Abstract
Exposure of cells to endoplasmic reticulum (ER) stress leads to activation of PKR-like ER kinase (PERK), eukaryotic translation initiation factor 2α (eIF2α) phosphorylation, repression of cyclin D1 translation, and subsequent cell cycle arrest in G1 phase. However, whether PERK is solely responsible for regulating cyclin D1 accumulation after unfolded protein response pathway (UPR) activation has not been assessed. Herein, we demonstrate that repression of cyclin D1 translation after UPR activation occurs independently of PERK, but it remains dependent on eIF2α phosphorylation. Although phosphorylation of eIF2α in PERK–/– fibroblasts is attenuated in comparison with wild-type fibroblasts, it is not eliminated. The residual eIF2α phosphorylation correlates with the kinetics of cyclin D1 loss, suggesting that another eIF2α kinase functions in the absence of PERK. In cells harboring targeted deletion of both PERK and GCN2, cyclin D1 loss is attenuated, suggesting GCN2 functions as the redundant kinase. Consistent with these results, cyclin D1 translation is also stabilized in cells expressing a nonphosphorylatable allele of eIF2α; in contrast, repression of global protein translation still occurs in these cells, highlighting a high degree of specificity in transcripts targeted for translation inhibition by phosphorylated eIF2α. Our results demonstrate that PERK and GCN2 function to cooperatively regulate eIF2α phosphorylation and cyclin D1 translation after UPR activation.
INTRODUCTION
Integral membrane proteins and secreted proteins are synthesized, folded, and posttranslationally modified in the endoplasmic reticulum (ER). When the integrity of protein folding in the ER is compromised and misfolded proteins accumulate, a signaling network referred to as the unfolded protein response pathway (UPR) is activated (Kaufman, 1999). Stresses that activate the UPR include disruption of proper protein glycosylation (glucose deprivation or treatment of cells with drugs that directly inhibit glycosylation such as tunicamycin), perturbations in ER calcium homeostasis (thapsigargin), perturbations in ER redox status (dithiothreitol [DTT]), and hypoxia (Kaufman, 1999; Koumenis et al., 2002).
Activation of the UPR triggers a checkpoint that provides cells with the opportunity to adapt and survive, or under conditions of chronic stress, to commit to a program of cell death. The proximal effectors of the mammalian UPR include the three homologous, ER-resident transmembrane protein kinases Ire1 (α and β) and PKR-like ER kinase (PERK); the ER-resident transmembrane protease caspase 12; and the ER-resident transmembrane bZIP transcription factor ATF6 (Cox et al., 1993; Shi et al., 1998; Tirasophon et al., 1998; Wang et al., 1998; Harding et al., 1999; Haze et al., 1999;
Nakagawa et al., 2000). Ire1 and PERK both have luminal, ER stress-sensing domains that regulate their dimerization and activation of their protein kinase activity. Activation of ATF6 occurs via a proteolytic cleavage that allows its translocation to the nucleus where it functions as an active transcription factor (Haze et al., 1999). Ire1 and ATF6 are primarily implicated in the transcriptional arm of the UPR, resulting in the induction of ER chaperones to remedy protein misfolding (Tirasophon et al., 1998).
In contrast, PERK contributes to the regulation of protein translation and cell adaptation to ER stress (Shi et al., 1998; Harding et al., 1999, 2000b; Cullinan et al., 2003). Two substrates/effectors of PERK have been identified. The first is eukaryotic translation initiation factor 2α (eIF2α) (Shi et al., 1998; Harding et al., 1999; Sood et al., 2000). PERK-dependent phosphorylation of eIF2α contributes to repression of protein translation after initiation of ER stress. The attenuation of translation by PERK has been attributed to cell survival because it is thought to limit the influx of new proteins into the ER and thereby limit buildup of misfolded proteins. A second substrate of PERK is the NF-E2-related factor 2 (Nrf2) transcription factor. Nrf2 is a direct substrate of PERK and has been demonstrated to function downstream of PERK in the regulation of cellular redox regulation and thereby contributes to cell adaptation (Cullinan and Diehl, 2004).
During ER stress, PERK-mediated translation inhibition is thought to contribute to cyclin D1 translation attenuation, provoking G1 arrest (Brewer et al., 1999). Arrest in G1 phase in response to ER stress is thought to provide the cell an opportunity to restore cellular homeostasis before committing to apoptosis (Brewer and Diehl, 2000). Although the precise role of cell cycle arrest during this stress response is undetermined, given that cell division places extreme energy demands on the cell, arrest may allow diversion of cell resources to mechanisms that contribute to restoring ER homeostasis.
Although activation of PERK is sufficient to trigger cyclin D1 loss, a dominant negative PERK allele cannot completely abrogate loss of cyclin D1, suggesting that PERK is not essential for regulation of cyclin D1 or G1 arrest during the UPR. We tested the hypothesis that PERK might be dispensable for loss of cyclin D1 translation during the UPR and explored compensatory mechanisms that could facilitate repression of cyclin D1 translation and consequent cell cycle arrest. We demonstrate that regulation of cyclin D1 translation and cell cycle progression, although not dependent on PERK, are dependent upon eIF2α phosphorylation. We further demonstrate that both PERK and GCN2 contribute to the ER stress-mediated regulation of eIF2α phosphorylation and cyclin D1 translation.
MATERIALS AND METHODS
Tissue Culture Conditions and Plasmids
Cell lines were maintained in MEF media (Dulbecco's modified Eagle's medium [Cellgro, Mediatech, Herndon, VA] supplemented with 10% fetal bovine serum [FBS] (Gemini), penicillin/streptomycin [Cellgro, Mediatech], nonessential amino acids [Invitrogen, Carlsbad, CA]), l-glutamine (Cellgro, Mediatech), and β-mercaptoethanol [Invitrogen]). PERK–/– fibroblasts used for our initial experiments were a gift from David Ron (Skirball Institute, New York University School of Medicine, New York, NY) or Douglas Cavener (Department of Biology, Pennsylvania State University, University Park, PA). The eIF2α S51A homozygous knockin murine embryonic fibroblasts (MEFs) were a gift from Donalyn Scheuner and Randal Kaufman (University of Michigan Medical Center, Ann Arbor, MI; Scheuner et al., 2001). PERK/GCN2–/– and PERK/PKR/GCN2–/– embryonic fibroblasts were a gift from Douglas Cavener (Zhang et al., 2002; Jiang et al., 2004). All MEF cell lines were immortalized via a standard 3T9 passage protocol, except for the cells received from David Ron, which were immortalized with SV40 Large T antigen. The plasmid encoding dominant negative PKR (K296D) was a gift from Randal Kaufman. Transfections were performed using LipofectAMINE Plus (Invitrogen) according to the manufacturer's instructions. A plasmid expressing green fluorescent protein (GFP) was cotransfected in all experiments to confirm equivalent transfection efficiency. To knockdown endogenous GCN2, hairpins were synthesized and cloned into pSuper retro (Oligo-Engine, Seattle, WA) (target sequence aaggcctgtcgaatgaaagt). Vectors encoding short hairpin RNA (shRNA) against firefly luciferase were a gift from P. Klein (Department of Pharmacology, University of Pennsylvania, Philadelphia, PA). To achieve knockdown, mouse fibroblasts were infected with retroviral supernatants as described previously (Brewer and Diehl, 2000). Forty-eight hours postinfection, cells were treated as indicated and harvested for analysis.
