Abstract
Inhibition of ubiquitin-proteasome pathway has been shown to be a promising strategy for the treatment of inflammation and cancer. Here, we show that proteasome inhibitors MG132, PSI-1, and lactacystin induce COX-2 expression via enhancing gene transcription rather than preventing protein degradation in the human alveolar NCI-H292 and A549, and gastric AGS epithelial cells. NF-IL6 and CRE, but not NF-κB elements on the COX-2 promoter were involved in the gene transcription event. The binding of CCAAT/enhancer binding protein (C/EBP)β and C/EBPδ to the CRE and NF-IL6 elements, as well as the recruitment of CBP and the enhancement of histone H3 and H4 acetylation on the COX-2 promoter was enhanced by MG132. However, it did not affect the total protein levels of C/EBPβ and C/EBPδ. MG132-induced DNA-binding activity of C/EBPδ, but not C/EBPβ was regulated by p38, PI3K, Src, and protein kinase C. Small interfering RNA of C/EBPδ suppressed COX-2 expression, further strengthening the role of C/EBPδ in COX-2 gene transcription. In addition, the generation of intracellular reactive oxygen species (ROS) in response to MG132 contributed to the activation of MAPKs and Akt. These findings reveal that the induction of COX-2 transcription induced by proteasome inhibitors requires ROS-dependent protein kinases activation and the subsequent recruitments of C/EBPδ and CBP.
INTRODUCTION
Cyclooxygenase (COX) is a rate-limiting enzyme that catalyzes the biosynthesis of prostaglandins and thromboxanes from arachidonic acid. These bioactive metabolites play an important role in the regulation of physiological process, such as mucosa secretion and smooth muscle contraction, and in the regulation of pathological condition, such as allergic diseases and rheumatoid arthritis (Smith, 1992; Goetzl et al., 1995). Two isoforms of cyclooxygenase, COX-1 and COX-2, have been identified (Fletcher et al., 1992; Hla and Neilson, 1992; Kraemer et al., 1992). COX-1 is constitutively expressed in most human tissues and functions as an housekeeping gene, whereas COX-2 is an immediate-early gene and is induced by oncogenes, growth factors, cytokines, endotoxins, and phorbol esters (Xie et al., 1994; Arias-Negrete et al., 1995; Inoue et al., 1995). Overexpression of COX-2 has been related to chronic inflammation, angiogenesis, and carcinogenesis (Tsuji et al., 2001). COX-2 expression is tightly regulated at both the transcriptional and the posttranscriptional levels. The cis-acting elements in the 5′-flanking promoter region of COX-2 contains a canonical TATA-box and multiple regulatory elements of NF-κB, NF-IL6 (CCAAT/enhancer-binding protein; C/EBP), and cAMP response element (CRE; Kosaka et al., 1994; Tazawa et al., 1994). The activation of intracellular signaling pathway induces the recruitment of specific transcription factors to these elements and triggers COX-2 expression.
Ubiquitin-proteasome system regulates diverse shortlived proteins degradation and homeostasis in eukaryotic cells. The 26S proteasome is a large multisubunit proteolytic enzyme complex playing a pivotal role in preventing accumulation of misfolded or damaged proteins involved in the cell cycle, apoptosis, and transcription, such as p53, p27(Kip1), cyclins, c-jun, IκB, Bax, and Bcl-2 family members (Ciechanover, 1998). Disorders in ubiquitin-dependent proteolysis have been implicated in the pathogenesis of a variety of human diseases, including cancer, inflammation, neurodegenerative diseases, and metabolic disorders (Schwartz and Ciechanover, 1999). Therefore, manipulating the ubiquitin-proteasome process has become a potential strategy for the treatment of these diseases. Recent reports have shown the inhibition of NF-κB activation and the induction of apoptosis by proteasome inhibitors in a broad range of cancer cells. These effects may contribute to the anti-inflammation and anti-tumor activity of proteasome inhibitors, which can thus serve as promising novel anticancer agents (Delic et al., 1998; Hideshima et al., 2001; Dai et al., 2003). Among them, the dipeptide boronic acid, PS-341 (Bortezomib) has been approved for the treatment of refractory multiple myeloma.
In addition to the regulation of protein turnover via ubiquitin-proteasome pathway, the proteasome inhibitor MG132 had been reported to activate activator protein-1 (AP-1) through the mitogenic activated protein kinases (MAPKs) pathway and induce the expressions of several inflammatory genes, such as monocyte chemoattractant protein-1 (MCP-1), IL-8, and IL-6 (Nakayama et al., 2001; Shibata et al., 2002; Joshi-Barve et al., 2003). However, the precise mechanism by which proteasome inhibitor triggers the expression of inflammatory genes is not fully clear. With this regard, the purpose of this study is to investigate the effect of proteasome inhibitors, MG132, PSI-1, and lactacystin on the transcriptional regulation of COX-2 expression.
In this study, we found the induction of COX-2 expression by proteasome inhibitors in human alveolar and gastric epithelial cells. Further exploration of the transcriptional regulation demonstrated that MG132 enhanced the bindings of C/EBPβ and C/EBPδ to the CRE and NF-IL6 elements on COX-2 promoter. Chromatin remodeling by recruitment of CBP to the promoter leading to an increase in histone H3 and H4 acetylation was also seen. We further demonstrated that reactive oxygen species (ROS) production in response to MG132 mediated activations of PI3K, p38, Src, and PKC which up-regulate the binding of C/EBPδ to the CRE and NF-IL6 elements.
MATERIALS AND METHODS
Materials
Goat polyclonal antibodies specific for COX-2, actin, and rabbit polyclonal antibodies specific for ubiquitin, C/EBPβ, C/EBPδ, MAPKs (p44/42 MAPK, p38, and JNK), Akt, and CBP were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). T4 polynucleotide kinase and rabbit polyclonal antibodies specific for the phosphorylated form of MAPKs, Akt, and C/EBPβ (Thr-235) were purchased from New England Biolabs (Beverly, MA). Anti-acetylated histone H3 and H4 antibodies were from Upstate Biotechnology (Lake Placid, NY). RPMI 1640, fetal calf serum (FCS), penicillin, and streptomycin were obtained from Invitrogen (Carlsbad, CA). Poly (dI/dC), horseradish peroxidase-labeled donkey anti-goat or anti-rabbit secondary antibody and the ECL detection reagent were from Amersham Pharmacia Biotechnology (Piscataway, NJ). [γ-32P]ATP (3000 Ci/mmol) was from Dupont-New England Nuclear (Boston, MA). Protein A-Sepharose, N-acetylcysteine (NAC), and glutathione (GSH) were from Sigma-Aldrich (St. Louis, MO). Proteasome inhibitor 1 (PSI-1), lactacystin, MG132, PD98059, SB203580, SP600125, LY294002, PP2, and Ro 31-8220 were from Calbiochem (San Diego, CA). The luciferase assay kit was from Promega (Madison, WI). SuperFect and plasmid purification kit were from Qiagen (Valencia, CA).
Plasmids
The COX-2 promoter construct pGS459 (–459/+9) was a generous gift from L. H. Wang (University of Texas, Houston, TX). The site-mutation of COX-2 promoter constructs (–327/+59, KBM, ILM, CRM) were provided by Inoue H (National Cardiovascular Center Research Institute, Osaka, Japan). The CRE-luc plasmid was from Stratagene (La Jolla, CA). The dominant-negative (DN) mutant of p38 (T180A/T182F) was from J. Han (Scripps Research Institute, San Diego, CA), and Akt KA was from Dr. Klippel A (Chiron, Emeryville, CA).
