Abstract
Founder analysis is a method for analysis of nonrecombining DNA sequence data, with the aim of identification and dating of migrations into new territory. The method picks out founder sequence types in potential source populations and dates lineage clusters deriving from them in the settlement zone of interest. Here, using mtDNA, we apply the approach to the colonization of Europe, to estimate the proportion of modern lineages whose ancestors arrived during each major phase of settlement. To estimate the Palaeolithic and Neolithic contributions to European mtDNA diversity more accurately than was previously achievable, we have now extended the Near Eastern, European, and northern-Caucasus databases to 1,234, 2,804, and 208 samples, respectively. Both back-migration into the source population and recurrent mutation in the source and derived populations represent major obstacles to this approach. We have developed phylogenetic criteria to take account of both these factors, and we suggest a way to account for multiple dispersals of common sequence types. We conclude that (i) there has been substantial back-migration into the Near East, (ii) the majority of extant mtDNA lineages entered Europe in several waves during the Upper Palaeolithic, (iii) there was a founder effect or bottleneck associated with the Last Glacial Maximum, 20,000 years ago, from which derives the largest fraction of surviving lineages, and (iv) the immigrant Neolithic component is likely to comprise less than one-quarter of the mtDNA pool of modern Europeans.
Introduction
It is generally agreed that many components of early European farming, including domesticated emmer wheat, barley, sheep and goats, were introduced into Europe from the Near East during the Neolithic, beginning some 9,000 years before the present (YBP) (Thorpe 1996). Nevertheless, there is uncertainty concerning the nature of the spread of these components into Europe. Two extreme hypotheses have been proposed. The replacement hypothesis suggests that the onset of agriculture was accompanied by extensive immigration by demic diffusion from the Near East, such that most of the gene pool of modern Europeans is derived from the newcomers (Diamond 1997; Barbujani et al. 1998; Chikhi et al. 1998b). Many archaeologists have rejected this view, in favor of a model based on trade and cultural diffusion (Dennell 1983; Barker 1985; Whittle 1996), which would have left the gene pool of prehistoric Europe essentially autochthonous. There is clearly a spectrum of possibilities between these two extremes, including demic diffusion involving a substantial minority of newcomers, perhaps practicing hypergamy (Cavalli-Sforza and Minch 1997), and pioneer colonization involving fewer newcomers and a more substantial contribution from the indigenous Mesolithic population (Zvelebil 1986, 1989; Sherratt 1994).
The study of the geographic distribution and diversity of genetic variation, known as the “phylogeographic approach” (Avise et al. 1987; Templeton et al. 1995), is emerging as a useful tool for the investigation of range expansions, migrations, and other forms of gene flow during prehistory. It is particularly suited to the study of nonrecombining-marker systems such as mtDNA, which is inherited down the female line and evolves rapidly, so that, provided that sufficient characters are assayed, the maternal genealogy can be well resolved. European mtDNAs fall into a number of distinct clusters, or haplogroups (Torroni et al. 1994a, 1996; Richards et al. 1998a; Macaulay et al. 1999). Most of these clusters are clades defined by particular control-region and/or coding-region motifs, although recurrent mutation, especially in the control region, can sometimes erase diagnostic elements of these motifs. The major clades are H–K, T, U3–U5, and V–X. As has been argued elsewhere (Richards et al. 1998a; Macaulay et al. 1999), the RFLP haplogroup U (Torroni et al. 1996) subsumes both haplogroup K and a number of other clusters (U1–U6), including several (U1, U2, and U6) found rarely in Europe but more frequently in the Near East and northern Africa (Macaulay et al. 1999). In addition, lineages are occasionally seen in Europe that belong to clusters more commonly found elsewhere, such as members of haplogroups M from eastern Eurasia (Ballinger et al. 1992; Passarino et al. 1992, 1996; Torroni et al. 1994b) or L1 and L2 from Africa (Chen et al. 1995; Watson et al. 1997).
In previous work, it was suggested that much of the extant European mtDNA lineages have their ancestry in Late Glacial expansions within Europe (Richards et al. 1996; Torroni et al. 1998), with only ∼10% dating to the earliest Upper Palaeolithic settlement of the continent (Richards et al. 1998a) and with ∼⩽20% dating to fresh immigrations during the early Neolithic. However, these estimates depend on a reliable determination of founder sequence types, since the undetected presence of ancestral heterogeneity in a colonizing population would result in an overestimation of the age. If this were the case, Europe could have been populated far more recently—for example, during the Neolithic—by a much more diverse founder population (Barbujani et al. 1998). A limitation of the initial analysis (Richards et al. 1996) was that it was based on a very small set of published Near Eastern sequences—42 from the Levant and the Arabian peninsula, mainly from the Bedouin (Di Rienzo and Wilson 1991; Richards and Sykes 1998). Although these sequences were difficult to assign, with certainty, to mtDNA clusters, since they encompassed only the first hypervariable segment (HVS-I) of the control region, they appeared to comprise mainly clusters J and T, pre-HV, a few sequences belonging to X, M, and L1/L2, and some probably belonging to cluster U. There were very few or no members of the major European cluster H, which occurs at a frequency of 40%–60% in most European populations, and there were no representatives of either its sister cluster V or of clusters I, W, K, or U5.
More data from the Near East have been published since this initial analysis, suggesting that the Bedouin may be unrepresentative of Near Eastern populations. Both Calafell et al. (1996) and Comas et al. (1996) have presented data from Turkey. These data suggest the presence of substantial frequencies of cluster H (although lower than that in Europe) (Torroni et al. 1998), as well as of I, W, K, and U4, in addition to clusters already identified in the Bedouin. Torroni et al. (1998) also analyzed a sample of Druze from Israel, using high-resolution RFLPs, and concluded that haplogroup H, but not haplogroup V, evolved first in the Near East and subsequently migrated into Europe. Here, we extend the Near Eastern database further, to a total of 1,234 individuals sampled from throughout the region, including ∼500 from the vicinity of the Fertile Crescent, where agriculture emerged from the increasingly sedentary Natufian populations at the end of the last Ice Age (Henry 1989).
We have formalized the procedure for founder analysis, investigated the extent of confounding recurrent gene flow between the putative source and derived populations, and developed criteria that take into account the effects of both gene flow and recurrent mutation. This has enabled us to provide an estimate of the contribution, to the present-day mtDNA pool, of immigration events at different times during Europe’s past.
Although previous genetic studies, using classical markers, have inferred a demic component to the spread of agriculture into Europe from the Near East (Menozzi et al. 1978; Ammerman and Cavalli-Sforza 1984; Sokal et al. 1991), the present study allows us for the first time to quantify that component realistically—at least for maternal lineages. Furthermore, the founder analysis using mtDNA allows us to trace lineages farther back into prehistory, through the Last Glacial Maximum (LGM), to the first settlement of Europe by anatomically modern humans, almost 50,000 YBP.
Subjects and Methods
Subjects
For the purposes of this analysis, the Near East was taken to include the whole of Turkey, the Fertile Crescent from Israel to western Iran, and the whole of the Arabian peninsula (see Kuhrt 1995, p. 1). The lower Nile (Egypt and northern Sudan) was also included, since this region is often treated historically with the Near East and since the HVS-I sequence data show that a large proportion of typically Near Eastern mtDNAs have penetrated the Nile Valley, where they coexist with sub-Saharan African mtDNAs (Krings et al. 1999). We sampled widely in the Near East, for several reasons. First, we wished to trace the ancestry of European lineages as far back as 50,000 YBP. We therefore needed as wide a source-population database as possible. Second, even though there is a particular concern with the origin of European Neolithic lineages in this work, we did not wish to focus exclusively on the core region for the origin of agriculture in the Fertile Crescent. This is because extensive gene flow within the Near East since the early Neolithic may well have dispersed founder sequence types at least as far afield as Egypt, the southern Caucasus, and Iran.
The Near Eastern populations analyzed for sequence variation in HVS-I of the mtDNA control region were as follows: 80 Nubians and 67 Egyptians (Krings et al. 1999); 29 Bedouin (Di Rienzo and Wilson 1991); 43 Yemeni Jews, including 5 from the study by Di Rienzo and Wilson (1991); 116 Iraqis, sampled from four regions of Iraq; 12 Iranians, sampled in Iran and Germany; 69 Syrians from Damascus; 146 Jordanians (45 from the Dead Sea region and 101 from the Amman region [V. Cabrera and N. Karadsheh, personal communication]); 117 Israeli Palestinians, including 8 from the study by Di Rienzo and Wilson (1991); 45 Israeli Druze (Macaulay et al. 1999); 218 Turks from Turkey, including 74 from the studies by Comas et al. (1996) and Calafell et al. (1996); 53 Kurds from eastern Turkey; 191 Armenians from Armenia; and 48 Azeris from Azerbaijan.
The European populations were analyzed by a modified version of the paleo-climatological model of Gamble (1986, 1999), as described elsewhere (Richards et al. 1998a): southeastern Europe—141 Bulgarians, including 30 from the study by Calafell et al. (1996), and 92 Romanians from Maramureş (65) and Vrancea (27); eastern Mediterranean—65 Greeks from Thessaloniki, 60 Sarakatsani from northern Greece, and 42 Albanians (Belledi et al. 2000); central Mediterranean—49 Italians from Tuscany (Francalacci et al. 1996; Torroni et al. 1998) and 48 from Rome, 90 Sicilians (42 from Troina and 48 from Trapani), and 115 Sardinians, including 69 from the study by Di Rienzo and Wilson (1991); western Mediterranean—54 Portuguese (Côrte-Real et al. 1996), 71 Spaniards (Côrte-Real et al. 1996), 92 Galicians (Salas et al. 1998) (156 Basques from northern Spain, including those from the studies by Bertranpetit et al. [1995] and Côrte-Real et al. [1996], were treated separately); Alps—70 Swiss (Pult et al. 1994), 49 South Germans from Bavaria (Richards et al. 1996), and 99 Austrians (Parson et al. 1998); north-central Europe—37 Poles, 83 Czechs, 174 Germans (Richards et al. 1996; Hofmann et al. 1997), and 38 Danes, including 33 from the study by Richards et al. (1996); Scandinavia—32 Swedes (Sajantila et al. 1996), 231 Norwegians, including 215 from the study by Opdal et al. (1998), and 53 Icelanders (Sajantila et al. 1995; Richards et al. 1996); northwestern Europe—71 French, comprising 47 from northeastern France and 24 from the CEPH database, 100 British (Piercy et al. 1993), 92 individuals from Cornwall, including 69 from the study by Richards et al. (1996), 92 individuals from Wales (Richards et al. 1996), and 101 individuals from western Ireland; northeastern Europe—25 Russians from the northern Caucasus, 36 Chuvash from Chuvashia (Russia), 163 Finns and Karelians, including 133 from the study by Sajantila et al. (1995) and 29 from the study by Richards et al. (1996), 149 Estonians, including 28 from the study by Sajantila et al. (1995) and 20 from the study by Sajantila et al. (1996), and 34 Volga-Finns (Sajantila et al. 1995); and northern Caucasus—106 northern Ossetians, 13 Chechens, 39 Kabardians, and 50 Adygei (Macaulay et al. 1999). Several published sequences containing ambiguities were excluded. Unattributed sequence data had previously been unpublished and were generated by the authors. HVS-I sequences were analyzed between nucleotide positions 16090 and 16365 (in the numbering system according to Anderson et al. [1981], which is used throughout this article), to be able to incorporate earlier data, but new sequences generally extended from ∼16050 to 16495, so that additional informative positions could be incorporated. The status at nucleotide position 16482 was checked in a number of previously analyzed samples, by use of the restriction enzyme DdeI, in order to assign H lineages including a transition at nucleotide position 16362 to the subcluster with HVS-I motif 16362-16482. In order to classify mtDNAs that did not harbor a diagnostic haplogroup motif in their HVS-I sequence, additional diagnostic markers were assayed, when this was possible. This screening mainly involved haplogroups H (7025 AluI), HV (14766 MseI and/or 00073), and U (by use of a mismatched 12308 HinfI [Torroni et al. 1996]). By use of restriction digestion with the enzymes HaeIII and Tsp509I, the former in conjunction with a mismatched primer, the status at nucleotide positions 11719 and 11251, respectively, was checked in 12 mtDNAs harboring the motif 16126C-16362C, which, until now, had been a cluster with an ambiguous position in the mtDNA phylogeny (Macaulay et al. 1999), being either pre-HV or pre-JT. All samples bore the 11719G (+11718HaeIII) mutation that is characteristic of HV (Saillard et al., in press), whereas none of them bore the 11251G (–11251Tsp509I) mutation that is characteristic of JT (Hofmann et al. 1997; Macaulay et al. 1999). Thus, these mtDNAs were shown to constitute an early branch in the pre-HV cluster.
mtDNA Classification
The mtDNA nomenclature has been described in detail by Richards et al. (1998a) and Macaulay et al. (1999). In brief, named clades of the phylogeny typically either refer to early branchings or are distinguished by an interesting geographic distribution. Major clades, by tradition called “haplogroups,” are denoted in terms of uppercase roman letters (e.g., H, J, etc.), and nested subclades are denoted by alternating positive integers and lowercase roman letters (e.g., J2, J1a, J1b1, etc.). Superclades, if not denoted by a single letter (e.g., M or N), are denoted by concatenating clade names (e.g., HV) whenever the smallest superclade comprising those clades is meant; the largest superclade containing those but no other named clades receives the prefix “pre-” (e.g., pre-HV). Possibly paraphyletic groups coalescing in an unresolved multifurcation that exclude the named clades deriving from this coalescence are marked by an asterisk (*) appended to the list of those named clades (e.g., HV*).
We use the term “sequence type” to refer to haplotypes of HVS-I sequences, and we use the term “lineage” to denote an individual subject’s sequence. Hence, a particular sequence type, such as that which, within HVS-I, matches the Cambridge reference sequence (CRS [Anderson et al. 1981]), might comprise several lineages, if several individuals in a population sample display the same sequence type. As before, we denote sequence types in terms of the positions at which they differ from the CRS, so that an HVS-I sequence type differing by a transition at nucleotide position 16311 is denoted “16311,” and a type differing by transitions at nucleotide positions 16145 and 16223 and a C→G transversion at nucleotide position 16176 is denoted “16145-16176G-16223.” The term “founder type” denotes a sequence type that has been carried from a source population to a derived population. “Founder cluster” refers to the cluster that has evolved from the founder type in the derived population.
