Abstract
Homing endonucleases, endonucleases capable of recognizing long DNA sequences, have been shown to be a tool of choice for precise and efficient genome engineering. Consequently, the possibility to engineer novel endonucleases with tailored specificities is under strong investigation. In this report, we present a simple and efficient method to select meganucleases from libraries of variants, based on their cleavage properties. The method has the advantage of directly selecting for the ability to induce double-strand break induced homologous recombination in a eukaryotic environment. Model selections demonstrated high levels of enrichments. Moreover, this method compared favorably with phage display for enrichment of active mutants from a mutant library. This approach makes possible the exploration of large sequence spaces and thereby represents a valuable tool for genome engineering.
INTRODUCTION
The use of rare-cutting double-stranded DNA endonucleases, called meganucleases, has emerged as the most successful means to enhance gene targeting (1), a powerful strategy for studying gene functions and for correcting genetic defects. In the wild, meganucleases are essentially represented by homing endonucleases, a family of DNA nucleases found in phage, bacteria, archaea and various eukaryotes (2–4). Homing endonucleases are encoded by self-propagating genetic elements, usually introns or inteins. These proteins recognize and cleave long DNA sequences (12–40 bp) within intron- or intein-less alleles, leaving recombinogenic double-stranded DNA breaks (DSBs). Repair of the DSB by homologous recombination with an intron or intein-containing gene results in the insertion (homing) of the intron or intein. Meganuclease-induced gene targeting was derived from this natural homing mechanism: a rare-cutting DNA endonuclease is used to cut a target at a chosen genomic locus, which provides significant enhancement of desired chromosomal DNA rearrangement by homologous recombination. In particular, the use of I-SceI (intron-encoded endonuclease I from Saccharomyces cerevisiae), one of the first homing endonucleases characterized (5,6), with its cognate homing site has rapidly become a standard strategy (7–17). Applications nevertheless remain limited to situations for which a natural homing endonuclease site is initially introduced at the chromosomal target locus. When such an engineering step is not practical, or simply not desired, DSB-induced gene targeting requires a dedicated cleavage agent that has to be custom-engineered.
Significant efforts are being made to find, design or engineer new endonucleases with desired cleavage specificities. A successful example is the fusion of Cys2–His2 type zinc-finger proteins (ZFP) with the catalytic domain of the FokI nuclease in order to generate functional sequence-specific endonucleases (18,19). Phage display selections and de novo design have been used to select a set of zinc fingers able to target (G/A/C)NN triplets, which can be combined to target a site of interest (20–23). Nevertheless, preserving a very narrow specificity is one of the major issues for genome engineering applications, and presently it is unclear whether ZFPs would fulfill the very strict requirements for many applications.
An alternative way to obtain custom meganucleases would be to change the specificity of natural homing endonucleases by molecular engineering. Several approaches have been undertaken by several teams to reach this goal, including construction of hybrid proteins, and screening or selection-based techniques. These techniques have been essentially based on bacterial genetic selection systems that link either DNA cleavage or DNA binding to cell survival or growth (24–27).
Being focused on DSB-induced gene targeting, we have developed two similar methods to identify active endonucleases that promote cleavage-induced homologous recombination in yeast cells. The use of eukaryotic cells is one advantage of our method over previous ones, in addition to the selection for chromosomal DNA rearrangements instead of DNA cleavage or binding. A first method places both the endonuclease gene and the DNA target site on separate replicative yeast plasmids, whereas a second one places the DNA targets on chromosomal loci. While both report the restoration of marker enzymes activity as a result of recombination events, they have distinct features and outcomes. The former one allows for high-throughput screening (HTS), and we have used it to isolate a large number of new highly specific I-CreI (intron-encoded endonuclease I from Chlamydomonas reinhardtii) based meganucleases cleaving novel target sites (28). The generation of collections of novel homing endonucleases, and the ability to combine them, could create a large catalog of engineered meganucleases. However, when a precise chromosomic site has to be targeted, one might need to explore large libraries, and HTS may prove insufficient. In such a case, the possibility to use a selection process that would isolate clones capable of cleaving a site of interest from large libraries of variants is invaluable.
The second method is a selection method. To assay its ability to select for variants of interests, we have processed a library based on I-CreI (29,30). This meganuclease is a homing endonuclease belonging to the LAGLIDADG family of homing endonucleases (the largest of the four homing endonucleases families). The 3D structure of I-CreI (4,31) is defined enough to allow the precise identification of amino acid residues interacting with the cognate DNA target. A semi rational approach aiming at randomizing only these relevant residues can thus be conducted. To further evaluate the results obtained with our new in vivo selection method, we performed similar selections using the well-known phage display strategy, which provided tremendous success with DNA-binding proteins such as zinc fingers (22,32). Our results show that this new in vivo selection method can deliver enrichment in the 105 range in a single run, and favorably compares with phage display in terms of output diversity. This method should facilitate meganuclease engineering and provide a valuable tool for genome surgery.
MATERIALS AND METHODS
Structure analyses
All analyses of protein structures were realized using Pymol. The structures from I-CreI correspond to pdb entry 1g9y. Residue numbering in the text always refers to these structures, except for residues in the second I-CreI protein domain of the homodimer where residue numbers were set as for the first domain.