Cell Culture
The human alveolar epithelial cell lines NCI-H292 and A549, and gastric epithelial cell line, AGS, were obtained from the American Type Culture Collection (Manassas, VA) and were cultured in the RPMI 1640 medium supplemented with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2. The cells were subcultured either in 12-well plates for transfection experiments, in 6-cm dishes for RNA extraction, in 10-cm dishes for cell extract preparations and chromatin immunoprecipitation experiments, or in 15-cm dishes for nuclear extraction.
Preparation of Total Cell Lysates and Nuclear Extracts
After pretreatment with various inhibitors for 30 min, cells were incubated with MG132 for the indicated time. Cells were then rapidly washed with phosphate-buffered saline (PBS) and lysed with ice-cold lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EGTA, 1 mM NaF, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 μg/ml leupeptin, 20 μg/ml aprotinin, 1 mM Na3VO4, 10 mM β-glycerophosphate, 5 mM Na-pyrophosphate, and 1% Triton X-100), as described previously (Chang et al., 2002).
Nuclear extracts were isolated as described previously (Chang et al., 2002). Briefly, cells were washed with ice-cold PBS and pelleted, and then the cell pellet was resuspended in a hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol [DTT], 0.5 mM PMSF, 1 mM NaF, and 1 mM Na3VO4), incubated for 15 min on ice, and then lysed by the addition of 0.5% Nonidet P-40 followed by vigorous vortexing for 10 s. The nuclei were pelleted and resuspended in extraction buffer (20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 1 mM NaF, and 1 mM Na3VO4), and the tube was vigorously shaken at 4°C for 15 min on a shaking platform. The nuclear extracts were then centrifuged, and the supernatants were aliquoted and stored at –80°C.
Western Blotting Analysis
After treatment with PSI-1, lactacystin, or MG132, total cell lysates or nuclear extracts were prepared and subjected to SDS-PAGE using 10% polyacrylamide gels. The proteins were transferred to a nitrocellulose membrane, which was then incubated successively at room temperature for 1 h with 0.1% milk in TTBS (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.05% Tween 20) for 1 h with indicated primary antibodies and for 30 min with horseradish peroxidase-labeled secondary antibody. After the each incubation, the membrane was washed extensively with TTBS. The immunoreactive bands were detected using enhanced chemiluminescence detection reagent and visualized using Hyperfilm-ECL (Amersham, Arlington Heights, IL). Quantitative data normalized with internal control were obtained using the computing densitometer and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
RT-PCR
Total RNA was isolated from NCI-H292 cells using TRIzol reagent (Invitrogen). The reverse transcription reaction was performed using 2 μg of total RNA that was reverse transcribed into cDNA using oligo(dT) primer, then amplified for 25 cycles using two oligonucleotide primers derived from a published COX-2 sequence (5′-TTCAAATGAGATTGTGGGAAAAT-3′ and 5′-AGATCATCTCTGCCTGAGTATCTT-3′) and two from a β-actin sequence (5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′ and 5′-CTAGAAGCATTTGCGGGGACGATGGAGGG-3′). Each PCR cycle was carried out for 30 s at 94°C, 30 s at 65°C, and 1 min at 70°C. The PCR products were subjected to electrophoresis on a 1% agarose gel. Quantitative data were obtained using the computing densitometer and ImageQuant software (Amersham Pharmacia Biosciences, Piscataway, NJ).
Coimmunoprecipitation
Cell lysates containing 400 μg of protein were incubated for 16 h at 4°C with 2 μg of anti-COX-2 or anti-ubiquitin antibodies, and then 40 μl of 50% protein A-agarose beads were added and mixed for 1 h at 4°C. The immunoprecipitates were collected and washed three times with lysis buffer without Triton X-100, and then Laemmli buffer was added and the samples were subjected to electrophoresis on 10% SDS-polyacrylamide gels. Western blot analysis was performed, as described above, using antibodies against COX-2 or ubiquitin.
Transient Transfection and Luciferase Activity Assay
NCI-H292 cells, grown in 12-well plates, were transfected with the human COX-2 pGS459 (–459/+9),–327/+59, KBM, ILM, CRM, or CRE/firefly luciferase (Luc) plasmid using SuperFect (Qiagen), according to the manufacturer's recommendations. Briefly, reporter DNA (1 μg) and β-galactosidase DNA (0.5 μg) were mixed with 0.75 μl of SuperFect in 0.9 ml of serum-free RPMI 1640. The plasmid pRK containing the β-galactosidase gene driven by the constitutively active SV40 promoter was used to normalize the transfection efficiency.
In experiments using DN mutants, cells were cotransfected with reporter (0.4 μg), β-galactosidase (0.1 μg), and 0.4 μg DN p38, Akt, or c-Src mutant or 0.4 μg pcDNA 3.1. After 10- to 15-min incubation at room temperature, the mixture was applied to the cells, and then 0.1 ml of FCS was added 8 h later. Twenty-four hours after transfection, the cells were treated with various inhibitors (as indicated) for 30 min, and then MG132 was added for 6 h. Cell extracts were prepared, and luciferase and β-galactosidase activities were measured. The luciferase activity was normalized to the β-galactosidase activity.
Electrophoretic Mobility Shift Assay
Oligonucleotides corresponding to the consensus sequences of NF-κB (5′-AGAGTGGGGACTACCCCCTCT-3′), NF-IL6 (5′-CGGCTTACGCAATTTTT-3′), and CRE (5′-TCATTTCGTCACATG-3′) of the human COX-2 promoter were synthesized, annealed, and end-labeled with [γ-32P]ATP using T4 polynucleotide kinase, and electrophoretic mobility shift assay (EMSA) was performed as described previously (Chen et al., 2000).
When supershift assays were performed, polyclonal antibodies specific for C/EBPβ, C/EBPδ, CREB-1, ATF-1, ATF-2, c-jun, and c-fos were added to the nuclear extracts for 30 min before the binding reaction, and the DNA/nuclear protein complexes were separated on a 4.5% polyacrylamide gel. Excess cold unlabeled NF-κB, NF-IL6, and CRE probe were used for competition assays.
DNA Affinity Protein-binding Assay (DAPA)
The biotinylated double-stranded oligonucleotides corresponding to the NF-IL6 sequences (5′-GCTTACGCAATTTTTTTAAG G-biotin-3′), and CRE sequences (5′-AAGAAACAGTCATTTCGTCACATGG-biotin-3′) of human COX-2 promoter were synthesized and annealed. Binding of transcription factors to the COX-2 promoter was assayed as described previously (Huang and Chen, 2005). The nuclear extract (400 μg) was precleared at room temperature for 1h with 20 μl of the 4% streptavidin-coated beads (Sigma) mixed with 50% slurry to reduce nonspecific binding. Precleared nuclear extract was incubated with 2 μg of biotinylated DNA oligonucleotides and 20 μl of 4% streptavidin agarose beads with 50% slurry in 400 μl PBS at room temperature for 1 h with shaking. Beads were then collected by centrifugation at 2000 rpm for 2 min and washed with cold PBS for three times. DNA/protein complexes bound to the beads were eluted with 30 μl of Laemmli sample buffer. Nuclear proteins were denatured by putting on dry bath at 95°C for 5 min and subjected to SDS-PAGE. Western blot analysis probed with specific anti-C/EBPβ or anti-C/EBPδ antibody was performed as described above. The quantitative data were obtained using the computing densitometer and ImageQuant software (Molecular Dynamics).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assay (ChIP) analysis was performed as described previously (Huang and Chen, 2005). DNA immunoprecipitated by C/EBPβ, C/EBPδ, CBP, or acetylated histone H3 or H4 antibodies was purified. The DNA was then extracted with phenol-chloroform. The purified DNA pellet was resuspended in H2O and subjected to PCR. To amplify the regions containing NF-IL6 or CRE element on the COX-2 promoter, PCR was performed with the following pairs of primers: NF-IL6, forward primer, 5′-GAAGCCAAGTGTCCTTCTGC-3′, and reverse primer, 5′-TTTTCCCTCCTCTCCCCTT A-3′; CRE, forward primer, 5′-TAAGGGGAGAGGAGGGAAAA-3′, and reverse primer, 5′-ACAATTGGTCGCTAACCGAG-3′. PCR products were then resolved by 1.5% agarose-ethidium bromide gel electrophoresis and visualized by UV.