The phylogenetic analysis was based on the construction of reduced median networks (Bandelt et al. 1995). These networks had to be further reduced, since we required a tree in order to perform founder analysis. For western-Eurasian mtDNA data, combined analyses of control-region and coding-region data have resulted in the basal part of the phylogeny becoming clear (Macaulay et al. 1999). Assignment to major clades on the basis of control-region data can therefore readily be achieved with the limited amount of additional RFLP typing described above. In the case of published data and other samples that could not be tested for additional markers, sequences could usually be unproblematically assigned to clusters by use of sequence matches or related types of known cluster. However, clear resolution within major clades can still remain a problem, particularly when the clades have little branching substructure. Several cases required special attention:
-
1.
A subset of haplogroup T (referred to as “T*” in the study by Richards et al. [1998a]) includes the nodes 16126-16294, 16126-16294-16296, 16126-16294-16296-16304, and 16126-16294-16304, which form a four-cycle in the network, with additional ambiguity surrounding nucleotide positions 16292 and 16153 (Richards et al. 1998a). Unfortunately, comparative coding-region data have not yet helped to resolve this four-cycle. A possible explanation for this is that the transition at nucleotide position 16294 destabilizes sites in its vicinity, where two runs of three cytosines are separated by an adenine. If this were the case, the nucleotide positions 16292 and 16296 might be unstable in this subcluster (as also has been noted by Malyarchuk and Derenko [1999]). Instability at nucleotide position 16296 is supported by its recurrence in T1 and by nucleotide position 16146 in T*, a slowly-evolving position that, nevertheless, occurs on both 16126-16294 and 16126-16294-16296 backgrounds; 16153 also occurs on both backgrounds (although it appears again, clearly resolved, on a 16126-16266-16294 background). The basic T* network therefore is probably best resolved into the path 16126-16294 (root), 16126-16294-16296, 16126-16294-16296-16304, and 16126-16294-16304 (since 16126-16294-16296 has higher diversity than does 16126-16294-16304). The difficulties with nucleotide position 16296 are acute, however, so we zero-weighted this character when constructing the T* network; this suggested that the position had mutated a minimum of 10 times within T*. This analysis also suggested that nucleotide position 16292, which might, for the same reason, also be thought likely to be destabilized has mutated only twice in T*. Position 16153 also appeared to have mutated only twice. Setting aside the status at nucleotide position 16296 enabled us to distinguish additional subclusters within T: T2 (16126-16294-16304), T3 (16126-16292-16294), T4 (16126-16294-16324), and T5 (16126-16153-16294). These clusters are worth delineating, since they represent some of the main founder clusters within T. We here update T* as the remainder of T when T1–T5 are excluded.
-
2.
Haplogroup U5 appears to suffer instability at nucleotide position 16192 (Macaulay et al. 1999; Finnilä et al. 2000), resulting in a four-cycle as above. We therefore assigned the 16189-16192-16270 type to subcluster U5b and assigned 16192-16311 to U5*. However, 16189-16192-16256-16270 was assigned to U5a1*, on the basis of the more stable nucleotide position 16256.
-
3.
Haplogroup K appears to suffer multiple hits at nucleotide position 16093. However, it was usually possible to resolve 16093 transitions on the basis of additional HVS-I information (resolving in favor of slower positions from the list in the study by Hasegawa et al. [1993]); therefore, this character was retained. Nevertheless, the 16093-16224-16311 type itself may well have evolved from 16224-16311 more than once, which would account for the very low age estimate for this cluster.
-
4. Haplogroup H needed particular processing. We did this by analyzing the data from cluster H site by site, constructing reduced median networks of all sequences containing the variant base at each site. Two criteria were then used to evaluate which of these aggregates formed valid clusters:
-
a. Connectivity.—If there was a starlike phylogeny with an extant central node and one-step connected derivatives, it was considered likely that the group of sequences formed a phylogenetic cluster.
-
b. Relative mutation rates of sites.—If ambiguity remained after criterion (a) was employed, the clusters were further resolved by cutting of links corresponding to positions with lower weights—that is, with higher individual mutation rates. The weights were assigned by counting the number of major clusters in which the variant base occurred, as a minimum estimate of the number of times that it has mutated in the data set.
-
a.
-
5.
The HVS-I CRS sequence type, along with common one-step derivatives resulting from transitions at fast sites such as 16129, 16189, 16311, and 16362, may belong to haplogroups H, HV, pre-HV, U, or R. For each region, the additional typing information available was sufficient to allow us to distribute untyped HVS-I CRS lineages among the haplogroups. This was done on the basis of their known frequency, with fully typed data, region by region. Typically, almost all lineages were allocated to haplogroup H within Europe. The HVS-I types 16129, 16189, 16311, and 16362 were treated in the same way where this was necessary, and types that included these variants were assigned to clusters by a favoring of slower-evolving positions.
Founder Analysis
Here we take an approach toward the identification of European founders that is more formal than that used elsewhere (Richards et al. 1996). Similar kinds of analysis have been performed by Stoneking and Wilson (1989), Stoneking et al. (1990), and Sykes et al. (1995), for the populations of the Pacific, and by Torroni et al. (1993a, 1993b) and Forster et al. (1996), for the populations of the Americas. However, as a consequence of both our larger data set and the closer genetic contact between the Near East and Europe, it has proved necessary here to incorporate a data-processing step, to allow for the high levels of recurrent mutation and back-migration.
We identified “candidate” founders by searching for
-
1.
identical sequence types in the Near East and Europe, and
-
2. inferred matches within the Near Eastern and European phylogeny, which are either
-
a. unsampled types with both European and Near Eastern derivatives, or
-
b. sequence types sampled only in the Near East and whose immediate derivatives include at least one European, or
-
c. sequence types sampled only in Europe and whose immediate derivatives include at least one Near Eastern individual.
-
a.
We then developed criteria for screening out recurrent mutation and back-migration. These criteria were designed to identify types that had most likely evolved in the Near East and to exclude those which had migrated there during the more recent past; the presence of derived types in the Near East was used to distinguish the former.
We applied three levels of stringency to identify founder candidates, and we also performed analyses on the candidate list itself (f0). Two of the levels, f1 and f2, were threshold levels, designed particularly to minimize the effects of recurrent mutation. Especially in the case of a shared frequent type, a parallel mutation in both regions, usually at a fast position, is likely to be reconstructed as a single event, so that the mtDNAs bearing the derived state seem to be more closely related than they are. A sequence match (either sampled or inferred) between populations—and, hence, a false founder candidate—can result. The threshold criteria aimed to reduce the impact of this effect, by requiring that matches should not be at the tips of the Near Eastern phylogeny: they are required to have either one (f1) or two (f2) branches deriving from them in the Near East. Furthermore, the derived types must connect to the founder candidate via Near Eastern (or shared) sequence types and not via sequence types found only in Europe. These criteria also provide a screen against recent back-migration into the Near East, since recently back-migrated types should also lack derivatives in the Near Eastern population.
A weakness of this approach for detection of back-migration is that it is dependent on the frequency of the founder cluster candidates in Europe. Clearly, for rarer types, the chance that back-migration or recurrent mutation will be detected is lower—and, for common types, it is higher—so that the criteria might be both too stringent for rare clusters and too weak for common clusters. We therefore introduced an alternative to f2, referred to as “fs,” in which the frequency of the cluster deriving from each candidate founder in Europe was used to scale the number of derivatives required in the Near East in order for the candidate to be counted as a founder type. To this end, we rescaled the (absolute) frequency of founder candidate clusters in Europe by taking logarithms to the base 10, rounding to the nearest integer, and then adding 1, allowing the outcome to be 1–4. This outcome was then used to designate the number of derivatives required in order for the candidate to qualify as a founder. In addition, to investigate the effect of sample size and differential back-migration into the more peripheral Near Eastern populations, we reapplied the fs criterion, excluding these populations (a procedure referred to as the “fs′” analysis).
Frequency Estimates
We estimated the posterior distribution of the proportion of a group of lineages in the population, given the sample, by using a binomial likelihood and a uniform prior on the population proportion. From this posterior distribution, we calculated a central 95% “credible region” (CR) (Berger 1985).
Dating and Age Classes
Having identified a list of founder types corresponding to each of these criteria, we measured the diversity in the clusters to which they have given rise within Europe, using the statistic ρ, the mean number of transitions from the founder sequence type to the lineages in the cluster (Forster et al. 1996). This is an unbiased estimate of the time to the most common ancestor of the cluster (TMRCA), measured in mutational units. This value was converted to an age estimate, by use of a mutation rate of 1 transition (between nucleotide positions 16090 and 16365) per 20,180 years (Forster et al. 1996), which closely approximates other rates used for HVS-I (Ward et al. 1991; Macaulay et al. 1997). If the underlying genealogy of a cluster is starlike, we can readily calculate the posterior distribution of its TMRCA, given the sequence type of the ancestor, assuming a uniform prior distribution for the TMRCA and a Poisson distribution for the mutational process. From the (gamma-distributed) posterior, we calculated a central 95% CR. We did this for all clusters, regardless of whether their phylogeny was starlike. When the phylogeny is markedly non-starlike, this is highlighted, since this method is expected to underestimate considerably the width of the true CR.
We employed two simple Procrustean models of demographic prehistory to partition the founder clusters, under each criterion, into migration events. The first, or “basic,” model assumes four major prehistoric migrations from the Near East to Europe: (i) early Upper Palaeolithic (EUP), 45,000 YBP; (ii) middle Upper Palaeolithic (MUP), 26,000 YBP; (iii) late Upper Palaeolithic (LUP), 14,500 YBP; and (iv) Neolithic, 9,000 YBP. It also employed a fifth class, at 3,000 YBP, in order to distinguish Neolithic from more-recent migration events.
These age classes were chosen by combining archaeological and paleo-climatological information (e.g., see Dansgaard et al. 1993; Strauss 1995) with an eyeballing of the ages of the more common founder clusters (see table 3 and fig. 1). These clusters appeared to fall roughly into at least three age classes, roughly corresponding to the beginning of the EUP, the LUP, and the Neolithic, with some clusters falling broadly between the LUP and the EUP. The MUP date of 26,000 YBP was chosen to allow immigrants arriving during ∼30,000–20,000 YBP to register, and it also corresponds to a slight climatic improvement. The EUP and LUP dates also correspond to more-substantial climatic ameliorations, especially the LUP dates, which are based on the rapid onset of the Bølling warm phase (Dansgaard et al. 1993).
Table 3.
Ages of the Major Founder Clusters Identified under Four Different Criteria
|
f0 |
f1 |
f2 |
fs |
||||||
| Haplogroup | Ancestral Sequence Typea | Proportion | Age(YBP) | Proportion | Age(YBP) | Proportion | Age(YBP) | Proportion | Age(YBP) |
| H | 0 | .212–.243 | 8,300–10,400 | .288–.322 | 11,500–13,600 | .367–.403 | 15,300–17,500 | .360–.396 | 15,000–17,200 |
| H | 16304 | .028–.042 | 10,100–16,500 | .032–.046 | 10,900–17,200 | .032–.046 | 10,900–17,200 | .032–.046 | 10,900–17,200 |
| H | 16293-16311b | .006–.014 | 5,400–16,300 | .010–.019 | 16,100–29,400 | .010–.019 | 16,100–29,400 | .010–.019 | 16,100–29,400 |
| H | 16291 | .008–.016 | 13,200–27,100 | .008–.016 | 13,200–27,100 | … | … | … | … |
| H | 16162 | .009–.017 | 5,400–14,700 | .009–.017 | 5,400–14,700 | … | … | … | … |
| H | 16362-16482 | .003–.009 | 4,600–19,400 | .004–.011 | 9,700–26,300 | .005–.011 | 10,000–26,200 | .004–.011 | 9,700–26,300 |
| H | 16093 | .008–.015 | 5,600–15,800 | .008–.015 | 5,600–15,800 | … | … | … | … |
| H | 16261 | .006–.014 | 6,500–18,200 | .006–.014 | 6,500–18,200 | .006–.014 | 6,500–18,200 | .006–.014 | 6,500–18,200 |
| H | 16354 | .005–.012 | 4,200–15,000 | .005–.012 | 4,200–15,000 | … | … | … | … |
| HV | 0b | .000–.003 | 0–30,200 | .001–.004 | 11,400–52,700 | .007–.014 | 22,000–40,700 | .046–.063 | 29,300–37,600 |
| V | 16298 | .035–.050 | 9,600–15,300 | .038–.054 | 11,100–16,900 | .039–.054 | 11,100–16,900 | … | … |
| I | 16129-16223b | .005–.012 | 3,800–14,500 | .005–.012 | 3,800–14,500 | .016–.026 | 26,900–40,300 | .013–.023 | 19,900–32,700 |
| I | 16129-16223-16311 | .002–.007 | 6,900–26,500 | .007–.014 | 13,900–29,400 | … | … | … | … |
| J | 16069-16126 | .047–.064 | 5,100–8,700 | .053–.071 | 6,900–10,900 | .054–.072 | 7,400–11,400 | .053–.071 | 6,900–10,900 |
| J1 | 16069-16126-16261 | .005–.012 | 1,000–8,000 | .005–.012 | 1,000–8,000 | .005–.012 | 1,000–8,000 | .005–.012 | 1,000–8,000 |
| J1a | 16069-16126-16145-16231-16261 | .005–.012 | 1,000–7,700 | .007–.014 | 2,600–10,800 | .007–.014 | 2,600–10,800 | .007–.014 | 2,600–10,800 |
| K | 16224-16311 | .036–.051 | 9,100–14,600 | .037–.052 | 9,300–14,800 | .040–.056 | 10,800–16,400 | .039–.054 | 10,000–15,500 |
| K | 16093-16224-16311 | .006–.013 | 1,400–8,600 | .007–.014 | 2,600–10,800 | .007–.014 | 2,600–10,800 | .006–.014 | 2,200–10,100 |
| T | 16126-16294 | .011–.020 | 3,100–9,700 | .011–.021 | 3,600–10,400 | .019–.030 | 10,900–19,200 | .017–.028 | 9,600–17,700 |
| T1 | 16126-16163-16186-16189-16294 | .018–.029 | 6,100–12,800 | .018–.029 | 6,100–12,800 | .018–.029 | 6,100–12,800 | .018–.029 | 6,100–12,800 |
| T2 | 16126-16294-16304 | .023–.036 | 9,200–16,100 | .024–.036 | 9,300–16,200 | .024–.036 | 9,300–16,200 | .024–.036 | 9,300–16,200 |
| U | 0 | .007–.014 | 13,400–28,300 | .007–.014 | 13,400–28,300 | .009–.017 | 15,500–29,600 | .049–.066 | 44,600–54,400 |
| U3 | 16343 | .002–.007 | 400–9,400 | .004–.009 | 5,200–19,900 | .005–.012 | 11,400–27,100 | .004–.009 | 5,200–19,900 |
| U4 | 16356 | .012–.021 | 4,800–12,200 | .016–.027 | 8,500–16,500 | .024–.037 | 16,100–24,700 | .024–.037 | 16,100–24,700 |
| U4 | 16134-16356 | .005–.011 | 13,900–32,400 | .005–.011 | 13,900–32,400 | … | … | … | … |
| U5 | 16270b | .015–.025 | 28,100–42,200 | .015–.025 | 28,000–42,200 | .041–.057 | 32,600–41,800 | … | … |
| U5a1c | 16192-16256-16270 | .021–.033 | 14,400–23,200 | .023–.036 | 15,600–24,300 | .023–.036 | 15,600–24,300 | .023–.036 | 15,600–24,300 |
| U5a1ac | 16256-16270 | .006–.014 | 2,200–10,100 | .007–.014 | 2,600–10,800 | .007–.015 | 3,800–12,800 | .007–.014 | 2,600–10,800 |
| U5b | 16189-16270 | .016–.026 | 13,100–22,900 | .017–.028 | 13,400–22,900 | … | … | … | … |
| W | 16223-16292 | .008–.016 | 8,200–19,500 | .010–.019 | 11,600–23,400 | .010–.019 | 11,600–23,400 | .010–.019 | 11,600–23,400 |
| X | 16189-16223-16278 | .007–.014 | 9,700–23,100 | .009–.017 | 12,000–24,700 | .011–.020 | 17,000–30,000 | .009–.017 | 12,000–24,700 |
| Overall (95% CR) | .689–.723 | .814–.842 | .897–.918 | .870–.894 | |||||
Major founder cluster (with a frequency, in either the f1, f2, or fs analyses, of >0.6%).
