Abstract
Alternative splicing of the bcl-x gene generates two transcripts: the anti-apoptotic bcl-xL isoform and the pro-apoptotic bcl-xS isoform. The ratio between the two isoforms is a key factor in development and in cancer progression. Here, we show that a short antisense chimeric peptide nucleic acid (PNA) oligonucleotide conjugated to a polypeptide containing eight Ser-Arg repeats (SR)8 can modulate splicing of bcl-x both in vitro and in vivo and induces apoptosis in HeLa cells. The PNA-SR oligo was targeted to a region of bcl-x that does not contain splicing regulatory sequences and was able to override the complex network of splicing enhancers and silencers that regulates the ratio between the two bcl-x isoforms. Thus, PNA-SR oligos are powerful tools that can potentially modulate splice site choice in endogenous genes independent of the presence of other splicing regulatory mechanisms on the target gene.
INTRODUCTION
Analysis of the human genome indicates that at least 65% of human genes are alternatively spliced (1), which contributes significantly to the complexity of the human proteome. The understanding of the mechanisms that regulate alternative splicing and the development of novel technologies to modulate splicing patterns thus have great therapeutic potential in the treatments of cancer, as well as genetic and infectious diseases.
In tumors, the alternative splicing pattern of several genes, including bcl-x, can undergo dramatic changes. Bcl-x is a member of the bcl-2 family of apoptosis regulators and alternative splicing of the bcl-x pre-mRNA generates two main transcripts, a long (bcl-xL) and a short (bcl-xS) isoform, that encode proteins with antagonistic functions (2). Bcl-xL inhibits apoptosis through heterodimerization with pro-apoptotic proteins (3), whereas bcl-xS has pro-apoptotic properties. A proper expression ratio between isoforms is essential in several physiological and developmental processes, such as thymic selection, mammary gland involution and neural reshaping (4,5).
Bcl-xL is over-expressed in numerous types of cancer including myelomas, lymphomas, hepatomas, neuroblastomas and breast cancers (6–10). This over-expression of bcl-xL is associated with decreased apoptosis in tumors, resistance to chemotherapeutic drugs and a poor clinical outcome. Since bcl-xS over-expression can induce apoptosis in tumoral cell lines (11), the ability to alter the bcl-xL/-xS ratio thus has great therapeutic potential for the treatment of cancer. Previous work has shown that antisense oligonucleotides targeted to the bcl-xL 5′ splice site can decrease the expression of bcl-xL while increasing bcl-xS expression, thus inducing cell death in response to apoptotic signals (12,13).
However promising, the use of antisense oligonucleotides targeted to splice junctions is not always a suitable approach to alter the splicing pattern of a gene. The blockage of a splice site can result in the activation of a nearby cryptic splice site, which often leads to the appearance of mRNAs which translate for truncated or modified proteins. Additionally, a splice site block can cause unspliced mRNAs to be produced that are subject to rapid nuclear degradation. It is also conceivable that several unrelated genes could be inadvertently targeted by the same antisense oligonucleotide because 5′ splice site sequences have a high degree of conservation.
A different approach to affect splicing regulation, developed by Cartegni and Krainer (14), utilizes a new class of compounds that mimic SR protein function. SR proteins regulate splicing by binding to exonic sequences named exonic splicing enhancers (ESEs) and recruiting components of the splicing machinery to adjacent 5′ and 3′ splice sites (15). The Ser-Arg (SR)-rich domains present at the C-termini of SR proteins mediate such protein–protein interactions. Chimeric antisense peptide nucleic acid (PNA) oligonucleotides (16) linked to a polypeptide containing 5–10 Ser-Arg repeats have been shown in vitro to activate splicing of substrates derived from the BRCA1 and SMN2 genes (14).
It is unclear whether splicing activation by the chimeric PNA-SR oligos is dependent on the presence of pre-existing splicing regulatory elements or on its targeted position relative to the splice sites. In both the SMN2 and BRCA1 substrates, the targeted sequence was a previously characterized ESE, located a few nucleotides downstream of a 3′ splice site, which had been mutated to abolish splicing (14). Furthermore, it is unknown whether the PNA-SR oligos can be delivered to cellular systems to activate splicing in complex endogenous transcripts.
In this work, we utilized an in vitro splicing substrate derived from the bcl-x gene to determine that PNA-SR oligos are suitable tools to activate splicing independent of the presence of pre-existing splicing enhancers and silencers in the target gene sequence. We also determined that a chimeric PNA-SR oligo can efficiently modulate splice site selection in the bcl-x gene in vivo and induce apoptosis in HeLa cells.
