitus (maturity-onset diabetes of the young subtype) is linked to DNA polymorphism on human chromosome 20q. Proc Natl Acad Sci USA 88:1484–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman RN (1989) Toward physiological understanding of glucose tolerance: minimal model approach. Diabetes 38:1512–1527 [DOI] [PubMed] [Google Scholar]
- Bergman RN, Ider YZ, Bowden CR, Cobelli C (1979) Quantitative estimation of insulin sensitivity. Am J Physiol 236:E667–E677 [DOI] [PubMed] [Google Scholar]
- Bergman RN, Phillips LS, Cobelli C (1981) Physiologic evaluation of factors controlling glucose tolerance in man: measurement of insulin sensitivity and β-cell glucose sensitivity from the response to intravenous glucose. J Clin Invest 68:1456–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangero J, Almasy L (1997) Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol 14:959–964 [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Hixson JE, Almasy L, Mitchell BD, Mahaney MC, Dyer TD, Stern MP, MacCluer JW, Blangero J (1997) A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nat Genet 15:273–276 [DOI] [PubMed] [Google Scholar]
- Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkanen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J (1998) A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet 20:284–287 [DOI] [PubMed] [Google Scholar]
- DeFronzo RA (1987) The triumvirate: β-cell, muscle, liver: a collusion responsible for NIDDM. Diabetes 37:667–687 [DOI] [PubMed] [Google Scholar]
- DeFronzo RA, Ferrannini E (1991) Insulin resistance: a multifaceted syndrome responsible for NIDDM, obesity, hypertension, dyslipidemia, and atherosclerotic cardiovascular disease. Diabetes Care 14:173–194 [DOI] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP (1999) Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Stern MP, Mitchell BD, Reinhart LJ, Shipman PA, Uresandi OC, Chung WK, Leibel RL, Hales CN, O'Connell P, Blangero J (1996) Quantitative variation in obesity-related traits and insulin precursors linked to the OB gene region on human chromosome 7. Am J Hum Genet 59:694–703 [PMC free article] [PubMed] [Google Scholar]
- Ehm MG, Karnoub MC, Sakul H, Gottschalk K, Holt DC, Weber JL, Vaske D, Briley D, Briley L, Kopf J, McMillen P, Nguyen Q, Reisman M, Lai EH, Joslyn G, Shepherd NS, Bell C, Wagner MJ, Burns DK, American Diabetes Association GENNID Study Group (2000) Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet 66:1871–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC, Hoffman MD, Teng K, Leppert MF, Hasstedt SJ (1999) A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes 48:1175–1182 [DOI] [PubMed] [Google Scholar]
- Froguel P, Vaxillaire M, Sun F, Velho G, Zouali H, Butel MO, Lesage S, Vionnet N, Clément K, Fougerousse F, Tanizawa Y, Weissenbach J, Beckmann JS, Lathrop GM, Passa P, Permutt MA, Cohen D (1992) Close linkage of glucokinase locus on chromosome 7p to early-onset non-insulin-dependent diabetes mellitus. Nature 356:162–164 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Schork NJ (1996) Genetic analysis of NIDDM: the study of quantitative traits. Diabetes 45:1–14 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Hauser ER, Valle T, Magnuson VL, Erdos MR, Langefeld CD, et al (1999) Type 2 diabetes: evidence for linkage on chromosome 20 in 716 Finnish affected sib pairs. Proc Natl Acad Sci USA 96:2198–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Langefeld CD, Magnuson VL, Ally DS, et al (2000) The Finland-United States Investigation of Non–Insulin-Dependent Diabetes Mellitus Genetics (FUSION) Study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67:1174–1185 (in this issue) [PMC free article] [PubMed] [Google Scholar]