Measurement of Intracellular ROS Generation
ROS generation was assessed using the oxidation-sensitive fluorescent probe 2′,7′-dichlorofluorescin diacetate (DCF). Briefly, NCI-H292 cells grown on glass coverslips in a 3.5-cm well were loaded with 10 μM DCF at 37°C for 1 h in the dark. Cells were replaced with serul-free phenol red-free DMEM and incubated with 25 μM MG132 or 200 μM H2O2 for 1 h in the present or absence of 20 mM NAC or GSH. Cells were observed and photographed with a fluorescence microscope (excitation and emission wavelength: 495 and 525 nm, respectively; Carl Zeiss, Oberkochen, Germany). The DCF fluorescence intensity were quantitated with ImageQuant software (Amersham Pharmacia Biosciences).
Small Interfering RNA Synthesis and Transfection
Complementary hairpin small interfering RNA (siRNA) oligonucleotides targeting C/EBPδ or scrambled siRNA sequence cloned in pSilencer 3.1-H1/neo (Ambion, Austin, TX) were kindly provided from Dr. W. C. Chang (National Cheng Kung University, Taiwan). These plasmids were transfected using SuperFect (Qiagen) as described above.
Statistical Analysis
Data were analyzed using Student's t test. P values < 0.05 were considered significant.
RESULTS
COX-2 Expression Induced by Proteasome Inhibitors and the Involvements of NF-IL6 and CRE Elements
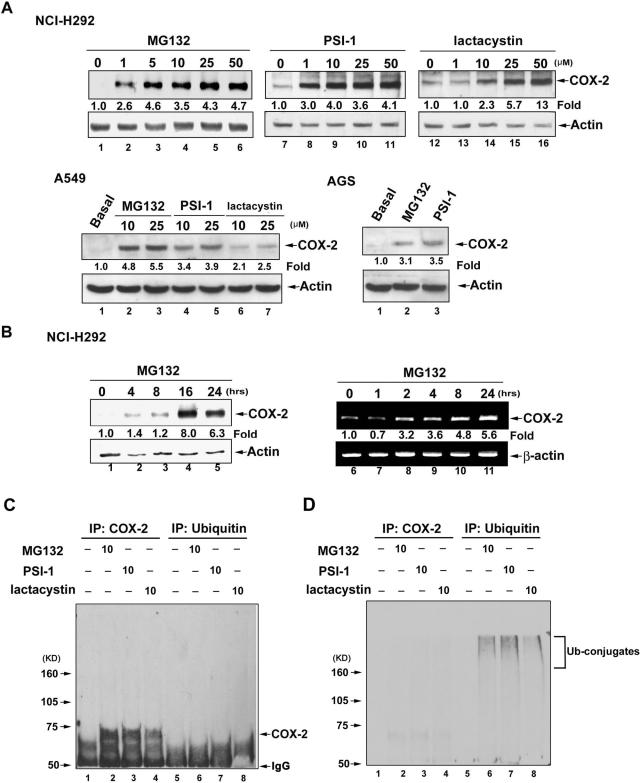
Three different proteasome inhibitors, MG132, PSI-1, and lactacystin induced COX-2 expression in a concentration-dependent manner in NCI-H292, A549, and AGS cells (Figure 1A). When NCI-H292 cells were treated with 25 μM MG132 for various time, the induction of COX-2 expression was increased significantly at 4 h and maximally at 16 h (Figure 1B, lanes 1–5). To further determine whether this induction occurred at the mRNA expression level, the time course of COX-2 mRNA expression induced by MG132 was examined by semiquantitative RT-PCR. COX-2 mRNA levels were significantly increased after treatment with MG132 for 2 h and maintained for 24 h (Figure 1B, lanes 6–11).
Figure 1.
Proteasome inhibitors stimulate COX-2 protein expression via enhancing gene transcription in NCI-H292, A549, and AGS epithelial cells. (A) Cells were treated with various concentrations of MG132, PSI-1, or lactacystin for 16 h, and then whole-cell lysates were prepared and subjected to Western blotting using antibodies specific for COX-2 or actin as described in Materials and Methods. (B) NCI-H292 cells were treated with 25 μM MG132 for the indicated time, and then whole-cell lysates were prepared and subjected to Western blotting using antibodies specific for COX-2 or actin, or mRNA was harvested and RT-PCR was performed to quantify COX-2 mRNA. Fold of induction of COX-2 protein or mRNA expression normalized with actin or β-actin was quantified using ImageQuant. (C and D) Cells were treated with MG132, PSI-1, or lactacystin for 16 h. Whole-cell lysates were immunoprecipitated with anti-COX-2 or anti-ubiquitin antibody, and then the precipitates were subjected to SDS-PAGE and Western blotting using anti-COX-2 (C) or anti-ubiquitin (D) antibody. Results are representative of three independent experiments.
It had been reported that proteasomal inhibition led to the stabilization of COX-2 in neuronal cells as ubiquitin conjugates, suggesting the contribution of ubiquitin/proteasome pathway to the upregulation of COX-2 protein level induced by proteasome inhibitors (Rockwell et al., 2000). To check whether COX-2 protein was also ubiquitinated in NCI-H292 cells, the association between COX-2 and ubiquitin after treatment with MG132, PSI-1, and lactacystin was examined. Anti-COX-2 or anti-ubiquitin antibody was used to precipitate COX-2 or ubiquitin from NCI-H292 cells, and then the immunoprecipitated proteins were subjected to Western blotting using anti-COX-2 or anti-ubiquitin antibody (Figure 1, C and D). No COX-2 was coimmunoprecipitated with ubiquitin after stimulation with MG132, PSI-1, and lactacystin (Figure 1C, lanes 6–8, and D, lanes 2–4). However, immunoprecipitated COX-2 and ubiquitin showed the respective expression of COX-2 (Figure 1C, lanes 2–4) and ubiquitin conjugates (Figure 1D, lanes 6–8).
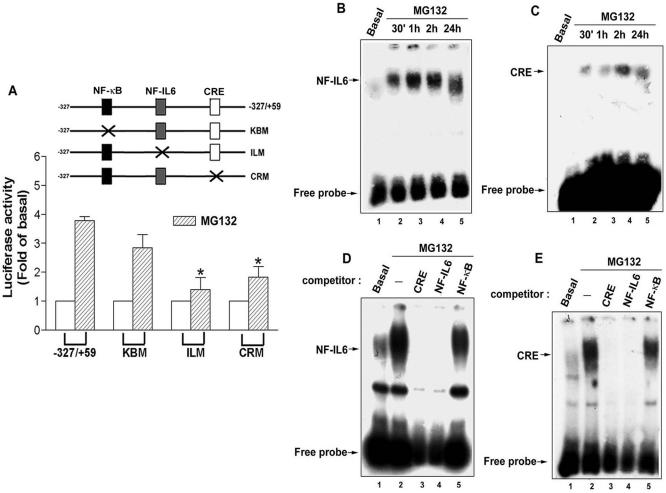
To identify which cis-acting element was involved in the regulation of MG132-induced COX-2 expression, cells were transfected with the human COX-2 promoter-luciferase constructs including–327/+59, KBM with κB site (–223/–214) mutation, ILM with NF-IL6 site (–132/–124) mutation, and CRM with CRE site (–59/–53) mutation, and then the promoter activity was examined by measuring the luciferase activity. Our results showed an attenuation of the MG132-induced COX-2 promoter activity using ILM and CRM (Figure 2A), demonstrating that the NF-IL6 and CRE elements are responsible for MG132-induced COX-2 transcription in NCI-H292 cells.