Markedly non-starlike phylogeny.
May be a reentrant into Europe, having evolved in Europe, migrated to the Near East, and remigrated to Europe; thus, the European sample may be a palimpsest of indigenous lineages and redispersed Near Eastern ones.
Figure 1.
Age ranges for major founder clusters—namely, those comprising ⩾40 lineages (which comprise 76% of the European data set), under the fs criterion. The proportion of lineages in each cluster is indicated. The 95% (50%) CRs for the age estimates of each cluster are shown by white (black) bars. The age classes used in the partition analysis are also indicated. Since the U* founder cluster (incorporating U5 under fs) is very non-starlike, its CRs are certainly underestimated. Although frequent, the cluster U5a1 is not shown, since it is probably of European origin, as discussed in the text.
With this latter point in mind, we also considered an “extended” model, which included a Mesolithic component during the dramatic rewarming following the Younger Dryas glacial interlude at 11,500 YBP (Dansgaard et al. 1993). This was stimulated by the suggestion, by Adams and Otte (1999), that recovery from this brief cold period, like that from the LGM, may have led to renewed population dispersals in Europe, possibly including some from Near Eastern refugia. Mesolithic events would, of course, be difficult to distinguish from both LUP and Neolithic expansions, but the possibility of a Mesolithic contribution should nevertheless be borne in mind.
Our partition analysis involves making the following assumptions: (i) each cluster can be assigned, in its entirety, to one of the proposed migration phases; (ii) each cluster expanded in Europe, immediately after the migration event, so that, as a result, the genealogy of each founder cluster is starlike, with a time depth closely approximating the time of the migration event; (iii) the mutation-rate estimate is accurate; (iv) the phylogenetic analysis has resolved all mutations; and (v) the founder analysis has correctly identified the sequence types of the founders. We determined the probabilities that each founder cluster took part in each of the migration events, on the basis of the age of the cluster. Then, given the proportion of the modern sample contained in each cluster, we estimated the proportion of the sample (and, by implication, the modern population of Europe) that is derived from each migration event. In detail, the migration-event times, tm (1⩽m⩽M, where M=5 for the basic model and M=6 for the extended model), were first scaled by the mutation rate μ; that is, τm=μtm. If we were to know from which event a cluster derived, then, under our assumption about the genealogy, the sampling distribution of ρ would be Poisson, with the parameter given by the total (scaled) length of the tree (which equals the number of samples in the cluster multiplied by the scaled time of the event); that is,
where ρi is the value of ρ for the ith cluster (1⩽i⩽I), ni is the sample size of the ith cluster, and aim=1 if the ith cluster is associated with the mth event and aim=0 otherwise. Then, the application of Bayes’s theorem, with an uninformative prior pr(aim=1)=M-1, yields the posterior probability that aim=1:
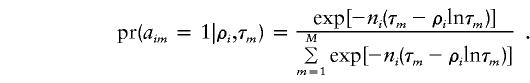 |
The
proportion of the total sample that is associated with the mth
event, Sm, is
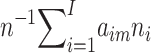 ,
where n is the total sample size. Exploiting the distribution
derived above for aim,
we evaluated the posterior mean of
Sm and the
root-mean-square deviation from the mean, to provide an
overall indication of the likely contribution of each migration to
the extant mtDNA pool. We performed the analysis on the three founder lists
identified on the basis of the criteria f1,
f2,
and fs, as well as on the basis of the f0 list.
The analysis was repeated with the migration dates varied by as much as
2,000 years, to establish that this did not greatly affect the
outcome.
,
where n is the total sample size. Exploiting the distribution
derived above for aim,
we evaluated the posterior mean of
Sm and the
root-mean-square deviation from the mean, to provide an
overall indication of the likely contribution of each migration to
the extant mtDNA pool. We performed the analysis on the three founder lists
identified on the basis of the criteria f1,
f2,
and fs, as well as on the basis of the f0 list.
The analysis was repeated with the migration dates varied by as much as
2,000 years, to establish that this did not greatly affect the
outcome.
Multiple dispersals of single sequence types are clearly a possibility, particularly for older types that are frequent in the Near East. Although most western-Eurasian mtDNA types are rare, one in particular, the root of haplogroup H (having the CRS in HVS-I and, hence, referred to as “H-CRS”), is very common, accounting for 16% of European lineages and for 6% of those from the Near East. Since this type is as much as 30,000 years old, it may have spread into Europe more than once. To allow for this possibility, we removed from the partition analysis the cluster derived from this type and then repeated the analysis. We then distributed the H-CRS cluster in Europe into the migrations, in proportion to the overall contribution of other lineages to each migration, while excluding the EUP, which occurred before this type had evolved. We term this the “fsr” analysis. No other Near Eastern type occurs at >2% of the total—except for the K type 16224-16311 (3.0%), which is <25,000 years old.
Members of haplogroups of eastern-Eurasian and African origin were excluded from these analyses, as “erratics”—that is, occasional migrants rather than parts of major range expansions. Few of these types occur more than once. We also excluded possible members of R1, R* (Macaulay et al. 1999), and N* (see below). These sets of lineages lack informative HVS-I markers, and, in the absence of additional RFLP typing, which was not possible for data assembled from the literature, they could not be unambiguously identified. However, they are extremely rare in Europe, amounting to <1% of the lineages.
Results
mtDNA in the Near East
Table 1 shows frequencies and age estimates of the main mtDNA haplogroups that occur in the Near East and Europe. These clusters are restricted primarily to Europe and the Near East (western Eurasia). Western-Eurasian lineages are found at moderate frequencies as far east as central Asia (Comas et al. 1998) and are found at low frequencies in both India (Kivisild et al. 1999a) and Siberia (Torroni et al. 1998), but, in these cases, only restricted subsets of the western-Eurasian haplogroups have been found, suggesting that they are most probably the result of secondary expansions from the core Near Eastern/European zone.
Table 1.
Estimated Frequencies and Ages of Major Haplogroups and Their Major Subclusters, in the Near East and Europe
|
Near
Eastern
Sample |
European
Sample |
||||||
| Haplogroup or Subclustera | Ancestral Sequence Type in HVS-I | No. of Lineages (95% CR for Proportion) | 95% CR for Age(YBP) | Phylogeny | No. of Lineages in European Sample (95% CR for Proportion) | 95% CR for Age(YBP) | Phylogeny |
| HV | CRS | 376 (.280–.331) | 24,300–29,000 | Starlike | 1,464 (.504–.541) | 20,700–22,800 | Starlike |
| H | CRS | 302 (.222–.270) | 23,200–28,400 | Starlike | 1,300 (.445–.482) | 19,200–21,400 | Starlike |
| V | 16298 | 6 (.002–.011) | 9,500–43,900 | Starlike | 128 (.039–.054) | 11,100–16,900 | Starlike |
| HV1 | 16067 | 29 (.016–.034) | 11,300–24,800 | Starlike | 9 (.002–.006) | 22,200–58,300 | Starlike |
| pre-HV | 16126-16362 | 44 (.027–.048) | 18,600–31,800 | Starlike | 12 (.002–.007) | 15,400–41,600 | Starlike |
| J | 16069-16126 | 116 (.079–.112) | 42,400–53,700 | Non-starlike | 261 (.083–.104) | 22,000–27,400 | Non-starlike |
| T | 16126-16294 | 121 (.083–.116) | 41,900–52,900 | Non-starlike | 229 (.072–.092) | 33,100–40,200 | Non-starlike |
| T1 | 16126-16163-16186-16189-16294 | 50 (.031–.053) | 16,700–28,400 | Starlike | 64 (.018–.029) | 6,100–12,800 | Starlike |
| U | CRS | 269 (.196–.242) | 50,400–58,300 | Starlike | 607 (.201–.232) | 53,600–58,900 | Starlike |
| K | 16224-16311 | 63 (.040–.065) | 15,500–25,500 | Non-starlike | 159 (.049–.066) | 12,900–18,300 | Non-starlike |
| U1a | 16189-16249 | 29 (.016–.034) | 17,000–33,100 | Starlike | 12 (.002–.007) | 20,500–49,900 | Starlike |
| U1b | 16249-16327 | 11 (.005–.016) | 14,000–40,800 | Starlike | 2 (.000–.003) | 2,400–56,200 | Starlike |
| U2 | 16129C-16189-16362 | 10 (.004–.015) | 14,000–42,300 | Non-starlike | 18 (.004–.010) | 23,600–48,000 | Starlike |
| U3 | 16343 | 62 (.039–.064) | 16,300–26,600 | Starlike | 26 (.006–.014) | 11,900–26,800 | Starlike |
| U4 | 16356 | 21 (.011–.026) | 16,300–35,500 | Starlike | 84 (.024–.037) | 16,100–24,700 | Non-starlike |
| U5 | 16270 | 22 (.012–.027) | 46,000–75,000 | Non-starlike | 257 (.081–.103) | 45,100–52,800 | Non-starlike |
| U7 | 16318T | 13 (.006–.018) | 23,900–53,600 | Non-starlike | 7 (.001–.005) | 11,900–45,400 | Non-starlike |
| N1b | 16145-16176G-16223 | 19 (.010–.024) | 8,900–24,900 | Starlike | 8 (.001–.006) | 21,100–59,300 | Starlike |
| I | 16129-16223 | 20 (.011–.025) | 32,300–58,400 | Non-starlike | 59 (.016–.027) | 27,200–40,500 | Non-starlike |
| W | 16223-16292 | 20 (.011–.025) | 18,000–38,400 | Starlike | 54 (.015–.025) | 17,100–28,400 | Starlike |
| X | 16189-16223-16278 | 36 (.021–.040) | 13,700–26,600 | Starlike | 42 (.011–.020) | 17,000–30,000 | Starlike |
Subclusters of haplogroups are indented. The less common haplogroups are not shown in this table; they account for 213 of the 1,234 Near Eastern lineages (95% CR = .153–.195) and for 68 of the 2,804 European lineages (95% CR = .019–.031). Also excluded are the northern-Caucasian samples.
The ages are estimates of the TMRCA of each cluster. Since these clusters are largely restricted to Europe and the Near East, they are likely to have originated in either one or the other region and to have subsequently dispersed into the other. In this case, there may be an overall reduction in the diversity of a cluster in the region that was settled, which gives an indication of the direction of gene flow, although this will not automatically be the case, depending on the diversity carried from the source. If this were the case, the older of the two age estimates would be a better estimate of the age of the cluster; the estimate for the younger population then would be rather meaningless. A founder analysis would be necessary to date the migration event.
As table 1 indicates, a number of the major haplogroups have greater diversities in the Near East than in Europe. This is the case for haplogroups H, J, and T, for which the central 95% CRs of their TMRCAs in the Near East and Europe do not overlap. Haplogroup U appears to be similar in age in both Europe and the Near East and has ancient geographically specific subclusters. In these two regions, haplogroups I, W, and X are also indistinguishable, possibly as a result of their low sample sizes. We also calculated haplogroup diversity in the northern-Caucasian samples; however, although high, these values cannot be very meaningfully converted into age estimates, since the cluster phylogenies in this region are markedly non-starlike, evidently displaying drift onto rare sequence types, often near the tips of the phylogenies. Although the Caucasian data are therefore difficult to interpret, the presence there of cluster distributions that are similar to those of Europe and the Near East should caution us that both Europe and the Near East could have been populated from a third region, perhaps closer to either the extant Caucasian population or other populations in eastern Europe. More-recent incursions from eastern Europe, particularly during the Bronze Age, are also likely to have taken place.
