Abstract
The Saccharomyces cerevisiae mitotic exit network (MEN) is a conserved signaling network that coordinates CDK inactivation, cytokinesis and G1 gene transcription. The MEN Cdc14p phosphatase is sequestered in the nucleolus and transiently released in early anaphase and telophase. Cdc14p mediates mitotic exit by dephosphorylating Cdk1p substrates and promoting Cdk1p inactivation. Cdc14p also regulates the localization of chromosomal passenger proteins, which redistribute from kinetochores to the mitotic spindle during anaphase. Here we present evidence that the MEN protein kinase complex Mob1p-Dbf2p localizes to mitotic nuclei and partially colocalizes with Cdc14p and kinetochore proteins. Chromatin immunoprecipitation (ChIP) experiments reveal that Mob1p, Dbf2p, and Cdc14p associate with centromere DNA and require the centromere binding protein Ndc10p for this association. We establish that Mob1p is essential for maintaining the localization of Aurora, INCENP, and Survivin chromosomal passenger proteins on anaphase spindles, whereas Cdc14p and the Mob1p-Dbf2p-activating kinase Cdc15p are required for establishing passenger protein localization on the spindle. Moreover, Mob1p, but not Cdc15p, is required for dissociating Aurora from the kinetochore region. These findings reveal kinetochores as sites for MEN signaling and implicate MEN in coordinating chromosome segregation and/or spindle integrity with mitotic exit and cytokinesis via regulation of chromosome passenger proteins.
INTRODUCTION
The Saccharomyces cerevisiae mitotic exit network (MEN) is a conserved signaling network that coordinates several events associated with mitotic exit, including the inactivation of cyclin dependent kinase (CDK), cytokinesis activation, initiation of G1 gene transcription, and formation of prereplication complexes (Seshan and Amon, 2004; Simanis, 2003). MEN is comprised of several regulatory proteins, including Tem1p GTPase, a two-component GTPase-activating factor (GAP) Bub1p-Bfa1p, a putative GTP exchange factor (GEF) Lte1p, Cdc14p protein phosphatase, four protein kinases Cdc5p, Cdc15p, Dbf2p, Dbf20p, and a Dbf2p-associated protein Mob1p. The equivalent signaling network in Schizosaccharomyces pombe is called the septation initiation network (SIN) and is required for coordinating cytokinesis with CDK inactivation (Simanis, 2003; Wolfe and Gould, 2004). Most MEN/SIN proteins are conserved from yeast to human, however the functions of MEN/SIN-related networks have not been resolved in other eukaryotes.
S. cerevisiae Cdc14p plays a key role in MEN signaling (reviewed in Seshan and Amon, 2004; Stegmeier and Amon, 2004). Cdc14p is a temporally and spatially regulated proline directed phosphatase that directly mediates mitotic exit by promoting CDK inactivation and dephosphorylating CDK substrates (Gray et al., 2003; Visintin et al., 1998). Cdc14p is sequestered in the nucleolus throughout most of the cell cycle by binding to the nucleolar RENT complex (Shou et al., 1999; Visintin et al., 1999). Its release from the nucleolus is essential for catalytic activity and appears to occur in two phases (Visintin et al., 2003; Seshan and Amon, 2004). The first phase of Cdc14p release takes place during early anaphase and is dependent on FEAR (Cdc fourteen early anaphase release) proteins, which include Cdc5p (a Polo-like kinase), Esp1p (separase), and the kinetochore protein Slk19p (Saunders, 2002; Stegmeier et al., 2002). Cdc5p and Esp1p are also required for dissociation of sister chromatids during the metaphase to anaphase transition (Alexandru et al., 2001; Stegmeier et al., 2002). Thus, FEAR proteins coordinate Cdc14p early anaphase release with anaphase initiation. The second phase of Cdc14p release occurs during telophase and is dependent on other MEN proteins, such as Cdc5p, Tem1p, Cdc15p, and Dbf2p (Shou et al., 1999; Visintin et al., 1999). Once released in telophase, Cdc14p disperses throughout the cell and concentrates on spindle pole bodies (SPBs) and the bud neck ring (the site of cytokinesis) before returning to the nucleolus (Yoshida et al., 2002; Pereira and Schiebel, 2003; Bembenek et al., 2005). Late mitotic substrates for Cdc14p include Cdh1p, Sic1p, and Swi5p, which are CDK substrates that regulate mitotic cyclin degradation, CDK inactivation, and G1 gene expression, respectively (Visintin et al., 1998).
Cdc14p was shown to regulate the kinetochore-associated chromosomal passenger proteins Sli15p, and Ipl1p kinase, which are homologues to mammalian INCENP and Aurora B kinase (Pereira and Schiebel, 2003; Vagnarelli and Earnshaw, 2004). Cdc14p binds Sli15p in two-hybrid assays and is important for Sli15p and Ipl1p kinase localization to the spindle midzone during anaphase. Significantly, the spindle localization of chromosome passenger proteins was reported to be dependent on FEAR proteins and independent of the MEN kinase Cdc15p. These data suggest that Cdc14p interacts with passenger proteins during early anaphase. The Schizosaccharomyces pombe Cdc14p ortholog also interacts with the Aurora kinase at mitotic kinetochores and contributes to proper chromosome segregation (Trautmann et al., 2004). The functional significance of the interactions between S. cerevisiae Cdc14p and passenger proteins is not well understood; however, recent evidence suggest that Cdc14p interacts with passenger proteins to influence spindle integrity and cytokinesis (Bouck and Bloom, 2005; Higuchi and Uhlmann, 2005).
Other MEN proteins appear to function upstream of Cdc14p with regard to mitotic exit (Simanis, 2003; Seshan and Amon, 2004). Tem1p GTPase functions on SPBs and is regulated by the putative GEF Lte1p and the two-component GAP Bub2p-Bfa1p. Cdc5p kinase promotes MEN activation by inhibiting Bub2p-Bfa1p GAP activity, leading to an accumulation of GTP-bound Tem1p (Tem1p-GTP; Hu and Elledge, 2002; Geymonat et al., 2003). Tem1p-GTP activates Cdc15p kinase, which phosphorylates and activates the Mob1p-Dbf2p kinase complex. Substrates for Mob1p-Dbf2p kinase are not known; however, Dbf2p kinase is required for the mass release of Cdc14p from the nucleolus during telophase (Shou et al., 1999).
A critical function for MEN signaling is Cdc14p release and activation. The early anaphase release of Cdc14p from the nucleolus is mediated by phosphorylation of the nucleolar RENT protein Net1p/Cfi1p by Cdc5p kinase (Shou et al., 2002). Cdc5p overexpression induces the premature release of some but not all Cdc14p from the nucleolus (Shou et al., 2002; Visintin et al., 2003). Furthermore, mutations in Cdc5p phosphorylation sites on Net1p weaken Net1p-Cdc14p association (Shou et al., 2002). Recent data suggest that FEAR proteins induce Cdc14p release by promoting Cdk1p phosphorylation of Net1p/Cfi1p (Azzam et al., 2004). The MEN-dependent mechanism for Cdc14p release during telophase remains a mystery. Current models for MEN signaling suggest that Mob1p-Dbf2p kinase complex functions late in MEN signaling and imply that Cdc14p release is regulated by Mob1p-Dbf2p kinase complex (Simanis, 2003; Seshan and Amon, 2004). However, Mob1p and Dbf2p were not observed in the nucleus, suggesting that Mob1p-Dbf2p regulates Cdc14p release indirectly.
Mob1p and Dbf2p kinase function together as a unit. Mob1p binding to Dbf2p is essential for kinase activation by Cdc15p (Mah et al., 2001). Mob1p-Dbf2p kinase activity varies throughout the cell cycle, with peak activity occurring during mitosis and minimal activity in G1 and S phase (Toyn and Johnston, 1994; Visintin and Amon, 2001). Dbf2p kinase activation is also dependent on other MEN proteins, which is consistent with the model that Mob1p-Dbf2p functions late in MEN signaling. Both Mob1p and Dbf2p localize to the cytoplasmic surfaces of SPBs and to the bud neck during late mitosis (Frenz et al., 2000; Luca et al., 2001; Visintin and Amon, 2001). The bud neck localization reflects a role for Mob1p-Dbf2p in regulating cytokinesis (Frenz et al., 2000; Luca et al., 2001; Hwa Lim et al., 2003). The SPB localization appears to be important for Mob1p-Dbf2p function, because mutations in the SPB protein Nud1p induce telophase arrest and abolish the SPB and bud neck localizations of Mob1p and other MEN proteins (Luca et al., 2001; Seshan and Amon, 2004). Curiously, bipolar SPB and bud neck localizations of Mob1p are aberrant in conditional cdc14 mutants, suggesting that some Mob1p-Dbf2p functions are dependent on Cdc14p activity (Luca et al., 2001). Some Mob1p also binds Mps1p, a kinase required for SPB duplication and mitotic checkpoint signaling (Luca and Winey, 1998). The functional significance of this interaction is not understood; however, certain mob1 mps1 double mutant cells display maintenance of ploidy defects (Luca and Winey, 1998). Similar interactions with Mps1p and Dbf2p have not been reported.