Figure 2.
Involvements of NF-IL-6 and CRE in the MG132-induced COX-2 promoter activity in NCI-H292 cells. (A) Cells were transfected with –327/+59, KBM, ILM, or CRM luciferase expression vector, and then treated with 25 μM MG132 for 6 h. Luciferase activity was measured as described in Materials and Methods. The data have been normalized with β-galactosidase activity and expressed as the mean ± SEM of three independent experiments performed in triplicate. * p < 0.05; compared with –327/+59. (B-E) Cells were treated with 25 μM MG132 for 30 min, 1 h, 2 h, or 24 h, and then nuclear extracts were prepared and DNA-protein binding activity measured by EMSA using NF-IL6 (B and D) and CRE (C and E) oligonucleotides probe as described in Materials and Methods. One hundred-fold molar excess of unlabeled NF-IL6, CRE, or NF-κB oligonucleotides was used as competitor (D and E). Results are representative of three independent experiments.
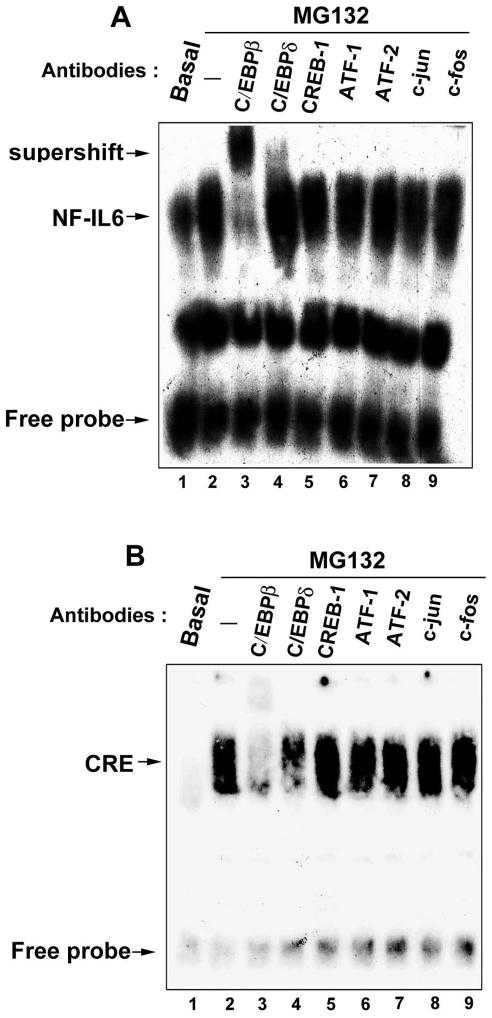
Because NF-IL6 and CRE elements were involved in the MG132-induced COX-2 gene transcription, their DNA-protein bindings were examined by EMSA. After stimulation with MG132 for 30 min, the NF-IL6 or CRE DNA-protein binding was rapidly increased and persistent for 24 h (Figure 2, B and C), and these effects were inhibited by the excess of cold NF-IL6 or CRE, but not NF-κB probe (Figure 2, D and E, lanes 3–5). To identify which transcription factor binds to the NF-IL6 and CRE elements after MG132 stimulation, supershift assays were performed in the presence of antibodies against C/EBPβ, C/EBPδ, CREB-1, ATF-1, ATF-2, c-jun, or c-fos. After incubation of nuclear extracts with anti-C/EBPβ and anti-C/EBPδ antibodies (Figure 3, A and B, lanes 3 and 4), the supershift was seen in the MG132-induced NF-IL6 DNA-protein binding (Figure 3A, lanes 3 and 4) and attenuation was seen in the CRE DNA-protein binding (Figure 3B, lanes 3 and 4). However, other antibodies did not induce supershift or attenuation of these two bindings (Figure 3, A and B, lanes 5–9). These results indicated that C/EBPβ and C/EBPδ were the components binding to the NF-IL6 and CRE sites induced by MG132. The weaker supershift of C/EBPδ in the NF-IL6 DNA-protein binding (Figure 3A, lane 4) was similar to the finding induced by growth hormone (Liao et al., 1999, Figure 2, lanes 7 and 8).
Figure 3.
Identification of transcription factors binding to the NF-IL6 and CRE sites. NCI-H292 cells were incubated with 25 μM MG132 for 1 h. Nuclear extracts were prepared and DNA-protein binding activity measured by EMSA using NF-IL6 (A) and CRE (B) oligonucleotides probe. Supershift assays were performed using 2 μg of the indicated antibodies as described in Materials and Methods. Results are representative of three independent experiments.
MG132 Induced the Bindings of C/EBPβ and C/EBPδ to Both the CRE and NF-IL6 Elements of the COX-2 Promoter
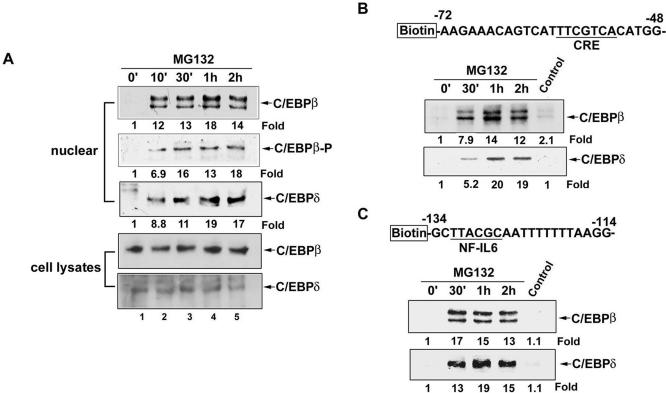
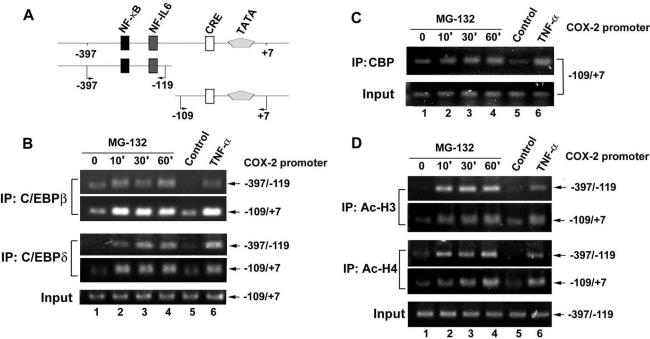
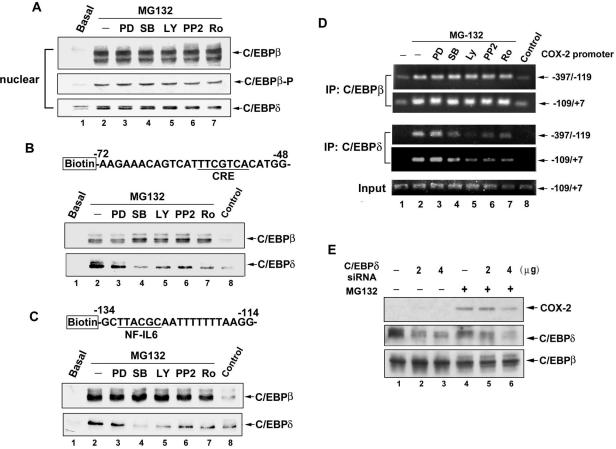
Phosphorylation and translocation to the nucleus have been reported to activate the C/EBP isoforms (Ramji and Foka, 2002). Therefore, whether MG132 induced translocation of C/EBPβ and C/EBPδ to the nucleus and whether phosphorylation occurred were examined. As shown in Figure 4A, C/EBPβ and C/EBPδ were translocated to the nuclear compartment after MG132 stimulation at 10 min, and this effect was lasted for 2 h. Phosphorylation of C/EBPβ at Thr-235 was also seen. However, the total level of C/EBPβ and C/EBPδ were not affected by MG132 (Figure 4A). In addition to the translocation and phosphorylation, the DNA-binding activity also plays an important role in the C/EBP-dependent transcription. The DNA-affinity protein-binding assay using specific biotinylated probe was performed to measure the in vitro binding of C/EBPβ and C/EBPδ to the respective CRE and NF-IL6 site on the COX-2 promoter. Using a biotinylated double-strand DNA fragment from –72 to –48 of proximal COX-2 promoter containing CRE consensus sequences, the increases in bindings of C/EBPβ and C/EBPδ to the CRE site were found after MG132 treatment for 30 min and sustained to 2 h (Figure 4B). Similar bindings of C/EBPβ and C/EBPδ to the DNA fragment from –134 to –114 of COX-2 promoter containing NF-IL6 site were also induced by MG132 (Figure 4C). To further confirm the interactions of these two transcription factors with the CRE and NF-IL6 elements in vivo, ChIP assay was performed using anti-C/EBPβ and anti-C/EBPδ antibodies. Two pairs of primers to amplify the COX-2 promoter regions from –397 to –119 containing NF-IL6 site and from –119 to +7 containing CRE sites were used (Figure 5A). In vivo bindings of C/EBPβ and C/EBPδ to these two fragments of COX-2 promoter occurred as early as 10 min and sustained to 60 min after MG132 treatment (Figure 5B, lanes 2–4). Similar bindings were also seen when cells were treated with TNF-α for 60 min (Figure 5B, lane 6).