Reverse Transcription (RT)-PCR
For detection of GCN2 and cyclin D1 mRNA by RT-PCR, total RNA was extracted from wild-type murine fibroblasts using TRIzol (Invitrogen) and purified by phenol-chloroform extraction. RT reactions were performed using Superscript II RT (Invitrogen) and oligo(dT) priming following the manufacturer's instructions. GCN2 was amplified using primers sense, 5′-atacccagatgtagttcccgaaa-3′, and antisense, 5′-atggaggatgtcacagcagccaggagag-3′; cyclin D1 was amplified using primers sense, 5′-atccggcccgaggagctg-3′, and antisense 5′-accacgctcccagcagc-3′; and hypoxanthine-guanine phosphoribosyl transferase (HPRT) was amplified as a control using primers sense, 5′-cctgctggattacattaaagcactg-3′, and antisense 5′-cctgaagtactcattatagtcaagg-3′.
Western Blot Analysis
For direct Western blot analysis, cells were lysed in EBC buffer (50 mM Tris-HCl, pH 8.0, 120 mM NaCl, 1 mM EDTA, 0.5% Igapel, 10 U/ml aprotinin, 5 μg/ml leupeptin, 1 mM DTT, 10 mM β-glycerophosphatase, 4 mM NaF, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). For phospho-eIF2α blots, cells were lysed in 1% SDS plus the protease/phosphatase inhibitors listed above. Total protein was resolved on denaturing polyacrylamide gels, transferred to nitrocellulose membranes (Millipore, Billerica, MA), and blotted with the indicated primary antibodies: cyclin D1 (Ab3; Calbiochem, San Diego, CA), β-tubulin (Sigma-Aldrich, St. Louis, MO), C/EBP homologous protein (Santa Cruz Biotechnology, Santa Cruz, CA), phospho-eIF2α (Ser51; Cell Signaling Technology, Beverly, MA), total eIF2α (Santa Cruz Biotechnology or Cell Signaling Technology), CDK4 (C-22; Santa Cruz Biotechnology), p27Kip1 (clone 57; BD Transduction Laboratories, Franklin Lakes, NJ), and heterogeneous nuclear ribonucleoprotein K (hnRNPK) (gift from Gideon Dreyfuss, Department of Biochemistry and Biophysics, University of Pennsylvania Medical Center, Philadelphia, PA). Sites of antibody binding were visualized by enhanced chemiluminescence detection (PerkinElmer Life and Analytical Sciences, Boston, MA).
Biosynthetic Labeling
Subconfluent cells were treated with 0.5 μg/ml tunicamycin for the indicated intervals, or left untreated, before being shifted to methionine/cysteine-free DMEM (Sigma-Aldrich) for the final 30 min of treatment. Cells were pulse labeled with medium (+/– tunicamycin) containing 150 μCi/ml trans-35S-label for the indicated intervals, lysed in NP-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 1% Igapel, 0.5% deoxycholate, 150 mM NaCl, 10 U/ml aprotinin, 5 μg/ml leupeptin, 1 mM DTT, 10 mM β-glycerophosphatase, 4 mM NaF, and 1 mM PMSF), and cyclin D1 was immunoprecipitated from whole cell lysates. Radiolabeled proteins were resolved on a denaturing polyacrylamide gel and visualized by autoradiography.
For quantification of newly synthesized radiolabeled proteins, 10-μl samples of whole cell lysate were spotted on Whatman paper and boiled in 10% trichloroacetic acid (TCA) supplemented with 0.1% l-methionine. Samples were then rinsed in water, 95% ethanol, and acetone before scintillation counting.
For pulse-chase analysis, cells were treated with tunicamycin and starved of methionine/cysteine for 30 min. Cells were then pulse-labeled in medium (+/– tunicamycin) containing 150 μCi/ml trans-35S-label for 30 min, washed with phosphate-buffered saline (PBS), and chased for the indicated interval in medium (+/– tunicamycin) supplemented with 200 μM cold methionine. Radiolabeled cyclin D1 was precipitated as described above.
Fluorescence-activated Cell Sorting (FACS) Analysis
For FACS analysis, cells were washed once with PBS, and pellets were suspended in 150 μl of PBS and 350 μl of 100% ethanol. Cells were fixed at –20°C overnight. Cells were incubated in propidium iodide (10 μg/ml in PBS) and scanned by flow cytometry using a Becton-Dickinson FACSCalibur (BD Biosciences, San Jose, CA) to determine DNA content.
Immunofluorescence
For detection of bromodeoxyuridine (BrdU)-positive cells, cells were plated on glass coverslips and treated as indicated. During the last 1.5 h of tunicamycin treatment, cells were pulsed with 10 μM BrdU. Cells were fixed and permeabilized with ice-cold methanol/acetone (1:1) for 10 min at –20°C. After treatment of cells with 1.5 M HCl, cells were stained with anti-BrdU antibody (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and fluorescein isothiocyanate-conjugated anti-mouse antibody (1:100; GE Healthcare). All incubations were performed in PBS containing 10% FBS. After a final wash in PBS, DNA was stained with Hoechst dye 33258 (Sigma-Aldrich). Cells were mounted with Vectashield (Vector Laboratories, Burlingame, CA) and visualized using a Nikon microscope fitted with the appropriate filters.