To elucidate the role of Mob1p-Dbf2p in MEN signaling and Cdc14p regulation, we investigated the subcellular localizations of wild-type and truncated Mob1p. Here, we present evidence that a fraction of Mob1p and Dbf2p colocalizes with Cdc14p in the nucleus and that all three proteins function at kinetochores and other nuclear structures during anaphase. We establish that Mob1p-Dbf2p complexes and Cdc15p kinase are important for maintaining chromosomal passenger proteins on the mitotic spindle during anaphase and mitotic exit. These data support models for cooperative regulation of chromosome passenger proteins by Cdc14p and Mob1p-Dbf2p, which may be important for coordinating chromosome segregation with cytokinesis and/or for coordinating spindle disassembly with other mitotic exit events. Significantly, these data establish kinetochores as major sites for MEN signaling during anaphase.
MATERIALS AND METHODS
Strain Construction and Culture Conditions
Yeast strains for this study are listed in Table 1. Yeast cells were cultured using standard growth media and conditions (Guthrie and Fink, 1991). Where designated, cells were synchronized in G1 with alpha factor, S phase with hydroxyurea, and M phase with nocodazole and benomyl, as described (Weiss and Winey, 1996).
Table 1.
Strain list
| Name | Genotype | Source |
|---|---|---|
| FLY060 | MATamps1-1 | Winey et al. (1991) |
| FLY331 | MATa/α GFP-MOB1-URA3::mob1Δ::HIS3/GFP-MOB1-URA3::mob1Δ::HIS3 | Luca et al. (2001) |
| FLY382 | MATamob1Δ::HIS3 cyh2 leu2-3, 112 [pRS315-CYH2-MOB1] | Luca et al. (2001) |
| FLY547 | MATacdc16-1 | This work |
| FLY558 | MATamob1-95 CDC14-GFP::Kanmx | This work |
| FLY724 | MATaDBF2-13Myc::Kanmx | This work |
| FLY1147 | MATaGFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1206 | MATaCDC14-CFP::Kanmx GFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1211 | MATaSLK19-CFP::Kanmx GFP-MOB1(79-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1213 | MATaSLK19-CFP::Kanmx GFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1217 | MATaOKP1-CFP::Kanmx GFP-MOB1(79-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1219 | MATaOKP1-CFP::Kanmx GFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1279 | MATaMOB1(1-314)::HIS3:URA3 | This work |
| FLY1605 | MATaMOB1(79-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1606 | MATaMOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1608 | MATaDBF2-GFP:Kanmx MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1609 | MATaDBF2-GFP:Kanmx MOB1(79-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1842 | MATamob1-77 BIR1-GFP::Kanmx | This work |
| FLY1846 | MATamob1-77 SLI15-GFP::HIS3 | This work |
| FLY1849 | MATacdc15-2 SLI15-GFP::HIS3 | This work |
| FLY1874 | MATa/α GFP-MOB1(79-314)-URA3::mob1Δ::HIS3/GFP-MOB1(79-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1875 | MATa/α GFP-MOB1(133-314)-URA3::mob1Δ::HIS3/GFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY1884 | MATamob1-77 IPL1-GFP::Kanmx | This work |
| FLY 1919 | MATα cdc16-1 GFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY 1920 | MATα cdc16-1 GFP-MOB1(79-314)-URA3::mob1Δ::HIS3 | This work |
| FLY 1923 | MATacdc16-1 GFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY 2004 | MATacdc15-2 IPL1-GFP::Kanmx | This work |
| FLY 2055 | MATaNIC96-RFP::Kanmx GFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY 2056 | MATα SIK1-RFP::Kanmx GFP-MOB1(133-314)-URA3::mob1Δ::HIS3 | This work |
| FLY 2069 | MATacdc14-1 IPL1-GFP::Kanmx | This work |
| FLY 2072 | MATaOKP1-CFP::Kanmx MOB1(79-314)-YFP:HIS3 | This work |
| FLY 2076 | MATaSLK19-CFP::Kanmx MOB1(79-314)-YFP:HIS3 | This work |
| IPY390 | MATaCHL4-13Myc::TRP1 ndc10-42 | Pot et al. (2003) |
| JSY59 | MATandc10-1 | This work |
| JSY96 | MATaMOB1-13Myc::Kanmx ndc10-1 | This work |
| JSY98 | MATaCDC14-13Myc::Kanmx ndc10-1 | This work |
| SBY322 | MATaipl1-321 | Biggins et al. (1999) |
Plasmid Construction
GFP-Mob1Δ78 and GFP-Mob1Δ132 centromeric and integrating plasmids were constructed by replacing the MOB1 ORF from pRS314-GFP-MOB1 (containing TRP1) and pRS306-GFP-MOB1 (containing URA3; Luca et al., 2001) with PCR-amplified MOB1Δ78 and MOB1Δ132 DNA. The plasmids were digested with BamHI and EcoRI to remove full-length MOB1 ORF. Untagged Mob1Δ78- and Mob1Δ132-integrating plasmids were constructed by removing the GFP ORF from pRS306-GFP-Mob1Δ78 and pRS306-GFP-Mob1Δ132 by digesting with XbaI and BamHI, filling in the 5′ overhangs with Klenow, and religating. For integration, the pRS306-derived MOB1 plasmids were digested with PmlI and integrated into the mob1Δ::HIS3 locus of FLY382, as previously described (Luca et al., 2001). Integration was confirmed by PCR. The localization patterns for GFP-Mob1Δ78 and GFP-Mob1Δ132 were the same regardless of whether the proteins were expressed from low copy plasmids or from single-copy integrants (unpublished data).
Microscopy
Differential interference contrast (DIC) and fluorescence microscopy were performed as described with a 100× oil immersion objective using a Leica DMR fluorescence microscope (Deerfield, IL; Luca et al., 2001). Images were captured and processed using OpenLab imaging software (Improvision, Lexington, MA), as previously described (Luca et al., 2001).
Immunoblots
SDS-PAGE and immunoblots were conducted as described (Weiss et al., 2002). Quantitative immunoblots were processed using enhanced chemifluorescence (ECF, Amersham Biosciences, Piscataway, NJ) and quantified using a STORM phosphorimager (Amersham Biosciences).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays and coimmunoprecipitation from yeast lysates were performed as in (Measday et al., 2002), with the following changes. For ChIP analysis, multiplex PCR was performed, with three sets of primers added to a single PCR reaction. The CEN1, CEN3, and PGK1 primer pairs used to amplify specific regions of DNA are described in Meluh and Koshland (1997). The expected sizes of PCR products are 302 base pairs (CEN1), 288 base pairs (PGK1), and 243 base pairs (CEN3). To equilibrate the amount of PCR products obtained, 0.35 μM primer was added for each of the CEN3 and PGK1 pairs, whereas 0.3 μM primer was used for the CEN1 pair. The amount of total chromatin added varied from 1/1000 to 1/500 of the available template, whereas that of immunoprecipitated chromatin template varied from 1/10 to 1/50 of the available template, depending on the linear range for PCR.
RESULTS
Mob1p Is Required for the Release of Cdc14p from Nucleoli during Mitotic Exit
It was previously established that several MEN proteins (Cdc5p, Tem1p, Cdc15p, Dbf2p) are required for Cdc14p release from the nucleolus during mitotic exit (Shou et al., 1999; Visintin et al., 1999). To test if Mob1p is also required for Cdc14p release during mitotic exit, we assayed Cdc14p-GFP localization in conditional mob1-77 and mob1-95 cells. Cells were grown asynchronously at permissive temperature (22°C) and shifted to restrictive temperature (34°C) for 2–3 h. At 34°C, mob1-77 and mob1-95 cells arrested in late anaphase as large budded cells with separated chromatin, as previously described (Luca and Winey, 1998). Cdc14-GFP was present in the nucleoli in all arrested mob1-77 and mob1-95 cells (n = 140 cells) and was not detectable on SPBs or bud necks (Supplementary Figure S1, shown for mob1-95). These results indicate that Mob1p is required for the complete release of Cdc14p from the nucleolus during mitotic exit.
Mob1p Localizes to the Nucleus during Mitosis
Mob1p and Dbf2p localize prominently to the cytoplasmic surface of SPBs and to the bud neck during mitotic exit and were not previously detected in the nucleus (Frenz et al., 2000; Luca et al., 2001). These data suggest that Mob1p-Dbf2p kinase complex indirectly regulates Cdc14p release from the nucleolus. Nevertheless, it is possible that a previously undetected fraction of Mob1p-Dbf2p translocates to the nucleus during mitosis and directly regulates Cdc14p. Indeed, the structurally related Mob2p-Cbk1p protein kinase complex, which functions in the RAM signaling network, translocates to the nucleus during mitotic exit (Weiss et al., 2002). Thus, to determine if any Mob1p localizes to the nucleus during mitosis, we examined Mob1p localization in cells expressing chromosomally tagged GFP-Mob1p (Figure 1). We did not observe nuclear GFP-Mob1p in most single or merged optical sections of mitotic cells. However, in 6% (n = 33) of large-budded anaphase cells, we observed extremely faint clouds of fluorescence that were proximal to the nuclear surface of each SPB (Figure 1, A, C, and D). The apparent nuclear GFP-Mob1p fluorescence was often more readily detectable in time-lapse movies of select anaphase cells (see Supplementary Movie 1). These data suggest that a small fraction of Mob1p localizes to the nucleus during mitosis.