Figure 4.
Kinetics of MG132-induced translocation of C/EBPβ and C/EBPδ and phosphorylation of C/EBPβ and their binding to CRE and NF-IL6 sites. (A) NCI-H292 cells were treated with 25 μM MG132 for 10 min, 30 min, 1 h, or 2 h, and then whole-cell lysates and nuclear extracts were prepared and subjected to Western blotting using antibodies against C/EBPβ, C/EBPδ, and phospho-C/EBPβ (Thr-235). (B and C) Cells were treated with 25 μM MG132 for the indicated time, and then nuclear extracts were prepared and mixed with biotinylated COX-2 promoter probe containing CRE (B) or NF-IL6 (C) sites or without probe (Control) and streptavidin-agarose beads. C/EBPβ or C/EBPδ in the complex was detected by Western blotting. Results are representative of three independent experiments.
Figure 5.
MG132 induced recruitment of C/EBPβ, C/EBPδ, and CBP to the COX-2 promoter and acetylation of histone H3 and H4 in vivo. (A) Schematic illustration of various regulatory elements on the COX-2 promoter (–397 to +7). (B–D) NCI-H292 cells were treated with 25 μM MG132 for the indicated time or 10 ng/ml TNF-α for 60 min, and then ChIP assays were performed using anti-C/EBPβ, anti-C/EBPδ (B), anti-CBP (C), anti-acetyl H3, or anti-acetyl H4 (D) antibody or without antibody (Control) to assay the precipitated COX-2 promoter regions (–397 to –119 or –109 to +7) as described in Materials and Methods. One percent of the chromatin was assayed to verify equal loading (Input). Results are representative of three independent experiments.
Eukaryotic chromatin is packaged into a highly compact structure composed with the basic nucleosomal unit, a complex of DNA tangled with the histone octamer. The N-terminal tails of core histones have several basic amino acid residues that are subject to modifications including acetylation. Histone acetylation is a crucial step for chromatin remodeling, which facilitate the accessibility of the binding of transcription factors to trigger gene transcription (Grunstein, 1997). CREB-binding protein (CBP), a transcriptional coactivator, possessing histone acetyltransferase activity (HAT) and had been reported to play an important role in C/EBP-mediated transcription through recruitment by C/EBPδ (Kovacs et al., 2003). We further examined whether MG132 increased the recruitment of CBP and the acetylation of histone H3 and H4 in the COX-2 promoter. In parallel with the bindings of C/EBPβ and C/EBPδ, a time-dependent increase in the recruitment of CBP to the COX-2 promoter containing CRE site (–109 to +7) was seen in response to MG132 stimulation or TNF-α treatment for 60 min (Figure 5C). Τhe acetylated H3 and H4 in both regions of COX-2 promoter were also increased by MG132 or TNF-α (Figure 5D).
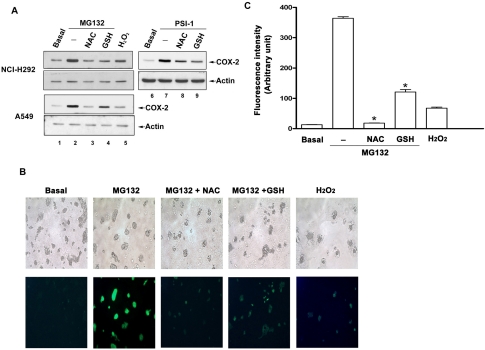
Inhibition of MG132-induced COX-2 Expression, ROS Production, and Kinases Activations by ROS Scavengers
Regulation of proinflammatory gene expressions by ROS-dependent signaling pathways had been reported (Rahman, 2002). We tested whether proteasome inhibitor-induced COX-2 expression was involved ROS; hence two ROS scavengers, NAC and GSH, were used. As shown in Figure 6A, MG132- or PSI-1-induced COX-2 expression in lung epithelial cells was attenuated by 20 mM NAC and GSH. H2 O2 also induced slight COX-2 expression (Figure 6, lane 5). To examine whether MG132 indeed elicited intracellular ROS generation when C/EBPβ and C/EBPδ were bound to the COX-2 promoter, fluorescence intensity of ROS was measured by using DCF after cells were treated with MG132 for 1 h. A dramatic increase in ROS production was seen by MG132, and this effect was abolished by NAC or GSH. H2 O2 also induced a weaker production of ROS in NCI cells (Figure 6, B and C).
Figure 6.
Effects of ROS scavengers on MG132-induced COX-2 expression and ROS production. (A) Cells were treated with 25 μM MG132, PSI-1, or 200 μM H2O2 for 16 h in the presence or absence of pretreatment with 20 mM NAC or GSH for 30 min. Whole-cell lysates were prepared and subjected to Western blotting using antibody specific for COX-2 or actin. (B) Cells were loaded with 10 μM DCF for 1 h and then replaced with fresh medium followed by incubation with 25 μM MG132 or 200 μM H2 O2 for 1 h in the present or absence of 20 mM NAC or GSH. Fluorescent images were observed and photographed with a fluorescence microscope as described in Materials and Methods. Results are representative of three independent experiments. (C) The fluorescence intensity were determined with ImageQuant software and represented as means of spots in three independent fields. * p < 0.05; compared with MG132 alone.