- Groop LC, Kankuri M, Schalin-Jantti C, Ekstrand A, Nikula-Ijas P, Widen E, Kuismanen E, Eriksson J, Franssila-Kallunki A, Saloranta C, Koskimies S (1993) Association between polymorphism of the glycogen synthase gene and non-insulin-dependent diabetes mellitus. N Engl J Med 328:10–14 [DOI] [PubMed] [Google Scholar]
- Hanis CL, Boerwinkle E, Chakraborty R, Ellsworth DL, Concannon P, Stirling B, Morrison VA, et al (1996) A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet 13:161–166 [DOI] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MI (1990) Noninsulin-dependent diabetes mellitus in black and white Americans. Diabetes Metab Rev 6:71–90 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M (1998) Genetic linkage analysis of complex genetic traits by using affected sibling pairs. Biometrics 54:1238–1246 [PubMed] [Google Scholar]
- Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, Lindner T, Yamagata K, Ogata M, Tomonaga O, Kuroki H, Kasahara T, Iwamoto Y, Bell GI (1997) Mutation in hepatocyte nuclear factor-1β gene (TCF2) associated with MODY. Nat Genet 17:384–385 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Imperatore G, Hanson RL, Pettitt DJ, Kobes S, Bennett PH, Knowler WC, Pima Diabetes Genes Group (1998) Sib-pair linkage analysis for susceptibility genes for microvascular complications among Pima Indians with type 2 diabetes. Diabetes 47:821–830 [DOI] [PubMed] [Google Scholar]
- Jacquard A (1974) The genetic structure of populations. Springer-Verlag, New York [Google Scholar]
- Kahn SE, Prigeon RL, McCulloch DK, Boyko EJ, Bergman RN, Schwartz MW, Neifing JL, Ward WK, Beard JC, Palmer JP, Porte D Jr (1993) Quantification of the relationship between insulin sensitivity and β-cell function in human subjects: evidence for a hyperbolic function. Diabetes 42:1663–1672 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M (1988) Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 5:471–472 [DOI] [PubMed] [Google Scholar]
- Lehtovirta M, Kaprio J, Forsblom C, Eriksson J, Tuomilehto J, Groop L (2000) Insulin sensitivity and insulin secretion in monozygotic and dizygotic twins. Diabetologia 43:285–293 [DOI] [PubMed] [Google Scholar]
- Lillioja S, Bogardus C (1988) Obesity and insulin resistance: lessons learned from the Pima Indians. Diabetes Metab Rev 4:517–540 [DOI] [PubMed] [Google Scholar]
- Mahtani MM, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, Chan G, Daly M, Forsblom C, Kanninen T, Kirby A, Kruglyak L, Munnelly K, Parkkonen M, Reeve-Daly MP, Weaver A, Brettin T, Duyk G, Lander ES, Groop LC (1996) Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet 14:90–94 [DOI] [PubMed] [Google Scholar]
- Majer M, Mott DM, Mochizuki H, Rowles JC, Pedersen O, Knowler WC, Bogardus C, Prochazka M (1996) Association of the glycogen synthase locus on 19q13 with NIDDM in Pima Indians. Diabetologia 39:314–321 [DOI] [PubMed] [Google Scholar]
- National Diabetes Data Group (1979) Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. Diabetes 28:1039–1057 [DOI] [PubMed] [Google Scholar]
- Ohlson LO, Larsson B, Björntorp P, Eriksson H, Svardsudd K, Welin L, Tibblin G, Wilhelmsen L (1988) Risk factors for type 2 (non-insulin dependent) diabetes mellitus: thirteen and one-half years of follow-up of the participants in a study of Swedish men born in 1913. Diabetologia 31:798–805 [DOI] [PubMed] [Google Scholar]