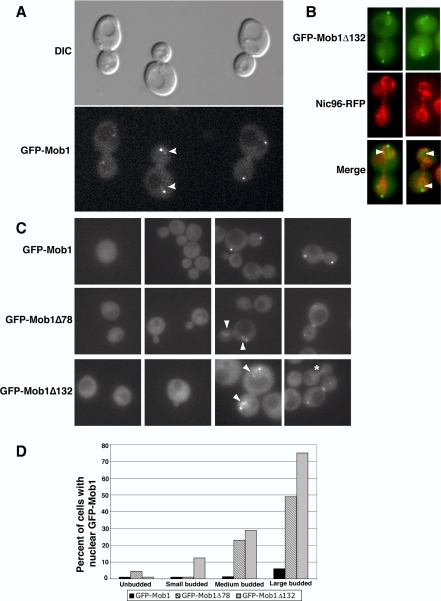
Figure 1.
Mob1p localizes to the nucleus. (A) Homozygous diploid cells expressing chromosomally tagged GFP-Mob1p (FLY331) were analyzed by DIC and fluorescence microscopy. Faint GFP fluorescence could be detected in nuclei of some anaphase cells (arrowheads). The bright fluorescence spots are GFP-Mob1p on SPBs. See corresponding Supplementary Movie 1. (B) Asynchronously growing haploid cells expressing GFP-Mob1Δ132 and Nic96p-RFP (FLY 2055) were analyzed by fluorescence microscopy. Mob1Δ132 was readily detectable within the RFP-marked nuclear envelope. (C) Asynchronously growing haploid GFP-Mob1 (FLY316), GFP-Mob1Δ78 (FLY1066) and GFP-Mob1Δ132 cells (FLY1147) were analyzed by fluorescence microscopy. Truncated GFP-Mob1p was readily detectable in the nuclei of large budded anaphase cells, in addition to previously established SPB and bud neck localizations. The arrowheads point to patches of nuclear Mob1p in anaphase cells. The asterisk (*) denotes a cell with Mob1Δ132 on what appears to be the anaphase spindle. The percentages of unbudded, small-, medium- (preanaphase), and large-budded cells with nuclear Mob1p were plotted from asynchronous populations of cells. n = 247, 191, and 161 for GFP-Mob1p, GFP-Mob1Δ78, and GFP-Mob1Δ132 cells, respectively.
N-terminally Truncated Mob1p Localizes More Prominently to Nuclei than Full-length Mob1p
To aid in analysis of the presumed nuclear fraction of Mob1p, we attempted to identify the protein domains that are important for establishing or maintaining Mob1p in the nucleus. We introduced plasmids encoding epitope-tagged or GFP-tagged C-terminal and N-terminal Mob1p deletion mutations into yeast and assayed protein expression and function by immunoblot and complementation. None of the C-terminally deleted Mob1p proteins were stable in yeast or complemented mob1Δ or conditional mob1 strains (unpublished data) and thus were not further analyzed in this study. Two N-terminally truncated forms of Mob1p, designated Mob1Δ78 and Mob1Δ132, were stably expressed in vivo and fully complemented mob1Δ and conditional mob1-77 cells (Figures 1 and 2 and unpublished data). These proteins lack the N-terminal 78 and 132 amino acids of Mob1p, which correspond to the nonconserved domain of Mob1p (Luca and Winey, 1998; Stavridi et al., 2003). The remaining region of Mob1p, which corresponds to amino acids 133–314, shares significant homology to human hMob1A (Stavridi et al., 2003).
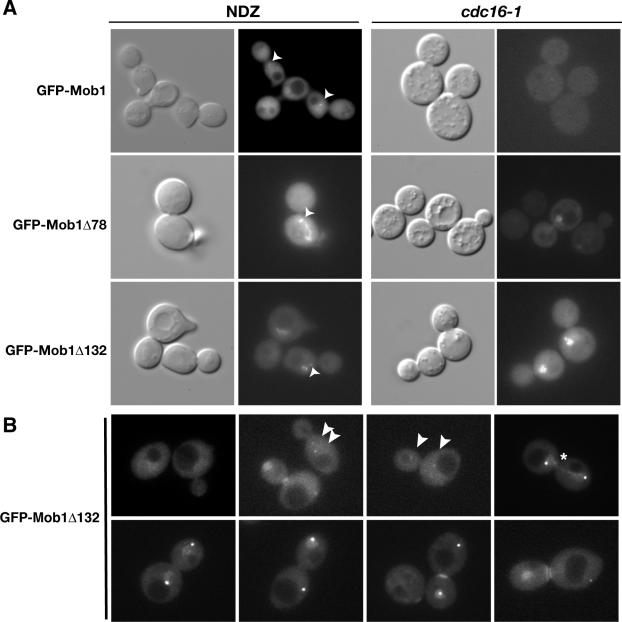
Figure 2.
Dynamics of truncated Mob1p localization. (A) Mob1Δ78 and Mob1Δ132 localize to nuclei in metaphase-arrested cells. GFP-Mob1p, GFP-Mob1Δ78, and GFP-Mob1Δ132 were analyzed in nocodazole (NDZ)-treated wild-type cells (left panels) and conditional cdc16-1 cells (right panels). The cells were synchronized in metaphase, as described in the text. The strains used in the NDZ experiments were FLY316, FLY1066, and FLY1147. The cdc16-1 cells (FLY547) contained pRS314-GFP-Mob1, pRS314-GFP-Mob1Δ78, or pRS314-GFP-Mob1Δ132. We obtained similar results for cdc16-1 cells expressing integrated Mob1p (unpublished data). (B) Representative images of GFP-Mob1Δ132 in homozygous diploid cells (FLY1875) from various stages of the cell cycle. The arrowheads point to GFP-Mob1Δ132 on early mitotic SPBs. Note that the second SPB in the last image on the bottom panel is out of focal plane. The pattern of GFP-Mob1Δ78 fluorescence was similar to GFP-Mob1Δ132 (Supplementary Movies 2–4). See Results for detailed description. All images in B are merges of 6 × 0.2-μm Z-sections.
To determine if the nonconserved N-terminal region of Mob1p is important for Mob1p localization, we analyzed the subcellular distributions of GFP-tagged Mob1Δ78 and Mob1Δ132 in vivo. DNA encoding GFP-Mob1Δ78 and GFP-Mob1Δ132 was integrated into mob1Δ strains and expressed under the control of the endogenous MOB1 promoter. As previously observed for full-length GFP-Mob1p (Luca et al., 2001), GFP-Mob1Δ78 and GFP-Mob1Δ132 localized to both SPBs and the bud neck in anaphase cells and localized diffusely to the cytoplasm in unbudded (G1 phase) and small budded (S phase) cells (Figure 1, C and D). Significantly, GFP-Mob1Δ78 and GFP-Mob1Δ132 also localized prominently to putative nuclear structures in ∼50 and 75% (n = 49 and 32) of large budded anaphase cells, respectively (Figure 1D). To confirm that truncated Mob1p localizes to the nucleus, we analyzed Mob1Δ78 and Mob1Δ132 in cells expressing a red fluorescence protein-tagged nuclear pore protein, Nic96p-RFP (provided by James Falvo and Erin O'Shea, UCSF). Indeed, a fraction of GFP-Mob1Δ78 and GFP-Mob1Δ132 localized within the confines of the RFP-labeled nuclear envelope during anaphase (Figure 1B, arrowheads; shown for Mob1Δ132). The nuclear localization of GFP-Mob1Δ78 and GFP-Mob1Δ132 often appeared as a diffuse cloud of fluorescence near each SPB; however, many anaphase cells contained one or more bright spots or patches within the more diffuse nuclear fluorescence (Figure 1C; see GFP-Mob1Δ132, third panel). Truncated Mob1p also transiently localized to an axial structure in the nucleus in ∼12% (n = 72) of the large budded anaphase cells (Figure 1C, see asterisk). The axial structure was distinct from the dividing nuclear envelope and the nucleoplasmic bridge that connects the dividing lobes of the nucleus, suggesting that the axial structure is probably the anaphase spindle (Supplementary Figure S2).
Truncated Mob1p also localized to preanaphase nuclei. GFP-Mob1Δ78 and GFP-Mob1Δ132 was detectable in nuclei in ∼25% (n = 100) of the medium budded preanaphase cells (Figure 1D). Moreover, GFP-Mob1Δ132 localized prominently to a crescent- or ring-shaped nuclear structure that was reminiscent of the preanaphase nucleolus in 19% of the medium budded cells (see Figure 5A). Collectively, these data suggest that Mob1p functions in the nucleus and that the N-terminal region of Mob1p is important for restricting the amount of Mob1p that localizes to the nucleus.
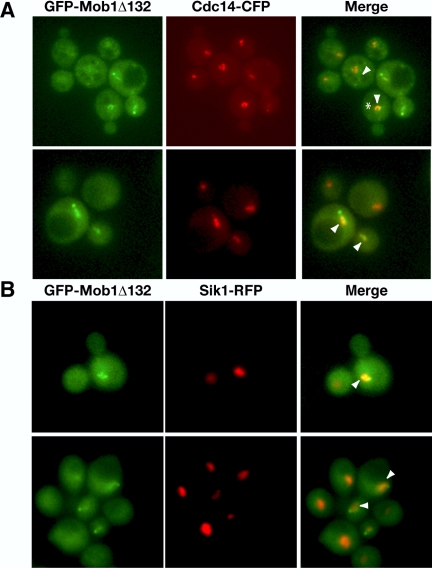
Figure 5.