Most of the major western-Eurasian clades (Macaulay et al. 1999, table 2) occur in the Near East at a frequency of ⩾1%. In addition to these, we here define U7 (HVS-I motif 16318T [Kivisild et al. 1999a]), HV1 (HVS-I motif 16067), and a clade in pre-HV (HVS-I motif 16126-16362). We subdivide the haplogroup N defined by 10873T (+10871MnlI) (Quintana-Murci et al. 1999), which encompasses almost all Eurasian mtDNAs (including haplogroups A, B, F, H–J, K, R, and T–Y) that do not fall into haplogroup M. A subcluster N1, characterized by 10238C (+10237HphI), can be identified (Kivisild et al. 1999b) that includes haplogroup I and that has distinct subclusters: N1a (tentative HVS-I motif 16147A/G–16172-16223-16248-16355), N1b (probable HVS-I motif 16145-16176G-16223), and N1c (probable HVS-I motif 16223-16265). Another N subcluster with HVS-I motif 16223-16257A-16261 has a predominantly eastern-Eurasian distribution. HV1, the specific clade of pre-HV, N1a–c, and U7 all occur at low frequency in the northern-Caucasian sample. If we enumerate named subclusters of mtDNA clades in the Near East, Europe, and the Caucasus, we also find more in the Near East than in either of the other two regions, again supporting a Near Eastern origin for the main clusters.
Table 2.
Founder Status of All Founder Candidate Sequence Types, under Five Different Criteria
|
Founder
Status, for
Criteriona |
||||||
| f0 | f1 | f2 | fs | fs′ | Haplogroup or Subcluster | HVS-I Sequence Typeb |
| • | • | • | • | • | H | 0 |
| • | • | • | • | H | 16092 | |
| • | H | 16092-16311 | ||||
| • | H | 16111 | ||||
| • | H | 16129 | ||||
| • | H | 16172 | ||||
| • | H | 16189 | ||||
| • | H | 16189-16311 | ||||
| • | H | 16212 | ||||
| • | H | 16244 | ||||
| • | H | 16284 | ||||
| • | H | 16289 | ||||
| • | H | 16311 | ||||
| • | H | 16362 | ||||
| • | H | 16192-16304 | ||||
| • | H | 16207-16304 | ||||
| • | H | 16210-16304 | ||||
| • | • | • | • | • | H | 16304 |
| • | H | 16304-16327 | ||||
| • | H | 16304-16362 | ||||
| • | H | 16256-16295-16352 | ||||
| • | • | • | • | • | H | 16256-16311-16352 |
| • | • | • | • | • | H | 16256-16352 |
| • | • | H | 16218-16328A | |||
| • | H | 16092-16293-16311 | ||||
| • | • | • | • | H | 16293-16311 | |
| • | • | H | 16291 | |||
| • | • | H | 16162 | |||
| • | • | • | H | 16162-16172 | ||
| • | H | 16148-16256-16319 | ||||
| • | • | H | 16256 | |||
| • | H | 16093-16293 | ||||
| • | • | H | 16293 | |||
| • | • | • | • | H | 16111-16362-16482 | |
| • | H | 16129-16362-16482 | ||||
| • | • | • | • | • | H | 16362-16482 |
| • | • | H | 16093 | |||
| • | • | • | H | 16318T | ||
| • | • | H | 16176 | |||
| • | H | 16209 | ||||
| • | • | H | 16213 | |||
| • | H | 16218 | ||||
| • | • | H | 16192 | |||
| • | H | 16222 | ||||
| • | • | H | 16240 | |||
| • | • | H | 16278 | |||
| • | • | • | • | • | H | 16261 |
| • | H | 16263 | ||||
| • | • | • | • | H | 16274 | |
| • | • | H | 16286 | |||
| • | • | H | 16287 | |||
| • | • | • | • | H | 16295 | |
| • | H | 16265-16298 | ||||
| • | • | • | • | H | 16298 | |
| • | • | H | 16354 | |||
| • | • | H | 16355 | |||
| • | • | • | • | • | H | 16234 |
| • | • | • | • | H | 16093-16265 | |
| • | • | • | • | H | 16265 | |
| • | • | • | H | 16150-16192 | ||
| • | • | H | 16248 | |||
| • | • | • | • | H | 16356 | |
| • | • | • | • | • | H | 16266 |
| • | • | • | • | H | 16266-16311 | |
| • | H | 16266-16311-16362 | ||||
| • | • | H | 16145 | |||
| • | H | 16188G | ||||
| • | • | • | • | • | H | 16288-16362 |
| • | • | • | • | • | H | 16357 |
| • | • | • | • | • | HV | 0 |
| • | • | • | • | HV | 16129 | |
| • | HV | 16129-16221 | ||||
| • | HV | 16172-16311 | ||||
| • | • | HV | 16221 | |||
| • | • | HV | 16311 | |||
| • | • | • | • | HV | 16362 | |
| • | • | • | • | • | HV1 | 16067 |
| • | • | • | HV1 | 16067-16311 | ||
| • | • | • | • | HV1 | 16067-16355 | |
| • | • | • | • | • | I | 16129-16223 |
| • | I | 16129-16172-16223-16311 | ||||
| • | • | I | 16129-16223-16311 | |||
| • | • | • | • | I | 16129-16223-16311-16362 | |
| • | • | • | • | I | 16223-16311 | |
| • | • | • | • | • | I | 16129-16148-16223 |
| • | • | • | • | I | 16129-16223-16311-16319 | |
| • | • | • | • | I | 16129-16223-16270-16311-16319-16362 | |
| • | • | • | • | • | J | 16069-16126 |
| • | J | 16069-16126-16145 | ||||
| • | J | 16069-16126-16148 | ||||
| • | J | 16069-16126-16189 | ||||
| • | J | 16069-16126-16193-16256-16300-16309 | ||||
| • | J | 16069-16126-16241 | ||||
| • | • | • | • | J | 16069-16126-16300 | |
| • | J | 16069-16126-16311 | ||||
| • | J | 16069-16126-16319 | ||||
| • | J1 | 16069-16126-16145-16172-16261 | ||||
| • | • | • | • | • | J1 | 16069-16126-16145-16261 |
| • | • | • | • | • | J1 | 16069-16126-16261 |
| • | J1a | 16069-16126-16145-16189-16231-16261 | ||||
| • | • | • | • | • | J1a | 16069-16126-16145-16231-16261 |
| • | • | • | • | • | J1b | 16069-16126-16145-16222-16261 |
| • | • | • | • | J1b | 16069-16126-16145-16222-16261-16274 | |
| • | J1b1 | 16069-16126-16145-16172-16222-16261 | ||||
| • | • | • | • | • | J2 | 16069-16126-16193 |
| • | J2 | 16069-16126-16193-16278 | ||||
| • | J2 | 16069-16126-16193-16311 | ||||
| • | K | 16093-16224-16234-16311 | ||||
| • | • | • | K | 16189-16224-16311 | ||
| • | • | K | 16224-16234-16311 | |||
| • | • | • | • | K | 16224-16291-16311 | |
| • | • | • | K | 16224-16304-16311 | ||
| • | • | • | • | • | K | 16224-16311 |
| • | • | K | 16093-16189-16224-16311 | |||
| • | K | 16093-16224 | ||||
| • | K | 16093-16224-16278-16311 | ||||
| • | • | • | • | • | K | 16093-16224-16311 |
| • | N | 16223 | ||||
| • | • | N1a | 16147A-16172-16223-16248-16320-16355 | |||
| • | • | • | • | N1a | 16147A-16172-16223-16248-16355 | |
| • | • | • | N1b | 16126-16145-16176G-16223 | ||
| • | • | • | N1b | 16128G-16145-16176G-16223-16311 | ||
| • | • | • | • | • | N1b | 16145-16176G-16223 |
| • | • | • | N1b | 16145-16176G-16223-16256 | ||
| • | • | • | • | • | N1c | 16201-16223-16265 |
| • | • | • | • | pre-HV | 16114-16126-16362 | |
| • | pre-HV | 16126 | ||||
| • | • | • | pre-HV | 16126-16264-16362 | ||
| • | • | • | • | • | pre-HV | 16126-16355-16362 |
| • | • | • | • | • | pre-HV | 16126-16362 |
| • | • | T | 16126-16189-16294-(16296) | |||
| • | T | 16126-16189-16294-(16296)-16298 | ||||
| • | T | 16126-16192-16294-(16296) | ||||
| • | • | • | • | • | T | 16126-16291-16294-(16296) |
| • | • | • | • | • | T | 16126-16294-(16296) |
| • | • | • | T | 16126-16294-(16296)-16311 | ||
| • | • | • | • | T | 16126-16294-(16296)-16362 | |
| • | • | • | • | • | T1 | 16126-16163-16186-16189-16294 |
| • | T2 | 16093-16126-16294-(16296)-16304 | ||||
| • | • | • | • | • | T2 | 16126-16294-(16296)-16304 |
| • | • | • | • | • | T3 | 16126-16146-16292-16294-(16296) |
| • | • | • | • | T3 | 16126-16189-16292-16294-(16296) | |
| • | • | T3 | 16126-16292-16294-(16296) | |||
| • | • | • | • | T4 | 16126-16294-(16296)-16324 | |
| • | • | • | • | • | T5 | 16126-16153-16294-(16296) |
| • | • | • | • | • | U | 0 |
| • | • | • | U | 16129-16189-16234 | ||
| • | • | • | U | 16189 | ||
| • | U | 16189-16234 | ||||
| • | • | • | U | 16189-16234-16324 | ||
| • | • | • | • | U | 16189-16362 | |
| • | • | • | U | 16311 | ||
| • | • | • | • | U | 16362 | |
| • | U1a | 16092-16189-16249 | ||||
| • | • | • | • | U1a | 16129-16189-16249-16288 | |
| • | U1a | 16129-16189-16249-16288-16362 | ||||
| • | • | • | • | • | U1a | 16189-16249 |
| • | U1a | 16189-16249-16288 | ||||
| • | U1a | 16189-16249-16362 | ||||
| • | • | • | • | • | U1b | 16111-16249-16327 |
| • | U2 | 16129C-16189-16256 | ||||
| • | • | • | • | U2 | 16129C-16189-16256-16362 | |
| • | • | • | • | • | U2 | 16129C-16189-16362 |
| • | • | • | • | U3 | 16093-16343 | |
| • | • | • | U3 | 16189-16343 | ||
| • | U3 | 16193-16249-16343 | ||||
| • | • | • | • | U3 | 16260-16343 | |
| • | • | • | • | U3 | 16301-16343 | |
| • | U3 | 16311-16343 | ||||
| • | • | • | • | • | U3 | 16343 |
| • | • | • | • | • | U3 | 16168-16343 |
| • | U4 | 16179-16356 | ||||
| • | U4 | 16223-16356 | ||||
| • | • | • | • | U4 | 16356 | |
| • | • | U4 | 16356-16362 | |||
| • | • | U4 | 16134-16356 | |||
| • | • | • | U5 | 16270 | ||
| • | • | • | U5 | 16270-16296 | ||
| • | • | • | U5a | 16187-16192-16270 | ||
| • | U5a1 | 16093-16192-16256-16270-16291 | ||||
| • | U5a1 | 16189-16192-16256-16270 | ||||
| • | • | • | • | U5a1 | 16192-16256-16270 | |
| • | • | • | • | U5a1 | 16192-16256-16270-16291 | |
| • | U5a1 | 16192-16256-16270-16311 | ||||
| • | • | • | U5a1a | 16189-16256-16270 | ||
| • | • | • | • | U5a1a | 16256-16270 | |
| • | U5a1a | 16256-16270-16295 | ||||
| • | • | U5b | 16189-16270 | |||
| • | U5b | 16189-16270-16311 | ||||
| • | • | U5b1 | 16144-16189-16270 | |||
| • | • | • | • | • | U7 | 16318T |
| • | • | • | • | • | U7 | 16309-16318C |
| • | • | • | • | • | U7 | 16309-16318T |
| • | V | 16216-16261-16298 | ||||
| • | V | 16239-16298 | ||||
| • | • | • | V | 16274-16298 | ||
| • | • | • | V | 16298 | ||
| • | V | 16298-16311 | ||||
| • | W | 16093-16223-16292 | ||||
| • | W | 16129-16223-16292 | ||||
| • | W | 16172-16223-16231-16292 | ||||
| • | • | • | • | W | 16223-16292 | |
| • | • | • | • | W | 16223-16292-16295 | |
| • | • | W | 16192-16223-16292-16325 | |||
| • | • | W | 16223-16292-16325 | |||
| • | X | 16093-16189-16223-16278 | ||||
| • | X | 16126-16189-16223-16278 | ||||
| • | • | • | X | 16189-16223-16248-16278 | ||
| • | • | • | X | 16189-16223-16265-16278 | ||
| • | • | • | X | 16189-16223-16274-16278 | ||
| • | • | • | • | • | X | 16189-16223-16278 |
| • | X | 16189-16223-16278-16293 | ||||
| • | • | • | • | X | 16189-16223-16278-16344 | |
| • | X | 16189-16278 | ||||
Founder clusters are indicated by bullets (•).
Parentheses denote that, as discussed in the Subjects and Methods section, nucleotide position 16296 in haplogroup T is unstable, making the state of this position in the founder sequence uncertain. Sequence types that are formally founders but whose clusters are empty in the European sample are not included.
The principal exception is cluster V, which seems to have expanded within Europe ∼13,000 YBP (Torroni et al. 1998). Cluster U5 is an additional unusual case. Although U5 occurs at ∼2% in the Near East, its phylogeography, as we discuss below, suggests that it evolved mainly within Europe during the past ∼50,000 years. Haplogroups V and U5 occur in the Near East at ∼11% and ∼19%, respectively, of their European frequencies, in most cases as occasional haplotypes that are derived from European lineages. These can be regarded as “erratics,” in the same way that African and eastern-Eurasian types can be regarded as such in Europe.