Yeast DNA target sequences
Both the wild-type natural I-CreI target (named C1234: tcaaaacgtcgtgagacagtttgg) and two 24 bp palindromes (called C1221: tcaaaacgtcgtacgacgttttga and C4334: ccaaactgtctcgagacagtttgg) corresponding to inverted repeats of both half-sites of this target were used in screening campaigns (our nomenclature for the target sites is based on describing the natural I-CreI cognate DNA target as four quarters of six bases). Three pGEM-T Easy vectors (Promega) were constructed that comprise our target sequences. Next, a 400 bp PvuII fragment was excised and cloned into the yeast vector pFL39-ADH-LACURAZ, described previously (33). Yeast reporter vectors were transformed into S.cerevisiae strain FYBL2-7B (MAT a, ura3Δ851, trp1Δ63, leu2Δ1, lys2Δ202).
Selection in yeast
A yeast strain, specific for selecting endonucleases cleaving the I-CreI related targets, was constructed with strain FYC2-6A (MAT α, trp1Δ63, leu2Δ1, his3Δ200). First, a LYS2 single-strand annealing (SSA) target was engineered by modifying the lys2 gene to introduce two direct repeats of 3240 bp separated by a 4.3 kb cassette containing a kanamycin resistance gene and the target site, using a plasmid harboring a lys2 gene containing a 5′ and 3′ deletion, and digested by NheI prior to transformation. The resulting intermediate yeast strain, resistant to aminoglycoside G418, was unable to grow on minimal media lacking lysine. Upon overexpression of I-CreI, however, the strain was able to grow on media without lysine (but sensitive to G418). Second, an ADE2 SSA target was added: the ade2 gene was disrupted by transforming the strain with a plasmid harboring an ade2 gene containing a 5′ and 3′ deletion, and linearized by StuI, thereby introducing two direct repeats of 1.1 kb separated by a 3.6 kb cassette containing a tryptophan selectable marker plus the target site. The final selection yeast strain was thus resistant to G418 and prototrophic for tryptophan, while unable to grow on minimal media lacking lysine and/or adenine (detailed protocol available upon request).
For selection, the I-CreI mutant library was used to transform yeast cells from the selection strain using the classical TRAFO method (34). Cells were plated onto a selective medium without leucine (selection of transformed cells), and containing galactose as a carbon source and to induce overexpression of I-CreI mutants. After three days of growth at 37°C, colonies were suspended in water and aliquots of the suspension were plated onto selective media lacking leucine, adenine and lysine (selection for the clones wherein both the ADE2 and LYS2 SSA reporters had recombined) and containing galactose. Eventually, isolated clones were arrayed into 96-well microtiter plates containing 100 µl/well of YPD medium, using a QPix2 colony picker (Genetix). All microplates were incubated overnight at 30°C, under agitation, and sterile glycerol was added (final glycerol concentration was 20%) for storage at −80°C.
Screening in yeast
Mating was performed using a colony gridder (QpixII, Genetix). Mutants were gridded on nylon filters covering YPD medium plates, using a high gridding density (∼20 spots/cm2). A second gridding process was performed on the same filters to spot a second layer consisting of six different reporter-harboring yeast strains for each variant. Membranes were placed on solid agar YPD rich medium, and incubated at 30°C for one night, to allow mating. Next, filters were transferred to synthetic medium, lacking leucine and tryptophan, with galactose (2%) as a carbon source and incubated for 5 days at 37°C, to select for diploids carrying the expression and target vectors. After 5 days, filters were placed on solid agarose medium with 0.02% X-Gal in 0.5 M sodium phosphate buffer, pH 7.0, 0.1% SDS, 6% dimethyl formamide, 7 mM β-mercaptoethanol, 1% agarose, and incubated at 37°C, to monitor β-galactosidase activity.
Construction of mutant libraries
I-CreI wt and I-CreI D75N open reading frames (ORFs) were synthesized, as described previously (33). Mutation D75N was introduced by replacing codon 75 with AAC. The diversity of the meganuclease library was generated by PCR using degenerate primers from Sigma harboring codon VVK (18 codons coding for amino acids A, D, E, G, H, K, N, P, Q, R, S or T) at position 44, 68 and 70, and as DNA template, the I-CreI gene. The final PCR product was digested with specific restriction enzymes, and cloned back into the I-CreI ORF digested with the same restriction enzymes (detailed protocol is available upon request), in pCLS542. In this 2 µm-based replicative vector marked with the LEU2 gene, I-CreI variants are under the control of a galactose inducible promoter (33). After electroporation in Escherichia coli, we obtained 7 × 104 clones, representing 12 times the theoretical diversity at the DNA level (183 = 5832). DNA was extracted and transformed into S.cerevisiae strain FYC2-6A (MATα, trp1Δ63, leu2Δ1, his3Δ200). Colonies (13 824) were picked using a colony picker (QpixII, genetix), and grown in 144 microtiter plates.
Phage display targets
For phage display selections and screening, specific 400 bp biotinylated DNA targets were produced by PCR using the corresponding pGEM-T vector as template and a pair of primers comprising a 5′-biotinylated oligonucleotide. Single-band PCR products were obtained (as examined on agarose gels) and working quantities for either selection or ELISA assays were determined empirically. In order to enrich our library with phage display I-CreI mutants that bind our DNA targets, several cycles of phage productions and selections were realized. S1234 contains a 24 bp I-SceI cleavage site (CACGCTAGGGATAACAGGGTAATA).