Mob1Δ132 colocalizes with Cdc14p. (A) Asynchronously growing cells expressing truncated GFP-Mob1Δ132 and CFP-tagged Cdc14p (FLY1206) were analyzed by fluorescence microscopy. GFP, CFP, and merged images are labeled accordingly. The arrowheads point to regions of significant colocalization. Note the colocalization of Mob1Δ132 and Cdc14p in the nucleolus of a preanaphase cell (*). Similar results were obtained for GFP-Mob1Δ78 (unpublished data). (B) Asynchronously growing cells expressing GFP-Mob1Δ132 and RFP-tagged nucleolar protein Sik1p (FLY2056) were analyzed by fluorescence microscopy. The arrowheads point to areas of colocalization in both preanaphase (top panels) and anaphase cells (bottom panels).
Mob1Δ78 and Mob1Δ132 Localize to the Nucleus before Anaphase
To confirm that truncated Mob1p localizes to the nucleus before anaphase, we monitored Mob1Δ78 and Mob1Δ132 localization in mitotic checkpoint-activated cells and in conditional anaphase promotion complex (APC) mutants. We synchronized cells in G1 with mating pheromone and released them into medium containing nocodazole (NDZ), which activates the spindle assembly checkpoint and induces a G2/M phase arrest. Full-length GFP-Mob1p was undetectable in the nuclei in >99% of the NDZ-arrested cells, but localized to both SPBs in 38% of the cells (n = 167; Figure 2A). In contrast, Mob1Δ78 and Mob1Δ132 localized prominently to nuclei in 57% and to SPBs in ∼43% of the NDZ-arrested cells (n = 167; Figure 2A). To determine if the preanaphase nuclear localization of truncated Mob1p was specific to mitotic checkpoint-activated cells, we assayed Mob1p localization in the conditional APC mutant, cdc16-1. When cdc16-1 cells were shifted to restrictive temperature, Mob1p, Mob1Δ78, and Mob1Δ132 was detectable in the nuclei of 0, 93, and 76% (n = 150, 218, and 215, respectively) of the metaphase-arrested cdc16-1 cells (Figure 2A, right panels). These data indicate that Mob1Δ78 and Mob1Δ132 can localize to nuclei before anaphase and suggest that Mob1p may be recruited to the nucleus before the early anaphase release of Cdc14p. Moreover, these data indicate that nuclear recruitment of Mob1p and presumably Dbf2p (see below) is not sufficient to initiate Cdc14p release from the nucleolus.
We analyzed images of asynchronous diploid cells and conducted time-lapse microscopy to investigate the timing of GFP-Mob1Δ78 and GFP-Mob1Δ132 disappearance from the nucleus at the end of mitosis. To enhance the detection and analysis of nuclear fractions of Mob1p, we monitored GFP-Mob1Δ78 and GFP-Mob1Δ132 in homozygous diploid cells, which are larger than haploid cells. As previously demonstrated (Luca et al., 2001), full-length Mob1p localizes to both SPBs during midanaphase and to the bud neck ring a few minutes before spindle disassembly (Supplementary Movie 1). Just as for full-length Mob1p, GFP-Mob1Δ78 and GFP-Mob1Δ132 localized to both SPBs during anaphase (Figure 2B, top row, panel 3; Supplementary Movies 1–4). During anaphase, nuclear Mob1Δ78 and Mob1Δ132 appeared proximal to the two SPBs and then localized briefly along the long axis of the dividing nucleus before separating to symmetrical clouds of nuclear fluorescence proximal to each SPB (Figure 2B and Supplementary Movies 2–4). Curiously, in some diploid cells (38% of large budded preanaphase cells, n = 40) truncated Mob1p localized to the SPBs before anaphase (Figure 2B, second panel in top row; Supplementary Movie 4). This phenomenon was rarely observed in haploid cells (<1% of cells, n > 500). In late anaphase and before spindle disassembly (which was detected by a sharp decrease in SPB to SPB distance), truncated Mob1p disappeared from the mother cell nucleus a few minutes before disappearing from the daughter cell nucleus. We did not observe the transient asymmetric nuclear localization of truncated Mob1p in haploid cells, perhaps because of the smaller cell size and reduced fluorescence signal in haploid cells. Collectively, these data indicate that truncated Mob1p localizes to the nucleus during mitosis in cycling cells and disappears from the nucleus shortly after or concurrent with cytokinesis. Of course it is possible that the relative timing of Mob1Δ78 and Mob1Δ132 nuclear localization differs from that of full-length Mob1p.
Deletion of the N Terminus of Mob1p Does not Affect Protein Stability
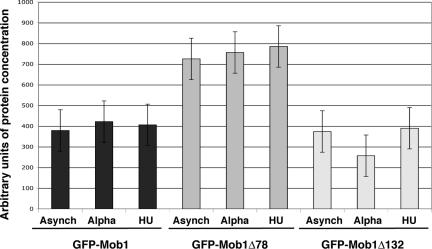
Our results establish that Mob1Δ78 and Mob1Δ132 localize more prominently to the nucleus than wild-type (full-length) Mob1p. The enhanced nuclear localizations of Mob1Δ78 and Mob1Δ132 might be caused by differences in protein stability. For example, truncated Mob1p may be more stable than wild-type Mob1p and consequently may accumulate more conspicuously in the nucleus. We therefore conducted quantitative immunoblots to compare the relative concentrations of GFP-tagged Mob1p, Mob1Δ78, and Mob1Δ132 from G1 and S phase-arrested and asynchronously growing cells. As a control for protein loading, we compared Mob1p protein expression levels to glucose-6-phosphate dehydrogenase (G6PDH), whose expression is not cell cycle regulated. Immunoblots revealed that GFP-Mob1Δ78 was about twofold higher in concentration than wild-type GFP-Mob1p (Figure 3). In contrast, cellular GFP-Mob1Δ132 levels were about the same as wild-type Mob1p. Thus, there was no correlation between total cellular levels of Mob1p and the degree of nuclear localization. In agreement, low-copy plasmids encoding full-length GFP-Mob1p, which induce higher expression than integrated GFP-Mob1p, did not enhance detection of nuclear fluorescence (unpublished data). These data indicate that the relative abundance of nuclear Mob1p is not influenced by changes in total cellular Mob1p protein concentration.
Figure 3.
Quantitative immunoblots of full-length and truncated Mob1p. (B) Quantitative immunoblots of full-length and truncated GFP-Mob1p were conducted as described in the Materials and Methods. The ratio of Mob1p to G6PDH levels (arbitrary units) from asynchronous, G1 and S phase-synchronized cells from three independent experiments were averaged and plotted. The SDs are denoted by error bars.
Mob1Δ78 or Mob1Δ132 Expression Increases the Nuclear Concentration of Dbf2p
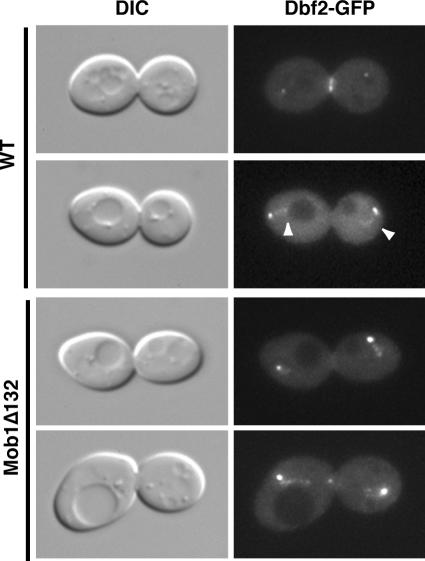
Mob1p associates with Dbf2p kinase and is necessary for kinase activity (Komarnitsky et al., 1998; Lee et al., 2001a). To determine if Dbf2p localizes to the nucleus during late mitosis, we monitored GFP-tagged Dbf2p in wild-type, Mob1Δ78, and Mob1Δ132 cells. As previously reported, Dbf2p-GFP localizes to SPBs and the bud neck ring in wild-type cells (Frenz et al., 2000). In addition, we detected nuclear Dbf2p-GFP in some wild-type anaphase cells (Figure 4). Nuclear Dbf2p-GFP was more readily detectable in Mob1Δ78 and Mob1Δ132 anaphase cells than in wild-type cells (Figure 4, shown for Mob1Δ132 cells). The overall pattern of nuclear fluorescence for Dbf2p-GFP was similar to that observed for GFP-Mob1p, GFP-Mob1Δ78, and GFP-Mob1Δ132 (Figures 1 and 2). These data indicate that the nuclear concentration of Dbf2p is enhanced in Mob1Δ78 and Mob1Δ132 cells and support the model that Mob1p and Dbf2p interact with each other in the nucleus, as they do in the cytoplasm.
Figure 4.
Dbf2p localizes to the nucleus during anaphase. DIC and fluorescence microscopy of wild-type (top panels) and Mob1Δ132 cells (bottom panels) expressing chromosomally tagged Dbf2-GFP (FLY626, FLY1608, respectively). Faint GFP fluorescence is detectable in nuclei of some anaphase wild-type cells (arrowheads). Nuclear Dbf2-GFP fluorescence is more prominent in Mob1Δ132 cells than in wild-type cells.