MATERIALS AND METHODS
Plasmid constructions
pBK-bclx was generated by amplifying human genomic DNA with the primer pairs MC35.21 (5′-AGATATCTAGAATGTCTCAGAGCAACCGGGAGCTG), MC36.11 (5′-ATAGCGAATTCCAATCACCCAACACAACAGAAAGAG), MC36.12 (5′-ATAGCGAATTCCCACCCACCTACATCACTCTCTGAG) and MC36.13 (5′-TAGATCTCGAGCACAGTCATGCCCGTCAGGAACCAG). The PCR products were then inserted in pBluescript KS+ under the T7 polymerase promoter. The construct pBK-bclx-PM was generated by PCR-mediated site directed mutagenesis of pBK-bclx utilizing the primers MC35.37 (5′-CACCCTAGCGAGAGTATATCAGAGCTTTGAACAG) and MC35.38 (5′-TGATATACTCTCGCTAGGGTGATGTGGAGCTGGGA).
In vitro pre-mRNA splicing assays and preparation of SR proteins
Capped, 32P-labeled run-off transcripts were synthesized by in vitro transcription using T7 RNA polymerase. HeLa cell and S100 extracts were prepared as described previously (17). In vitro splicing reactions were performed in a total volume of 25 µl, containing 15 µl of HeLa cell nuclear extract or S100 as described previously (17). The reaction mixtures were incubated at 30°C for 2 h. RNAs recovered from the reaction mixtures were separated on an 8 M urea–6% polyacrylamide gel and visualized with a Kodak 200R Image Station. Splicing ratios were calculated adjusting for the relative length of the spliced and unspliced RNA products. SR proteins were prepared from HeLa cells and calf thymus as described previously (18), except for SF2/ASF that was graciously donated by Dr Adrian Krainer, Cold Spring Harbor Laboratory. SR proteins were normalized for splicing activity on a β-globin substrate as described previously (19). Approximately 200 ng of each SR protein, purified from calf thymus, or 500 ng of a total SR protein preparation from HeLa cells was added to the splicing mixture as indicated in Figure 2. All PNA, PNA-SR and SR peptides were synthesized and high-performance liquid chromatography purified by Oswald Scientific.
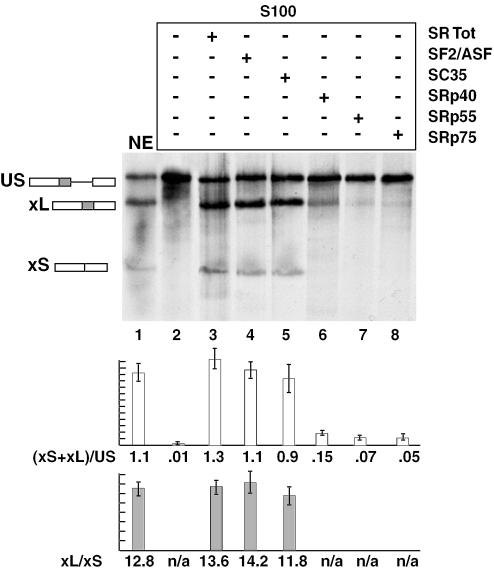
Figure 2.
In vitro splicing of pBK-bclx in HeLa S100 extract complemented with SR proteins. pBK-bclx pre-mRNAs were spliced in in vitro splicing reactions containing HeLa S100 extract (lanes 2–8) or HeLa nuclear extract (lane 1). Equal amounts of each purified SR protein were added where indicated. Single SR proteins were normalized in activity so that they had equivalent function in activation of splicing of a substrate containing the first intron of the human β-globin gene when added to a S100 extract (data not shown). Splicing ratios and error bars have been obtained from three independent experiments.
Transfection of PNA-SR oligos and RT–PCR
HeLa cells were maintained at below 80% confluence in D-MEM (Gibco BRL) supplemented with 5% calf serum and antibiotics. Cells were seeded 24 h before treatment at 40% confluence in a 24-well plate. At the time of transfection, growth media was removed and replaced with fresh media not containing serum or antibiotics. In addition, a mixture of Lipofectamine 2000 (Invitrogen) and oligos was added to the cells. After 5 h, growth media containing the transfection mixture was replaced with fresh media containing serum and antibiotics. Cells were harvested 48 h after transfection. Total RNA was extracted with Trizol (Invitrogen), reverse transcribed utilizing a poly(dT) oligonucleotide, and PCR amplified with primers MC21.50 (5′-GCGCTGAGGGAGGCAGGCGAC) and MC36.13 for 32 cycles. PCR products were visualized and quantified with a Kodak 200R Image Station. Splicing ratios were calculated adjusting for the relative length of the spliced and unspliced PCR products.