Cluster H is the most frequent cluster in the Near East, as it is in Europe; nevertheless, it is present at a frequency of only 25% (95% CR = .222–.270) in the Near East, compared with 46% (95% CR = .445–.482) in Europeans as a whole. It occurs in the northern Caucasus at a frequency of ∼25% (95% CR = .200–.318). It is almost absent in certain populations, such as the Arabians and the Saami (Sajantila et al. 1995). The age estimate for H in the Near East is 23,200–28,400 years. This is significantly older than its age estimate in Europe (19,200–21,400 years) and perhaps gives an indication of the TMRCA of haplogroup H.
A similar picture emerges with regard to the sister haplogroups, T and J, which both date to ∼50,000 YBP in the Near East but more-recent dates in Europe. Cluster J reaches its highest frequencies in Arabia (25% in the Bedouin and Yemeni [95% CR .165–.361]), alongside equally exceptional frequencies of the specific clade within pre-HV (22% [95% CR .142–.331]), perhaps as a result of the same founder effects or low population sizes that appear to have excluded or eliminated cluster H from the Arabian peninsula. Arabia is by far the most distinctive region in the Near East, and it is notable that the Bedouin and Yemeni populations would appear to have a common origin, as judged on the basis of their striking similarity in unusual cluster frequencies. Haplogroup U, which is >50,000 years old in the Near East and which harbors both specific European (U5), northern-African (U6 [Rando et al. 1998; Macaulay et al. 1999]), and Indian (U2i [Kivisild et al. 1999a]) components, each dating to ∼50,000 YBP, occurs in both Arabia and the northern Caucasus and, indeed, throughout the Near East.
There is in the Near East a moderate frequency of clusters originating in Africa (even when Egypt and Nubia—where the frequency of lineages of African origin is obviously higher—are excluded from the Near Eastern sample): ∼1% L1, ∼1% L2, and <3% African L3* (distinguished from the Eurasian haplogroups M and N, at nucleotide positions 10400 and 10873, respectively [Quintana-Murci et al. 1999]). The cluster M1, usually found in eastern Africa (Passarino et al. 1998; Quintana-Murci et al. 1999), also occurs at <1%. Thus, sub-Saharan African input in the Near East amounts to ∼5%, rather less than our estimates of gene flow from Europe. There are also <1% northern-African U6 mtDNAs.
There are even fewer eastern-Eurasian lineages represented, amounting to ∼2% in total: 3 individuals with haplogroup A, 4 with B, 7 with C (or pre-C), 2 with F, 1 from N*, 1 with Y, and 10 additional potential members of the eastern-Eurasian haplogroup M, some of which may be D (Torroni et al. 1993b). As in the case of Africa, these are probably attributable to fairly recent gene flow. Most of them would imply incursions from central/eastern Asia, and their occurrence in Turkey, Greece, Bulgaria, and the Caucasus, as well as in both the Saami and northeastern Europe, implies that they may be the result of historically attested migrations into these areas.
Back-Migration from Europe
Recent back-migration can be estimated by an examination of the presence, in the Near East, of clusters that are most likely to have evolved within Europe. Haplogroup U5 is very ancient (∼50,000 years old) in both Europe and the Near East, but it occurs more sporadically in the Near East and is absent from Arabia. In the Near East, it is largely restricted to peripheral populations (Turks, Kurds, Armenians, Azeris, or Egyptians): only three individuals from the core Near Eastern regions (namely, the Fertile Crescent and Arabia) harbor U5 sequence types; of these, one is the root sequence type, whereas the other two are members of the highly derived subcluster U5a1a (for the nomenclature for U5, seetable 2table 2). Overall, 8 of 22 Near Eastern U5 types are members of this highly derived subcluster, and an additional 6 are members of the next-most-derived subcluster, U5a1*. There are four members of U5b, one member of U5a*, and only three members of U5*. Moreover, these Near Eastern types are frequently derivatives of European intermediate types: one Egyptian type is derived from a Basque type, and many Armenian and Azeri types are derived from European and northern-Caucasian types. Therefore, whereas the U5 root sequence type (16270) could conceivably have originated in the Near East and have spread to Europe ∼50,000 YBP, with recurrent back-migration ever since, a European origin for the U5 cluster seems just as probable. In either case, the U5 cluster itself would have evolved essentially in Europe. U5 lineages, although rare elsewhere in the Near East, are especially concentrated in the Kurds, Armenians, and Azeris. This may be a hint of a partial European ancestry for these populations—not entirely unexpected on historical and linguistic grounds—but may simply reflect their proximity to the Caucasus and the steppes. Of the Near Eastern lineages, 1.8% (95% CR = .012–.027) are members of U5, in contrast to 9.1% (95% CR = .081–.103) in Europe; in the core region of Syria-Palestine through Iraq, the proportion falls to 0.5% (95% CR = .002–.015). Overall, this suggests the presence of as much as 20% of back-migrated mtDNA in the Near East but only ∼6% in the core region.
It seems likely that haplogroup V also originated within Europe and subsequently spread eastward (Torroni et al. 1998), although its lower diversity provides less opportunity to differentiate lineages by their ages. A slightly lower figure for back-migration is obtained when V is used: 0.5% (95% CR = .002–.011) of samples in the Near East (in Turkey, Azerbaijan, and Syria) versus 4.6% (95% CR = .039–.054) in Europe, suggesting a value of ∼11% back-migrants overall. Again, two-thirds of these back-migrants are in either Turkey or the southern Caucasus, which reduces the estimate for the core region to ∼8%. Given the small sample sizes involved and the resulting uncertainties in the estimates, these values are in good agreement with the figure estimated when U5 is used, especially since haplogroup V is both rarer in eastern Europe (whence much of the back-migration is likely to have originated) than in western Europe (Torroni et al. 1998) and of more recent origin than U5. Hence, the scale of back-migration is considerable. It needs to be taken into account as a major factor in the founder analysis and also suggests that it will be worthwhile to compare a founder analysis based only on the core regions versus a founder analysis based on the Near Eastern data as a whole.
Founder Analysis
Identification of founders
A total of 2,736 of 2,804 lineages in Europe could be assigned to haplogroups of western-Eurasian origin; of the remaining 68, “erratic” lineages, there were likely members of African (19), north-African (6), and eastern-Eurasian (22) clusters, the remainder being either members of R (7), ambiguous between (African) L3* and (Eurasian) N* (11), or unclassified (3).Table 2Table 2 shows all of the candidate types for European founders, as well as their founder status under the various founder criteria. There were 210 founder-candidate types (referred to as “f0”). Of these, 134 were types shared by Europe and the Near East, and the remaining 76 were inferred matches. A total of 134 founders were identified by use of the f1 criterion; 58 by the more stringent, f2 criterion; and 106 by the more flexible, fs criterion. Under the fs criterion only, the root types of both haplogroup V and haplogroup U5 were excluded as founders. U5 is very likely to be of indigenous European origin (see above). Within U5, types that qualified as founders could have back-migrated into the Near East sufficiently long ago to have contributed to subsequent dispersals into Europe (as, e.g., the root types of U5a1 or U5a1a), or they may represent cases in which the founder criteria have not winnowed out simple back-migrants. U5a1 and U5a1a lineages in Europe may, therefore, have been derived from either indigenous European or redispersing Near Eastern types. (Although this may be true for U5a1a, U5a1 is an implausible founder cluster, since its “Near Eastern” distribution is accounted for primarily by the southern Caucasus, where only a few derived types occur. Since related derived types are also quite common in the northern Caucasus, U5a1 seems likely to have arrived from Europe via the northern Caucasus, fairly recently. This being the case, the fs′ analysis would provide a better estimate for the EUP component than would be provided by fs.) Haplogroup V is also thought likely to have evolved in Europe (Torroni et al. 1998), and, again, a number of the Near Eastern V sequence types could be identified as derivatives of European types. This outcome suggests that the fs criterion indeed performs better than the threshold criteria f1 and f2. The fs′ analysis, performed by applying the fs criterion when the more peripheral Near Eastern populations (Egyptians, Turks, Kurds, Armenians, and Azeris) are excluded, resulted in 72 founders.
The 95% CRs of the ages of the more common founders under each criterion are given in table 3. Figure 1 shows the major founders and also indicates the age classes of the migration models. There are two major founders associated with the Neolithic (the root types of J and T1), several with the LUP (the root types of T, T2, and K and the H-16304 type), and several with somewhat earlier dates through the LUP and MUP (the root types of H, U4, I, and HV); and the root type of U is associated with the EUP. Note that, although, in the partition analysis, the H-CRS founder would be firmly associated with the LUP, it is in fact somewhat older than the Bølling rewarming, suggesting an earlier MUP immigration as well. It is also worth noting that, although several 95% CRs overlap the Mesolithic, only one of the 50% CRs does. The figure therefore provides some provisional support for the age classes in the basic model—but rather little support for the extended model with a Mesolithic migration.
Partition analyses
Table 4 shows the results of the partitioning analysis. For the f0 results first, with no allowance for back-migration, the first point to note is a value of 16% for recent gene flow. This is similar to the values that we estimated for back-migration into the Near East when haplogroups U5 and V were used (above). The value falls to 2%–6% in the subsequent analyses, in which most of the recently migrated lineages are reapportioned into the earlier dispersal events.
Table 4.
Percentage, of Extant European mtDNA Pool, Derived, in Each Migration Event, from Near Eastern Founder Lineages
|
Mean
± Root-Mean-Square Error, of Contribution, for
Criteriona(%) |
||||||
| Migration Event | f0 | f1 | f2 | fs | fs′ | fsr |
| Basic model: | ||||||
| Bronze Age/recent | 16.3 ± 1.2 | 5.9 ± 1.2 | 2.6 ± .7 | 4.0 ± .9 | 2.7 ± .7 | 7 |
| Neolithic | 48.5 ± 3.5 | 21.8 ± 3.1 | 12.4 ± 1.6 | 13.3 ± 2.0 | 11.9 ± 1.9 | 23 |
| LUP | 25.1 ± 3.5 | 58.8 ± 3.4 | 63.7 ± 2.6 | 58.8 ± 2.8 | 55.4 ± 1.9 | 36 |
| MUP | 5.8 ± 1.5 | 9.3 ± 2.1 | 12.8 ± 2.3 | 14.6 ± 2.2 | 11.0 ± .9 | 25 |
| EUP | 1.8 ± 1.0 | 1.7 ± 1.0 | 6.0 ± .7 | 6.9 ± .5 | 16.5 ± .5 | 7 |
| Extended model: | ||||||
| Bronze Age/recent | 15.2 ± 1.2 | 5.1 ± 1.2 | 2.4 ± .8 | 3.6 ± .9 | 2.5 ± .7 | 6 |
| Neolithic | 41.5 ± 3.0 | 16.5 ± 2.8 | 10.1 ± 2.5 | 10.7 ± 2.6 | 9.7 ± 2.5 | 18 |
| Mesolithic | 18.5 ± 4.1 | 45.2 ± 5.9 | 9.6 ± 4.6 | 10.8 ± 4.0 | 9.5 ± 4.0 | 19 |
| LUP | 15.4 ± 3.6 | 20.3 ± 5.8 | 56.9 ± 4.5 | 51.2 ± 4.0 | 48.6 ± 3.5 | 23 |
| MUP | 5.3 ± 1.5 | 8.8 ± 2.1 | 12.6 ± 2.3 | 14.4 ± 2.2 | 10.8 ± .9 | 25 |
| EUP | 1.7 ± 1.0 | 1.6 ± 1.0 | 6.0 ± .7 | 6.8 ± .5 | 16.5 ± .5 | 7 |
Calculated as described in the Subjects and Methods section. No error estimates are shown for fsr because the (intuitively large) uncertainty introduced by the repartitioning process is hard to assess. Some (2.4%) of lineages (“erratics”) were not assigned to founders and account for the remainder for each criterion; they are either the result of recent east Eurasian or African admixture or are rare types that could not be classified.
When the f0 partition analysis and the basic model were used, the age class with the most lineages was the Neolithic. This was also the case with the extended model, although the Neolithic contribution fell slightly, and a large component was attributed to the putative Mesolithic dispersal. However, the f1 analysis gave a quite different picture. When the basic model was used, the Neolithic contribution fell considerably, and the LUP rose, to become the majority component. For the extended model, much of the LUP contribution and some of the Neolithic contribution were taken up by the Mesolithic migration, which became the most significant migration (for the only time) under this criterion. For the more stringent f2 analysis, under the basic model, the Neolithic component fell further, and the LUP rose again. This pattern was repeated under the extended model.
Under f0, f1, and f2, the EUP component was 2%–6% and was contributed essentially by subsets of haplogroup U5. However, the other categories were rather unstable. We therefore applied the frequency-scaled criterion, fs. Although the number of founders identified by use of this criterion was closer to the number identified for f1 than to that identified for f2, the result of the partition analysis (under both the basic model and the extended model) was closer to that for f2. We based our subsequent analyses on the fs criterion.
We varied the dates of the basic model used for the partition analyses, to ensure that the outcomes were not crucially dependent on the value used. The analysis was most sensitive to the dates assigned to the Neolithic and the LUP: these are closest to each other in time and, hence, most easily confounded. However, even the placement of the LUP at 17,000 YBP had only a minor effect: the Neolithic contribution rose by <3%, the MUP fell slightly, and the LUP was more or less unchanged (data not shown).
It is possible to summarize the most likely contributing founders to each migration (see fig. 1). In the fs analyses, the principal Neolithic founder clusters were members of haplogroup J (in particular, the clusters based on the root sequence types of J and of J1a), T1, U3, and a few subclusters of H and W. The main contributors to the LUP expansions were the major subclusters of haplogroup H (including those derived from the H-CRS, 16304, and 16362-16482), K, T*, T2, W, and X. The main components of the MUP were HV*, U1, possibly U2, and U4, and the main component of the EUP was U. In the extended analysis, the Mesolithic component arose mainly from the reallocation of parts of haplogroup T.
Robustness of outcomes
To investigate the effect of sample size and differential back-migration into the more peripheral Near Eastern populations (Egyptians, Turks, Kurds, and southern Caucasians), we performed the fs′ analysis by excluding these populations and, using the remaining 577 Near Eastern samples, repeating the founder analysis. As table 4 shows, the results are remarkably similar to those derived from use of the complete data set, with the exception of the EUP category, which grows slightly at the expense of the others (since several haplogroups, including U4 and W, lose founder status and, hence, gain time depth within Europe). This suggests that our sample size is likely to be adequate and that most important founders have been identified.