Mob1Δ78 and Mob1Δ132 Colocalize with Cdc14p
If Mob1p-Dbf2p complex localizes to the nucleus during mitosis to regulate Cdc14p release or activity, then some Mob1p-Dbf2p should colocalize with Cdc14p in the nucleus. Thus, we analyzed GFP-Mob1Δ78 and GFP-Mob1Δ132 in cells expressing CFP-tagged Cdc14p and found that truncated Mob1p partially colocalized with nuclear Cdc14p in all (100%; n = 100) anaphase cells (Figure 5A, shown for Mob1Δ132). Moreover, when Mob1Δ132 was detectable in preanaphase medium budded cells, it always (100%, n = 100) colocalized with nucleolar Cdc14p (Figure 5A, see asterisk). We confirmed that Mob1Δ132 colocalizes with the nucleolus in some preanaphase cells and anaphase cells by monitoring GFP-Mob1Δ132 in cells expressing an RFP-tagged nucleolar protein, Sik1p (Figure 5B, see arrowheads). Significantly, some nuclear Mob1Δ78 and Mob1Δ132 appeared distinct from Cdc14p, suggesting that Mob1p also associates with other nuclear proteins. Control experiments using single tagged strains revealed that CFP and RFP fluorescence were undetectable when the microscope and camera settings were optimized for GFP fluorescence and vice versa (unpublished data). These data are consistent with the model that Mob1p-Dbf2p directly regulates Cdc14p release/activity during anaphase or functions cooperatively with Cdc14p.
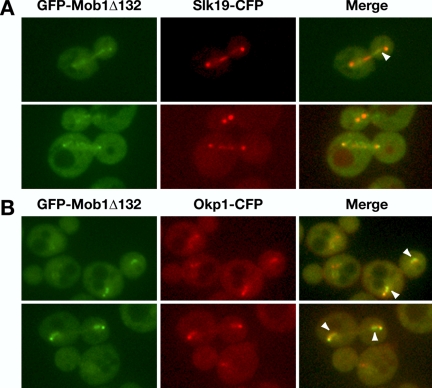
Mob1Δ78 and Mob1Δ132 Colocalize with Kinetochore Proteins
It was recently demonstrated that Cdc14p associates with and regulates kinetochore-associated chromosome passenger protein localization (Pereira and Schiebel, 2003; Trautmann et al., 2004). To determine if Mob1p also transiently associates with kinetochores, we monitored Mob1Δ132 localization in cells expressing CFP-tagged kinetochore proteins, Slk19p and Okp1p. Kinetochore proteins localize very close to SPBs during anaphase and thus are not easily distinguished from the SPB-associated Mob1p. Some kinetochore proteins, such as Slk19p and Okp1p, partially redistribute to the spindle during anaphase (Zeng et al., 1999; Bouck and Bloom, 2005; Figure 6). We observed that some SPB-independent pools of Mob1Δ132 partially colocalized with Slk19p-CFP and Okp1p-CFP during anaphase in the nuclear region just inside of the SPBs and along portions of the anaphase spindle (Figure 6, A and B; see arrowheads). Intriguingly, some nuclear Mob1Δ132 appeared distinct from Okp1p and Slk19p. We obtained similar data using cells expressing Mob1Δ78-YFP and Slk19-CFP or Okp1p-CFP (Supplementary Figure S3 and unpublished data). These data are consistent with the model that some Mob1p-Dbf2p kinase regulates Cdc14p and/or contributes to kinetochore regulation.
Figure 6.
Mob1Δ132 partially colocalizes with kinetochore proteins. Asynchronously growing cells expressing truncated GFP-Mob1Δ132 and CFP-tagged Slk19p (FLY1213) and Okp1p (FLY1219) were analyzed by fluorescence microscopy. GFP, CFP, and merged images are labeled accordingly. The arrowheads point to regions of significant colocalization. Similar results were obtained for GFP-Mob1Δ78 (unpublished data).
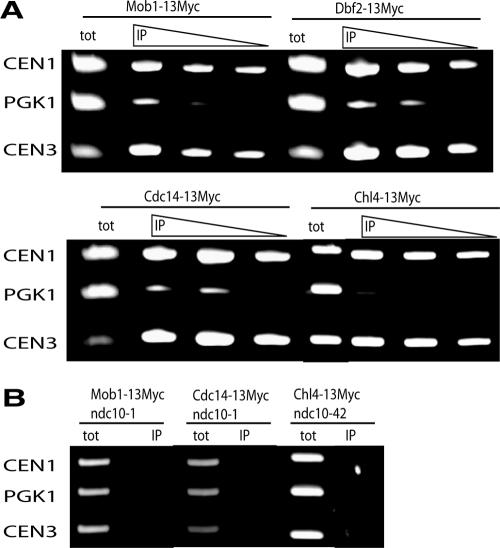
Full-length Mob1p, Dbf2p, and Cdc14p Associate with Centromere DNA
Centromere ChIP is a standard method to determine if a protein (at least transiently) associates with kinetochores. We therefore conducted centromere ChIP experiments to test if wild-type (full-length) Mob1p and Dbf2p associate with kinetochores (Figure 7). We treated cells expressing chromosomally Myc-tagged full-length Mob1p or Dbf2p with formaldehyde to cross-link protein-DNA complexes and immunoprecipitated the complexes using anti-Myc antibody, as described (Pot et al., 2003). We reversed the cross-links, purified the precipitated DNA, and conducted PCR using primers specific to centromeric (CEN1, CEN3) and noncentromeric DNA (PGK1). As a positive control, we conducted parallel ChIP assays for a central centromere-binding protein (Chl4p) and Cdc14p, the latter of which was shown to interact with the kinetochore-associated chromosome passenger protein and INCENP orthologue Sli15p (Pereira and Schiebel, 2003). The ChIP assays revealed that full-length Mob1p, Dbf2p, Cdc14p, and Chl4p precipitate CEN1 and CEN3 DNA, indicating that each of these proteins associate with kinetochores (Figure 7A). To determine the specificity of Mob1p and Cdc14p for centromere binding, we conducted ChIP experiments in conditional kinetochore mutant, ndc10-1 (Figure 7B). At 37°C, all kinetochore proteins dissociate from centromere DNA in ndc10 cells (McAinsh et al., 2003). Mob1p and Cdc14p failed to precipitate centromere DNA from extracts of ndc10-1 cells, indicating that the Mob1p and Cdc14p centromere DNA associations are kinetochore-dependent. These data indicate that Mob1p, Dbf2p, and Cdc14p physically associate with kinetochores.
Figure 7.
Mob1p, Dbf2p, and Cdc14p precipitate centromere DNA. (A) Centromere chromatin immunoprecipitation (ChIP) was conducted, as described in Materials and Methods. Myc-tagged-Mob1p, Dbf2p, Cdc14p, and Chl4p were immunoprecipitated from asynchronous cells. Dilutions of immunoprecipitated material (IP) were used as template for PCR to test for coprecipitating centromere DNA (CEN1, CEN3). tot, total cell extract used as positive control for PCR template. (B) ChIP of Myc-tagged Mob1p, Cdc14p, and Chl4p from conditional ndc10-1 or ndc10-42 cells.
Mob1p, Cdc15p, and Cdc14p Are Required for Regulating Passenger Protein Localization during Anaphase
ChIP experiments suggest that Mob1p-Dbf2p functions at kinetochores, but do not reveal the role of Mob1p-Dbf2p at kinetochores. Previous data regarding Cdc14p function might provide insight to the nuclear role(s) of Mob1p-Dbf2p. Cdc14p was demonstrated to interact with the chromosome passenger protein Sli15p and is required for the anaphase spindle localization of both Sli15p and Ipl1p (Pereira and Schiebel, 2003). This Cdc14p function was proposed to be independent of other MEN proteins because Sli15p prominently localizes to the mitotic spindle in conditional cdc15-1 cells (Pereira and Schiebel, 2003). Because Cdc15p kinase, which is an activator of Mob1p-Dbf2p kinase complex, did not appear to be required for regulating chromosome passenger protein localization during anaphase, these data led to the hypothesis that FEAR-dependent but not MEN-dependent Cdc14p activity is required for regulating chromosome passenger proteins (Pereira and Schiebel, 2003; Stegmeier and Amon, 2004).
Nevertheless, in light of the centromere/kinetochore associations of Mob1p and Dbf2p, it seemed plausible that Mob1p-Dbf2p and Cdc14p might function together at kinetochores to regulate the kinetochore-associated chromosomal passenger proteins. Thus, we tested if Ipl1p-GFP, Sli15p-GFP, and Bir1p-GFP (aurora, INCENP, and survivin orthologues) localization was aberrant in conditional mob1-77 cells. At restrictive temperature (37°C), Ipl1p-GFP localized to SPBs and kinetochore structures near the nuclear periphery but was absent from the spindle midzone in 95% (n = 160) of late anaphase-arrested mob1-77 cells (Figure 8A). Bir1p-GFP and Sli15p-GFP were absent from kinetochores and the spindles in 100% (n = 160) and 86% (n = 191) of arrested cells, respectively. When the arrested mob1-77 cells were shifted back to permissive temperature, Ipl1p-GFP, Bir1p-GFP, and Sli15p-GFP rapidly localized to the mitotic spindle within a few minutes (Figure 8A). These data suggest that Mob1p is required to establish and/or maintain chromosomal passenger proteins on the spindle during late mitosis. Furthermore, these data support the model that Mob1p, Dbf2p, and Cdc14p associate with kinetochores to regulate the localization of chromosome passenger proteins during anaphase.
Figure 8.