Phage production
The Ulib2 N75 library was subcloned by NcoI and EagI digestion and ligation into a phagemid vector (35). TG1 E.coli cells transformed with the phagemid library or collected after a round of selection were grown in 50 ml 2YTAG (2YT medium containing 100 µg/ml ampicilin and 2% glucose), at 37°C. Once the culture reached an OD600 of 0.5, helper phage M13KO7 (Amersham) was added to a 5 ml aliquot, at a phage:bacteria ratio of 20:1 and this sample was kept at 37°C during 30 min, without agitation. Cells were then recovered by centrifugation (3100 g for 10 min at room temperature), suspended in 25 ml of 2YTAK (2YT medium containing 100 µg/ml ampicilin and 25 µg/ml kanamycin) and grown overnight at 30°C to allow production of phage particles. Cells were removed from the culture by centrifugation (3100 g for 20 min at 4°C) and 0.2 vol of 20% PEG6000 2.5 M NaCl was added to precipitate phages (1 h on ice). A phage pellet was obtained by centrifugation (3100 g for 20 min at 4°C) and dissolved in 1 ml phosphate-buffered saline (PBS). The solution was cleaned (centrifugation at 1000 g for 5 min) and phage particles precipitated as before. Phage were finally suspended in 250 µl PBS.
Phage selection
Phage particles were diluted in 1 ml TMC (10 mM Tris buffer at pH 8, containing 150 mM NaCl, 3% powder milk and 25 mM CaCl2) and incubated for 1 h at room temperature. A sample of biotinylated DNA targets was then added to the phage solution. After 1 h at room temperature, streptavidin beads (Dynal, 100 µl or 200 µl for the first selection round) washed three times previously in TMC and blocked for 1 h in the same buffer, were added to the phage/DNA mix. After 15 min at room temperature, these beads were collected (on the vial wall using a magnet), washed 10 times in TMC containing 0.1% Tween-20, and once more in PBS. They were eventually suspended in 0.5 ml of 100 mM Triethanolamine, pH 12, and incubated for 10 min, to release phage particles. The supernatant was collected and immediately neutralized, adding 0.5 ml of 1 M Tris, pH 8. Aliquots of the input and output phage solutions were serially diluted and used to evaluate the corresponding numbers of phage particles, from which enrichment levels can be monitored. 2YT (4 ml) and exponentially growing TG1 cells (5 ml) were added to the rest of the output phage solution and the whole was incubated for 30 min at 37°C without agitation. The (phage-infected) cells were then plated on 2YTAG plates, which were incubated overnight at 30°C. All grown colonies were recovered and suspended in 2YTAG, adjusting the final OD600 to 100 and adding glycerol to a final concentration of 15%, and the resulting cell sample was stored at −80°C.
Screening by phage display
TG1 E.coli cells transformed with the phagemid library or collected after a round of selection were grown to form isolated colonies on agar plates. Colonies were then tooth-picked into 100 µl 2YTAG in 96-well microtiter plates, which were incubated overnight at 37°C, under agitation. All plates were replicated using a transfer device, before sterile glycerol was added (final glycerol concentration was 20%) for storage at −80°C. The replicate plates were incubated with agitation at 37°C for 2.5 h. 2YTAG containing 2 × 109 pfu of helper phage M13KO7 was added and the plates were further incubated for 30 min at 30°C (without agitation), then spun at 1700 r.p.m. for 15 min. Cell pellets were suspended in 150 µl 2YTAK, and cells were grown overnight at 30°C to produce phage particles in the culture medium. After centrifugation (1700 r.p.m. for 15 min at 4°C), 20 µl of supernatant was used in phage ELISAs. Microtiter plates (Maxisorp, Nunc) were coated with biotinylated BSA (Sigma) (1 h incubation at 37°C with 100 µl/well of PBS containing 2 µg/ml BSA). After three washes with PBS containing 0.1% Tween20 (PBST), streptavidin (Promega) was bound to the coated plates (1 h incubation at room temperature with 100 µl/well of PBS containing 10 µg/ml streptavidin). The plates were washed as before and specific biotinylated DNA targets were added (100 µl/well of PCR products at ∼250 pM in PBS). After 1 h incubation at room temperature and washing (as before), plates were saturated with TMC (300 µl/well of 10 mM Tris buffer at pH 8, containing 150 mM NaCl, 3% powder milk and 25 mM CaCl2). TMC was discarded 1 h later and plates were filled with 80 µl TMC and 20 µl/well of culture supernatant containing the phage particles. After 1 h at room temperature, microplates were extensively washed (10 times) with PBST. Anti-M13-horse radish peroxidase (HRP) conjugated antibody (Pharmacia) was diluted 1/5000 in TMC, and added to the microplates (100 µl/well). Plates were incubated for 1 h at room temperature and then washed (10 times) with PBST. Finally, TMB (Sigma) was added (100 µl/well), providing a substrate of the HRP, and after 5 min the HRP reaction was quenched adding 50 µl/well of 1 M H2SO4. The absorbance at 450 nm was then read for all plates (BioRad Microplate Reader 550). Positive binders were identified as those with an absorbance signal higher than a threshold set as the background level (measured with either no phage or phage displaying non-functional endonucleases) plus three times the standard deviation of this background signal.