- Pratley RE, Thompson DB, Prochazka M, Baier L, Mott D, Ravussin E, Sakul H, Ehm MG, Burns DK, Foroud T, Garvey WT, Hanson RL, Knowler WC, Bennett PH, Bogardus C (1998) An autosomal genomic scan for loci linked to prediabetic phenotypes in Pima Indians. J Clin Invest 101:1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich SS (1990) Mapping genes in diabetes. Diabetes 39:1315–1319 [DOI] [PubMed] [Google Scholar]
- Rubenstein AH, Clark JL, Melani F, Steiner DF (1969) Secretion of proinsulin C-peptide by pancreatic β cells and its circulation in blood. Nature 224:697–699 [Google Scholar]
- Self SG, Liang K-Y (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under non-standard conditions. J Am Stat Assoc 82:605–610 [Google Scholar]
- Sluiter WJ, Erkelens DW, Terpstra P, Weitsma WD, Doorenbos H (1976) Glucose tolerance and insulin release, a mathematical approach. II. Approximation of the peripheral insulin resistance after oral glucose loading. Diabetes 25:245–249 [DOI] [PubMed] [Google Scholar]
- St Jean PL, Mitchell BD, Hsueh W-C, Burns DK, Ehm MG, Wagner MJ, Bell CJ, Aburomia R, Nanthakumar E, Shuldiner AR (1999) Type 2 diabetes loci in the Old Order Amish. Diabetes Suppl 48:A46 [Google Scholar]
- Steil GM, Volund A, Kahn SE, Bergman RN (1993) Reduced sample number for calculation of insulin sensitivity and glucose effectiveness from the minimal model: suitability for use in population studies. Diabetes 42:250–256 [DOI] [PubMed] [Google Scholar]
- Stern MP, Duggirala R, Mitchell BD, Reinhart LJ, Shivakumar S, Shipman PA, Uresandi OC, Benavides E, Blangero J, O'Connell P (1996) Evidence for linkage of regions on chromosomes 6 and 11 to plasma glucose concentrations in Mexican Americans. Genome Res 6:724–734 [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Ferrer J, Clarke WL, Habener JF (1997) Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet 17:138–139 [DOI] [PubMed] [Google Scholar]
- Thompson DB, Janssen RC, Ossowski VM, Prochazka M, Knowler WC, Bogardus C (1995) Evidence for linkage between a region on chromosome 1p and the acute insulin response in Pima Indians. Diabetes 44:478–481 [DOI] [PubMed] [Google Scholar]
- Valle T, Tuomilehto J, Bergman RN, Ghosh S, Hauser ER, Eriksson J, Nylund SJ, Kohtamäki K, Toivanen L, Vidgren G, Tuomilehto-Wolf E, Ehnholm C, Blaschak J, Langefeld CD, Watanabe RM, Magnuson V, Ally DS, Hagopian WA, Ross E, Buchanan TA, Collins F, Boehnke M (1998) Mapping genes for NIDDM: design of the Finland-United States Investigation of NIDDM Genetics (FUSION) Study. Diabetes Care 21:949–958 [DOI] [PubMed] [Google Scholar]
- Vaxillaire M, Boccio V, Philippi A, Vigouroux C, Terwilliger J, Passa P, Beckmann JS, Velho G, Lathrop GM, Froguel P (1995) A gene for maturity onset diabetes of the young (MODY) maps to chromosome 12q. Nat Genet 9:418–423 [DOI] [PubMed] [Google Scholar]
- Vionnet N, Stoffel M, Takeda J, Yasuda K, Bell GI, Zouali H, Lesage S, Velho G, Iris F, Passa P, Froguel P, Cohen D (1992) Nonsense mutation in the glucokinase gene causes early-onset non-insulin-dependent diabetes mellitus. Nature 356:721–722 [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Valle T, Hauser ER, Ghosh S, Eriksson J, Kohtamäki K, Ehnholm C, Tuomilehto J, Collins FS, Bergman RN, Boehnke M (1999) Familiality of quantitative metabolic traits in Finnish families with non-insulin-dependent diabetes mellitus. Hum Hered 49:159–168 [DOI] [PubMed] [Google Scholar]
- World Health Organization (1985) Report of the expert committee on diabetes. WHO tech rep 727. World Health Organization, Geneva [Google Scholar]