Western blot analysis
HeLa cells were transfected as described above and 24 h post-transfection, cells were scraped from six-well plates into the media, washed twice with phosphate-buffered saline and resuspended in 80 µl of SDS–polyacrylamide gel electrophoresis sample buffer. Samples were subjected to electrophoresis on a SDS–12% polyacrylamide gel and transferred to a PVDF membrane (Immobilon™ P, Millipore). Membranes were immunoblotted using a rabbit anti-bcl-x polyclonal antibody S18 (Santa Cruz Biotech) or a rabbit anti-hnRNP H/H′ polyclonal kindly provided by Dr D. L. Black (University of California, Los Angeles) and visualized by chemiluminescence (ECL™-system, Pierce).
Annexin-V/propidium iodide staining
HeLa cells were transfected as described above with 5 µM of PNA-(SR)8, PNA oligo control or (SR)8 polypeptide per well. Twenty-four hours post-transfection, cells were stained with Annexin-V and propidium iodide using the Annexin-V-Fluos Staining Kit (Roche) following the manufacturer's instructions. Annexin-V and propidium iodide staining were then detected by microscopy using a FITC filter (Zeiss inverted microscope-Axiovert 40 CFL).
RESULTS
SF2/ASF and SC35 activate bcl-x splicing
A combination of exonic silencers, exonic enhancers and intronic regulatory elements has been shown to regulate alternative splicing of the bcl-x gene in response to a variety of stimuli (Figure 1A) (20,21). To date, hnRNPs H and F are the only splicing factors found to bind one of those elements to regulate splicing (B2G in Figure 1A) (21).
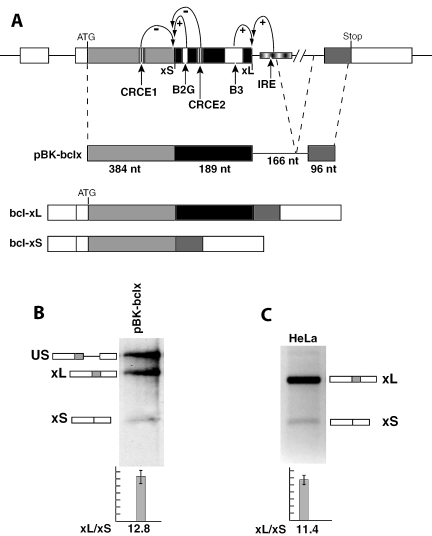
Figure 1.
Alternative splicing of bcl-x. (A) Schematic representation of the bcl-x gene and of the bcl-xL and bcl-xS alternatively spliced transcripts. Cis-regulatory sequences that act as splicing enhancers or silencers are indicated. The in vitro splicing construct used in this study, pBK-bclx, is schematically represented. (B) In vitro splicing of pBK-bclx. Radiolabeled pre-mRNA substrates were incubated in HeLa nuclear extract. RNA precursors and spliced products are indicated by the schematics. The ratio of bcl-xL to -xS observed is shown. (C) Endogenous bcl-x expression in HeLa cells. The ratio of bcl-xL/-xS in HeLa cells was determined by RT–PCR. Splicing ratios and error bars have been obtained from three independent experiments.
To gain insight into the regulation of bcl-x splicing, we constructed a minimal bcl-x splicing substrate containing the coding regions of exons 2 and 3 with a shortened intervening intron (Figure 1A). All the exonic splicing regulatory sequences previously characterized (20,21) are present in the pBK-bclx splicing substrate, whereas only part of the Intronic responsive element (IRE in Figure 1A) is present (22). Addition of further intronic sequences to include the entire IRE did not alter the bcl-xL/-xS splicing ratio (data not shown). In vitro splicing in HeLa nuclear extracts of the pBK-bclx substrate generated two spliced products corresponding to the bcl-xL and bcl-xS isoforms. The 12.8 ratio between the two isoforms (Figure 1B) is remarkably similar to the 11.4 bcl-xL/-xS ratio of the endogenous gene in HeLa cells (Figure 1C).