RESULTS
PERK Is Not Essential for Inhibition of Cyclin D1 Translation during the UPR
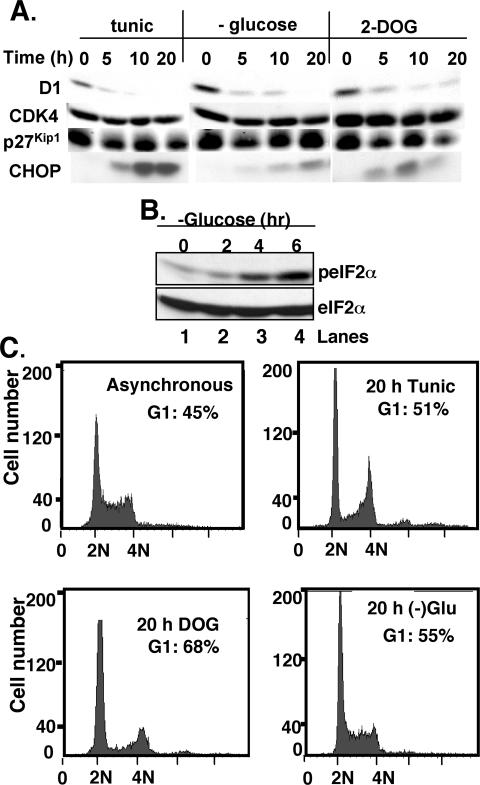
Cyclin D1 is regulated at the translational level after UPR activation with tunicamycin (Brewer et al., 1999; Brewer and Diehl, 2000). To ascertain that UPR induction and D1 loss are not a pleiotropic consequence of tunicamycin treatment, we assessed cyclin D1 accumulation and cell cycle progression after glucose restriction, a physiological trigger of the UPR (Yang et al., 2000; Fernandez et al., 2002). NIH-3T3 cells cultured in either glucose-free media, complete media supplemented with the nonhydrolysable glucose analogue 2-deoxyglucose (2-DOG), or as a control, tunicamycin, were harvested, and levels of cyclin D1 were assessed by immunoblot. Tunicamycin treatment resulted in the rapid loss of cyclin D1 by 5 h (Figure 1A, top). Rapid loss of cyclin D1 was also apparent in cells cultured in glucose-free media or 2-DOG (Figure 1A, top). Induction of CHOP (Figure 1A, bottom) and phosphorylation of eIF2α (Figure 1B) were noted under all three conditions, consistent with induction of the unfolded protein response. Cells treated as described above were also analyzed by FACS; after 20 h of stress, an accumulation of cells with a 2N DNA content was noted, consistent with G1 arrest (Figure 1C). No alterations in the accumulation of p27Kip1 (1A, second panel), CDK4 (third panel), or cyclin E (our unpublished data) were noted under these conditions. These data demonstrate that although tunicamycin treatment is a nonphysiological stress, the checkpoint it mimics is closely associated with that trigged by glucose restriction, and both stresses trigger cyclin D1 loss and growth arrest.
Figure 1.
Glucose restriction triggers UPR activation and cyclin D1 loss. (A) Cell lysates prepared from asynchronously proliferating NIH-3T3 cells treated with 0.5 μg/ml tunicamycin (lanes 1–4), glucose free-media (lanes 5–8), or 10 mM 2-deoxyglucose (lanes 9–12) were subjected to immunoblot analysis with antibodies directed toward cyclin D1 (top), CDK4 (second panel), p27Kip1 (third panel), or CHOP (bottom). (B) Lysates prepared from cells cultured in glucose-free media for the indicated intervals were subjected to Western analysis with antibodies directed toward either phosphorylated (top) or total eIF2α. (C) Untreated NIH-3T3 cells or those cultured in the presence of tunicamycin, 2-DOG, or glucose-free media for 20 h were fixed and stained with propidium iodide and analyzed by flow cytometry. The percentage of cells with a 2N DNA content is presented in the inset.
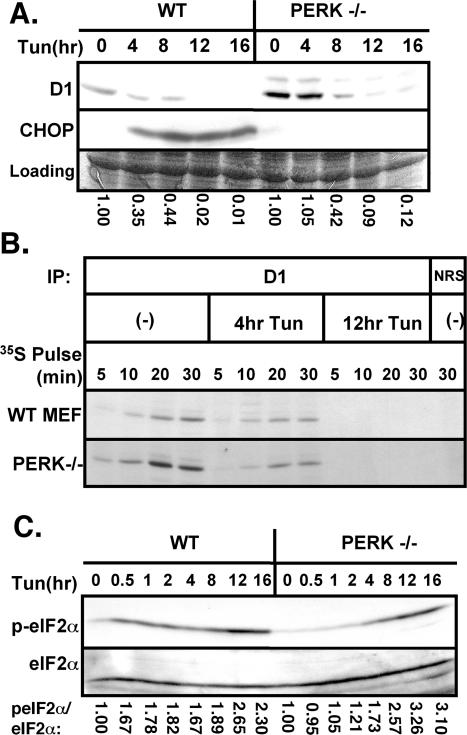
PERK is a primary effector of translational repression during ER stress (Harding et al., 1999; Zhang et al., 2002), and previous work has demonstrated that activation of PERK promotes inhibition of cyclin D1 translation and subsequent G1 phase arrest (Brewer and Diehl, 2000). However, cyclin D1 translational repression is not completely attenuated in cell lines expressing a dominant negative PERK (PERKΔC), suggesting the possibility that PERK-independent mechanisms might contribute to cyclin D1 translational inhibition (Brewer and Diehl, 2000). To directly address this idea, wild-type or PERK–/– fibroblasts were treated with 0.5 μg/ml tunicamycin for 0, 4, 8, 12, or 16 h. Western analysis revealed reduced cyclin D1 accumulation after exposure of PERK–/– fibroblasts to tunicamycin (Figure 2A, lanes 6–10), although loss was attenuated in comparison with wild-type mouse fibroblasts (lanes 1–5). In agreement with previous results (Harding et al., 2000a), we noted that CHOP induction was PERK dependent (Figure 2A, middle).
Figure 2.
PERK is not essential for repression of cyclin D1 translation during UPR activation. (A) Wild-type or PERK–/– fibroblasts were treated with 0.5 μg/ml tunicamycin for the indicated intervals. Equivalent amounts of total protein were subjected to Western blot analysis with antibodies specific for cyclin D1 or CHOP. (B) Lysates prepared from either wild-type or PERK–/– MEFs that had been pulsed for the indicated intervals with [35S]methionine after treatment with 0.5 μg/ml tunicamycin for 0, 4, or 12 h were subject to precipitation with normal rabbit antiserum (NRS) or a cyclin D1-specific mAb. Precipitated proteins were resolved by SDS-PAGE and visualized by autoradiography. (C) Lysates prepared as described in A were subjected to immunoblot analysis using antibodies directed toward serine 51 phosphorylated eIF2α or total eIF2α. The ratio of phosphorylated eIF2α relative to total eIF2α was determined by densitometry and was set to 1 in untreated cells within each cell type.