Multiple migrations
To try to address the problem of possible multiple dispersals of lineages bearing the H-CRS, we partitioned the fs data into migration classes, with the H-CRS cluster omitted (the fsr analysis). We then repartitioned the H-CRS cluster (out of the LUP class, where it is placed when a single migration is assumed) into other feasible age categories (i.e., recent, Neolithic, and MUP; EUP is earlier than the estimated age of the H-CRS). The results are shown in table 4. The Neolithic component rises to 23% (in the basic model), and lineages are also partitioned more evenly between LUP and MUP. Therefore the main implications of reexpansion of the H-CRS, when this crude extrapolation from more–easily characterized lineages are used, would be a moderate rise in the Neolithic and MUP contributions and a concomitant fall in the LUP.
Regional analyses
To examine the data for regional patterns, we performed the analysis region by region, using the fs criterion. The results are shown in table 5. Strikingly, although the level of recent gene flow surviving under this criterion is similar for most populations, at 5%–9%, the eastern-Mediterranean region (samples from Thessaloniki, Sarakatsani, and Albanians) has a very high value, 20%. This may reflect the heavy historical gene flow known between Greece and other populations of the eastern Mediterranean.
Table 5.
Percentage, of Extant European mtDNA Pool, in Each Migration Event, by Region[Note]
|
Mean
± Root-Mean-Square Error,
for |
||||||||||
| Southeastern Europe (n = 166) | Eastern Mediterranean (n = 233) | Central Mediterranean (n = 302) | Alps (n = 218) | North-Central Europe (n = 332) | Western Mediterranean (n = 217) | Basque Country (n = 156) | Northwestern Europe (n = 456) | Scandinavia (n = 316) | Northeastern Europe (n = 407) | |
| Migration Event: | ||||||||||
| Bronze Age/recent | 8.2 ± 3.3 | 19.5 ± 3.7 | 4.6 ± 1.6 | 6.9 ± 2.7 | 8.9 ± 3.5 | 6.3 ± 2.5 | 5.4 ± 2.6 | 4.6 ± 1.5 | 7.4 ± 3.3 | 5.5 ± 3.0 |
| Neolithic | 19.7 ± 6.0 | 10.7 ± 5.0 | 9.2 ± 5.1 | 15.1 ± 5.4 | 17.1 ± 5.6 | 12.0 ± 4.2 | 6.7 ± 4.7 | 21.7 ± 4.5 | 11.7 ± 4.5 | 18.0 ± 4.5 |
| LUP | 44.1 ± 6.5 | 43.0 ± 4.9 | 49.1 ± 5.8 | 54.4 ± 5.8 | 52.2 ± 12.1 | 56.3 ± 5.0 | 58.8 ± 4.7 | 52.7 ± 4.5 | 52.8 ± 4.7 | 43.3 ± 4.4 |
| MUP | 14.6 ± 4.9 | 13.2 ± 4.7 | 21.1 ± 4.4 | 14.7 ± 4.8 | 11.8 ± 11.7 | 14.4 ± 4.8 | 13.0 ± 3.4 | 12.2 ± 2.9 | 12.4 ± 4.0 | 16.0 ± 3.5 |
| EUP | 10.9 ± 3.2 | 11.2 ± 3.6 | 12.6 ± 3.1 | 7.5 ± 3.0 | 8.6 ± 3.0 | 4.5 ± 3.3 | 13.6 ± 2.6 | 7.5 ± 2.6 | 14.1 ± 1.8 | 14.6 ± 2.6 |
| Erratics | 2.6 | 2.4 | 3.3 | 1.4 | 1.5 | 6.5 | 2.6 | 1.3 | 1.6 | 2.7 |
Note.— Partitioning is under the five-migration model and the fs criterion.
With respect to their Neolithic components, the regions fall into several groups. The southeastern, north-central, Alpine, northeastern, and northwestern regions of Europe have the highest components (15%–22%). The Mediterranean zone has a consistently lower (9%–12%) Neolithic component, suggesting that Neolithic colonization along the coast had a demographic impact less than that which resulted from the expansions in central Europe. Scandinavia has a similarly low value, and the Basque Country has the lowest value of all, only 7%.
The LUP values are, by contrast, higher toward the west: the western Mediterranean, the Basque Country, and the northwestern, north-central, Scandinavian, and Alpine regions of Europe have 52%–59% LUP, with the central-Mediterranean region having a value of almost 50%. The MUP values are perhaps highest in the Mediterranean zone, especially the central Mediterranean region. The EUP values are highest in Scandinavia, the Basque Country, and northeastern Europe.
Discussion
Assumptions of the Founder Analysis
Several previous studies have applied the basic principles of founder analysis to human mtDNA variation in America (Torroni et al. 1992, 1993a) and in the Pacific (Stoneking and Wilson 1989; Stoneking et al. 1992; Sykes et al. 1995; Richards et al. 1998b). The use of Y-chromosome variation for founder analysis of data from America also has begun (Ruiz Linares et al. 1996; Karafet et al. 1999). Both situations are thought to be likely to be amenable to such an analysis, in that they have relatively well-defined source regions, only one major dispersal event, and probably minimal postsettlement gene exchange with those regions (although they undoubtedly are more complex than usually is supposed; see Terrell 1986).
Europe clearly presents a more difficult case. The time depth is such that it is unclear whether the Near East represents a suitable source population stretching back prior to the LGM. Settlement seems likely to have occurred in multiple waves from the east and to have been subsequently obscured by millennia of recurrent gene flow. There may well have been significant levels of gene flow throughout Eurasia, from the Upper Palaeolithic to the present, particularly during the Holocene (the “Holocene filter”), which would obscure the signals of earlier dispersals. The problem is particularly acute for the Near East, since the latter forms the junction between three continents. Therefore, it is important to take into account recurrent gene flow when a founder analysis of Europe is performed.
Sample size may also be an issue. Despite a source-population sample (n = 1,234) much larger than has been used in all previous studies, there are reasons to be cautious. Both the higher diversity and degree of substructure in the Near East, in comparison with Europe, and the greater number of potential founder lineages raise the possibility that some founders may be missed in the sampling.
Our aim was to identify the principal founder lineages that have entered Europe and to date the times of their entry, in order to quantify the contribution that the main episodes of new settlement during European prehistory have made to the modern mtDNA pool. As regularly has been pointed out (e.g., see Barbujani et al. 1998), the divergence time estimated on the basis of the genetic diversity of the population as a whole will not, in general, indicate the time of settlement. This is because some of the preexisting diversity of the source population is expected to be carried into the derived population, so that some of the earlier branches in the genealogy will have been generated in the former rather than in the latter. The founder methodology is intended to take into account the presence of ancestral heterogeneity in the founding population. The principle is to sample extensively from the likely source population and to identify matching lineages between the source population and the derived population. The diversity in the derived population can then be corrected to allow for the preexisting diversity generated before the founder event. However, several assumptions that are involved when this is attempted should be made explicit:
-
1.
We assume that the Near East was the source region for most of the genetic variation extant in Europe. For the Neolithic, this assumption is readily justified on archaeological grounds (Henry 1989; Harris 1996); it is much less secure as one goes farther back in time, although archaeologists have argued in favor of a Near Eastern origin for the EUP, and it is even possible that the Aurignacian industry may have spread from the Levant and Anatolia (Gilead 1991; Mellars 1992; Olszewski and Dibble 1994; Bar-Yosef 1998). Analyses of classical genetic markers have also indicated expansions from the Near East, albeit also from eastern Europe (Cavalli-Sforza et al. 1994). The raw age estimates for the major clusters in Europe and the Near East are consistent with this assumption, since they indicate that the clusters are at least as old—and, in some cases, considerably older—in the Near East compared with Europe. However, we cannot rule out the possibility that significant dispersals may have originated not in the Near East but in either the northern Caucasus or eastern Europe, as has been suggested for the MUP (Soffer 1987). Given the high levels of drift that have occurred in the northern Caucasus (which have resulted in markedly non-starlike phylogenies for most haplogroups), our present sample size of 208 is insufficient for realistic estimation of the age of the various haplogroups.
-
2.
We assume that the Near East and Europe can be meaningfully considered as well-separated populations. This overlooks the extreme proximity of Greece and Turkey, for example. In fact, the historical evidence for gene flow between Europe and the Near East provides strong grounds for assuming that there is at least some back-migration from Europe across the Bosporus—or, farther east, across the Caucasus into the Near East—throughout the past 10,000 years. Candidates include the Philistine migrations from the Aegean into the Levant during the Bronze Age (Kuhrt 1995; Tubb 1998); the expansion of Greek, Phrygian, and Armenian speakers into western Anatolia, central Anatolia, and Armenia, respectively, ∼1,200 b.c. (Redgate 1998); and the importation of European as well as African slaves by the Islamic caliphs of Syria and Iraq during the medieval period (Lewis 1998). Recurrent gene flow would raise the number of matches between the two regions—and would reduce the estimated divergence times. That said, the level of recurrent gene flow has certainly not been large enough to equilibrate the European and Near Eastern mtDNA pools. However, the question of back-migration is one of the major challenges for this analysis, a challenge that we have addressed by demanding further evidence—that is, more evidence than merely the existence of a shared node in the phylogeny—of a Near Eastern origin for any founder candidate.
-
3.
A further assumption for the founder methodology is the infinite-alleles model—that is, that recurrent mutation is not a disturbing factor. In fact, parallel mutation and back-mutation are an important force in mtDNA, especially in the control region (Hasegawa et al. 1993), a force that, again, will artificially raise the number of matches between the two regions, thereby acting to reduce the divergence times. Our founder criteria again endeavor to address this issue by attributing an independent origin to recently derived shared types.
-
4.
The method assumes that most of the founding lineages have survived in the source population and that they have all been sampled. For a region as genetically diverse as the Near East, this assumption may be problematic, even with the sample of >1,200 presented here. Failure to identify important founders would increase the estimated age of the founder events. A recent founder analysis of Icelandic mtDNA makes this point well (Helgason et al. 2000). The sparseness of Near Eastern data previously available was a handicap for previous studies of the settlement of Europe (Richards et al. 1996; Barbujani et al. 1998; Renfrew 1998). However, several lines of reasoning suggest that we may have identified the majority of important founders by use of the present, larger-scale analysis. First, one argument against equating the Icelandic settlement with the European (Neolithic) founder scenario is that in the former case there likely was punctuated displacement of some unknown population(s), whereas in the latter case the expansion (in both range and head count) affected a large territory in the source, as well as in the settled areas, so that founders should be much easier to identify. Second, by employing a phylogenetic approach to the identification of founder candidates, rather than simply screening for sequence matches, we identify many candidate nodes in the network, even if they are unsampled. Third, although we have identified a large number of founders, we also have identified a similarly large number of likely back-migrants, using our screening criteria. In comparison with major founder types, back-migrants are likely to be rare in the source population, having typically arrived in the population more recently and at low frequency; this suggests that further searching would tend to uncover more back-migrants than genuine founder lineages. Furthermore, the outcome of our partitioning analysis was very little affected when we excluded a large proportion (∼50%) of the Near Eastern sample: Egyptians, Turks, Kurds, Armenians, and Azeris. This implies both that the sample is adequate and that the presence of peripheral and intermediate populations in the Near East has not unduly skewed the results.
-
5.
It is an implicit assumption of the founder analysis that each founder type is involved in only a single migration. If the same founder type were involved in two migrations, the estimated age of the founder in the derived population would be sometime between the age of the two events. However, if there were more than two migrations, the problem would become more acute. In practice, this is only likely to be a problem in the case of the CRS-derived founder cluster in haplogroup H. When only HVS-I data, a minimum of diagnostic RFLP sites, and, when available, HVS-II data are used, this part of the mtDNA genealogy is not well resolved. The H-CRS sequence type, which is as much as 30,000 years old, represents 6% of the Near Eastern lineages and 16% of the European lineages, and the H-CRS cluster in the fs analysis amounts to 38% of the European lineages. Other founder types were either infrequent in the Near East or insufficiently old to have contributed to more than two consecutive dispersals. Our response to this difficulty therefore focused on the H-CRS founder, exploring how the outcome was effected by repartitioning this cluster in the proportions exhibited by the remainder of the data.
-
6.
With regard to the formation of the distinct populations, the method also assumes a model in which only a limited number of types are transferred, during each successive phase of settlement. The outcome in the present day is a genetic palimpsest. This contrasts with the model of sequential population splits, which is often assumed in classical population genetics (e.g., see Cavalli-Sforza et al. 1994). Simulations (not shown) suggest that, as the initial size of the derived population approaches that of the source population, the founder methodology will overestimate the divergence time of clusters in the derived population, since the number of founders would become too large to be adequately sampled. Several features of worldwide mtDNA diversity patterns imply support for strong founder effects during colonization—for example, during the late-Pleistocene movement of anatomically modern humans out of Africa (Watson et al. 1997; Quintana-Murci et al. 1999) and during the colonization of Oceania from Indonesia (Redd et al. 1995; Sykes et al. 1995; Richards et al. 1998b).
-
7.
These assumptions are, of course, in addition to the usual problems associated with genetic dating. These problems include the rate of mutation and the dependence of the variance on the demographic history, as reflected in the shape of the genealogy. It has been argued elsewhere that the mutation rate is well supported (Macaulay et al. 1997), and, since we are usually dating very starlike mtDNA phylogenies when dealing with European founder candidates, the demographic-variance issue may not be a major problem for the dating of most pan-European clusters. For some minor non-starlike clusters, such as I, J1b, and J2 in Europe, the variance of these time estimates is certainly underestimated. Furthermore, it is quite possible that the phylogeny of a small cluster could appear starlike and yet that the underlying genealogy could be markedly structured, since the phylogeny may resolve only a small fraction of the underlying genealogy; in such a case, although ρ would be an unbiased estimator of the TMRCA of the sample, the variance would again be underestimated.