RESULTS
Constructions of selection strains and reporter plasmids
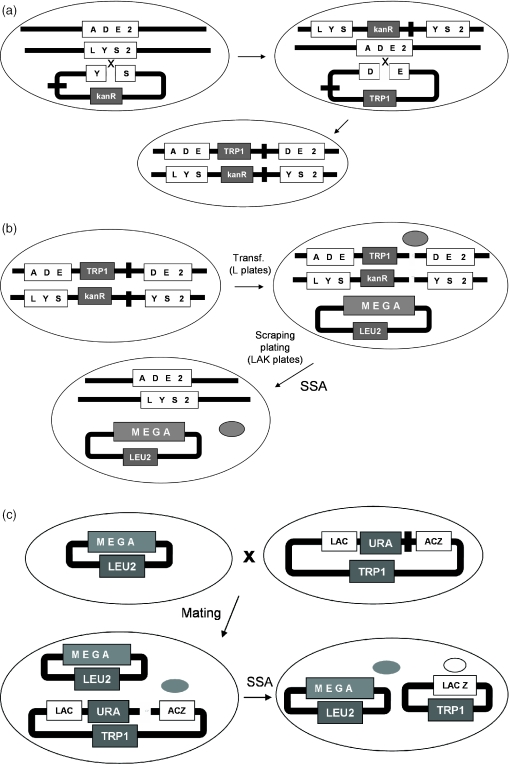
The principle of our in vivo yeast selection is based on the restoration of an auxotrophic marker. In this case, a marker of choice (named selection marker) of a yeast strain is disrupted by an insert containing a transformation marker and the site of interest flanked by two direct repeats (Figure 1a), thereby creating a strain named selection strain. To obtain a very low background owing to spontaneous recombination, each selection strain was disrupted at two different loci (namely ADE2 and LYS2). Upon cleavage of the site of interest by an active meganuclease expressed in the cell, an intact selection marker is obtained by SSA, permitting the selection of recombinant clones on suitable plates (Figure 1b). Five strains were built, following this procedure, corresponding to targets C1234, C1221, C4334 and two related palindromic targets named H1221 and H4334 (Figure 3).
Figure 1.
Selection and screening principle. (a) Selection strain construction: Strain FYC2-6A is transformed with a linearized plasmid harboring a deleted version of the gene LYS2, the target site and marker KanR. After selection on selective medium containing aminoglycoside G418, the resulting strain is transformed with a linearized plasmid harboring a deleted version of the gene ADE2, the target site and marker TRP1. The final strain phenotype is thus Trp+, KanR, Ade−, Lys−. (b) Selection process: a selection strain is transformed with a plasmid harboring a LEU2 marker and expressing the library. After selection of transformants on selective medium without leucine (L plates), colonies are resuspended and a fixed amount of cells are plated on medium selecting for intact selection markers and the presence of the expression vector (LAK plates containing selective medium without leucine, adenine and lysine). SSA, event allowing the restoration of the selection markers. (c) Screening process, the strain harboring the expression vector is mated to a strain harboring a reporter based on lacZ gene disruption. Upon induction, a mutant is produced, cleaves the target site and allows the restoration of the marker by SSA. Expression of the LacZ gene can be directly visualized on X-gal containing plates. Gray ball represents the meganuclease (mega), white ball represents the β-galactosidase.
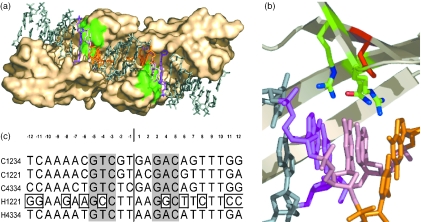
Figure 3.
Mutant library and targets (a) Localization of the area of the binding interface (bottom view) chosen for randomization (green) and interacting base pairs (−3, orange; −4, pink; −5, magenta). (b) Zoom showing residues 44, 68 and 70 chosen for randomization (green), D75 (red) and interacting base pairs (−3, orange; −4, pink; −5, magenta). (c) Sequences of the target used for selection and screening. C1234, wild-type target C1221; and C4334, palindromic site derived from C1234. H1221 and H4334, palindromic sites related to C1221. Boxes highlight bases not found in sites C1221 or C4334.
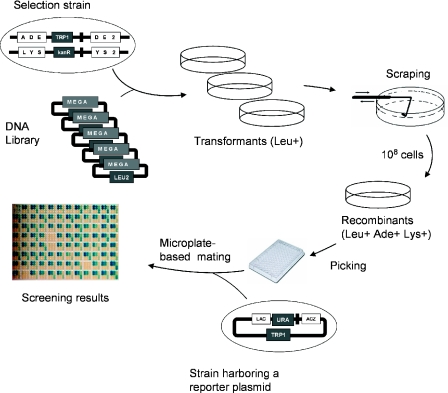
We have also developed a similar method, wherein DSB-induced recombination results in the restoration of a functional LacZ gene (Figure 1c), instead of a chromosomal auxotrophic marker. This method can be used for direct HTS (28), but also downstream of selection experiments: after selection, the activity of the resulting clones can still be screened by mating them with a strain harboring the LacZ reporter plasmid (Figure 1c). The general scheme of the experiment is described in Figure 2.
Figure 2.
General scheme of the yeast selection and screening method.