The loss of cyclin D1 in tunicamycin treated PERK–/– cells suggested that translation inhibition can occur in a PERK-independent manner. To test this notion, wild-type or PERK–/– fibroblasts were treated with 0.5 μg/ml tunicamycin for 4 or 12 h, or left untreated, and subsequently pulse labeled with [35S]methionine for 5, 10, 20, or 30 min (Figure 2B). Cyclin D1 was immunoprecipitated from whole cell lysates, resolved on a denaturing polyacrylamide gel, and visualized by autoradiography. Similar to wild-type cells, a significant reduction in radiolabeled cyclin D1 was detected in PERK–/– fibroblasts treated with tunicamycin compared with those that were left untreated (Figure 2B).
PERK is a member of the eIF2α kinase family, the other members of which include the heme-regulated kinase (HRI); the interferon-inducible, RNA-dependent protein kinase (PKR), and GCN2, which is activated by nutrient deprivation (Kaufman, 1999). We reasoned that in the absence of PERK, another eIF2α kinase might function redundantly to promote eIF2α phosphorylation and resultant inhibition of protein synthesis during ER stress, as demonstrated by recent data (Jiang et al., 2003, 2004). Wild-type or PERK–/– fibroblasts were treated with tunicamycin for the indicated intervals, or left untreated. Western blot analysis with a phosphospecific antibody to serine 51 of eIF2α revealed that eIF2α is indeed phosphorylated in fibroblasts lacking PERK (Figure 2C, top), although with delayed kinetics in comparison with wild-type fibroblasts. Immunoblotting with an antibody that recognizes both nonphosphorylated and phosphorylated eIF2α confirmed that equivalent amounts of total eIF2α were loaded (Figure 2C, bottom). The kinetics of eIF2α phosphorylation in PERK–/– fibroblasts correlate with the kinetics of cyclin D1 translational repression, suggesting that PERK-independent regulation of cyclin D1 may still be eIF2α dependent. Thus, PERK-independent mechanisms can promote eIF2α phosphorylation and inhibition of cyclin D1 protein synthesis during UPR activation.
Inhibition of cyclin D1 Is Dependent on eIF2α Phosphorylation
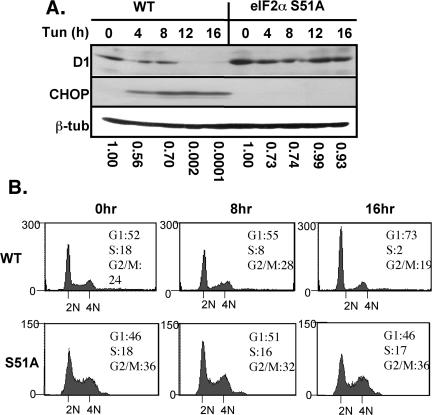
If UPR-dependent inhibition of cyclin D1 remains dependent upon eIF2α phosphorylation in the absence of PERK, we expected cyclin D1 translation to be stabilized if phosphorylation of eIF2α was prevented. To address this hypothesis, wild-type cells or those harboring a homozygous knockin of a nonphosphorylatable eIF2α allele (S51A) (Scheuner et al., 2001) were treated with tunicamycin for 0, 4, 8, 12, or 16 h. Western blot analysis revealed that cyclin D1 protein levels are stabilized in eIF2α S51A fibroblasts (Figure 3A, lanes 6–10) in comparison with wild-type fibroblasts (lanes 1–5). Levels of β-tubulin were assessed by immunoblotting to confirm equivalent amounts of protein (Figure 3A, bottom). CHOP induction was also ablated in the eIF2α S51A fibroblasts (Figure 3A, middle).
Figure 3.
Translational inhibition of cyclin D1 and consequent cell cycle arrest are dependent upon eIF2α phosphorylation. (A) Wild-type or eIF2α S51A MEFs treated with 0.5 μg/ml tunicamycin for the indicated intervals were subject to Western blot analysis with antibodies specific for cyclin D1, CHOP, or β-tubulin. (B) FACS analysis of wild-type or eIF2α S51A MEFs treated with 0.5 μg/ml tunicamycin for the indicated intervals.
The continued synthesis of cyclin D1 in the eIF2α S51A fibroblasts suggested that these cells should be refractory to UPR-induced G1 arrest (Brewer et al., 1999). Asynchronous populations of wild-type or eIF2α S51A fibroblasts were treated with 0.5 μg/ml tunicamycin for 0, 8, or 16 h, and cell cycle progression was monitored by FACS. Although wild-type fibroblasts lost a significant portion of S-phase cells with a concomitant increase in G1-phase cells by 16 h of UPR activation (Figure 3B, top), eIF2α S51A fibroblasts failed to arrest in G1 phase and maintained a steady S-phase population even after prolonged exposure to ER stress (Figure 3B, bottom), consistent with the stabilization of cyclin D1 protein synthesis in these cells.
To further investigate the functional consequences of cyclin D1 stabilization in eIF2α S51A fibroblasts, we evaluated the phosphorylation status of the primary cyclin D1 substrate, the retinoblastoma tumor suppressor protein pRb. Serine 780 of Rb is a specific target of the cyclin D1/CDK4 holoenzyme (Kitagawa et al., 1996). ER stress inhibited Rb phosphorylation in wild-type cells, whereas in eIF2α S51A cells, levels of phospho-serine 780 remained elevated even at 16 h of UPR activation, consistent with the stabilization of cyclin D1 and the cyclin D1-dependent kinases in these cells (our unpublished data). We conclude that phosphorylation of eIF2α is essential for the specific inhibition of cyclin D1 translation after UPR activation. Furthermore, inhibition of cyclin D1 translation is essential for cell cycle arrest after initiation of an ER stress response.
