Abstract
Bietti crystalline corneoretinal dystrophy (BCD) is an autosomal recessive retinal degeneration characterized by multiple glistening intraretinal dots scattered over the fundus, degeneration of the retina, and sclerosis of the choroidal vessels, ultimately resulting in progressive night blindness and constriction of the visual field. Although BCD has been associated with abnormalities in fatty-acid metabolism and absence of fatty-acid binding by two cytosolic proteins, the genetic basis of BCD is unknown. We report linkage of the BCD locus to D4S426 (maximum LOD score [Zmax] 4.81; recombination fraction [θ] 0), D4S2688 (Zmax=3.97; θ=0), and D4S2299 (Zmax=5.31; θ=0), on chromosome 4q35-4qtel. Multipoint analysis confirmed linkage to the region telomeric of D4S1652 with a Zmax of 5.3 located 4 cM telomeric of marker D4S2930.
Bietti crystalline corneoretinal dystrophy (BCD [MIM 210370]) is a form of autosomal recessive retinitis pigmentosa (RP) characterized by the presence of crystals in the corneal limbus, posterior pole of the eye, and circulating lymphocytes. Clinically, progressive night blindness and constriction of the visual fields occurs around the 3d and 4th decade of life, followed by visual loss rapidly progressing to legal blindness in the 5th or 6th decade. Multiple glistening intraretinal dots occur over the fundus, associated with degeneration of the retina and sclerosis of the choroidal vessels (Bietti 1937). Although BCD is felt to be rare worldwide, it accounted for ∼3% (3/104 index patients) of all nonsyndromic RP and for ∼10% of nonsyndromic autosomal recessive RP in a recent series from Europe (F.L.M., unpublished data). BCD also appears to be more common in individuals of Asian descent (Hu 1983; Kaiser-Kupfer et al. 1994), especially in those from China, where the gene frequency has been estimated to be .005. Wilson et al. (1989) described, in the retina, crystals resembling cholesterol or cholesterol esters. Complex lipid inclusions also occur in the cornea, conjunctiva, fibroblasts, and circulating lymphocytes, suggesting that BCD may result from a systemic abnormality of lipid metabolism. More recently, histopathologic studies of the eye demonstrated advanced panchorioretinal atrophy, with crystals and complex lipid inclusions seen in choroidal fibroblasts. (Kaiser-Kupfer et al. 1994)
A number of findings point to the existence of systemic abnormalities of lipid metabolism in BCD (Lee et al. 1999). Abnormally high levels of triglycerides and cholesterol storage are seen in cultured cells from patients with BCD, whereas metabolism of labeled fatty-acid precursors into n-3 PUFA is decreased in BCD. A deficiency or dysfunction of a fatty-acid–binding protein could cause decreased uptake and transfer between lipid pools. In addition, deficient elongation and desaturation of fatty-acid precursors could cause the deposit, in crystalline form, of lipids within disease cells. Lymphocytes from patients with BCD have been shown to lack two fatty-acid–binding activities (Lee et al. 1998), but the relationship between the disease and these two absent binding activities remains unclear, especially since the disease is inherited as a single autosomal recessive trait. To compliment the biochemical approach to defining the gene causing BCD, linkage analysis of this disease in 10 families was carried out. Linkage was established with markers on chromosome 4q35-4qtel, including D4S426 (maximum LOD score [Zmax] 4.81; recombination fraction [θ] 0), D4S2688 (Zmax=3.97; θ=0), and D4S2299 (Zmax=5.31; θ=0).
Initially, a genomewide linkage screen for the BCD locus was performed in 10 families of Chinese (033001 and 033014), Japanese (033002–03307), and European (033010 and 033011) ethnic origin. Samples from four additional affected individuals were included in homozygosity mapping. Forty-nine individuals, 16 of whom were affected and 33 of whom were unaffected, were enrolled in this study. This study was approved by the National Eye Institute Institutional Review Board, and the patients admitted to the study gave their informed consent. Detailed ocular, medical, and family histories were obtained from each available family member. The affected family members underwent a complete physical examination. Fundus photography was performed on all affected patients. Clinical diagnosis was based both on the presence of superficial corneal crystals seen at the corneoscleral limbus (by slit-lamp biomicroscopy) and of abundant small sparkling yellow white crystals in the posterior pole and on atrophy of retinal pigment epithelium, sclera, and choroid (on funduscopic examination). Diagnosis was confirmed by the presence of lipid inclusions in lymphocytes in families 1, 10, and 11.