Model selections
To assess the efficiency of our selection strategy, we transformed the selection strain harboring the wild-type I-CreI site with a fixed amount of expression vector encoding an irrelevant homing endonuclease (I-DmoI) spiked with different amounts of expression vector encoding I-CreI N75, using ratios of 10−3, 10−4 and 10−5. Previous experiments comparing a direct selection of recombinant clones (with restored selection markers) to a first selection of clones harboring an expression vector (transformants) followed by scraping and re-plating on medium selecting for recombinants demonstrated significantly greater enrichments with the two-step method (data not shown). Consequently, selection strains transformed with the different ratios were first plated on medium lacking leucine. Colonies were subsequently scraped off the plates, and 107 cells per selection were plated on medium selecting for Leu+ Ade+ yeast cells, referred to as recombinant clones. Only ratios of 10−4 and 10−3 showed a significant difference in the number of recombinant clones compared with the control (Table 1). However, screening of 16 selected clones for each ratio showed the presence of I-CreI N75 expressing clones, even for the 10−5 ratio (Table 1). PCR and restriction assays confirmed that these positive clones were harboring the I-CreI N75 ORF. This result demonstrates that our selection technique can lead to enrichment factors in the 105 range in a single round.
Table 1.
Results from model selections
| Ratio | Transformants | Recombinants | Positives |
|---|---|---|---|
| 0 | 298 000 | 45 | 0/16 |
| 10−5 | 310 000 | 51 | 4/16 |
| 10−4 | 305 000 | 275 | 10/16 |
| 10−3 | 308 000 | 2540 | 16/16 |
Ratio, ratio of I-CreI-N75 encoding plasmid to I-DmoI encoding plasmid. Transformants, number of colonies obtained after transformation. Recombinants, number of colonies obtained after plating of 107 transformed cells on selection plates. Positive, number of clones positive by screening against C1234.
Design and construction of Ulib2 N75
To demonstrate further the efficiency of the selection method, we decided to create a library of I-CreI variants, to subsequently enrich for active mutants. I-CreI is a dimeric homing endonuclease that cleaves a 22 bp pseudo-palindromic target (Figure 3). Analysis of I-CreI structure bound to its natural target shows that in each monomer, eight residues establish direct interactions with seven bases (31). Residues Q44, R68, R70 contact three consecutive base pairs at position 3–5 (and −3 to −5; Figure 3a and b). As a first attempt to generate active I-CreI mutants with modified specificities and to validate our selection strategies, we chose to randomize this area of the I-CreI DNA-binding interface and select the resulting libraries against the I-CreI wild-type target (named here C1234). Since expression in yeast leads to homodimers, we also selected our library against the two palindromic sites derived from the I-CreI wild-type target (C1221 and C4334) and two related palindromic sites (H1221 and H4334) (Figure 3c). Our I-CreI scaffold was first mutated from D75 to N to decrease probable energetic strains caused by the replacement of the basic residues R68 and R70 in the library that satisfy the hydrogen-acceptor potential of the buried D75 in the I-CreI structure (Figure 3b). The D75N mutation did not affect the protein structure, but decreased the toxicity of I-CreI in overexpression experiments (data not shown). This scaffold mutant, named QRR, was assayed by screening and was shown to cleave efficiently targets C1234 and C1221, and to a lesser extent C4334 and H4334. Target H1221 is not cleaved at all (Figure 4b). Next, positions 44, 68 and 70 (Figure 3b) were randomized using codon VVK coding for amino acids A, D, E, G, H, K, N, P, Q, R, S or T, leading to a small library with a theoretical diversity of 5832 (at the DNA level), that was cloned into a vector suitable for yeast expression. Sequencing of 80 clones picked at random led to 79 different mutants (scaffold QRR was not present in this sample), and a representation of all possible residues at the three randomized positions, demonstrating a good diversity in the library (data not shown).
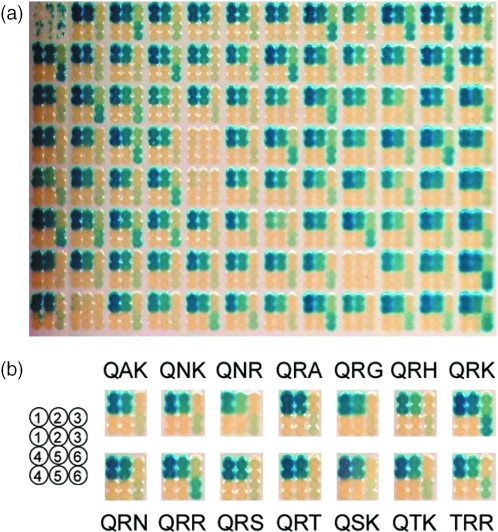
Figure 4.
Screening results of clones from C1234 yeast selection on. (a) Raw results for 95 clones (well A1 is a negative control). (b) Profiles of mutants selected against C1234. Target legend: 1, C1234; 2, C1221; 3, C4334; 4, negative control; 5, H1221; and 6, H4334.
In vivo yeast selection of Ulib2 N75 for cleavage
Next, library Ulib2 N75 was transformed into the five different selection strains and selected as above. About 106 transformant colonies were obtained in each case, and 108 transformant cells were plated onto selection plates to retrieve enough recombinants for further analysis. Recombinant clones (95) from each selection were picked and assayed by mating using our screening assay. The ORF of several clones was also PCR-amplified and sequenced. Table 2 shows for each selection the number of clones able to cleave the site used for selection and the sequence and frequency of each mutant, identified by residues 44, 68 and 70. As expected, the selection performed on the wild-type site C1234 was very efficient and led to a vast majority of active clones, with the scaffold protein (QRR) being the second most frequent clone (15%) behind mutant QTK (17%). Among the 78 sequences obtained from this selection, 13 new active mutants were identified. Selection performed on the palindromic left site (C1221) was equally efficient and mostly led to the same mutants (Table 2). However QRR was found dominant (19%) among the 84 sequences obtained. The right palindromic site of I-CreI wild-type site (C4334) is less efficiently cleaved by I-CreI and mutant QRR (Figure 4b). Strikingly, selection of Ulib2 N75 against this target yielded mainly one mutant, QRK (95%) and only a few QRR (3%). Screening results showed that clone QRK displays a slightly higher activity against C4334 (Figure 4b). Selection against H4334 led to only two populations: QRR (66%) and QRK (34%). The screening assay did not show any significant difference of activity against C4334 for these two clones (Figure 4b). Finally, selection against H1221 yielded only few recombinants that turned out negative in the screening assay.