Migration into the Near East
We have employed a novel method to identify and quantify back-migration from Europe and the Near East. We have done this by identifying two European haplogroups (i.e., U5 and V) that appear to have evolved in situ. Extrapolating from the frequency of these clusters in the Near East has provided us with estimates for back-migration in general. These are strikingly high. We estimate that 10%–20% of extant Near Eastern lineages have a European ancestry, although this estimate falls to 6%–8% for the core zone of the Fertile Crescent. This contrasts with estimates of ∼5% for sub-Saharan African lineages and only ∼2% for lineages originating in eastern Eurasia. It emphasizes the importance of taking back-migration from Europe into account when colonization times are estimated by founder analysis.
Distribution of European Colonization Times
We first performed a naive founder analysis, using all founder candidates (f0). Although this analysis makes no allowance whatsoever for back-migration, the result from the partition analysis is interesting for what it does not show. Even this analysis attributes only 49% of maternal lineages to the Neolithic expansion, contradicting extreme Neolithic demic-diffusion–replacement views such as those of Chikhi et al. (1998a). Even when we allow for multiple dispersals of the H-CRS, by repartitioning this cluster within Europe, the analysis yields a Neolithic component of only slightly >50% (data not shown).
The Neolithic components in the f1, f2, and fs analyses were 22%, 12%, and 13%, respectively; the fsr value reaches 23% when possible multiple migrations of the H-CRS are allowed. This robustness to differing criteria for the exclusion of back-migration and recurrent mutation suggests that the Neolithic contribution to the extant mtDNA pool is probably on the order of 10%–20% overall. Our regional analyses support this, with values of ∼20% for southeastern, central, northwestern, and northeastern Europe. The principal clusters involved seem to have been most of J, T1, and U3, with a possible H component. This would suggest that the early-Neolithic LBK expansions through central Europe did indeed include a substantial demic component, as has been proposed both by archaeologists and by geneticists (Ammerman and Cavalli-Sforza 1984; Sokal et al. 1991). Incoming lineages, at least on the maternal side, were nevertheless in the minority, in comparison with indigenous Mesolithic lineages whose bearers adopted the new way of life. This does not exclude the possibility that acculturation occurred principally in southeastern Europe and that there was considerable replacement in central Europe. The Mesolithic component is even higher along the Mediterranean coastline, where archaeologists have suggested Neolithic pioneer colonization of uninhabited coastal areas by boat and a developing patchwork of coexisting Mesolithic and Neolithic communities for several millennia (Zilhão 1993, 1998). The Neolithic component here is ∼10%. It is similar in Scandinavia, where, again, the development of the Neolithic was very late and the impact of newcomers likely was slight. It is lowest of all, as might be expected, in the Basque Country (7%), although the presence of a number of rare European types at elevated frequency in the Basques points to the action of genetic drift in the region, as well as to a lack of Neolithic settlement. It is worth noting that the consistency between these results and the evidence of archaeology provides additional support for the founder methodology. Our analyses provide little support for Mesolithic dispersals into Europe after the Younger Dryas glacial interlude, suggesting that, if they occurred at all, they probably were limited to ∼10% of the total.
The new analyses confirm that the greatest impact on the modern mtDNA pool was migration during the LUP. The regional analyses lend some support to the suggestion that much of western and central Europe was repopulated largely from the southwest when the climate improved, as has been suggested, on both archaeological (Housley et al. 1997) and genetic (Torroni et al. 1998) grounds, by previous studies. The LUP component is highest in western and central Europe and is slightly lower to the north and east. However, allowing for multiple dispersals modifies the picture somewhat. The LUP component is on the order of 45%–60% of extant lineages in the fs analysis but falls to ∼36% if the H-CRS is repartitioned to allow for possible multiple expansions. The lineages involved include much of the most common haplogroup, H, as well as much of K, T, W, and X.
Whether there were migrations of H into Europe from the east during the LUP is unclear, but, despite the assumptions of the founder analysis, such immigrations seem unlikely to constitute the main LUP component, for several reasons. First, there is little or no archaeological evidence for expansions from the Near East during the Late Glacial period, and there is strong evidence for major demographic expansions from core areas in southwestern Europe and probably also central and southeastern Europe (Jochim 1987; Soffer 1987; Housley et al. 1997). Second, haplogroup V, the sister cluster of H within HV, appears to have evolved within Europe, possibly in the southwest, and to have expanded with the LUP component (Torroni et al. 1998). Finally, the LUP component is most common in the west (the western-Mediterranean region, the Basque Country, and northwestern Europe), substantiating its western origin.
Although a Near Eastern refugium giving rise to fresh European immigration after the LGM is not impossible, the dates for the major founders of H (especially the H-CRS, 16304, and 16362-16482) can be readily explained if bottlenecks in Europe at the LGM were sufficiently dramatic to partially erase preexisting diversity. The fact that the overall age of the European H-CRS cluster in the f2 and fs analyses is somewhat greater than the LUP date of, for example, haplogroup V and the H–16304, K, and T founder clusters, might also support this suggestion. Fortunately, this hypothesis is testable. We can date the H-CRS founder cluster in mainland Italy, where continuity would be expected between MUP populations and LUP populations (Bietti 1990; Leighton 1999). In this region, the Late Glacial–expansion cluster V (Torroni et al. 1998) is rare, and a founder analysis suggests a date of ∼24,000 YBP (95% CR = 16,400–32,900) for the H-CRS cluster—markedly older than the age for the continent overall. This is supported by the regional analysis shown in table 5, in which the central-Mediterranean region has the greatest MUP component outside the Caucasus, where continuity may also be anticipated (Dolukhanov 1994). It seems plausible, then, that many founders of haplogroup H—and, possibly, founders from other haplogroups dating to the LUP, such as much of K, T, W, and X—may have (a) arrived prior to the LGM, (b) suffered reductions in diversity, as a result of population contractions at the onset of the LGM, and (c) subsequently reexpanded.
As we move back in time, the picture becomes less clear. The value for the MUP is rather low in the basic fs analysis, at ∼10%–15%, and is highest along the Mediterranean, especially in the central-Mediterranean region. However, after allowance is made for multiple expansions of the H-CRS, it rises to ∼25% overall. The contributing clusters are mainly HV*, I, U4, and (in the repartitioned version) H. For the first settlement of Europe, at least, the picture seems to be clearer. The regional EUP component varies 5%–15% and comprises mainly haplogroup U5. The values are highest in southern and eastern Europe, as well as in Scandinavia and the Basque Country.
These analyses allow us to quantify the effects that various prehistoric processes have had on the composition of the modern mtDNA pool of Europe. They suggest that <10% of extant lineages date back to the first colonization of Europe by anatomically modern humans and that ∼20% arrived during the Neolithic. Most of the other lineages seem most likely to have arrived during the MUP and to have reexpanded during the LUP. Given the uncertainties associated with the analyses, we should not rule out the possibility of a Mesolithic migration, but we have found virtually no evidence supporting this idea. The results of our study are consistent with the archaeological evidence but, nevertheless, are interesting for the low values obtained for the demic component of the Neolithic expansion. Classical analyses, which were the first that used genetic data to predict colonization from the Near East (Ammerman and Cavalli-Sforza 1984; Sokal et al. 1991; Cavalli-Sforza et al. 1994), have often been interpreted as implying a majority Neolithic input, but the identification of relatively few markers showing northwest-southeast clines (e.g., see Sokal et al. 1989) seems to be consistent with the mtDNA picture. Indeed, Cavalli-Sforza and Minch (1997) also have recently interpreted the low proportion of variance associated with the first principal component for classical markers (26%) as implying a minority contribution from the Neolithic newcomers. It remains to be seen whether similar results will be obtained by performing such analyses on Y-chromosome data, preliminary analyses of which have indicated northwest-southeast clines (Semino et al. 1996; Casalotti et al. 1999). Nevertheless, particularly in view of the possibility of hypergamy (in which case, the mtDNA picture might somewhat underestimate the overall Neolithic genetic contribution; see Cavalli-Sforza and Minch 1997), it seems that a consensus may be within reach.
Finally, it is important to bear in mind that these values indicate the likely contribution of each prehistoric expansion to the composition of the present-day mtDNA pool. Extrapolating from this information to details of the demography at the time of the migration, although of course highly desirable for the reconstruction of archaeological processes, is unlikely to be straightforward.
Acknowledgments
We thank Peter Forster, Heinrich Härke, Agnar Helgason, Martin Jones, Stephen Oppenheimer, Colin Renfrew, and Stephen Shennan for critical advice; David Goldstein for support; Vicente Cabrera and Naif Karadsheh for access to their unpublished Jordanian data; Peter Forster and Anna Katharina Forster for providing Polish and Danish samples; Pierre-Marie Danze, Anne Cambon-Thomsen, and CEPH for the French samples; Heinrich Härke and Andrej B. Belinskij for help with collecting the Russian samples; Sarah Webb, Kersten Griffin, and members of the Stavanger Archaeological Museum for help with collecting the Norwegian samples; Steve Jones for collecting the Syrian samples; Savita Shanker for her work at the University of Florida DNA Sequencing Core Laboratory; and Mark Stoneking for forwarding the Nile Valley sequences. This research received support from The Wellcome Trust and from Italian Consiglio Nazionale delle Ricerche grant 99.02620.CT04 (to A.T.), by the Faculty of Biological Sciences, University of Urbino (support to A.T.), by the Ministero dell’Università e Ricerca Scientifica e Tecnologica: Grandi Progetti Ateneo (support to R.S.), by the Progetti Ricerca Interesse Nazionale 1999 (support to R.S. and S.S.-B.), and by the Progetto Finalizzato C.N.R. “Beni Culturali” (Cultural Heritage, Italy). N.A.-Z. was supported by an International Center for Genetic Engineering and Biotechnology fellowship. The Romanian contribution to this work was supported by the EC Commission, Directorate General XII (50%), and by the Romanian Ministry of Research and Technology (50%), within the framework of the Cooperation in Science and Technology with Central and Eastern European Countries (supplementary agreement ERBCIPDCT 940038 to the contract ERBCHRXCT 920032 coordinated by Prof. Alberto Piazza, University of Turin, Turin). M.R. was supported by a Wellcome Trust Bioarchaeology fellowship, V.M. was supported by a Wellcome Trust Research Career Development fellowship, and H.-J.B. was supported by a Deutscher Akademischer Austauschdienst travel grant.