DNA was extracted either directly from blood or from transformed lymphoblastoid cell lines, by standard phenol-chloroform protocols (Smith et al. 1989). A genomewide scan was performed with 106 fluorescently labeled microsatellite markers (ABI Prism Linkage Mapping Set, version 2). For multiplexing PCR, each reaction was carried out in a 5-μl mixture containing 40 ng of genomic DNA, different combinations of 10 μM fluorescent dye–labeled primer pairs, 10× GeneAmp PCR Buffer II, 250 μM GeneAmp dNTP mix, 2.5 mM MgCl2, 0.2 U Taq DNA (AmpliTaq Gold Enzyme). Amplification was done by using an 800 Catalyst Molecular Biology Labstation (PE Biosystems). Initial denaturation was 12 min at 95°C; followed by 10 cycles of 15 s at 94°C, 15 s at 55°C, and 30 s at 72°C; then 20 cycles of 15 s at 89°, 15 s at 55°, and 30 s at 72°; and, last, with a 20-min extension cycle at 72°C and final hold at 4°C. PCR products from each DNA sample were pooled and mixed with a loading cocktail containing formamide, Gs-400HD ROX standards (PE Biosystems), and loading dye. The product was loaded onto 5% acrylamide gel and run in an ABI 377 Prism DNA Sequencer. The data were analyzed by ABI GENESCAN 3.1 and ABI GENOTYPER 2.1 software (PE Biosystems). Two independent individuals who were blinded to each other and to any phenotypic or family information read all gels, with conflicts resolved by a third independent reader. Data producing conflicts that could not be unambiguously resolved were discarded or, in an area of interest, repeated. Family relationships were confirmed by observation of Mendelian inheritance with 382 autosomal microsatellite markers from the ABI Linkage Mapping Set MD-10.
Initially a genomewide linkage screen was carried out using the ABI Medium Density Marker panel, with microsatellite markers distributed at an average of 10-cM intervals. Homozygosity mapping with pooled affected and unaffected siblings from each ethnic group gave no suggestion of linkage, over all chromosomes. Standard two-point linkage analysis was then carried out for all markers. Two-point linkage analysis was performed using the MLINK program of the LINKMAP program package, version 5.1 (Lathrop and Lalouel 1984), Zmax scores were calculated using ILINK, and LINKMAP was used for multipoint analysis. BCD was analyzed as a fully penetrant autosomal recessive trait. The gene frequency of BCD was set at .005 (Hu 1983). For screening, equal allele frequencies were assumed for all markers. When an initial LOD score of 3.6 was obtained with D4S426 at θ=0, five additional markers from the Généthon database (D4S3047–4cM–D4S3032–5cM–D4S3051–2cM–D4S2921/D4S426–1cM–D4S2930) and four polymorphic markers, positioned immediately telomeric of D4S2930, by physical mapping on the Stanford Human Genome Center YAC contig (D4S2688, D4S2283, D4S1652, and D4S2299), were analyzed to confirm the linkage. To our knowledge, these are the most telomeric polymorphic markers currently identified on chromosome 4q.
For fine mapping, the appropriate ethnic-specific allele frequencies for markers D4S426, D4S3047, D4S3032, D4S3051, D4S2921, D4S2930, D4S2688, D4S2283, D4S1652, and D4S2299 were approximated on the basis of 14 Chinese, 12 Japanese, and 15 European unrelated and unaffected individuals (28, 24, and 30 chromosomes, respectively). As seen in table 1, two-point linkage analysis gave evidence for linkage to markers on chromosome 4q35, with Zmax values of 4.81 with D4S426, at θ=0, 3.97 with D4S2688, at θ=0, and 5.31 with D4S2299, at θ=0. Suggestive LOD scores were also seen with other markers in the region, including D4S2921 (Zmax=2.70; θ=.04), D4S2930 (Zmax=2.75; θ=.06), and D4S2283 (Zmax=2.37; θ=.05). These markers, however, showed obligate recombinants. Multipoint linkage analysis carried out with overlapping sets of three markers from the Généthon database confirmed linkage to this region (Zmax=5.3) located 4 cM telomeric of marker D4S2930 (data not shown). Multipoint analysis carried out with VITESSE, using, in this interval, all mapped markers from the Généthon database simultaneously but discounting consanguinity loops, also confirmed linkage (Zmax=3.36) in the same region. Heterogeneity was examined using both the two-point linkage data from D4S3051, D4S2921, D4S426, and D4S2930 and the multipoint linkage data from these markers. Neither the M test for the three ethnic groups nor the admixture test (HOMOG) gave significant support for genetic heterogeneity. The admixture test was also performed for any marker in the genome screen with a Zmax>1 at any recombination fraction. No significant evidence was obtained for linkage with or without heterogeneity to any region outside chromosome 4q35-tel.