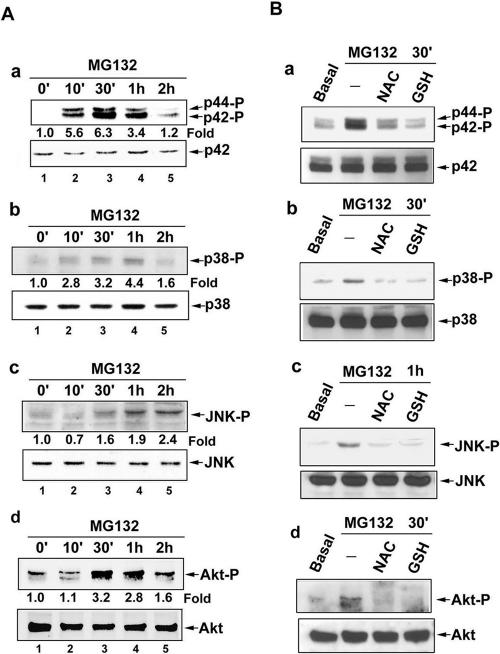
Time-dependent activations of ERK, p38, JNK, and Akt were induced by MG132 (Figure 7A). Consistently, these effects were blocked by NAC and GSH (Figure 7B), indicating that MG-132-induced ROS generation mediated activation of these protein kinases.
Figure 7.
Effects of ROS scavengers on MG132-induced MAPKs and Akt activations. Cells were treated with 25 μM MG132 for the indicated time (A), or pretreated with 20 mM NAC or GSH for 30 min before incubation with 25 μM MG132 for 30 min (B). Whole-cell lysates were prepared and subjected to Western blotting using antibodies specific for phosphorylated and unphosphorylated form of ERK, p38, JNK, or Akt. The immunoblots shown are representative of three independent experiments.
Effects of Various Inhibitors on MG132-induced COX-2 Expression
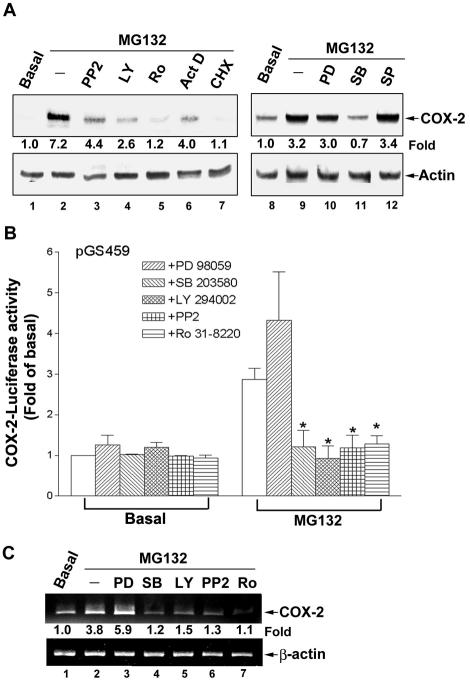
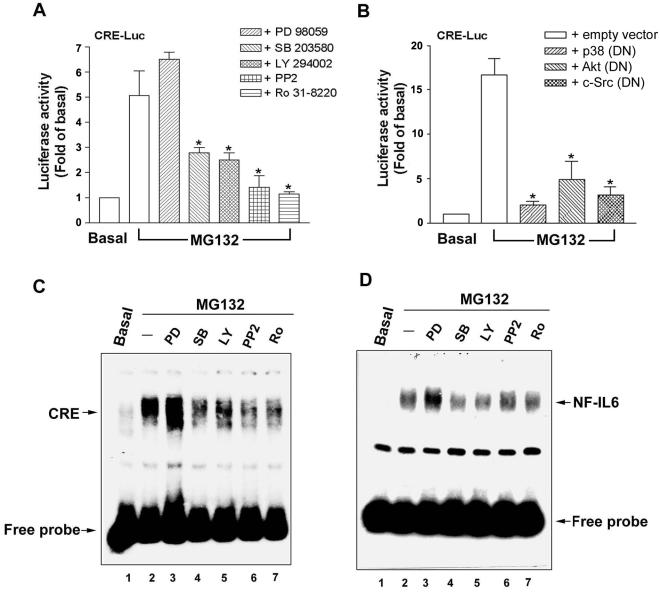
Both transcriptional (actinomycin D) and translational (cycloheximide) inhibitors blocked the MG132-induced COX-2 protein expression (Figure 8A, lanes 6 and 7), confirming the regulation at transcriptional level. Above results have demonstrated that MG132-induced COX-2 transcription was regulated by the binding of C/EBPs to the CRE and NF-IL6 elements. Phosphorylation plays a major role in the activation of C/EBP isoforms (Ramji and Foka, 2002). Therefore, which kinase involved in the regulation of COX-2 expression induced by MG132 was examined. As shown in Figure 8A, the COX-2 expression was inhibited by 10 μM PP2 (a Src kinase inhibitor), 10 μM LY294002 (a PI3K inhibitor), 1 μM Ro 31-8220 (a PKC inhibitor; Figure 8A, lanes 3–5), and 30 μM SB203580 (a p38 inhibitor; Figure 8B, lane 11), but not by PD98059 (a MEK inhibitor) and SP600125 (a JNK inhibitor; Figure 8A, lanes 10 and 12), indicating the involvements of PI3K, p38, Src, and PKC, but not p42/p44 and JNK. In accordance with the results of COX-2 protein expression, the induction of COX-2 promoter activity was attenuated by SB203580, LY294002, PP2, or Ro 31-8220, but not by PD98059 (Figure 8B). MG132-induced COX-2 mRNA expression was also inhibited by the inhibitors of p38, PI3K, Src, and PKC (Figure 8C, lanes 4–7). These results demonstrated that PI3K, p38, Src, or PKC plays a role in the MG132-mediated COX-2 expression.
Figure 8.
The effects of various inhibitors on the MG132-induced COX-2 expression and promoter activity. (A) NCI-H292 cells were pretreated with 10 μM PP2, 10 μM LY294002 (LY), 1 μM Ro 31-8220 (Ro), 200 ng/ml actinomycin D (Act D), 500 nM cycloheximide (CHX), 30 μM PD98059 (PD), 30 μM SB203580 (SB), or 25 μM SP600125 (SP) for 30 min before incubation with 25 μM MG132 for 16 h. Whole-cell lysates were prepared and subjected to Western blotting using antibodies specific for COX-2 and actin. Fold of induction of COX-2 protein expression normalized with actin was quantified using ImageQuant. (B) Cells transfected with pGS459 luciferase expression vector were pretreated with 30 μM PD98059, 30 μM SB203580, 10 μM PP2, 10 μM LY294002, or 1 μM Ro 31-8220 for 30 min before incubation with 25 μM MG132 for 6 h, and then luciferase activity was measured. The data were normalized with β-galactosidase activity and expressed as the mean ± SEM of three independent experiments performed in triplicate. * p < 0.05; compared with MG132 alone. (C) Cells were pretreated with various inhibitors as described in B for 30 min before incubation with 25 μM MG132 for 2 h, and then mRNA was harvested and RT-PCR was performed to quantify COX-2 mRNA. Results are representative of three independent experiments.