Members of the SR protein family are arguably the best characterized regulators of splicing. Single SR proteins have been shown to have similar, yet distinct substrate specificities and thus can differentially promote splicing of a given substrate or splice site. Nevertheless, SR proteins also operate as a constitutive part of the splicing machinery and are essential for the splicing of all transcripts (15,23,24). To determine whether specific SR proteins were involved in the splicing regulation of the bcl-x gene, the pBK-bclx splicing substrate was incubated in HeLa S100 cytoplasmic extracts complemented with single SR proteins. S100 extracts are depleted of SR proteins, thus splicing reactions performed with such extracts are not efficient. However, complementation with SR proteins has been shown to restore the ability of S100 extracts to activate splicing (23,25). Splicing of pBK-bclx was only slightly activated by complementation by SRp40, SRp55 or SRp75 (Figure 2, lanes 6–8). In contrast, complementation by a mixture of all SR proteins, SF2/ASF or SC35 activated splicing of the pBK-bclx substrate to levels similar to that obtained with nuclear extract (Figure 2, lanes 3–5).
Addition of SF2/ASF or SC35 to the splicing deficient S100 extracts did not significantly alter the bcl-xL/-xS ratio, 14.2 and 11.8, respectively (versus 12.8 when incubated with nuclear extract). This indicates that SF2/ASF and SC35 may play a constitutive role in the splicing of the bcl-x gene, but they do not selectively direct splicing toward one of the two alternative 5′ splice sites. We also cannot exclude the hypothesis that a SR protein different from the ones tested is specifically promoting splicing at one of the two alternative 5′ splice sites.
A chimeric PNA-SR oligo activates bcl-xS in vitro
To enhance the splicing of the bcl-xS isoform, a chimeric PNA oligo tethered to eight Ser-Arg (SR) repeats was made complementary to a site 20–31 nt upstream of the bcl-xS 5′ splice site (Figure 3A). The addition of the PNA-SR oligo to the in vitro splicing reactions in HeLa nuclear extract caused a dramatic dose-dependent switch in the bcl-xL/-xS ratio from 12.8 to 0.3 (Figure 3B, lanes 1–4). The switch in isoform ratio occurred at a compound concentration of 0.5–5 µM. Splicing was unaffected by a control PNA oligo lacking the (SR)8 domain (Figure 3B, lanes 5–7) or by the (SR)8 peptide alone (Figure 3B, lanes 8–9), demonstrating that both the antisense PNA targeting domain and the SR domain were required.
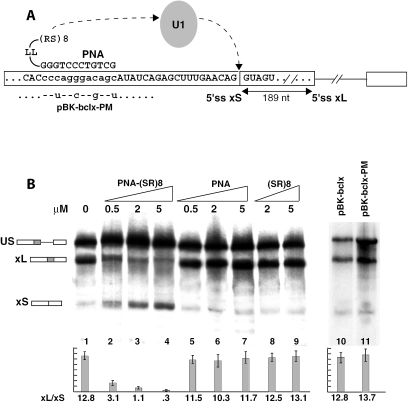
Figure 3.
A PNA oligo conjugated to an SR domain switches splicing to bcl-xS in vitro. (A) Schematic representation of the PNA-SR oligonucleotide, the location of its target sequence on the bcl-x transcript and likely mechanism of activation of the -xS splice site through recruitment of U1 snRNP. The mutations in the pBK-bclx-PM substrate are indicated. (B) In vitro splicing reactions. Increasing amounts of PNA-(SR)8 oligo, the control PNA oligo or the SR peptide alone were added to in vitro splicing reactions containing HeLa nuclear extract and the pBK-bclx substrate. The splicing profiles of pBK-bclx and pBK-bclx-PM are compared (lanes 10–11). Splicing ratios and error bars have been obtained from three independent experiments.
Regulatory sequences that either upregulate or downregulate splicing are often found in close proximity to the splice site they regulate (26). The absence of an effect by the PNA oligo control indicates that no other regulatory factors bind in the same region targeted by the antisense oligonucleotide. Nevertheless, to verify that no regulatory sequence is present in the region targeted by the PNA-SR antisense oligo, we mutated the splicing substrate (pBK-bclx-PM in Figure 3A). The wild-type pBK-bclx and mutant pBK-bclx-PM substrates splice with similar efficiency (12.8 and 13.7, respectively), thus confirming that no ESE or ESS sequences are present in the region targeted (Figure 3B). Additionally, the PNA-SR oligo, the PNA alone and the (SR)8 peptide alone all had no effect on the splicing of the mutant pBK-bclx-PM substrate (data not shown).