Chromosome passenger proteins are not maintained on anaphase spindles in mob1-77, cdc15-2, or cdc14-1 cells at 37°C. (A) The patterns of Ipl1-GFP, Bir1-GFP, and Sli15p-GFP were analyzed in late anaphase-arrested mob1-77 cells. Asynchronously growing cells (FLY1884, FLY1842, FLY1846) were shifted to 37°C for 2–3 h and analyzed by DIC and fluorescence microscopy. Ipl1-GFP, Bir1-GFP, and Sli15p-GFP were not maintained on anaphase spindles in late anaphase-arrested mob1-77 cells. Wild-type cells (wt) are shown for comparison. The mob1-77 cells rapidly recruited all three chromosomal passenger proteins to anaphase spindles when returned to permissive temperature (22°). (B) Sli15p-GFP and Ipl1p-GFP were not present on anaphase spindles in arrested cdc15-2 cells (FLY1849 and FLY2004). When returned to permissive temperature (22°), the cells rapidly recruited Sli15p and Ipl1p to the spindles before mitotic exit. (C) Ipl1p-GFP failed to localize to anaphase spindles in cdc14-1 cells (FLY2069), as previously described (Pereira and Schiebel, 2003). When returned to permissive temperature (22°C), the cells rapidly recruited Ipl1p to the spindles before mitotic exit. We obtained similar data for Sli15p localization (unpublished data). (D) The percentage of arrested cdc15-2, mob1-77, and cdc14-1 cells with Ipl1p-GFP on the kinetochore region was plotted (n = 200 for each strain).
Considering that Cdc15p is critical for Mob1p-Dbf2p activity and localization (Lee et al., 2001a; Luca et al., 2001; Mah et al., 2001), it seemed counterintuitive that mob1-77 and cdc15-1 cells would arrest with different phenotypes with respect to Sli15p localization. It is possible that the mutant protein encoded by the cdc15-1 allele, which was used in the previous study (Pereira and Schiebel, 2003), is partially active at restrictive temperature. The residual Cdc15p activity may persist in cdc15-1 cells at restrictive temperature and allow chromosome passenger proteins to localize to the mitotic spindle. We therefore used a different conditional allele of CDC15, cdc15-2, to investigate the role of Cdc15p in regulating passenger protein localization. We shifted asynchronously growing cdc15-2 cells to restrictive temperature and assayed Sli15p-GFP and Ipl1p-GFP localization after 2–3 h. We found that Sli15p and Ipl1p were absent from the mitotic spindle in more than 90% (n = 250) of the late anaphase-arrested cdc15-2 cells (Figure 8B). On release from the restrictive temperature, the cdc15-2 cells rapidly recruited Ipl1p and Sli15p to the spindle (Figure 8B). We obtained similar results for cdc14-1 cells, in agreement with Pereira and Schiebel (2003) (Figure 8C; shown for Ipl1-GFP). At 37°C, 88% (n = 200) and 91% (n = 100) of cdc14-1 cells arrested without Ipl1p or Sli15p on the spindle. Thus, like Mob1p and Cdc14p, Cdc15p is required for establishing or maintaining Sli15p on the mitotic spindle.
Intriguingly, some Ipl1p remained in the kinetochore region in 99% (n = 157) of the mob1-77 cells at restrictive temperature (Figure 8, A and D, see arrowheads). This was in sharp contrast to cdc15-2 and cdc14-1 cells, where only 12 and 61% cells (n = 260 and 200) had detectable Ipl1p in the kinetochore region when shifted to restrictive temperature (Figure 8D). Given that Cdc15p kinase is the presumed activator of Mob1p-Dfb2p kinase, it is curious that conditional mob1-77 and cdc15-2 cells display such dramatic differences in Ipl1p localization in the kinetochore region. This phenomenon might be explained if the diminished Mob1p-Dbf2p kinase activity in cdc15-2 cells is sufficient to regulate Ipl1p dissociation from kinetochore region but not sufficient to initiate or maintain Ipl1p on the anaphase spindle. Nevertheless, these data indicate that Mob1p is critical for dissociating Ipl1p from the kinetochore region and may suggest that Mob1p-Dbf2p functions independently of other MEN proteins.
Mob1p Is Required for Maintaining Passenger Proteins on the Anaphase Spindle
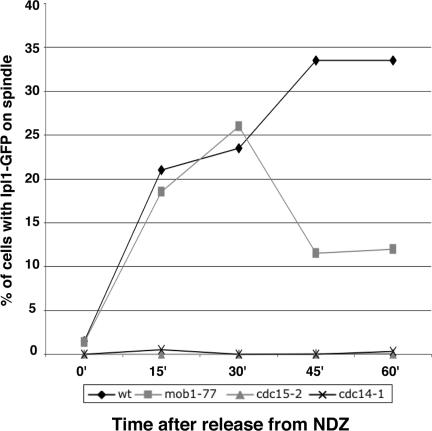
To determine if the chromosome passenger proteins localize to the spindle before the late anaphase arrest of conditional MEN mutants, we analyzed Ipl1p-GFP in synchronized mob1-77, cdc15-2, cdc14-1, and wild-type cells. We synchronized cells in G1 at permissive temperature (22°C), released them from the G1 block, and incubated them in NDZ-containing medium at restrictive temperature (34 or 37°C) to arrest them in metaphase. Once the cells were metaphase-arrested, we transferred them to drug-free medium at restrictive temperature and monitored Ipl1p localization as the cells progressed to the late anaphase arrest. Figure 9 shows the data from a representative experiment. The cells did not contain any Ipl1p-associated spindles in NDZ-containing medium (t = 0). Within 30 min after removing NDZ, 21% (n = 150) of mob1-77 cells contained Ipl1p on the spindle, which closely mirrored the percent of Ipl1p-associated spindles in similarly treated wild-type cells (Figure 9). After 30 min, the number of mob1-77 cells with Ipl1p on the spindles dropped continuously, reaching 5–10% by 2 h (unpublished data). We obtained similar results for Sli15p and Bir1p localization (Supplementary Figure S4). In contrast to mob1-77 cells, Ipl1p failed to localize to the anaphase spindles in all similarly treated cdc14-1 and cdc15-2 cells (Figure 9). These data indicate that Mob1p is not required for establishing chromosome passenger proteins on the mitotic spindle, as are Cdc14p and Cdc15p, but rather Mob1p is necessary for maintaining passenger proteins on the spindle. Thus, at least one nuclear role for Mob1p-Dbf2p kinase complex is to regulate the localization of kinetochore and spindle-associated chromosome passenger proteins during anaphase.
Figure 9.
Ipl1p transiently localizes to anaphase spindles in mob1-77 cells, but not in cdc15-2 or cdc14-1 cells. Ipl1p-GFP was analyzed in wild-type, mob1-77, cdc15-2, and cdc14-1 cells after release from NDZ-induced metaphase arrest, as described in the text. On release from metaphase block (t = 0), the percentage of cells that displayed Ipl1p-GFP on the anaphase spindle was scored over time. The presented data are from a representative experiment.
DISCUSSION
It was previously established that Mob1p and Dbf2p localize to the cytoplasmic surface of SPBs during anaphase and to a ring at the bud neck during cytokinesis (Frenz et al., 2000; Luca et al., 2001). In this work, we discovered that Mob1p and Dbf2p also localize to nuclei during mitosis and are required for maintaining chromosomal passenger proteins on the spindle during anaphase. By monitoring truncated Mob1p, which localizes more prominently to the nucleus than does full-length Mob1p, we established that nuclear Mob1p-Dbf2p complex transiently colocalizes with nucleolar Cdc14p before anaphase and with released Cdc14p and kinetochore proteins during anaphase. We independently confirmed the kinetochore association of Mob1p, Dbf2p, and Cdc14p by ChIP analysis and demonstrated that all three MEN proteins are required for the anaphase spindle localization of the Aurora chromosomal passenger protein complex. These data suggest that Mob1p-Dbf2p and Cdc14p functionally interact in the nucleus and establish kinetochores as sites for MEN signaling. These results evoke several models for nuclear Mob1p-Dbf2p function, including regulation of spindle integrity or cytokinesis via kinetochore or chromosomal passenger proteins, mitotic checkpoint regulation, or regulation of Cdc14p release from the nucleolus.
Kinetochore Function for MEN
One role for Mob1p-Dbf2p at kinetochores is to regulate the localization of kinetochore-associated passenger proteins during anaphase. Chromosomal passenger proteins are a conserved group of interacting proteins that localize to kinetochores before metaphase and move to the central spindle in anaphase (Terada, 2001; Lens and Medema, 2003; Vagnarelli and Earnshaw, 2004). Mob1p-Dbf2p may control chromosomal passenger proteins directly or via Cdc14p. Cdc14p appears to promote the redistribution of passenger proteins to the anaphase spindle by dephosphorylating Sli15p (Pereira and Schiebel, 2003). Thus, the early anaphase localization of Sli15p in conditional mob1 and cdc15 mutants is almost certainly mediated by Cdc14p upon FEAR-activation (Stegmeier et al., 2002; Pereira and Schiebel, 2003). Cdc15p and Mob1p-Dbf2p are not required for FEAR and therefore may contribute to passenger protein localization by ensuring that a small amount of Cdc14p remains active in the nucleus throughout anaphase. Alternatively, Mob1p-Dbf2p might function cooperatively with or separately from Cdc14p to regulate the chromosomal passenger proteins. The differences in Ipl1p localization in conditional MEN mutants support separate roles for MEN in regulating passenger proteins (Figure 8). The more severe failure of Ipl1p to dissociate from kinetochore regions in conditional mob1 mutants than in cdc14 mutants argues that Mob1p-Dbf2p regulates passenger protein localization distinctly from Cdc14p. Moreover, coprecipitation experiments from asynchronous cells have not revealed a direct association between Mob1p and Cdc14p complexes (unpublished data). Nevertheless, physiologically relevant interactions between Mob1p-Dbf2p and other nuclear proteins may be difficult to demonstrate because of the small concentrations and transient nature of nuclear Mob1-Dbf2p.