Table 2.
Results from selections of I-CreI variants in yeast
| Target | C1234 | C1221 | C4334 | H1221 | H4334 |
|---|---|---|---|---|---|
| Recombinants | 8.6 × 105 | 7.8 × 105 | 3.9 × 103 | 1.6 × 106 | 2.3 × 104 |
| Cutters (%) | 96 | 98 | 68 | 0 | 97 |
| Mutants | QTK 13 | QRR 16 | QRK 59 | none | QRR 57 |
| QRR 12 | QRN 12 | QRR 2 | QRK 29 | ||
| QRK 8 | QRT 12 | TRR 1 | |||
| QRN 8 | QRA 8 | ||||
| QRT 8 | QRK 8 | ||||
| QRA 7 | QTK 7 | ||||
| TRR 4 | QRG 4 | ||||
| QAK 3 | QSK 4 | ||||
| QNK 3 | QAK 3 | ||||
| QRG 3 | QGK 3 | ||||
| QSK 3 | QRQ 3 | ||||
| QNR 2 | TRR 2 | ||||
| QRH 2 | QNK 1 | ||||
| QRS 2 | QQK 1 |
Recombinants, number of recombinant clones obtained after plating 108 transformants on selection medium. Cutters, % of clones (out of 95) found to be positive against at least one target by screening assay after selection. Mutants, residues 44, 68 and 70 and the number of times these sequences were found (clones with unreadable sequences not mentioned).
In vitro phage display-based selection of Ulib2 N75 for binding
To compare the efficiency of our selection procedure with a standard phage display-based selection, Library Ulib2 N75 was also cloned into a phage display vector and electroporated in E.coli. After rescue, the library was panned against streptavidin-coated magnetic microbeads preincubated with a biotinylated 400 bp double-strand DNA harboring I-CreI-derived sites. Selections were iterated three times and 80 clones from each output of each round were assayed for binding against all targets using a standard phage ELISA procedure. The first round of selection against C1234 and C1221 yielded 60 and 71% of binders, respectively (Table 3). Selection against C4334 yielded only 5% of positives, giving weak signals. As a control, a selection performed on an irrelevant target yielded only one clone, capable of binding to C1234 but not against the sequence used for selection. Round 2 and 3 on targets C1234 and C1221 yielded almost exclusively binders whereas round 2 and 3 on C4334 led to a moderate increase in the proportion of binders. Thirty-two clones from each round of each selection were picked and sequenced (Table 3). Overall, the phage display selection outputs were similar to the yeast outputs. Most of the frequent yeast mutants were also found in phage display (QRN, QRR, QRK, QRT, QRA, QRS and QAK). Two striking exceptions were QTK, the most frequent clone of the yeast C1234 selection, absent from the phage display output, and QQK representing 35 and 31% of the final phage selection against C1234 and C1221, respectively, and not found in the C1234 yeast selection (although present at 1% in the C1221 yeast selection). Among the nine mutants isolated by phage display, only QHK was not present in the yeast output, but eight mutants found by yeast selection were not found by phage display.
Table 3.
Results from selections of I-CreI variants by phage display
| Target | C1234 | C1221 | C4334 | S1234 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Round | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| Binders (%) | 60 | 98 | 99 | 71 | 99 | 100 | 5 | 46 | 90 | 1 | 1 | 1 |
| Mutants | QRN 8 | QRN 13 | QRN 16 | QRN 14 | QRN 23 | QRN 17 | QQK 3 | QRR 15 | QRR 29 | QRA 1 | QRT 1 | QRR 1 |
| QRT 6 | QQK 7 | QQK 10 | QQK 4 | QQK 9 | QQK 9 | QRT 1 | QRK 2 | QRK 1 | ||||
| QQK 5 | QRT 3 | QRS 3 | QRT 4 | QRT 3 | QRT 3 | QRA 1 | QRN 1 | |||||
| QRA 2 | QRS 3 | QAK 2 | QRA 4 | QRR 1 | ||||||||
| QRR 1 | QHK 2 | QRR 2 | ||||||||||
| QRS 1 | QRK 1 | QRS 2 | ||||||||||
| QRK 1 | ||||||||||||
| QAK 1 | ||||||||||||
| QHK 1 | ||||||||||||
Binders, % of clones (out of 80) leading to positive signal by ELISA. Mutants, residues 44, 68 and 70 for clones positive against at least one of the targets by ELISA and number of times this sequence was found among 32 clones picked at random for each round and each target. The sequences of ELISA-negative clones are not shown. For S1234 sequence, see Materials and Methods.
In both techniques, selection against a difficult target like C4334 led to mutants QRK and QRR. However QRR was dominant in phage display whereas QRK represents 94% of the yeast output.