The PNA-SR oligo can modulate bcl-x splicing in vivo
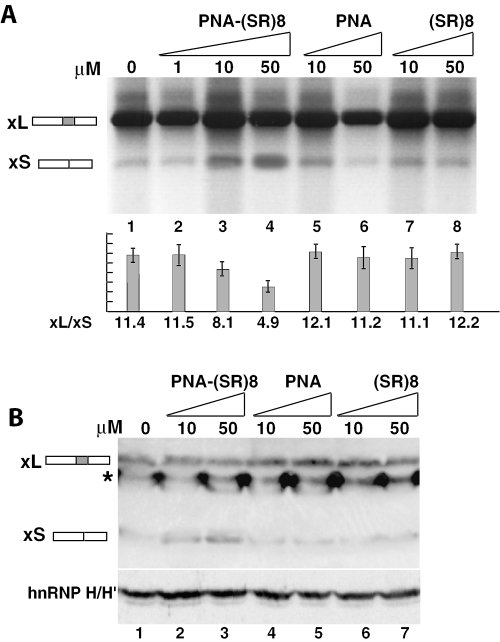
Since the chimeric PNA-SR oligo was able to dramatically and specifically switch splicing to the bcl-xS isoform in vitro, we investigated the possibility that this oligo would also alter the bcl-xL/-xS ratio in HeLa cells. The PNA-SR oligo was transfected into HeLa cells by cell scraping, spontaneous uptake and Lipofectamine. Transfection by Lipofectamine caused a dose-dependent switch in the bcl-xL/-xS ratio, at both the RNA and protein levels, when cells were transfected with a 10 µM solution of the PNA-SR oligo (Figure 4A, lanes 1–4 and Figure 4B, lanes 1–3), while no change was observed when cells were transfected by spontaneous uptake or cell scraping at concentrations up to 50 µM (data not shown). Using Lipofectamine, the bcl-xL/-xS mRNA ratio decreased from 11.4 in untreated cells to 4.9 when the PNA-SR oligo was added to a final concentration of 50 µM. The PNA-SR oligo effect on the bcl-xL/-xS ratio was highly specific since neither the PNA oligo alone nor the (SR)8 peptide alone affected bcl-x splicing (Figure 4A, lanes 5–8 and Figure 4B lanes 4–7). This is the first demonstration that PNA-SR oligos can modulate splicing in vivo.
Figure 4.
The PNA-SR oligo specifically alters the bcl-xL to -xS ratio in vivo. (A) HeLa cells were transfected with Lipofectamine and the indicated amounts of PNA-(SR)8 oligo, the PNA alone or the SR peptide. After 48 h, total RNA was isolated, reverse transcribed, and amplified by PCR. Splicing ratios and error bars have been obtained from three independent experiments. (B) Western blot of protein extracts derived from transfected HeLa cells. Cell lysates were analyzed with a bcl-x specific polyclonal antibody (upper panel) or a polyclonal antibody specific for hnRNP H/H′ as a control (lower panel). The asterisk symbol indicates a non-specific band recognized by the bcl-x antibody.
Modulation of splicing by PNA-SR oligos induces apoptosis in HeLa cells
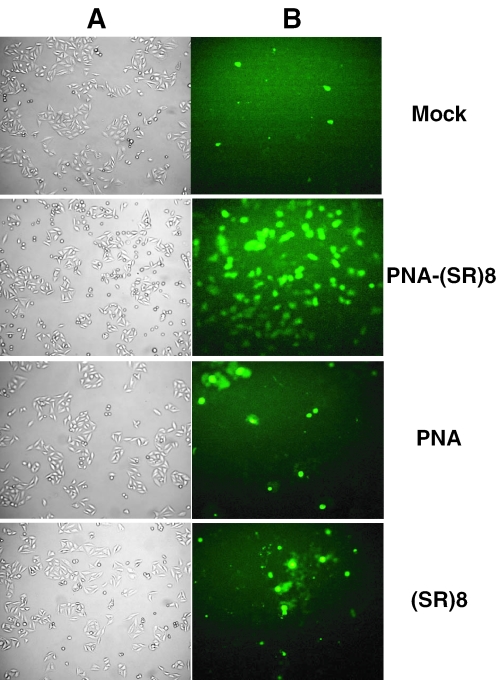
The short isoform (bcl-xS) of the bcl-x gene is known to have pro-apoptotic properties. To study the effect that alteration of the bcl-xL/-xS ratio by the PNA-SR oligo had on the transfected cells, we utilized Annexin-V and propidium iodide staining to detect apoptosis-mediated cell death and necrosis. During apoptosis, cells break up the phospholipid symmetry of their plasma membrane and expose phosphatidylserine on the external side of the lipid bilayer. Annexin-V preferentially binds to negatively charged phospholipids like phosphatidylserine and thus can be used to detect cells undergoing apoptosis. Following transfection with the bcl-x-specific PNA-(SR)8 oligo, cells showed a much higher level of apoptosis than cells that were transfected with the PNA alone or (SR)8 peptide control (Figure 5). The increased level of apoptosis was evident 24 h after transfection, whereas no increase in the level of necrotic cells was detected (data not shown). The chimeric PNA-SR oligo thus specifically triggers apoptosis in HeLa cells.