Mob1p-Dbf2p and Cdc14p may also transiently associate with kinetochores to maintain genomic stability during mitosis. In agreement, mutations in several MEN genes, including MOB1 and CDC14, increase genomic instability, although the mechanisms for MEN-associated genomic instability are not known (Hartwell and Smith, 1985; Luca and Winey, 1998; Dai et al., 2003). Mob1p-Dbf2p and Cdc14p might contribute to kinetochore assembly or chromosome attachment to the spindle. In support, the S. pombe Cdc14p homologue interacts with Aurora kinase at kinetochores before anaphase and regulates chromosome attachment to the spindle (Trautmann et al., 2004). Experiments with truncated Mob1p suggest that Mob1p and Dbf2p enter nuclei before anaphase, which is consistent with a preanaphase role for Mob1p-Dbf2p at kinetochores (Figures 2 and 5 and unpublished data). Nevertheless, current data suggest that Cdc14p remains sequestered in the nucleolus before anaphase and thus cannot be essential for mitotic spindle assembly or establishing chromosome attachment to the spindle. It is perhaps more likely that kinetochore-associated Mob1p, Dbf2p, and Cdc14p regulate later mitotic events, such as kinetochore disassembly or spindle disassembly during mitotic exit. These functions are more in line with the general role of MEN in coordinating multiple mitotic exit events, such as CDK inactivation, cytokinesis, and initiation of G1-specific transcription (Seshan and Amon, 2004). Accordingly, kinetochore-associated Mob1p-Dbf2p and Cdc14p might contribute to spindle disassembly at the end of mitosis by regulating the interactions of essential kinetochore- or spindle-associated proteins, such as the passenger proteins.
It is tempting to speculate that kinetochore-associated MEN proteins coordinate chromosome segregation and/or spindle disassembly with cytokinesis by regulating chromosomal passenger proteins. Indeed, mammalian chromosomal passengers Aurora B kinase, survivin, and INCENP are implicated in the coordination of chromosome segregation with cytokinesis (Terada, 2001; Tanaka et al., 2002; Higuchi and Uhlmann, 2003). Depletion or expression of dominant negative forms of passenger proteins induces cytokinesis defects in mammalian cells (Tatsuka et al., 1998; Vagnarelli and Earnshaw, 2004). The S. cerevisiae orthologues, Ipl1p, Bir1p, and Sli15p, are important for maintaining bipolar attachment of chromosomes to the mitotic spindle, but have not been directly implicated in cytokinesis. The functions of yeast passenger proteins on anaphase spindles are not fully established; however, current evidence suggests that Ipl1p and Bir1p regulate spindle disassembly (Buvelot et al., 2003; Bouck and Bloom, 2005). Moreover, Bir1p is required for the transport of the kinetochore protein Ndc10p to the plus-ends of interpolar microtubules at the spindle midzone (Bouck and Bloom, 2005). The function of Ndc10p on the spindle midzone is not known; however, conditional ndc10 mutants display anaphase spindle and cytokinesis defects (Bouck and Bloom, 2005). Thus, MEN may regulate spindle disassembly and cytokinesis by controlling the localization and activity of passenger proteins and Ndc10p.
Checkpoint Regulation of Mob1p-Dbf2p
Kinetochores are major sites for mitotic checkpoint regulation. Thus, the kinetochore localization of Mob1p-Dbf2p and Cdc14p might implicate MEN in mitotic checkpoint signaling. In support, at least one of the MEN proteins (Mob1p) interacts with Mps1p kinase, a component of the spindle assembly checkpoint (SAC) that localizes to kinetochores and SPBs (Luca and Winey, 1998; Castillo et al., 2002; Liu et al., 2003). Moreover, both Mps1p and Ipl1p kinase are components of the spindle tension checkpoint (Biggins and Murray, 2001). Although no current evidence supports an essential role for Mob1-Dbf2p in mitotic checkpoint signaling, checkpoint activation might result in the direct inhibition of Mob1p-Dbf2p kinase by Mps1p, as part of a mechanism to prevent premature mitotic exit and cytokinesis during spindle malfunction. In agreement, Mob1p is hyperphosphorylated in checkpoint-activated cells (unpublished data) and Dbf2p kinase activity is inhibited in an Mps1p-dependent manner upon SAC activation (Fesquet et al., 1999). Bub2p is also required for SAC-mediated inhibition of Dbf2p, suggesting that the SAC inhibits Mob1p-Dbf2p indirectly via preventing Tem1p activation (Fesquet et al., 1999). Nevertheless, there are likely to be multiple mechanisms for MEN inhibition upon checkpoint activation.
Mob1p-Dbf2p Regulation of Cdc14p Release
Another important function for nuclear Mob1p-Dbf2p may be to regulate Cdc14p release from the nucleolus. Previous work indicates that Dbf2p is not required for Cdc14p early anaphase release (FEAR) from the nucleolus (Shou et al., 1999; Visintin et al., 1999); however, our colocalization data are consistent with the model that Mob1p-Dbf2p plays a direct role in the release of Cdc14p from the nucleolus during telophase (Seshan and Amon, 2004). Mob1p-Dbf2p may phosphorylate Cdc14p, Net1p/Cfi1p, or another member of the nucleolar RENT complex to help mediate Cdc14p release during telophase. Alternatively, Mob1p-Dbf2p may trigger Cdc14p release indirectly by activating or functioning cooperatively with other regulators of Cdc14p release, such as Cdc5p or Cdc15p kinases. Indeed, Cdc5p is required for both Cdc14p early anaphase and telophase release. Cdc5p phosphorylates Net1p/Cfi1p, which decreases its capacity to bind Cdc14p (Shou et al., 2002; Visintin et al., 2003), and stimulates MEN activation by inhibiting the Bub2p-Bfa1p complex (Lee et al., 2001b; Geymonat et al., 2003; Lew and Burke, 2003). Cdc15p kinase is not required for FEAR, but is essential for Cdc14p telophase release (Shou et al., 1999; Visintin et al., 1999). Overexpression of truncated Cdc15p promotes Cdc14p release and leads to hyperphosphorylation of Net1p/Cfi1p (Bardin et al., 2003). Thus, nuclear Mob1p-Dbf2p may function downstream or in concert with Cdc5p or Cdc15p to regulate Cdc14p telophase release. Given that Cdc15p is an in vitro activator of Mob1p-Dbf2p and has not been observed in the nucleus, it may be more likely that Cdc15p overexpression induces Cdc14p release by activating Mob1p-Dbf2p kinase.
Regulation of Nuclear Localization of Mob1p-Dbf2p Complex
Experiments with N-terminally truncated Mob1p indicate that recruitment and retention of Mob1p-Dbf2p in the nucleus is tightly regulated, although the regulatory details remain unknown. Mob1p nuclear recruitment is not dependent on Ipl1p or Mps1p (Supplementary Figure S5), and nuclear import or retention is unlikely to be stimulated by Cdc14p, because truncated Mob1p can localize to the nucleus in preanaphase cells. The observation that truncated Mob1p localizes more prominently to the nucleus than full-length Mob1p suggests that the nonconserved N terminus of Mob1p, which lacks known protein motifs, is important for limiting the amount of protein in the nucleus. Deletion of the N-terminal 132 amino acids does not significantly affect the total cellular Mob1p concentration, although it is formally possible that it stabilizes the nuclear fraction of Mob1p. Alternatively, the Mob1p N-terminal domain may be subject to posttranslational modifications or interactions that limit its nuclear import and/or promote efficient Mob1p nuclear export. In support, Mob1p is phosphorylated in the N-terminal region (Ficarro et al., 2002; unpublished data). We did not detect any nuclear enhancement of full-length Mob1p in cells harboring a conditional allele of the Crm1 nuclear export factor (xpo1-1; unpublished data), suggesting that nuclear accumulation of Mob1p-Dbf2p kinase is not mediated by inhibition of Crm1-dependent nuclear export. Nevertheless, at this time we cannot rule out that there are Crm1-independent nuclear export mechanisms that control Mob1p nuclear accumulation.
It is likely that the nuclear Mob1p-Dbf2p and Cdc14p are subject to feedback regulations. Most models for MEN suggest that Mob1p-Dbf2p functions upstream of Cdc14p because Cdc14p telophase release is dependent on Dbf2p (Seshan and Amon, 2004). Intriguingly, Cdc14p is important for Mob1p-Dbf2p activity and localization. Activation of Dbf2p by Cdc5p overexpression is dependent on Cdc14p (Visintin et al., 2003), and the bipolar and bud neck localizations of Mob1p are Cdc14p-dependent (Luca et al., 2001). Given that Mob1p-Dbf2p and Cdc14p partially colocalize in the nucleus during anaphase, it is possible that they function together in a common complex. To date, coprecipitation experiments have failed to yield supporting evidence for a direct interaction between Mob1p-Dbf2p and Cdc14p or between Mob1p-Dbf2p and chromosome passenger proteins (unpublished data). Nevertheless, our data indicate that Mob1p-Dbf2p complex is critical for ensuring that Cdc14p is released from the nucleolus during late mitosis and for regulating the localization of chromosomal passenger proteins. These results provide insight to the identity of Mob1p-Dbf2p targets and predict that mammalian Mob1p and Dbf2p homologues will regulate spindle integrity and/or cytokinesis via passenger proteins.