DISCUSSION
The first in vivo selection strategy applied to homing endonucleases was developed to study the sequence requirements for I-CreI homing site recognition and cleavage (24). Although it allowed testing the importance of residues involved in direct DNA contacts, this method was based on a negative selection, aimed at retrieving clones unable to cleave their targets. A second E.coli-based strategy was designed to provide a selection for functional homing endonucleases, and was validated with three LAGLIDADG homing endonucleases (I-SceI, PI-SceI and I-ScaI) (25). However, the need to lower background growth level was reported to be important, if this selection strategy was to be used for finding functional endonucleases within protein libraries. More recently, in vivo selection from a randomized (but directed) library of homing endonuclease variants was performed (27). However, instead of selecting for cleavage, a two-hybrid strategy was used that selects for binding to the target DNA.
We wanted to design a method able to isolate mutants on the basis of their ability to trigger homologous recombination induced by DSBs inside eukaryotic cells, thereby directly selecting the mutants for their final desired property instead of indirectly selecting them for binding, or unrepaired cleavage. We used the high efficiency of SSA in yeast to derive a selection and a screening assay based on the restoration of a marker (auxotrophy for selection, reporter for screening) triggered by a meganuclease-induced DSB at the target site. Model selection experiments using spiked mixes revealed two striking features of the method. Firstly, we obtained a very low background since the selection done in the absence of I-CreI led to only 45 colonies for 107 transformants, i.e. in the 10−5 range. This characteristic is essentially owing to the presence of two SSA markers per strain. Indeed, preliminary experiments demonstrated a background of 10−3 to 10−4 for strains harboring only one marker (data not shown). Secondly, 25% of the clones picked after selection, using an input ratio I-CreI N75/I-DmoI of 10−5, were found positive in our screening assay. This demonstrates that the in vivo selection can provide enrichment factors in the 105 range, which can be considered as remarkable. For comparison, model phage display selections done with antibody fragments usually reach enrichment factors in the 102 to 103 range per round.
Next, we used the same method to select active homing endonucleases from a small library of I-CreI variants. As expected, the wild-type target C1234 and its left palindrome C1221 yielded a high percentage of cutters whereas H1221, showing a small degree of identity with C1234, led to background clones only. Interestingly, successful selections on C1234 and C1221 led to a high diversity of mutants, unlike selections on C4334 and H4334, two targets slowly cut by I-CreI N75 (also named QRR), where two clones only, QRK and QRR, survived the selection. Thus, many mutants are able to cut efficiently C1234 and C1221, but only the scaffold clone QRR and a closely related mutant, QRK, cut C4334 and H4334 efficiently enough to be enriched. It should be noted that many mutants selected against C1234 cleave very efficiently this target but no longer show any detectable signal with C4334, unlike scaffold clone QRR, demonstrating how an activity can be lost if not under selection pressure. We could observe a good correlation between our selection and screening assay since the only two clones present after selection on H4334, QRR and QRK are the only two showing a high signal against H4334 in the screening assay (Figure 4b).
To compare our results with an established method in terms of efficiency, and to see whether an in vivo recombination-based strategy would lead to a strong bias, we performed in vitro phage display selections of our library against magnetic beads displaying the I-CreI-derived sites. Despite some striking differences, the overall outputs of these two methods were very similar. This result can be considered as remarkable since the two techniques are very different. Indeed, the main difference resides in the principle of selection, the yeast technique selects for cutters whereas phage display is based on binding. Other differences could induce strong bias. For instance, successful display on the surface of a phage particle is conditioned by the ability of the protein to be efficiently produced in the prokaryotic environment of E.coli cytoplasm, exported in the periplasm and finally incorporated in the phage capsid. However, yeast selection uses a eukaryotic protein synthesis machinery and therefore is less prone to expression problems. Such differences between these two techniques might explain why mutant QTK performs well in yeast selection but is absent from the phage display output.
Being performed in vitro, a phage display selection strongly depends on the conditions used such as buffer composition, temperature and number of washes. Finding the right conditions can thus be complex. In contrast, only a few parameters, such as temperature (but in the range tolerated by yeast) or induction (glucose or galactose) are operator-dependent in the yeast selection method. However, the in vivo environment is very relevant since it closely mimics the conditions where the meganuclease will have to operate as a final product. Another advantage of an in vivo selection process is a built-in depletion for cytotoxic mutants since they are lost during transformation and recombinants selection.
Overall, yeast selection favorably compared with phage display in terms of efficiency and diversity of the selection output, and does not seem to induce strong bias. Its strong enrichment capacity and simplicity of use qualifies this technique as a tool of choice for homing endonuclease engineering. We have recently devised a method based on HTS that could deliver a large number of novel meganucleases with new specificities (28). While this method was demonstrated to be very fast and powerful, it imposes a restriction on the size of the libraries that can be analyzed, ∼104 clones. Consequently it only applies in situations where structural information can help to precisely choose the residues of the homing endonuclease to be tested and severely limits the number of these residues to be randomized. The in vivo selection method described in this article allows the handling of much larger sequence spaces. Indeed, the only size limitation is due to the initial transformation of the library into the selection strain. The average transformation efficiency in yeast using the classical heat-shock based yeast transformation technique (34) is 106 transformants/µg of DNA. A simple upscale of this technique can lead to 108 transformants in one experiment, and repeating this step can lead to library sizes in the 109 range, allowing the handling of libraries 105 times bigger than those used in direct HTS. However, this transformation step has to be done for each selection strain. We are currently removing this limitation by developing a protocol using a reusable library strain mated to different selection strains by a highly efficient liquid mating method.