Functional Cooperativity among eIF2α Kinases
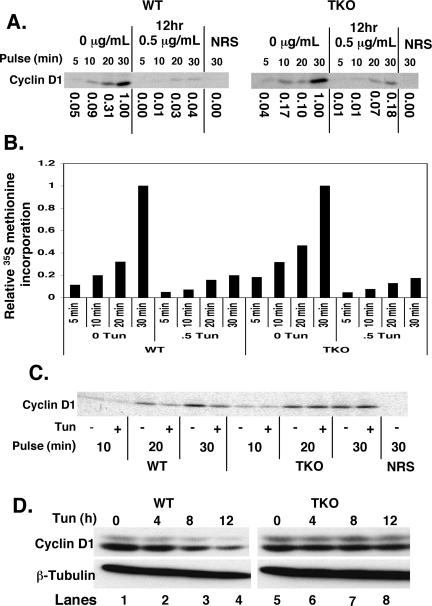
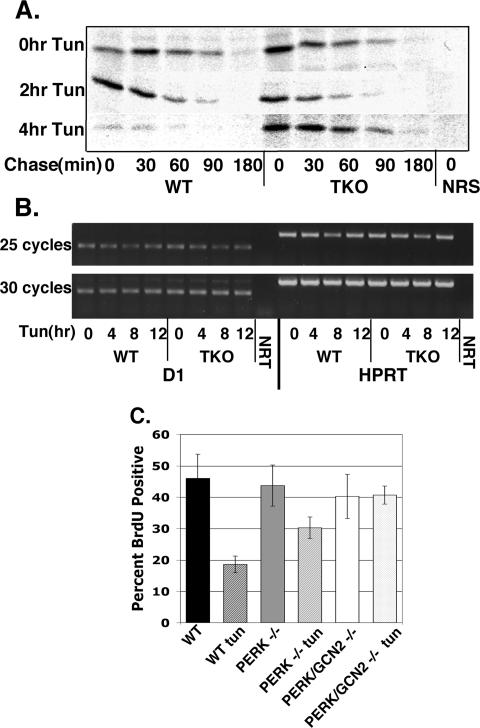
The results presented above demonstrate that during the UPR, regulation of cyclin D1 translation is determined by the phosphorylation status of eIF2α. The fact that cyclin D1 translation is repressed in PERK null cells demonstrates that other eIF2α kinases can function in its absence. Of the four known eIF2α kinases, GCN2 and PKR are the most likely to serve in this capacity because expression of the fourth eIF2α kinase, HRI, is restricted to erythroid cells. We assessed cyclin D1 protein synthesis in fibroblasts isolated from mice lacking PERK, GCN2, and PKR (TKO cells) (Jiang et al., 2004). Wild-type or TKO cells were treated with 0.5 μg/ml tunicamycin for 12 h or left untreated. Cells were subsequently pulsed with [35S]methionine for 5, 10, 20, or 30 min; samples were normalized to total protein; and cyclin D1 levels were assessed after precipitation with a cyclin D1-specific monoclonal antibody (mAb) (Brewer et al., 1999). As demonstrated previously (Brewer et al., 1999; Brewer and Diehl, 2000), cyclin D1 synthesis is inhibited by tunicamycin treatment in wild-type fibroblasts (Figure 4A). In contrast, we observed that cyclin D1 synthesis remained elevated in TKO fibroblasts (Figure 4A). Although cyclin D1 synthesis is reduced in TKO cells after tunicamycin treatment (5-fold), this repression of cyclin D1 synthesis was significantly reduced relative to that observed in wild-type cells (∼25-fold) (Figure 4A). Analysis of global protein translation revealed a fivefold decrease in [35S]methionine incorporation into newly synthesized polypeptides in both wild-type and TKO cells (Figure 4B). Similar results were obtained with the eIF2α S51A fibroblasts (our unpublished data). These results demonstrate that eIF2α-independent mechanisms contribute to a “bulk” or global translation inhibition after UPR activation; the phosphorylation of eIF2α further contributes specificity to translation inhibition further reducing synthesis of specific transcripts such as cyclin D1.
Figure 4.
Continued cyclin D1 protein synthesis after treatment of cells lacking PERK/GCN2/PKR with tunicamycin. (A) PERK/GCN2/PKR triple knockout or wild-type fibroblasts were treated with the 0.5 μg/ml tunicamycin before pulse label with [35S]methionine. Cyclin D1 synthesis was assessed after metabolic labeling by precipitation with a cyclin D1-specific mAb or normal rabbit antiserum (NRS). (B) Total protein synthesis was monitored by precipitation of equivalent concentrations of total cellular lysates with TCA. Precipitated proteins were spotted onto filter paper, and new protein was quantified by scintillation counting. (C) TKO or wild-type fibroblasts were treated with 0.5 μg/ml tunicamycin for 6 h before a pulse with [35S]methionine for the indicated intervals. Protein concentrations were normalized to TCA-precipitable counts, and equivalent counts for each pulse length were immunoprecipitated for cyclin D1. (D) TKO or wild-type fibroblasts were treated with 0.5 μg/ml tunicamycin. Cyclin D1 and β-tubulin levels were assessed by Western analysis after separation of cell lysates on a denaturing polyacrylamide gel.
To illustrate the specificity of eIF2α phosphorylation for specific transcripts such as cyclin D1, we performed an additional in vivo labeling experiment in which newly synthesized cyclin D1 protein levels were normalized to newly synthesized total protein, as determined by [35S]methionine incorporation. Wild-type (WT) and TKO cells were treated with 0.5 μg/ml tunicamycin for 6 h or left untreated. Cells were subsequently pulsed with [35S]methionine for 10, 20, or 30 min, and new protein synthesis was determined by scintillation counting of TCA-precipitable counts. Protein content of samples from each time point was normalized to TCA-precipitable counts, and cyclin D1 was immunoprecipitated. As illustrated in Figure 4C, cyclin D1 synthesis is repressed in wild-type cells beyond the level of bulk, eIF2α-independent total translational repression. In contrast, TKO cells, lacking eIF2α phosphorylation, do not repress cyclin D1 translation further than the level of bulk repression, allowing cyclin D1 levels to remain elevated in these cells.
Although bulk eIF2α-independent translation inhibition impacts total protein synthesis, we predicted that it would not be sufficient to eliminate D1. This was evident when equivalent concentrations of total protein were resolved and processed for immunoblot with either a β-tubulin antibody or cyclin D1 (Figure 4D).
To eliminate the possibility that cyclin D1 accumulation in TKO cells relative to wild-type cells reflects decreased protein degradation, we conducted pulse-chase experiments in both cell lines under normal conditions or after tunicamycin treatment. WT and TKO cells were left untreated or treated with 0.5 μg/ml tunicamycin for 2 or 4 h. Cells were pulse labeled with [35S]methionine for 30 min followed by a “chase” in media containing excess cold methionine 0, 30, 60, 90, or 180 min. The rate of cyclin D1 turnover remained unchanged in either cell line upon treatment with tunicamycin (Figure 6A).
Figure 6.