Figure 5.
Annexin-V staining of HeLa Cells transfected with the PNA-SR oligo. HeLa cells were transfected with 5 µM PNA-(SR)8, PNA oligo control, (SR)8 peptide or the transfection mix alone (Mock). Twenty-four hours after transfection, cells were stained with Annexin-V and analyzed by (A) phase contrast and (B) fluorescence microscopy. Fluorescence indicates cells undergoing apoptosis.
DISCUSSION
Utilizing the bcl-x gene as a model, we have shown that chimeric antisense PNA oligos conjugated to an SR domain can modulate splicing patterns independent of the existence of an SR-dependent-exonic splicing enhancer and that they can be efficiently delivered in vivo to trigger apoptosis in HeLa cells. SR proteins are important regulators of alternative splicing and two members of this family, SF2/ASF and SC35, up-regulate splicing of the bcl-x substrate. Nevertheless, SF2/ASF and SC35 did not promote specific splice site selection in this alternative 5′ splice site system.
Although SR proteins do not appear to be involved in regulating splice site choice in bcl-x, we showed that a chimeric oligo mimicking SR protein activity (PNA-SR) targeted upstream of the bcl-xS 5′ splice site did redirect the splicing machinery, inducing a dramatic switch in the ratio between the bcl-xL and bcl-xS isoforms. The use of PNA-SR compounds was developed previously by Cartegni and Krainer (14) who utilized in vitro splicing substrates derived from the SMN2 and BRCA1 genes. In that study, the PNA-SR compounds were targeted to a sequence immediately downstream of the 5′ splice site to restore inclusion of a skipped exon (14). In contrast, the sequence we targeted in bcl-x is located upstream of a 5′ splice site and the PNA-SR oligo causes that splice site to become preferentially utilized over a downstream 5′ splice site. The different kinds of splicing regulation induced, as well as the different positioning of the oligos in these systems, underscore the versatility of this technique.
Control PNA oligos not carrying the SR peptide have been shown to activate splicing when targeted to the BRCA1 and SMN2 substrates, although requiring a PNA oligo concentration 5–10 times higher to achieve comparable effects to the PNA-SR oligo (14). For bcl-x, we found that the PNA alone control oligo had no effect in vitro, even at a concentration of 5 µM. The discrepancy in the effect of the control PNA oligos between the bcl-x and the BRCA1 and SMN2 substrates can be explained, at least for the SMN2 substrate, with the fact that binding of the control PNA to the SMN2 substrate masks the binding site for hnRNP A1, a negative regulator of splicing (27), thus de-repressing splicing.
Transfection of antisense oligonucleotides to block the use of the bcl-xL 5′ splice site has been shown to efficiently alter the bcl-xL/-xS splicing ratio at a concentration of 1 µM (12,13). Additionally, antisense oligos that contain protein binding sites as part of a non-hybridizing 5′ tail targeted upstream of the bcl-xL 5′ splice site have been shown to alter the splicing ratio in the nM range (28). Here, we showed that a detectable change in the bcl-xL/-xS ratio in vivo is observed upon transfection of cells with the PNA-SR oligo at a concentration higher than 10 µM. Rather than blocking the bcl-xL 5′ splice site as previous studies have done, we activated splicing to bcl-xS and thus a high concentration of PNA-SR oligo may be needed to overcome the active mechanism repressing use of the bcl-xS 5′ splice site. Furthermore, it is possible that the chimeric PNA-SR oligonucleotide cannot efficiently cross the cellular and/or the nuclear membranes. Conjugation of the PNA-SR oligo with translocation peptides or to several positively charged residues (29) may be used to facilitate the absorption through the membranes and the nuclear localization of these oligos.