In conclusion, this in vivo yeast selection and our previously described robot-assisted HTS are two key processes that greatly facilitate engineering of new custom-made meganucleases and should lead to a set of valuable tools for genome engineering.
Acknowledgments
We would like to thank Julianne Smith for critical reading of the manuscript. This work has been partly supported by Grant 0212508Q The French Agency for Innovation (ANVAR), and grant 04W107 from the French Ministry of Research, under label EUREKA Σ!3294. Funding to pay the Open Access publication charges for this article was provided by The French Agency for Innovation (ANVAR).
Conflict of interest statement. None declared.
REFERENCES
- 1.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 2.Belfort M., Roberts R.J. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jurica M.S., Stoddard B.L. Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevalier B.S., Monnat R.J., Jr, Stoddard B.L. The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nature Struct. Biol. 2001;8:312–316. doi: 10.1038/86181. [DOI] [PubMed] [Google Scholar]
- 5.Jacquier A., Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 6.Perrin A., Buckle M., Dujon B. Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron–exon junctions. EMBO J. 1993;12:2939–2947. doi: 10.1002/j.1460-2075.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouet P., Smih F., Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choulika A., Perrin A., Dujon B., Nicolas J.F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segal D.J., Carroll D. Endonuclease-induced, targeted homologous extrachromosomal recombination in Xenopus oocytes. Proc. Natl Acad. Sci. USA. 1995;92:806–810. doi: 10.1073/pnas.92.3.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smih F., Rouet P., Romanienko P.J., Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5015. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puchta H., Dujon B., Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl Acad. Sci. USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taghian D.G., Nickoloff J.A. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen-Tannoudji M., Robine S., Choulika A., Pinto D., El Marjou F., Babinet C., Louvard D., Jaisser F. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol. Cell. Biol. 1998;18:1444–1448. doi: 10.1128/mcb.18.3.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donoho G., Jasin M., Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol. Cell. Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott B., Richardson C., Winderbaum J., Nickoloff J.A., Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Posfai G., Kolisnychenko V., Bereczki Z., Blattner F.R. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rong Y.S., Golic K.G. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 18.Smith J., Berg J.M., Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27:674–681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 20.Pabo C.O., Peisach E., Grant R.A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 21.Segal D.J., Barbas C.F., III Custom DNA-binding proteins come of age: polydactyl zinc-finger proteins. Curr. Opin. Biotechnol. 2001;12:632–637. doi: 10.1016/s0958-1669(01)00272-5. [DOI] [PubMed] [Google Scholar]
- 22.Isalan M., Klug A., Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreier B., Fuller R.P., Segal D.J., Lund C., Blancafort P., Huber A., Koksch B., Barbas C.F., III Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 24.Seligman L.M., Stephens K.M., Savage J.H., Monnat R.J., ,Jr Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics. 1997;147:1653–1664. doi: 10.1093/genetics/147.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruen M., Chang K., Serbanescu I., Liu D.R. An in vivo selection system for homing endonuclease activity. Nucleic Acids Res. 2002;30:e29. doi: 10.1093/nar/30.7.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seligman L.M., Chisholm K.M., Chevalier B.S., Chadsey M.S., Edwards S.T., Savage J.H., Veillet A.L. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30:3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimble F.S., Moure C.M., Posey K.L. Assessing the plasticity of DNA target site recognition of the PI-SceI homing endonuclease using a bacterial two-hybrid selection system. J. Mol. Biol. 2003;334:993–1008. doi: 10.1016/j.jmb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Arnould S., Chames P., Perez C., Lacroix E., Duclert A., Epinat J.-C., Stricher F., Petit A.S., Patin A., Guillier S., Rolland S., Prieto J., Blanco F.J., Bravo J., Montoya G., Serrano L., Duchateau P., Pâques F. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006 doi: 10.1016/j.jmb.2005.10.065. in press. [DOI] [PubMed] [Google Scholar]
- 29.Thompson A.J., Yuan X., Kudlicki W., Herrin D.L. Cleavage and recognition pattern of a double-strand-specific endonuclease (I-creI) encoded by the chloroplast 23S rRNA intron of Chlamydomonas reinhardtii. Gene. 1992;119:247–251. doi: 10.1016/0378-1119(92)90278-w. [DOI] [PubMed] [Google Scholar]
- 30.Durrenberger F., Thompson A.J., Herrin D.L., Rochaix J.D. Double strand break-induced recombination in Chlamydomonas reinhardtii chloroplasts. Nucleic Acids Res. 1996;24:3323–3331. doi: 10.1093/nar/24.17.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurica M.S., Monnat R.J., Jr, Stoddard B.L. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 32.Rebar E.J., Greisman H.A., Pabo C.O. Phage display methods for selecting zinc finger proteins with novel DNA-binding specificities. Methods Enzymol. 1996;267:129–149. doi: 10.1016/s0076-6879(96)67010-4. [DOI] [PubMed] [Google Scholar]
- 33.Epinat J.C., Arnould S., Chames P., Rochaix P., Desfontaines D., Puzin C., Patin A., Zanghellini A., Paques F., Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gietz R.D., Woods R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 35.Chames P., Hufton S.E., Coulie P.G., Uchanska-Ziegler B., Hoogenboom H.R. Direct selection of a human antibody fragment directed against the tumor T-cell epitope HLA-A1-MAGE-A1 from a nonimmunized phage-Fab library. Proc. Natl Acad. Sci. USA. 2000;97:7969–7974. doi: 10.1073/pnas.97.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]