Table 1.
Summed Two-Point Linkage Results for BCD in All Families
|
LOD Score at θ = |
|||||||||
| Markers(Distancea) | 0 | .01 | .05 | .1 | .2 | .3 | .4 | Zmax | θ |
| D4S3047 | −206.07 | −6.08 | −1.85 | .47 | .61 | .37 | .11 | .61 | .20 |
| D4S3032 (4) | −99.52 | .59 | 1.45 | 1.31 | .91 | .46 | .12 | 1.52 | .07 |
| D4S3051 (5) | −101.09 | −.03 | .96 | 1.14 | .87 | .50 | .16 | 1.16 | .09 |
| D4S2921 (2) | 1.04 | 2.37 | 2.69 | 1.97 | 1.32 | .70 | .21 | 2.70 | .04 |
| D4S426 | 4.81 | 4.66 | 4.08 | 2.60 | 1.66 | .84 | .25 | 4.81 | .00 |
| D4S2930 (1) | − 99.15 | 2.01 | 2.75 | 1.82 | 1.26 | .66 | .20 | 2.75 | .06 |
| D4S2688 | 3.97 | 3.84 | 3.31 | 2.70 | 1.63 | .81 | .24 | 3.97 | .00 |
| D4S2283 | −98.66 | 1.61 | 2.37 | 2.20 | 1.43 | .66 | .16 | 2.37 | .05 |
| D4S1652 | −99.55 | 1.13 | 1.94 | 1.85 | 1.24 | .61 | .15 | 1.97 | .06 |
| D4S2299 | 5.31 | 5.14 | 4.51 | 3.76 | 2.34 | 1.18 | .39 | 5.31 | .00 |
Distance (in cM) from preceding marker.
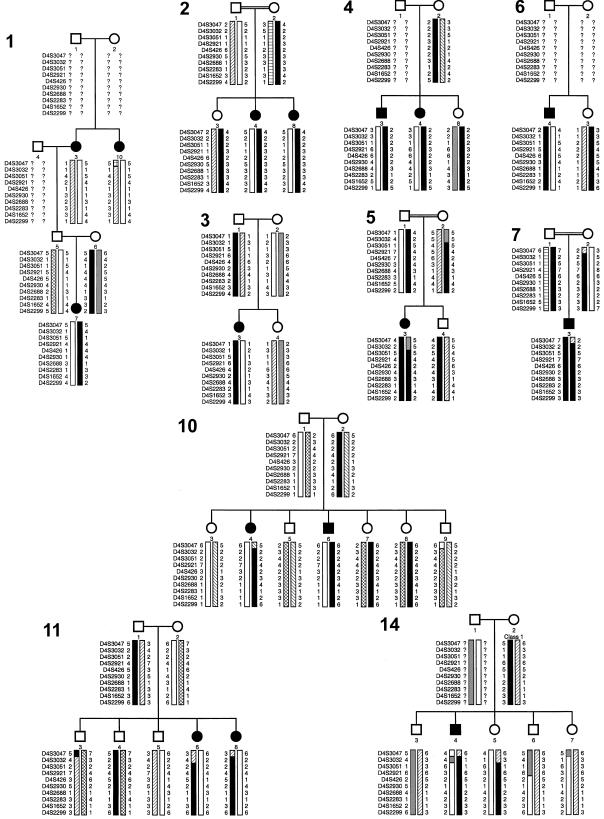
Visual inspection of the haplotypes for markers in this region supported the linkage-analysis data (fig. 1). In family 10, affected individuals 4 and 6 have inherited the same parental haplotypes for markers D4S2921–D4S2299, but a recombination event has taken place, at D4S3047, in individual 4, with markers D4S3032 and D4S3501 being uninformative. In family 14, affected individual 4 and unaffected sibling 5 have inherited the same parental haplotypes for markers D4S1652–D4S3051, a result that excludes that part of the interval. Although they have the same alleles for D4S2299, their mother is not informative for this marker, so this does not represent an obligate recombination event. She is also homozygous for D4S426, so this marker does not show an obligate recombination either. D4S2688 is not excluded because the father, whose DNA is not available for testing, could theoretically be uninformative at that locus.
Figure 1 .