Nevertheless, our data shown in Figure 5 indicate that the PNA-SR oligo can induce apoptosis in HeLa cells by altering the bcl-xL/-xS ratio. Apoptosis is detected by 24 h after transfection of the cells with a 5 µM solution of the PNA-SR oligo. This concentration of PNA-SR oligo is much lower than the one utilized to observe a marked shift in the bcl-xL/-xS ratio at the RNA and protein levels (Figure 4). It is possible that only a sub-population of cells is transfected with high efficiency by the PNA-SR oligo, and thus the non-transfected cells dilute the effect of the splicing ratio switch. Furthermore, cells that are transfected with high efficiency presumably undergo apoptosis and degrade their mRNAs and proteins at much higher levels than the untransfected healthy cells, thus interfering with a direct analysis of the bcl-xL/-xS ratio switch.
PNAs have a neutral peptide-like backbone, form highly stable and specific base pairing with RNA, are resistant to nucleases, do not trigger RNase H activity and can be easily conjugated with any polypeptide (16). Furthermore, PNA oligos have been successfully delivered into cells in culture and into animal tissues (30). All these characteristics make these compounds ideal for their use as therapeutic agents. Not only can PNA-SR oligos modify the expression of single protein isoforms, but they also have the potential to generate novel mRNAs coding for new proteins. This would further increase the diversity of the proteome and possibly lead to new therapeutic tools.
In particular, it is conceivable that PNA-SR oligos could be targeted to activate cryptic splice sites that closely match the consensus sequence but are normally not used in vivo. The splicing machinery often does not recognize such splice sites because splicing enhancers within close proximity are often required for a splice site to be active. Accordingly, we have previously shown that even a perfect consensus 5′ splice site cannot splice without the presence of a nearby ESE in a viral transcript (31). Targeting a PNA-SR oligo in proximity of a cryptic splice site could activate such site, thus generating novel gene products.
An example for such applications could be the bcl-2 gene, an inhibitor of apoptosis. Despite its high homology at the nucleotide level with bcl-x, the bcl-2 gene is not alternatively spliced into a short and a long isoform like bcl-x. The bcl-xS 5′ splice site sequence AG/GUAGUG is similar to the corresponding sequence present in the bcl-2 gene CG/GUGGUG that is never utilized, but is close to the consensus 5′ splice site AG/GURAGU. Therefore, it may be possible to use a PNA-SR oligo targeted to that region of bcl-2 to create a novel bcl-2 isoform that encodes a protein that is likely to have pro-apoptotic properties. PNA-SR oligos have the potential to change not only known splicing patterns, but also to create novel gene products in cellular and possibly animal systems.
Acknowledgments
The authors thank Dr Alan Zahler (University of California, Santa Cruz) and Dr Adrian Krainer (Cold Spring Harbor Laboratory) for SR protein preparations as well as Dr Doug Black (University of California, Los Angeles) for hnRNP H/H′ antibodies. The authors also thank Dr Mark Kantorow and Dr Maria Marchetti (Florida Atlantic University) for their helpful comments and technical help. This work was supported by NIH/NIAID grants R01AI052820 and R01AI052820-02S1 to M.C. Funding to pay the Open Access publication charges for this article was provided by NIH/NIAID grant R01AI052820.
Conflict of interest statement. None declared.