Effects of Various Kinase Inhibitors on the Binding of C/EBPβ and C/EBPδ to the CRE and NF-IL6 Sites
Because translocation of C/EBPβ and C/EBPδ to the nucleus and phosphorylation of C/EBPβ at Thr-235 induced by MG132 were seen (Figure 4A), the effects of various kinase inhibitors were examined. As shown in Figure 9A, the translocations and phosphorylation were not affected by PD98059, SB203580, LY294002, PP2, and Ro 31-8220, indicating that PI3K, p38, Src, and PKC activations were not involved in these events. However, the in vitro binding of C/EBPδ, but not C/EBPβ to the CRE and NF-IL6 sites was attenuated by SB203580, LY294002, PP2, and Ro 31-8220 (Figure 9, B and C). ChIP assays further confirmed these findings. Binding of C/EBPδ to the fragments containing CRE and NF-IL6 sites were inhibited by SB203580, LY294002, PP2, and Ro 31-8220. However, that of C/EBPβ was not affected (Figure 9D, lanes 4–7). These results suggested the involvement of C/EBPδ, but not C/EBPβ in the MG132-induced COX-2 expression. To further confirm the role of C/EBPδ, siRNA of C/EBPδ was used. As shown in Figure 9E, the expression of C/EBPδ as well as MG132-induced COX-2 expression was attenuated by siRNA of C/EBPδ (Figure 9E).
Figure 9.
The effects of various kinase inhibitors on the MG132-induced translocation of C/EBPβ and C/EBPδ and their binding to the CRE and NF-IL6 sites. NCI-H292 cells were pretreated with 30 μM PD98059 (PD), 30 μM SB203580 (SB), 10 μM PP2, 10 μM LY294002 (LY), or 1 μM Ro 31-8220 (Ro) for 30 min before incubation with 25 μM MG132 for 1 h, and then nuclear extracts were prepared and subjected to Western blotting using antibodies specific for C/EBPβ, C/EBPδ, or phosphorylated form of C/EBPβ (A) or mixed with biotinylated COX-2 promoter probe containing CRE (B) or NF-IL6 (C) sites or without probe (Control) and streptavidin-agarose beads. C/EBPβ or C/EBPδ in the complex was detected by Western blotting. In D, after cells were treated, ChIP assays were performed using anti-C/EBPβ, or anti-C/EBPδ antibody or without antibody (Control) to assay the precipitated COX-2 promoter regions (–397 to –119 or –109 to +7) as described in Materials and Methods. One percent of the chromatin was assayed to verify equal loading (Input). (E) NCI-H292 cells were transfected with pSilencer 3.1 plasmids expressing scrambled (–), or 2 or 4 μg C/EBPδ siRNA. After 48 h, cells were incubated with 25 μM MG132 for 16 h, and then whole-cell lysates were prepared and subjected to Western blot using antibodies specific for anti-C/EBPβ, anti-C/EBPδ, and anti-COX-2. Results are representative of three independent experiments.
Because CRE element was involved in the MG132-induced COX-2 transcription, the effects of various kinase inhibitors on the CRE-luc activity were also examined. As shown in Figure 10, A and B, CRE-luc activity induced by MG132 was attenuated by LY294002, SB203580, PP2, or Ro 31-8220, as well as the DN mutants of p38, Akt, and c-Src We further examined the effects of these inhibitors on MG132-induced CRE and NF-IL6 DNA-protein complex formation. The formations of these two complexes were inhibited by SB203580, LY294002, PP2, and Ro 31-8220, but not PD98059 (Figure 10, B and C, lanes 3–7). All these data suggested that MG132 acts through PI3K, p38, Src, and PKC to induce C/EBPδ binding to both CRE and NF-IL6 sites on the COX-2 promoter, resulting in the initiation of COX-2 transcription.
Figure 10.
The effects of various kinase inhibitors on MG132-induced CRE promoter activity and CRE- or NF-IL6-specific DNA-protein complex formation. (A and B) NCI-H292 cells transfected with CRE luciferase expression vector were pretreated with 30 μM PD98059, 30 μM SB203580, 10 μM PP2, 10 μM LY294002, or 1 μM Ro 31-8220 for 30 min (A) or cotransfected with the empty vector or the DN mutants of p38 (DN), Akt (DN), or c-Src (DN; B) before incubation with 25 μM MG132 for 6 h, and then luciferase activity was measured. The data were normalized with β-galactosidase activity and expressed as the mean ± SEM of three independent experiments performed in triplicate. * p < 0.05 compared with MG132 alone. (C and D) Cells were pretreated with various inhibitors as described in (A) before incubation with 25 μM MG132 for 1 h. Nuclear extracts were prepared and the CRE (B) or NF-IL6 (C) DNA-protein complex formation was measured by EMSA. Results are representative of three independent experiments.
DISCUSSION
Proteasome inhibitors control the process of apoptosis and cell cycle by blocking the ubiquitin-dependent pathway (Zhang et al., 2004). In addition, the proteasome inhibitor MG132 act through activation of intracellular signaling pathways to up-regulate acute phase genes and induce accumulation of heat shock protein (Meriin et al., 1998). In this study, proteasome inhibitors MG132, PSI-1, and lactacystin were found to induce COX-2 expression in different epithelial cells. Although proteasome inhibition has been reported to lead to the stabilization of COX-2 as ubiquitin conjugates (Rockwell et al., 2000), our data did not show the ubiquitination of COX-2 by MG132 (Figure 1, C and D). Furthermore, RT-PCR, reporter assay and EMSA revealed that MG132 acts via transcriptional regulation to induce COX-2 gene expression. MG132 enhances bindings of C/EBPβ and C/EBPδ to the CRE and NF-IL6 elements on the COX-2 promoter. Recruitment of CBP and chromatin remodeling through acetylations of histone H3 and H4 are also involved. ROS play a critical role in the MG132-induced activations of PI3K/Akt, p38, Src, and PKC, which up-regulate the binding of C/EBPδ to the CRE and NF-IL6 elements on the COX-2 promoter, resulting in COX-2 expression.
COX-2 expression is regulated by the binding of specific transcription factors to their cis-acting elements on the COX-2 promoter (Tanabe and Tohnai, 2002). Our promoterluciferase assay and EMSA demonstrated the participations of NF-IL6 and CRE elements, but not NF-κB in MG132-induced COX-2 transcription. The involvements of NF-IL6 and CRE sites in the COX-2 expression have also been reported in many types of cells stimulated with cytokines or phorbol ester (Inoue et al., 1995; Potter et al., 2000; Zhu et al., 2002; Tamura et al., 2003). Typically, the family of C/EBPs binds to the NF-IL6 element, and CREB-1, ATF-1, c-jun, c-fos, and ATF-2, which belong to the AP-1 family is associated with the CRE site (Tang et al., 2001; Schroer et al., 2002; Nie et al., 2003). C/EBPδ binding to and increasing the COX-2 gene promoter activity mainly through CRE have been reported (Inoue et al., 1995). Our results showed that only C/EBPβ and C/EBPδ, but not CREB-1, ATF-1, c-fos, or c-jun bind to the CRE site in response to MG132 stimulation (Figure 3B). Similar observation of the binding of C/EBP, but not CREB-1, ATF-1, or ATF-2 to the CRE site induced by conditioned medium was also reported in human endometrial stromal cells (Tamura et al., 2003).