UPR activation does not affect cyclin D1 stability or transcription. (A) TKO or wild-type fibroblasts were treated with tunicamycin for 0, 2, or 4 h before a 30-min pulse with [35S]methionine. Cells were “chased” in media containing excess cold methionine for the indicated intervals. Cyclin D1 was precipitated with a D1 mAb and visualized by autoradiography. (B) TKO or wild-type fibroblasts were treated with 0.5 μg/ml tunicamycin for the indicated intervals. Total RNA was purified, reverse transcribed, and messages specific for cyclin D1 or HPRT were amplified by PCR. NRT indicates no RT input control. (C) Wild-type, PERK–/– and PERK/GCN2–/– fibroblasts proliferating on glass coverslips were left untreated or treated with tunicamycin (0.5 μg/ml) for 16 h. During the last 1.5 h, cells were pulsed with BrdU; BrdU incorporation was determined by immunofluorescence. Bars represent SD determined from three independent experiments.
We also considered the possibility that elevated cyclin D1 levels in TKO cells reflects differential mRNA accumulation. WT and TKO cells were treated with tunicamycin for 4, 8, or 12 h or left untreated. Total RNA was isolated, reverse transcribed, and amplified by PCR using primers specific for cyclin D1, or HPRT as a control. We detected no difference in cyclin D1 transcript level in WT or TKO cells over the course of tunicamycin treatment (Figure 6B). Similar results were observed by Northern analysis (our unpublished data).
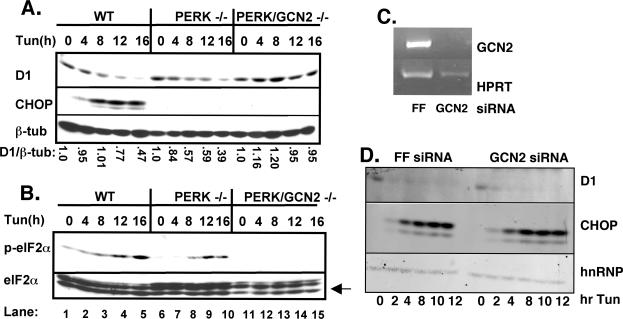
We next considered whether GCN2 or PKR plays the primary role in regulating protein synthesis cooperatively with PERK or whether they might participate in an equivalent manner. Because GCN2 responds to nutrient limitation, we first concentrated on this protein kinase. Fibroblasts isolated from wild-type, PERK–/–, or PERK/GCN2–/– embryos were treated with tunicamycin for 0, 4, 8, 12, or 16 h. Western analysis revealed that cyclin D1 protein is lost in both wild-type and PERK–/– fibroblasts, with slightly delayed kinetics in the PERK–/– (Figure 5A, top). In contrast, cyclin D1 levels were maintained in PERK/GCN2–/– fibroblasts (Figure 5A, top). β-Tubulin was assessed to confirm equivalent loading of total cellular protein (Figure 5A, bottom), and levels of CHOP were assessed (Figure 5A, middle) to verify UPR activation.
Figure 5.
GCN2 cooperates with PERK in the inhibition of cyclin D1 translation after ER stress. Whole cell lysates prepared from wild-type, PERK–/–, or PERK/GCN2–/– cells treated with 0.5 μg/ml tunicamycin for the indicated intervals were resolved on a denaturing polyacrylamide gel and subject to Western blot analysis with antibodies specific for cyclin D1, CHOP, or β-tubulin (A) and phospho-eIF2α, total eIF2α, or CHOP (B). (C and D) Wild-type fibroblasts were infected with short hairpin vectors targeting GCN2. Forty-eight hours postinfection, cells were treated with 2 μg/ml tunicamycin for the indicated intervals and levels of cyclin D1, CHOP, and hnRNPK were determined by Western analysis, and knockdown of GCN2 was assessed by RT-PCR.
The kinetics of eIF2α phosphorylation in PERK–/– fibroblasts correlates with the kinetics of cyclin D1 loss in these cells. The maintenance of cyclin D1 levels in PERK/GCN2–/– fibroblasts suggests that GCN2 might function as the redundant eIF2α kinase during UPR activation. To explore this possibility further, we evaluated the phosphorylation status of eIF2α in PERK/GCN2–/– fibroblasts. Whole cell lysates (see above) were subject to immunoblot analysis using a phosphospecific antibody to serine 51 of eIF2α. In agreement with previous data (Figure 2C; Jiang et al., 2003, 2004), eIF2α is phosphorylated in PERK–/– fibroblasts with delayed kinetics (Figure 5B, top, lanes 5–10) in comparison with wild-type cells (lanes 1–5). However, eIF2α phosphorylation is completely abrogated in PERK/GCN2–/– fibroblasts (Figure 5B, top, lanes 11–15). Western analysis with an antibody that recognizes both nonphosphorylated and phosphorylated eIF2α confirmed equivalent amounts of total eIF2α(Figure 5B, bottom). CHOP was induced in wild-type cells but not in the cell lines lacking PERK. GCN2 is not considered the primary eIF2α kinase activated by ER stress and thus is unlikely to contribute to cyclin D1 translational regulation in cells expressing functional PERK. To test this hypothesis, we infected wild-type murine fibroblasts with retrovirus encoding GCN2-specific short hairpins (Paddison et al., 2002; Paddison and Hannon, 2002). Because antibodies that recognize GCN2 are not available, we confirmed GCN2 knockdown by RT-PCR (Figure 5C). The kinetics and magnitude of cyclin D1 loss after initiation of ER stress were essentially identical in cells infected with either a control virus (short hairpin targeting luciferase) or GCN2 (Figure 5D). Likewise, knockdown of GCN2 attenuated cyclin D1 loss in a PERK–/– background, confirming data from the PERK/GCN2-deficient MEFs (our unpublished data). These results demonstrate that although GCN2 compensates for PERK loss, in wild-type cells it does not make substantial contributions to translational regulation after ER stress.
Although previous work has determined that PKR is not essential for translational repression during the UPR (Harding et al., 1999), these experiments were not performed in a PERK null background. Because mice harboring deletion of both PERK and PKR are not currently available, we used a documented PKR dominant negative allele (Donze et al., 1995) to assess its contribution to the regulation of cyclin D1 translation after UPR activation. PERK–/– cells were transiently transfected with an empty vector or a plasmid encoding a dominant negative PKR (K296D) and subsequently treated with tunicamycin. Inactivation of PKR did not inhibit cyclin D1 loss during an ER stress response (our unpublished data). These results suggest that unlike GCN2, PKR may not function as a redundant eIF2α kinase to promote loss of cyclin D1 during UPR activation, and this conclusion is supported by our demonstration that eIF2α phosphorylation (Figure 5B) and cyclin D1 loss are abrogated in PERK/GCN2–/– double null fibroblasts (Figure 5A).