Supplementary Material
Acknowledgments
We thank Zachary Kern and Manali Mody for technical assistance; Sue Biggins, Tricia Melloy, James Falvo, and Erin O'Shea for yeast strains; Erfei Bi, Phong Tran, Sue Biggins, and members of the Luca lab for helpful discussions. We also thank Vivien Measday for critically reading this manuscript. This work was supported by the National Institutes of Health (GM60575 to F.C.L.). J.S. was supported in part by the Degussa-Hermann-Schlosser Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–04–0337) on September 21, 2005.
Abbreviations used: MEN, mitotic exit network; SPB, spindle pole body; ChIP, chromatin immunoprecipitation.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alexandru, G., Uhlmann, F., Mechtler, K., Poupart, M. A., and Nasmyth, K. (2001). Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459–472. [DOI] [PubMed] [Google Scholar]
- Azzam, R., Chen, S. L., Shou, W., Mah, A. S., Alexandru, G., Nasmyth, K., Annan, R. S., Carr, S. A., and Deshaies, R. J. (2004). Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305, 516–519. [DOI] [PubMed] [Google Scholar]
- Bardin, A. J., Boselli, M. G., and Amon, A. (2003). Mitotic exit regulation through distinct domains within the protein kinase Cdc15. Mol. Cell. Biol. 23, 5018–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek, J., Kang, J., Kurischko, C., Li, B., Raab, J. R., Belanger, K. D., Luca, F. C., and Yu, H. (2005). Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle 4, 951–961. [DOI] [PubMed] [Google Scholar]
- Biggins, S., and Murray, A. W. (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15, 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., Severin, F. F., Bhalla, N., Sassoon, I., Hyman, A. A., and Murray, A. W. (1999). The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck, D. C., and Bloom, K. S. (2005). The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA 102, 5408–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot, S., Tatsutani, S. Y., Vermaak, D., and Biggins, S. (2003). The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo, A. R., Meehl, J. B., Morgan, G., Schutz-Geschwender, A., and Winey, M. (2002). The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p. J. Cell Biol. 156, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W., Huang, X., and Ruan, Q. (2003). Polo-like kinases in cell cycle checkpoint control. Front. Biosci. 8, d1128–d1133. [DOI] [PubMed] [Google Scholar]
- Fesquet, D., Fitzpatrick, P. J., Johnson, A. L., Kramer, K. M., Toyn, J. H., and Johnston, L. H. (1999). A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 18, 2424–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro, S. B., McCleland, M. L., Stukenberg, P. T., Burke, D. J., Ross, M. M., Shabanowitz, J., Hunt, D. F., and White, F. M. (2002). Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305. [DOI] [PubMed] [Google Scholar]
- Frenz, L. M., Lee, S. E., Fesquet, D., and Johnston, L. H. (2000). The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J. Cell Sci. 113, 3399–3408. [DOI] [PubMed] [Google Scholar]
- Geymonat, M., Spanos, A., Walker, P. A., Johnston, L. H., and Sedgwick, S. G. (2003). In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 278, 14591–14594. [DOI] [PubMed] [Google Scholar]
- Gray, C. H., Good, V. M., Tonks, N. K., and Barford, D. (2003). The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 22, 3524–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. R. (1991). Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology 194, 1–933. [PubMed] [Google Scholar]
- Hartwell, L. H., and Smith, D. (1985). Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, T., and Uhlmann, F. (2003). Cell cycle: passenger acrobatics. Nature 426, 780–781. [DOI] [PubMed] [Google Scholar]
- Higuchi, T., and Uhlmann, F. (2005). Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 433, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, F., and Elledge, S. J. (2002). Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle 1, 351–355. [PubMed] [Google Scholar]
- Hwa Lim, H., Yeong, F. M., and Surana, U. (2003). Inactivation of mitotic kinase triggers translocation of MEN components to mother-daughter neck in yeast. Mol. Biol. Cell 14, 4734–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky, S. I., Chiang, Y. C., Luca, F. C., Chen, J., Toyn, J. H., Winey, M., Johnston, L. H., and Denis, C. L. (1998). Dbf2 protein kinase binds to and acts through the cell cycle-regulated Mob1 protein. Mol. Cell Biol. 18, 2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. E., Frenz, L. M., Wells, N. J., Johnson, A. L., and Johnston, L. H. (2001a). Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 11, 784–788. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., Jensen, S., Frenz, L. M., Johnson, A. L., Fesquet, D., and Johnston, L. H. (2001b). The Bub2-dependent mitotic pathway in yeast acts every cell cycle and regulates cytokinesis. J. Cell Sci. 114, 2345–2354. [DOI] [PubMed] [Google Scholar]
- Lens, S. M., and Medema, R. H. (2003). The survivin/Aurora B complex: its role in coordinating tension and attachment. Cell Cycle 2, 507–510. [DOI] [PubMed] [Google Scholar]
- Lew, D. J., and Burke, D. J. (2003). The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37, 251–282. [DOI] [PubMed] [Google Scholar]
- Liu, S. T., van Deursen, J. M., and Yen, T. J. (2003). The role of mitotic checkpoint in maintaining genomic stability. Curr. Top. Dev. Biol. 58, 27–51. [DOI] [PubMed] [Google Scholar]
- Luca, F. C., Mody, M., Kurischko, C., Roof, D. M., Giddings, T. H., and Winey, M. (2001). Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell Biol. 21, 6972–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca, F. C., and Winey, M. (1998). MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 9, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah, A. S., Jang, J., and Deshaies, R. J. (2001). Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98, 7325–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh, A. D., Tytell, J. D., and Sorger, P. K. (2003). Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19, 519–539. [DOI] [PubMed] [Google Scholar]
- Measday, V., Hailey, D. W., Pot, I., Givan, S. A., Hyland, K. M., Cagney, G., Fields, S., Davis, T. N., and Hieter, P. (2002). Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., and Koshland, D. (1997). Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11, 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., and Schiebel, E. (2003). Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302, 2120–2124. [DOI] [PubMed] [Google Scholar]
- Pot, I., Measday, V., Snydsman, B., Cagney, G., Fields, S., Davis, T. N., Muller, E. G., and Hieter, P. (2003). Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell 14, 460–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, W. S. (2002). The FEAR factor. Mol. Cell 9, 207–209. [DOI] [PubMed] [Google Scholar]
- Seshan, A., and Amon, A. (2004). Linked for life: temporal and spatial coordination of late mitotic events. Curr. Opin. Cell Biol. 16, 41–48. [DOI] [PubMed] [Google Scholar]
- Shou, W., Azzam, R., Chen, S. L., Huddleston, M. J., Baskerville, C., Charbonneau, H., Annan, R. S., Carr, S. A., and Deshaies, R. J. (2002). Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex. BMC Mol. Biol. 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., Seol, J. H., Shevchenko, A., Baskerville, C., Moazed, D., Chen, Z. W., Jang, Charbonneau, J. H., and Deshaies, R. J. (1999). Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97, 233–244. [DOI] [PubMed] [Google Scholar]
- Simanis, V. (2003). The mitotic exit and septation initiation networks. J. Cell Sci. 116, 4261–4262. [DOI] [PubMed] [Google Scholar]
- Stavridi, E. S., Harris, K. G., Huyen, Y., Bothos, J., Verwoerd, P. M., Stayrook, S. E., Pavletich, N. P., Jeffrey, P. D., and Luca, F. C. (2003). Crystal structure of a human mob1 protein. Toward understanding mob-regulated cell cycle pathways. Structure (Camb). 11, 1163–1170. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., and Amon, A. (2004). Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38, 203–232. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., Visintin, R., and Amon, A. (2002). Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108, 207–220. [DOI] [PubMed] [Google Scholar]
- Tanaka, T. U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E. Stark, M. J., and Nasmyth, K. (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317–329. [DOI] [PubMed] [Google Scholar]
- Tatsuka, M., Katayama, H., Ota, T., Tanaka, T., Odashima, S., Suzuki, F., and Terada, Y. (1998). Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 58, 4811–4816. [PubMed] [Google Scholar]
- Terada, Y. (2001). Role of chromosomal passenger complex in chromosome segregation and cytokinesis. Cell Struct. Funct. 26, 653–657. [DOI] [PubMed] [Google Scholar]
- Toyn, J. H., and Johnston, L. H. (1994). The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 13, 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann, S., Rajagopalan, S., and McCollum, D. (2004). The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev. Cell 7, 755–762. [DOI] [PubMed] [Google Scholar]
- Vagnarelli, P., and Earnshaw, W. C. (2004). Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma 113, 211–222. [DOI] [PubMed] [Google Scholar]
- Visintin, R., and Amon, A. (2001). Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12, 2961–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., Craig, K., Hwang, E. S., Prinz, S., Tyers, M., and Amon, A. (1998). The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2, 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin, R., Hwang, E. S., and Amon, A. (1999). Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398, 818–823. [DOI] [PubMed] [Google Scholar]
- Visintin, R., Stegmeier, F., and Amon, A. (2003). The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol. Biol. Cell 14, 4486–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E., and Winey, M. (1996). The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E. L., Kurischko, C., Zhang, C., Shokat, K., Drubin, D. G., and Luca, F. C. (2002). The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158, 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., Goetsch, L., Baum, P., and Byers, B. (1991). MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 114, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, B. A., and Gould, K. L. (2004). Fission yeast Clp1p phosphatase affects G(2)/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 23, 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., Asakawa, K., and Toh-e, A. (2002). Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr. Biol. 12, 944–950. [DOI] [PubMed] [Google Scholar]
- Zeng, X., Kahana, J. A., Silver, P. A., Morphew, M. K., McIntosh, J. R., Fitch, I. T., Carbon, J., and Saunders, W. S. (1999). Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.