Haplotypes for 10 markers from 4q35-4qtel, for 10 families with BCD. Marker order is shown to the left of each generation. Shading indicates haplotypes, with alleles of uncertain origin shaded to maximize the critical region. Families 2, 5, and 7 are all first-cousin matings, and, in these families, haplotypes consistent with identity by descent are shaded in solid black.
Families 033002, 033005, and 033007 are all first-cousin matings, so markers closely linked to the BCD locus are likely to be homozygous. In family 2, the only markers in the region that are homozygous are D4S3032, D4S426, D4S2688, and D4S2299. Because DNA samples are not available from all individuals who make up the consanguinity loop, the origin of these alleles cannot be traced unambiguously, and the presence of closely linked flanking markers suggests that this might represent identity by state rather than identity by descent. In family 5, individual 3 is not homozygous for markers D4S3047 and D4S3032, a result that limits the candidate region to the area telomeric of D4S3032. Similarly, in family 7, individual 3 is not homozygous for D4S3047.
The explanation most consistent with the aforementioned data is that the BCD locus resides on chromosome 4q, at a point telomeric of marker D4S1652. Although it remains a formal possibility, it seems unlikely that these results with highly significant two-point and multipoint LOD scores are spurious, produced by a fortuitous combination of alleles. Markers in the D4S3051–D4S1652 region recombine with BCD in families 033002 (Japanese) and 033014 (Chinese). Recently, an autosomal dominant retinopathy phenotypically similar to BCD has been described (Richards et al. 1991), suggesting that more than one genetic locus might exist for the BCD phenotype. However, although they certainly do not exclude locus heterogeneity, these data do not statistically support it. Thus, whereas locus heterogeneity with one locus in the D4S3051–D4S1652 interval also remains a formal possibility, our current results suggest a single BCD locus telomeric of D4S1652. Although, without having identified a distal flanking marker, it is difficult to estimate the size of this interval, within 8 cM the multipoint LOD scores drop by 1. This telomeric region has not been mapped, but if the properties and gene density were found to be similar to the genome averages, this might correspond to 8×106 bp containing 100–300 genes. Studies to refine this interval and to identify candidate genes in this chromosomal region are currently under way.
Acknowledgment
We would like to acknowledge support from the Swiss National Science Foundation (grant 32-053750-98) the families who donated samples to make this work possible.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Généthon database, http://www.genethon.fr/
- Online Mendelian Inheritance in Man (OMIM), http://www3.ncbi.nlm.nih.gov/Omim/ (for BCD [MIM 210370]) [PubMed]
- Stanford Human Genome Center, http://shgc-www.stanford.edu/
References
- Bietti G (1937) Ueber familiaeres vorkommen von “retinitis punctata albescens” (verbunden mit “dystrophia marginalis cristallinea corneae”), glitzern des glaskoerpers und anderen degenerativen augenveraenderungen. Klin Mbl Augenheilk 99:737–757 [Google Scholar]
- Hu DN (1983) Ophthalmic genetics in China. Ophthal Paediatr Genet 2:39–45 [Google Scholar]
- Kaiser-Kupfer MI, Chan CC, Markello TC, Crawford MA, Caruso RC, Csaky KG, Guo J, Gahl WA (1994) Clinical biochemical and pathologic correlations in Bietti's crystalline dystrophy. Am J Ophthalmol 118:569–582 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lee J, Jiao X, Gahl WA, Markello TC, Guo J, Chader GJ. The metabolism of fatty acids in human Bietti crystallin dystrophy. Invest Ophthalmol Vis Sci (in press) [PubMed] [Google Scholar]
- Lee J, Jiao X, Hejtmancik JF, Kaiser-Kupfer M, Chader GJ (1998) Identification, isolation, and characterization of a 32-kDa fatty acid-binding protein missing from lymphocytes in humans with Bietti crystalline dystrophy (BCD). Mol Genet Metab 65:143–154 [DOI] [PubMed] [Google Scholar]
- Richards BW, Brodstein DE, Nussbaum JJ, Ferencz JR, Maeda K, Weiss L (1991) Autosomal dominant crystalline dystrophy. Ophthalmology 98:658–665 [DOI] [PubMed] [Google Scholar]
- Smith RJH, Holcomb JD, Daiger SP, Caskey CT, Pelias MZ, Alford BR, Fontenot DD, Hejtmancik JF (1989) Exclusion of Usher syndrome gene from much of chromosome 4. Cytogenet Cell Genet 50:102–106 [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Weleber RG, Klein ML, Welch RB, Green WR (1989) Bietti's crystalline dystrophy: a clinicopathologic correlative study. Arch Ophthalmol 107:213–221 [DOI] [PubMed] [Google Scholar]