To explore the possibility that PERK/GCN2–/– fibroblasts are refractory to UPR-dependent cell cycle arrest, asynchronous populations of wild-type, PERK–/–, or PERK/GCN2–/– fibroblasts were treated with tunicamycin for 16 h or left untreated. Cell cycle arrest was monitored by determining the fraction of cells able to incorporate BrdU during the final 1.5 h of tunicamycin treatment. UPR activation resulted in a significant reduction in S-phase populations in both wild-type and PERK–/– fibroblasts (Figure 6C). In contrast, the percentage of BrdU-positive PERK/GCN2–/– cells did not change over this time course (Figure 6C). These data are in agreement with Western blot analysis and support the theory that a redundant eIF2α kinase, GCN2, is necessary for inhibition of cyclin D1 translation and subsequent cell cycle arrest during UPR activation in the absence of PERK.
DISCUSSION
Activation of the UPR leads to the rapid repression of protein translation (Kaufman, 1999). Among the proximal targets of translation inhibition is the cell cycle regulatory protein cyclin D1. Cyclin D1 loss after exposure of cells to ER stress is sufficient to trigger arrest in G1 phase of the cell cycle (Brewer et al., 1999; Brewer and Diehl, 2000). G1 arrest during the UPR likely contributes to cellular adaptation by serving as a period during which cells attempt to reestablish homeostatic conditions or commit to apoptosis. Previous work revealed that overexpression of wild-type PERK, which results in its activation without an ER stress-promoting agent, inhibits cyclin D1 translation and induces cell cycle arrest (Brewer and Diehl, 2000). The capacity of PERK to regulate cyclin D1 protein synthesis was linked to phosphorylation of the PERK substrate eIF2α. The fact that cyclin D1 translational repression is not completely attenuated in cell lines expressing a dominant negative PERK (PERKΔC) suggests that PERK-independent mechanisms may contribute to repression of cyclin D1 translation (Brewer and Diehl, 2000). Indeed, we have found that UPR activation still triggers loss of cyclin D1 in cells harboring a targeted deletion of PERK. Thus, although PERK can regulate cyclin D1 translation (Brewer and Diehl, 2000), alternative pathways or PERK-like kinases must be used in cells deficient for PERK.
Cyclin D1 Loss Requires Phosphorylation of eIF2α
Translational regulation is most commonly achieved through modulation of eIF2α phosphorylation or via modulation of the activity of the cap binding complex eIF4F (Sonenberg and Gingras, 1998; Sonenberg and Dever, 2003). To ascertain that regulation of cyclin D1 during an ER stress response reflects regulation of eIF2α, we used cells isolated from mice harboring the nonphosphorylatable eIF2α S51A (Scheuner et al., 2001). Indeed, cyclin D1 protein levels were maintained in eIF2α S51A knockin fibroblasts after ER stress, demonstrating that cyclin D1 loss was absolutely dependent upon eIF2α phosphorylation. Furthermore, eIF2α S51A fibroblasts failed to arrest in G1 phase during ER stress, providing further support for the requirement for eIF2α phosphorylation in cyclin D1 regulation.
Because PERK is a member of the eIF2α kinase family, we considered it likely that another family member might cooperate and thereby promote eIF2α phosphorylation and attenuation of cyclin D1 synthesis (Dever et al., 1993). Two candidates for a redundant eIF2α kinase include PKR and GCN2, which are activated by double-stranded RNA and nutrient deprivation, respectively. Our data suggest that GCN2 but not PKR compensates in the absence of PERK to trigger inhibition of cyclin D1 translation. In fibroblasts lacking both PERK and GCN2, cyclin D1 protein levels are maintained, and these cells are refractory to UPR-triggered cell cycle arrest. In addition, phosphorylation of eIF2α was completely abrogated in these cells. When a dominant negative PKR was overexpressed in PERK–/– fibroblasts, there was no rescue of cyclin D1 protein levels; the kinetics of cyclin D1 loss matched those of cells transfected with empty vector. Consistent with our results, the activation of additional eIF2α kinases during the UPR has been reported previously (Jiang et al., 2003, 2004).
A striking implication of our results lies within the continued inhibition of protein translation upon treatment of eIF2αA/A or TKO cells with ER stress-inducing agents. Although phosphorylation of eIF2α has been considered the major effector of translation inhibition during ER stress, these results demonstrate that other pathways of regulation are indeed present. These results demonstrate that eIF2α inactivation is not absolutely necessary for global translation attenuation. Yet, eIF2α phosphorylation is absolutely required for effective inhibition of cyclin D1 synthesis and induction of the prosurvival transcription factor ATF4. Thus, our results suggest that although eIF2α phosphorylation may contribute to decreased protein load during stress, perhaps the more important output lies within its modulation of a specific subset of transcripts whose increased or decreased translation specifically contributes to a cells ability to adapt to stress. Although we have not elucidated the mechanism of translation inhibition that occurs in cells wherein regulation of eIF2α is eliminated, the most probable mechanism likely involves inactivation of the eIF4F complex that binds to the 7-methyl guanosine mRNA cap. Indeed, we have noted decreased phosphorylation, and thus decreased activity, of 4BP1, a cellular inhibitor of eIF4E, after exposure of cells to tunicamycin (our unpublished data).
Cross-Talk among the eIF2α Kinases
Our data demonstrate that both PERK and GCN2 contribute to the inhibition of translation initiation after ER stress. Deletion of both kinases is necessary and sufficient to prevent eIF2α phosphorylation, loss of cyclin D1, and subsequent cell cycle arrest after induction of the UPR. Although cross-talk between signaling cascades is not without precedence, a question remains regarding how a cell communicates the accumulation of misfolded proteins in the ER to the cytoplasmic GCN2 kinase in the absence of the ER-resident kinase PERK. Many of the cellular stresses that activate the UPR could potentially interfere with cellular homeostasis on multiple levels and activate additional signaling pathways. For example, glucose deprivation, which results in improper protein glycosylation and thus UPR activation, likely influences ATP levels in the cell. The mammalian target of rapamycin mTOR has been identified as a sensor of cellular ATP levels (Dennis et al., 2001). Therefore, it is possible that during ER stress, mTOR responds to the decreased ATP concentration in the cell, and as a consequence, either directly or indirectly activates GCN2 (Cherkasova and Hinnebusch, 2003). In the same way, as yet unidentified proteins that detect ER stress-dependent alterations in cell homeostasis (e.g., Ca2+ and ATP) could activate GCN2. Alternatively, other proximal ER-resident UPR effecters besides PERK (e.g., Ire1) might signal to GCN2 to promote eIF2α phosphorylation, directly or indirectly. Identification of the effector of GCN2 activation remains an important question to be addressed.