Electronic-Database Information
The URL for data in this article is as follows:
- Authors' data available online, http://www.stats.ox.ac.uk/~macaulay/founder2000/index.html
References
- Adams J, Otte M (1999) Did Indo-European languages spread before farming? Curr Anthropol 40:73–77 [Google Scholar]
- Ammerman AJ, Cavalli-Sforza LL (1984) The Neolithic transition and the genetics of populations in Europe. Princeton University Press, Princeton, NJ [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Avise J, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC (1987) Intraspecific phylogeography: the molecular bridge between population genetics and systematics. Annu Rev Ecol Syst 18:489–522 [Google Scholar]
- Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen K-H, Wallace DC (1992) Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics 130:139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbujani G, Bertorelle G, Chikhi L (1998) Evidence for Paleolithic and Neolithic gene flow in Europe. Am J Hum Genet 62:488–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker G (1985) Prehistoric farming in Europe. Cambridge University Press, Cambridge [Google Scholar]
- Bar-Yosef O (1998) On the nature of transitions: the Middle to Upper Palaeolithic and the Neolithic Revolution. Camb Archeol J 8:141–163 [Google Scholar]
- Belledi M, Poloni ES, Casalotti R, Conterio F, Mikerezi I, Tagliavini J, Excoffier L (2000) Maternal and paternal lineages in Albania and the genetic structure of Indo-European populations. Eur J Hum Genet 8:480–486 [DOI] [PubMed] [Google Scholar]
- Berger JO (1985) Statistical decision theory and Bayesian analysis. Springer-Verlag, New York [Google Scholar]
- Bertranpetit J, Sala J, Calafell F, Underhill PA, Moral P, Comas D (1995) Human mitochondrial DNA variation and the origin of Basques. Ann Hum Genet 59:63–81 [DOI] [PubMed] [Google Scholar]
- Bietti A (1990) The Late Upper Palaeolithic of Italy: an overview. J World Prehist 4:95–155 [Google Scholar]
- Calafell F, Underhill P, Tolun A, Angelicheva D, Kalaydjieva L (1996) From Asia to Europe: mitochondrial DNA sequence variability in Bulgarians and Turks. Ann Hum Genet 60:35–49 [DOI] [PubMed] [Google Scholar]
- Casalotti R, Simoni L, Belledi M, Barbujani G (1999) Y-chromosome polymorphisms and the origins of the European gene pool. Proc R Soc Lond B Biol Sci 266:1959–1965 [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Cavalli-Sforza LL, Minch E (1997) Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet 61:247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-S, Torroni A, Excoffier L, Santachiara-Benerecetti AS, Wallace DC (1995) Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am J Hum Genet 57:133–149 [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Destro-Bisol G, Bertorelle G, Pascali V, Barbujani G (1998a) Clines of nuclear DNA markers suggest a largely Neolithic ancestry of the European gene pool. Proc Natl Acad Sci USA 95:9053–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Destro-Bisol G, Pascali V, Baravelli V, Dobosz M, Barbujani G (1998b) Clinal variation in the nuclear DNA of Europeans. Hum Biol 70:643–657 [PubMed] [Google Scholar]
- Comas D, Calafell F, Mateu E, Pérez-Lezaun A, Bertranpetit J (1996) Geographic variation in human mitochondrial DNA control region sequence: the population history of Turkey and its relationship to the European populations. Mol Biol Evol 13:1067–1077 [DOI] [PubMed] [Google Scholar]
- Comas D, Calafell F, Mateu E, Pérez-Lezaun A, Bosch E, Martínez-Arias R, Clarimon J, Facchini F, Fiori G, Luiselli D, Pettener D, Bertranpetit J (1998) Trading genes along the Silk Road: mtDNA sequences and the origin of central Asian populations. Am J Hum Genet 63:1824–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côrte-Real HBSM, Macaulay VA, Richards MB, Hariti G, Issad MS, Cambon-Thomsen A, Papiha S, Bertranpetit J, Sykes BC (1996) Genetic diversity in the Iberian peninsula determined from mitochondrial sequence analysis. Ann Hum Genet 60:331–350 [DOI] [PubMed] [Google Scholar]
- Dansgaard W, Johnsen SJ, Clausen HB, Dahl-Jensen D, Gundestrup NS, Hammer CU, Hvidberg CS, Steffensen JO, Sveinbjornsdottir AE, Jouzel J, Bond G (1993) Evidence for general instability of past climate from a 250-kyr ice-core record. Nature 364:218–220 [Google Scholar]
- Dennell R (1983) European economic prehistory: a new approach. Academic Press, London [Google Scholar]
- Diamond JM (1997) The language steamrollers. Nature 389:544–546 [Google Scholar]
- Di Rienzo A, Wilson AC (1991) Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci USA 88:1597–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolukhanov P (1994) Environment and ethnicity in the ancient Middle East. Avebury, Aldershot [Google Scholar]
- Finnilä, S, Hassinen IE, Ala-Kokko L, Majamaa K (2000) Phylogenetic network of the mtDNA haplogroup U in northern Finland based on sequence analysis of the complete coding region by conformative-sensitive gel electrophoresis. Am J Hum Genet 66:1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt H-J (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Francalacci P, Bertranpetit J, Calafell F, Underhill PA (1996) Sequence diversity of the control region of mitochondrial DNA in Tuscany and its implications for the peopling of Europe. Am J Phys Anthropol 100:443–460 [DOI] [PubMed] [Google Scholar]
- Gamble C (1986) The Palaeolithic settlement of Europe. Cambridge University Press, Cambridge [Google Scholar]
- ——— (1999) The Palaeolithic societies of Europe. Cambridge University Press, Cambridge [Google Scholar]
- Gilead I (1991) The Upper Palaeolithic Period in the Levant. J World Prehist 5:105–154 [Google Scholar]
- Harris DR (1996) The origins and spread of agriculture and pastoralism in Eurasia. University College London Press, London [Google Scholar]
- Hasegawa M, Di Rienzo A, Kocher TD, Wilson AC (1993) Toward a more accurate time scale for the human mitochondrial DNA tree. J Mol Evol 37:347–354 [DOI] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir S, Gulcher JR, Ward R, Stefánsson K (2000) mtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet 66:999–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry DO (1989) From foraging to agriculture. University of Pennsylvania Press, Philadelphia [Google Scholar]
- Hofmann S, Jaksch M, Bezold R, Mertens S, Aholt S, Paprotta A, Gerbitz KD (1997) Population genetics and disease susceptibility: characterization of central European haplogroups by mtDNA gene mutations, correlations with D loop variants and association with disease. Hum Mol Genet 6:1835–1846 [DOI] [PubMed] [Google Scholar]
- Housley RA, Gamble CS, Street M, Pettitt P (1997) Radiocarbon evidence for the Lateglacial human recolonisaton of northern Europe. Proc Prehist Soc 63:25–54 [Google Scholar]
- Jochim M (1987) Late Pleistocene refugia in Europe. In: Soffer O (ed) The Pleistocene Old World. Plenum Press, New York, pp 317–331 [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz K, Harihara S, de Knijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of New World Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS, Mastana SS, Mir MR, Ferak V, Villems R (1999a) Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol 9:1331–1334 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha S, Villems R (1999b) The place of the Indian mitochondrial DNA variants in the global network of maternal lineages and the peopling of the Old World. In: Papiha S, Deka R, Chakraborty R (eds) Genomic diversity: applications in human population genetics. Plenum Press, New York, pp 135–152 [Google Scholar]
- Krings M, Salem AH, Bauer K, Geisert H, Malek A, Chaix L, Simon C, Welsby D, Di Rienzo A, Utermann G, Sajantila A, Pääbo S, Stoneking M (1999) mtDNA analysis of Nile River Valley populations: a genetic corridor or a barrier to migration? Am J Hum Genet 64:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhrt A (1995) The ancient Near East. Vol 1: 3000–330 b.c. Routledge, London [Google Scholar]
- Leighton R (1999) Sicily before history. Duckworth, London [Google Scholar]
- Lewis B (1998) The multiple identities of the Middle East. Weidenfeld & Nicholson, London [Google Scholar]
- Macaulay VA, Richards MB, Forster P, Bendall KA, Watson E, Sykes BC, Bandelt H-J (1997) mtDNA mutation rates—no need to panic. Am J Hum Genet 61:983–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk BA, Derenko MV (1999) Molecular instability of the mitochondrial haplogroup T sequences at nucleotide positions 16292 and 16296. Ann Hum Genet 63:489–497 [DOI] [PubMed] [Google Scholar]
- Mellars PA (1992) Archaeology and the population-dispersal hypothesis of modern human origins in Europe. Philos Trans R Soc Lond B Biol Sci 337:225–234 [DOI] [PubMed] [Google Scholar]
- Menozzi P, Piazza A, Cavalli-Sforza LL (1978) Synthetic maps of human gene frequencies in Europeans. Science 201:786–792 [DOI] [PubMed] [Google Scholar]
- Olszewski DI, Dibble HL (1994) The Zagros Aurignacian. Curr Anthropol 35:68–75 [Google Scholar]
- Opdal SH, Rognum TO, Vege A, Stave A, Dupuy BM, Egeland T (1998) Increased number of substitutions in the D-loop of mitochondrial DNA in the sudden infant death syndrome. Acta Paediatr 87:1039–1044 [DOI] [PubMed] [Google Scholar]
- Parson W, Parsons TJ, Scheithauer R, Holland MM (1998) Population data for 101 Austrian Caucasian mitochondrial DNA D-loop sequences: application of mtDNA sequence analysis to a forensic case. Int J Legal Med 111:124–132 [DOI] [PubMed] [Google Scholar]
- Passarino G, Semino O, Bernini LG, Santachiara-Benerecetti AS (1996) Pre-caucasoid and caucasoid genetic features of the Indian population, revealed by mtDNA polymorphisms. Am J Hum Genet 59:927–934 [PMC free article] [PubMed] [Google Scholar]
- Passarino G, Semino O, Pepe G, Shrestha SL, Modiano G, Santachiara-Benerecetti AS (1992) mtDNA polymorphisms among Tharus of eastern Terai (Nepal). Gene Geogr 6:139–147 [PubMed] [Google Scholar]
- Passarino G, Semino O, Quintana-Murci L, Excoffier L, Hammer M, Santachiara-Benerecetti AS (1998) Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am J Hum Genet 62:420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piercy R, Sullivan KM, Benson N, Gill P (1993) The application of mitochondrial DNA typing to the study of white caucasian genetic identification. Int J Legal Med 106:85–90 [DOI] [PubMed] [Google Scholar]
- Pult I, Sajantila A, Simanainen J, Georgiev O, Schaffner W, Pääbo S (1994) Mitochondrial DNA sequences from Switzerland reveal striking homogeneity of European populations. Biol Chem Hoppe Seyler 375:837–840 [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt H-J, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence for an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 [DOI] [PubMed] [Google Scholar]
- Rando JC, Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM, Bandelt H-J (1998) Mitochondrial DNA analysis of northwest African populations reveals genetic exchanges with European, near-Eastern, and sub-Saharan populations. Ann Hum Genet 62:531–550 [DOI] [PubMed] [Google Scholar]
- Redd AJ, Takezaki N, Sherry ST, McGarvey ST, Sofro ASM, Stoneking M (1995) Evolutionary history of the COII/tRNALys intergenic 9 base pair deletion in human mitochondrial DNAs from the Pacific. Mol Biol Evol 12:604–615 [DOI] [PubMed] [Google Scholar]
- Redgate AE (1998) The Armenians. Blackwell, Oxford [Google Scholar]
- Renfrew C (1998) The origins of world linguistic diversity: an archaeological perspective. In: Jablonski NG, Aiello LC (eds) The origins and diversification of language. California Academy of Sciences, San Francisco, pp 171–192 [Google Scholar]
- Richards M, Côrte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, Hedges R, Bandelt H-J, Sykes B (1996) Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet 59:185–203 [PMC free article] [PubMed] [Google Scholar]
- Richards MB, Macaulay VA, Bandelt H-J, Sykes BC (1998a) Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet 62:241–260 [DOI] [PubMed] [Google Scholar]
- Richards M, Oppenheimer S, Sykes B (1998b) mtDNA suggests Polynesian origins in eastern Indonesia. Am J Hum Genet 63:1234–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M, Sykes B (1998) Reply to Barbujani et al. Am J Hum Genet 62:491–4929463340 [Google Scholar]
- Ruiz Linares A, Nayar K, Goldstein DB, Hebert JM, Seielstad MT, Underhill PA, Lin AA, Feldman MW, Cavalli-Sforza LL (1996) Geographic clustering of human Y-chromosome haplotypes. Ann Hum Genet 60:401–408 [DOI] [PubMed] [Google Scholar]
- Saillard J, Magalhães PJ, Schwartz M, Rosenberg T, Nørby S. Mitochondrial DNA variant 11719G is a marker for the mtDNA haplogroup cluster HV. Hum Biol (in press) [PubMed] [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus ML, Aula P, Beckman L, Tranebjaerg L, Geddedahl T, Isseltarver L, Dirienzo A, Paabo S (1995) Genes and languages in Europe—an analysis of mitochondrial lineages. Genome Res 5:42–52 [DOI] [PubMed] [Google Scholar]
- Sajantila A, Salem AH, Savolainen P, Bauer K, Gierig C, Pääbo S (1996) Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc Natl Acad Sci USA 93:12035–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo A (1998) mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet 6:365–375 [DOI] [PubMed] [Google Scholar]
- Semino O, Passarino G, Brega A, Fellous M, Santachiara-Benerecetti AS (1996) A view of the neolithic demic diffusion in Europe through two Y chromosome-specific markers. Am J Hum Genet 59:964–968 [PMC free article] [PubMed] [Google Scholar]
- Sherratt A (1994) The transformation of early agrarian Europe: the later Neolithic and Copper Ages. In: Cunliffe B (ed) The Oxford illustrated prehistory of Europe. Oxford University Press, Oxford [Google Scholar]
- Soffer O (1987) Upper Paleolithic connubia, refugia, and the archaeological record from eastern Europe. In: Soffer O (ed) The Pleistocene Old World. Plenum Press, New York, pp 333–348 [Google Scholar]
- Sokal RR, Harding RM, Oden NL (1989) Spatial patterns of human gene frequencies in Europe. Am J Phys Anthropol 80:267–294 [DOI] [PubMed] [Google Scholar]
- Sokal RR, Ogden NL, Wilson C (1991) Genetic evidence for the spread of agriculture in Europe by demic diffusion. Nature 351:143–144 [DOI] [PubMed] [Google Scholar]
- Stoneking M, Jorde LB, Bhatia K, Wilson AC (1990) Geographic variation in human mitochondrial DNA from Papua New Guinea. Genetics 124:717–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M, Sherry ST, Redd AJ, Vigilant L (1992) New approaches to dating suggest a recent age for the human mtDNA ancestor. Philos Trans R Soc Lond B Biol Sci 337:167–175 [DOI] [PubMed] [Google Scholar]
- Stoneking M, Wilson AC (1989) Mitochondrial DNA. In: Hill AVS, Serjeantson S (eds) The colonization of the Pacific: a genetic trail. Oxford University Press, Oxford, pp 215–245 [Google Scholar]
- Strauss LG (1995) The Upper Palaeolithic of Europe: an overview. Evol Anthropol 4:4–16 [Google Scholar]
- Sykes B, Leiboff A, Low-Beer J, Tetzner S, Richards M (1995) The origins of the Polynesians—an interpretation from mitochondrial lineage analysis. Am J Hum Genet 57:1463–1475 [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Routman E, Phillips CA (1995) Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics 140:767–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell JE (1986) Prehistory in the Pacific Islands. Cambridge University Press, Cambridge [Google Scholar]
- Thorpe IJ (1996) The origins of agriculture in Europe. Routledge, London [Google Scholar]
- Torroni A, Bandelt H-J, D’Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savantaus M-L, Bonné-Tamir B, Scozzari R (1998) mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet 62:1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus M-L, Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Lott MT, Cabell MF, Chen Y-S, Lavergne L, Wallace DC (1994a) mtDNA and the origin of Caucasians: identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am J Hum Genet 55:760–776 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Miller JA, Moore LG, Zamudio S, Zhuang J, Droma T, Wallace DC (1994b) Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol 93:189–199 [DOI] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993a) Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Yang C-C, Szathmary EJE, Williams RC, Schanfield MS, Troup GA, Knowler WC, Lawrence DN, Weiss KM, Wallace DC (1992) Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Sukernik RI, Schurr TG, Starikovskaya YB, Cabell MF, Crawford MH, Comuzzie AG, Wallace DC (1993b) mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet 53:591–608 [PMC free article] [PubMed] [Google Scholar]
- Tubb JN (1998) Canaanites. British Museum Press, London [Google Scholar]
- Ward RH, Frazier BL, Dew-Jager K, Pääbo S (1991) Extensive mitochondrial diversity within a single Amerindian tribe. Proc Natl Acad Sci USA 88:8720–8724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt H-J (1997) Mitochondrial footprints of human expansions in Africa. Am J Hum Genet 61:691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle A (1996) Europe in the Neolithic. Cambridge University Press, Cambridge [Google Scholar]
- Zilhão J (1993) The spread of agro-pastoral economies across Mediterranean Europe: a view from the Far-West. J Mediterr Archeol 6:5–63 [Google Scholar]
- ——— (1998) On logical and empirical aspects of the Mesolithic-Neolithic transition in the Iberian peninsula. Curr Anthropol 39:690–698 [Google Scholar]
- Zvelebil M (1986) Mesolithic prelude and neolithic revolution. In: Zvelebil M (ed) Hunters in transition: Mesolithic societies of temperate Eurasia and their transition to farming. Cambridge University Press, Cambridge, pp 5–15 [Google Scholar]
- ——— (1989) On the transition to farming in Europe, or what was spreading with the Neolithic: a reply to Ammerman (1989). Antiquity 63:379–383 [Google Scholar]