REFERENCES
- 1.Leipzig J., Pevzner P., Heber S. The Alternative Splicing Gallery (ASG): bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boise L.H., Gonzales-Garcia M., Postema C.E., Ding L., Lindsten T., Turka L.A., Mao X., Nunez G., Thompson C.B. bcl-x a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 3.Kelekar A., Thompson C.B. Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 4.Reed J.C. Splicing and dicing apoptosis genes. Nat. Biotechnol. 1999;17:1064–1065. doi: 10.1038/15048. [DOI] [PubMed] [Google Scholar]
- 5.Motoyama N., Wang F., Roth K.A., Sawa H., Nakayama K., Nakayama K., Negishi I., Senju S., Zhang Q., Fujii S., et al. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 6.Olopade O.I., Adeyanju M.O., Safa A.R., Hagos F., Mick R., Thompson C.B., Recant W.M. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J. Sci. Am. 1997;3:230–237. [PubMed] [Google Scholar]
- 7.Xerri L., Parc P., Brousset P., Schlaifer D., Hassoun J., Reed J.C., Krajewski S., Birnbaum D. Predominant expression of the long isoform of Bcl-x (Bcl-xL) in human lymphomas. Br. J. Haematol. 1996;92:900–906. doi: 10.1046/j.1365-2141.1996.423958.x. [DOI] [PubMed] [Google Scholar]
- 8.Takehara T., Liu X., Fujimoto J., Friedman S.L., Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34:55–61. doi: 10.1053/jhep.2001.25387. [DOI] [PubMed] [Google Scholar]
- 9.Tu Y., Renner S., Xu F., Fleishman A., Taylor J., Weisz J., Vescio R., Rettig M., Berenson J., Krajewski S., et al. BCL-X expression in multiple myeloma: possible indicator of chemoresistance. Cancer Res. 1998;58:256–262. [PubMed] [Google Scholar]
- 10.Reeve J.G., Xiong J., Morgan J., Bleehen N.M. Expression of apoptosis-regulatory genes in lung tumour cell lines: relationship to p53 expression and relevance to acquired drug resistance. Br. J. Cancer. 1996;73:1193–1200. doi: 10.1038/bjc.1996.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dole M.G., Clarke M.F., Holman P., Benedict M., Lu J., Jasty R., Eipers P., Thompson C.B., Rode C., Bloch C., et al. Bcl-xS enhances adenoviral vector-induced apoptosis in neuroblastoma cells. Cancer Res. 1996;56:5734–5740. [PubMed] [Google Scholar]
- 12.Taylor J.K., Zhang Q.Q., Wyatt J.R., Dean N.M. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat. Biotechnol. 1999;17:1097–1100. doi: 10.1038/15079. [DOI] [PubMed] [Google Scholar]
- 13.Mercatante D.R., Bortner C.D., Cidlowski J.A., Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. Analysis of apoptosis and cell death. J. Biol. Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 14.Cartegni L., Krainer A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nature Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- 15.Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen P.E. Antisense peptide nucleic acids. Curr. Opin. Mol. Ther. 2000;2:282–287. [PubMed] [Google Scholar]
- 17.Mayeda A., Krainer A.R. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- 18.Zahler A.M. Purification of SR protein splicing factors. Methods Mol. Biol. 1999;118:419–432. doi: 10.1385/1-59259-676-2:419. [DOI] [PubMed] [Google Scholar]
- 19.Caputi M., Zahler A.M. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massiello A., Salas A., Pinkerman R.L., Roddy P., Roesser J.R., Chalfant C.E. Identification of two RNA cis-elements that function to regulate the 5′ splice site selection of Bcl-x pre-mRNA in response to ceramide. J. Biol. Chem. 2004;279:15799–15804. doi: 10.1074/jbc.M313950200. [DOI] [PubMed] [Google Scholar]
- 21.Garneau D., Revil T., Fisette J.F., Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 22.Li C.Y., Chu J.Y., Yu J.K., Huang X.Q., Liu X.J., Shi L., Che Y.C., Xie J.Y. Regulation of alternative splicing of Bcl-x by IL-6, GM-CSF and TPA. Cell Res. 2004;14:473–479. doi: 10.1038/sj.cr.7290250. [DOI] [PubMed] [Google Scholar]
- 23.Zahler A.M., Neugebauer K.M., Lane W.S., Roth M.B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 24.Caputi M., Kendzior R.J., Jr, Beemon K.L. A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes Dev. 2002;16:1754–1759. doi: 10.1101/gad.997502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krainer A.R., Mayeda A., Kozak D., Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 26.Blencowe B.J. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 27.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nature Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 28.Villemaire J., Dion I., Elela S.A., Chabot B. Reprogramming alternative pre-messenger RNA splicing through the use of protein-binding antisense oligonucleotides. J. Biol. Chem. 2003;278:50031–50039. doi: 10.1074/jbc.M308897200. [DOI] [PubMed] [Google Scholar]
- 29.Sazani P., Kang S.H., Maier M.A., Wei C., Dillman J., Summerton J., Manoharan M., Kole R. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 2001;29:3965–3974. doi: 10.1093/nar/29.19.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pooga M., Soomets U., Hallbrink M., Valkna A., Saar K., Rezaei K., Kahl U., Hao J.X., Xu X.J., Wiesenfeld-Hallin Z., et al. Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo. Nat. Biotechnol. 1998;16:857–861. doi: 10.1038/nbt0998-857. [DOI] [PubMed] [Google Scholar]
- 31.Caputi M., Freund M., Kammler S., Asang C., Schaal H. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J. Virol. 2004;78:6517–6526. doi: 10.1128/JVI.78.12.6517-6526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]