UPR-dependent cell cycle arrest is considered an opportunity for cells to restore cellular homeostasis (Niwa and Walter, 2000). What are the requirements for a cell to be able to reenter the cell cycle? Certainly, contributions from chaperone proteins to help remedy protein misfolding are essential (Kaufman, 1999). However, the cellular stress threshold below which a cell can reestablish homeostasis and above which it will initiate apoptosis remains a curious phenomenon. There may in fact be a link between cell cycle arrest and initiation of apoptosis during the UPR (Brewer et al., 1999). Given that the UPR has been implicated in the development and progression of a variety of diseases (Kaufman et al., 2002), understanding the biochemical events that occur during ER stress will reveal molecules that can be targeted to potentially treat and to ideally cure individuals who suffer from UPR-based pathologies.
Acknowledgments
We thank D. Cavener for providing the PERK–/–, PERK/GCN2–/–, and PERK/GCN2/PKR–/– MEFs; H. Harding and D. Ron for the PERK–/–fibroblasts; D. Scheuner and R. Kaufman for providing the eIF2α S51A knockin fibroblasts; and R. Woolery for technical assistance. This work was supported by grants from the National Institutes of Health (P01 CA104838) and the Abramson Family Cancer Research Institute (to J.A.D.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–03–0268) on September 21, 2005.
References
- Brewer, J. W., and Diehl, J. A. (2000). PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl. Acad. Sci. USA 97, 12625–12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, J. W., Hendershot, L. M., Sherr, C. J., and Diehl, J. A. (1999). Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. USA 96, 8505–8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova, V. A., and Hinnebusch, A. G. (2003). Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 17, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. S., Shamu, C. E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Cullinan, S. B., and Diehl, J. A. (2004). PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following ER stress. J. Biol. Chem. 279, 20076–20087. [DOI] [PubMed] [Google Scholar]
- Cullinan, S. B., Zhang, D., Hannink, M., Arvisais, E., Kaufman, R. J., and Diehl, J. A. (2003). Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell Biol. 23, 7198–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, P. B., Jaeschke, A., Saitoh, M., Fowler, B., Kozma, S. C., and Thomas, G. (2001). Mammalian TOR: a homeostatic ATP sensor. Science 294, 1102–1105. [DOI] [PubMed] [Google Scholar]
- Dever, T. E., Chen, J. J., Barber, G. N., Cigan, A. M., Feng, L., Donahue, T. F., London, I. M., Katze, M. G., and Hinnebusch, A. G. (1993). Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc. Natl. Acad. Sci. USA 90, 4616–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, O., Jagus, R., Koromilas, A. E., Hershey, J. W., and Sonenberg, N. (1995). Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 14, 3828–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, J., Bode, B., Koromilas, A., Diehl, J. A., Krukovets, I., Snider, M. D., and Hatzoglou, M. (2002). Translation mediated by the internal ribosome entry site of the cat-1 mRNA is regulated by glucose availability in a PERK kinase-dependent manner. J. Biol. Chem. 277, 11780–11787. [DOI] [PubMed] [Google Scholar]
- Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., and Ron, D. (2000a). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding, H. P., Zhang, Y., Bertolotti, A., Zeng, H., and Ron, D. (2000b). Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904. [DOI] [PubMed] [Google Scholar]
- Harding, H. P., Zhang, Y., and Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274. [DOI] [PubMed] [Google Scholar]
- Haze, K., Yoshida, H., Yanagi, H., Yura, T., and Mori, K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H. Y., Wek, S. A., McGrath, B. C., Lu, D., Hai, T., Harding, H. P., Wang, X., Ron, D., Cavener, D. R., and Wek, R. C. (2004). Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24, 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H. Y., Wek, S. A., McGrath, B. C., Scheuner, D., Kaufman, R. J., Cavener, D. R., and Wek, R. C. (2003). Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-κB in response to diverse cellular stresses. Mol. Cell. Biol. 23, 5651–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, R. J. (1999). Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211–1233. [DOI] [PubMed] [Google Scholar]
- Kaufman, R. J., Scheuner, D., Schroder, M., Shen, X., Lee, K., Liu, C. Y., and Arnold, S. M. (2002). The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell Biol. 3, 411–421. [DOI] [PubMed] [Google Scholar]
- Kitagawa, M., et al. (1996). The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15, 7060–7069. [PMC free article] [PubMed] [Google Scholar]
- Koumenis, C., Naczki, C., Koritzinsky, M., Rastani, S., Diehl, A., Sonenberg, N., Koromilas, A., and Wouters, B. G. (2002). Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol. Cell. Biol. 22, 7405–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., Zhu, H., Morishima, N., Li, E., Xu, J., Yankner, B. A., and Yuan, J. (2000). Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403, 98–103. [DOI] [PubMed] [Google Scholar]
- Niwa, M., and Walter, P. (2000). Pausing to decide. Proc. Natl. Acad. Sci. USA 97, 12396–12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison, P. J., Caudy, A. A., and Hannon, G. J. (2002). Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA 99, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison, P. J., and Hannon, G. J. (2002). RNA interference: the new somatic cell genetics? Cancer Cell 2, 17–23. [DOI] [PubMed] [Google Scholar]
- Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R. J. (2001). Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176. [DOI] [PubMed] [Google Scholar]
- Shi, Y., Vattem, K. M., Sood, R., An, J., Liang, J., Stramm, L., and Wek, R. C. (1998). Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell Biol. 18, 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg, N., and Dever, T. E. (2003). Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 13, 56–63. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N., and Gingras, A. C. (1998). The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10, 268–275. [DOI] [PubMed] [Google Scholar]
- Sood, R., Porter, A. C., Ma, K., Quilliam, L. A., and Wek, R. C. (2000). Pancreatic eukaryotic initiation factor-2alpha kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to endoplasmic reticulum stress. Biochem. J. 346, 281–293. [PMC free article] [PubMed] [Google Scholar]
- Tirasophon, W., Welihinda, A. A., and Kaufman, R. J. (1998). A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12, 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. Z., Harding, H. P., Zhang, Y., Jolicoeur, E. M., Kuroda, M., and Ron, D. (1998). Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17, 5708–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R., Wek, S. A., and Wek, R. C. (2000). Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell Biol. 20, 2706–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., McGrath, B., Li, S., Frank, A., Zambito, F., Reinert, J., Gannon, M., Ma, K., McNaughton, K., and Cavener, D. R. (2002). The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22, 3864–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]