C/EBPβ and C/EBPδ belong to the CCAAT/enhancer binding protein family, which contain a transactivation domain and a highly conserved basic-leucine zipper for dimerization with other members (Ramji and Foka, 2002). C/EBPβ and C/EBPδ regulate the expressions of an array of genes involved in cell proliferation, inflammation, and immunity (Ramji et al., 1993; Poli, 1998; Hu et al., 2000). The activity of C/EBP isoforms has been demonstrated to be regulated at several levels, including gene transcription, translation, protein-protein interactions, nuclear localization, and phosphorylation-mediated changes in DNA-binding activity (Ramji and Foka, 2002). In the present study, the total protein levels of C/EBPβ and C/EBPδ were not affected by the treatment with MG132, indicating that protein stability changes do not account for the enhanced binding of C/EBPβ and C/EBPδ to the COX-2 promoter. The binding of C/EBPβ to the promoter has been reported to be stimulated by phosphorylation at several amino acid residues, including Ser-105, Ser-299, and Thr-235 (Nakajima et al., 1993; Trautwein et al., 1993; Chinery et al., 1997). The DNA-binding of C/EBPδ has also been found to be regulated by phosphorylation (Ray and Ray, 1994; Milosavljevic et al., 2002); however, which kinase mediates this effect is not understood. We found the increases in the phosphorylation of nuclear C/EBPβ at Thr-235 and the nuclear translocation and DNA-binding of C/EBPβ and C/EBPδ after stimulation with MG132. However, only the binding of C/EBPδ to the CRE and NF-IL6 sites was attenuated by the inhibitors of p38, PI3K, c-Src, and PKC. The differential regulation of the binding of C/EBPδ to the COX-2 promoter by p38, PI3K, Src, and PKC remains to be investigated. However, recent study showed that the specific inhibitor of p38 repressed the transcriptional activity of C/EBPδ. An important domain overlapping with a docking site for p38 was found, and deletion of this domain led to a decrease in its transcriptional activity (Svotelis et al., 2005). Inhibitors of p38, PI3K, c-Src, and PKC also blocked the MG-132-induced COX-2 mRNA and protein expressions, implying that the C/EBPδ recruitment to COX-2 promoter is the major regulatory mechanism in this event. This assumption was further confirmed by the findings that knockdown of endogenous C/EBPδ expression by siRNA attenuated MG132-induced COX-2 expression (Figure 9). To our knowledge, this is the first report to demonstrate that C/EBPδ regulates COX-2 expression induced by MG132.
CBP and p300, transcriptional coactivators possessing HAT activity, and histones play important roles in orchestrating multiple transcription factors and functions as an adaptor between the transcription factors and the general transcriptional machinery. Therefore, CBP is believed to activate gene transcription by recruiting basal transcription factors (TFIIB, TBP, and RNA polymerase II holoenzyme), modifying chromatin structure via histone acetylation and recruiting other HATs, such as P/CAF (Imhof et al., 1997; Marmorstein, 2001). We revealed that MG132 enhanced recruitment of CBP to the COX-2 promoter containing CRE site. The acetylations of histone H3 and H4 on the COX-2 promoter were increased as well. Therefore, CBP-dependent acetylation of histones and recruitment of basal transcription machinery may be involved in the MG132-induced COX-2 gene transcription. It has been reported that acetylation of histone H3 and H4 is a prerequisite for TNF-α production in monocytes and macrophages (Lee et al., 2003), and acetylation of histone H4 mediated oxidative stress-regulated proinflammatory gene, such as IL-8 expression in human pulmonary epithelial cells (Gilmour et al., 2003). It is probable that recruitment of transcriptional coactivators accompanied with histone acetylation is also necessary for the proinflammatory gene expression.
Previous studies have shown that MG132 triggered IL-8 release and MCP-1 expression through JNK-dependent pathway in human hepatocytes and rat mesangial cells (Nakayama et al., 2001; Joshi-Barve et al., 2003) and induced IL-6 production through MEK/ERK activation in human umbilical vein endothelial cells (Shibata et al., 2002). The mechanism why MG132 induces activation of these kinases is not well elucidated. ROS production served as a pivotal linkage in enhancing the proinflammatory gene expression through the activation of stress kinases (JNK, MAPK, p38, PI3K/Akt; Rahman, 2002). Here, we found that MG132 indeed elevated intracellular ROS production. MAPKs and Akt activations and COX-2 induction were both abrogated by the ROS scavengers, NAC and GSH, indicating that MG132 acts through ROS generation to activate downstream protein kinases, resulting in the induction of COX-2 expression. Whether MG132-induced ROS generation and kinase activation depends on its proteasome inhibitory action remains to be determined. However, proteasome inhibitors have been reported to induce ROS production through mitochondrial dysfunction and ER stress (Ling et al., 2003; Fribley et al., 2004).
In conclusion, proteasome inhibitors MG132, PSI-1, and lactacystin induced COX-2 expression in three different epithelial cells. The induction of COX-2 expression in response to proteasome inhibition is regulated at transcriptional level and via the ROS-mediated pathway involving activation of p38, PI3K, PKC, or c-Src, which enhances binding of C/EBPδ to the CRE and NF-IL6 sites. This coordinated with the recruitment of CBP and enhanced acetylations of histone H3 and H4 on the COX-2 promoter, resulting in the transactivation of COX-2 expression.
Acknowledgments
This work was supported by a research grant from the National Science Council of Taiwan, NSC 94-2320-B002-098.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–08–0778) on September 29, 2005.
Abbreviations used: NF-κB, nuclear factor κB; NF-IL6, nuclear factor for interleukin 6; IL, interleukin; PI3K, phosphatidylinositol 3-kinase; RT-PCR, reverse transcription-polymerase chain reaction; TNF, tumor necrosis factor.
References
- Arias-Negrete, S., Keller, K., and Chadee, K. (1995). Proinflammatory cytokines regulate cyclooxygenase-2 mRNA expression in human macrophages. Biochem. Biophys. Res. Commun. 208, 582–589. [DOI] [PubMed] [Google Scholar]
- Chang, Y. J., Holtzman, M. J., and Chen, C. C. (2002). Interferon-gammainduced epithelial ICAM-1 expression and monocyte adhesion. Involvement of PKC-dependent c-Src tyrosine kinase activation pathway. J. Biol. Chem. 277, 7118–7126. [DOI] [PubMed] [Google Scholar]
- Chen, C. C., Sun, Y. T., Chen, J. J., and Chiu, K. T. (2000). TNF-alpha-induced cyclooxygenase-2 expression in human lung epithelial cells: involvement of the phospholipase C-gamma 2, PKC-alpha, tyrosine kinase, NF-kappa B-inducing kinase, and I-kappa B kinase 1/2 pathway. J. Immunol. 165, 2719–2728. [DOI] [PubMed] [Google Scholar]
- Chinery, R., Brockman, J. A., Dransfield, D. T., and Coffey, R. J. (1997). Antioxidant-induced nuclear translocation of CCAAT/enhancer-binding protein beta. A critical role for protein kinase A-mediated phosphorylation of Ser299. J. Biol. Chem. 272, 30356–30361. [DOI] [PubMed] [Google Scholar]
- Ciechanover, A. (1998). The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17, 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Y., Rahmani, M., and Grant, S. (2003). Proteasome inhibitors potentiate leukemic cell apoptosis induced by the cyclin-dependent kinase inhibitor flavopiridol through a SAPK/JNK- and NF-kappaB-dependent process. Oncogene 22, 7108–7122. [DOI] [PubMed] [Google Scholar]
- Delic, J., Masdehors, P., Omura, S., Cosset, J. M., Dumont, J., Binet, J. L., and Magdelenat, H. (1998). The proteasome inhibitor lactacystin induces apoptosis and sensitizes chemo- and radioresistant human chronic lymphocytic leukaemia lymphocytes to TNF-alpha-initiated apoptosis. Br. J. Cancer 77, 1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, B. S., Kujubu, D. A., Perrin, D. M., and Herschman, H. R. (1992). Structure of the mitogen-inducible TIS10 gene and demonstration that the TIS10-encoded protein is a functional prostaglandin G/H synthase. J. Biol. Chem. 267, 4338–4344. [PubMed] [Google Scholar]
- Fribley, A., Zeng, Q., and Wang, C. Y. (2004). Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol. Cell. Biol. 24, 9695–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, P. S., Rahman, I., Donaldson, K., and MacNee, W. (2003). Histone acetylation regulates epithelial IL-8 release mediated by oxidative stress from environmental particles. Am. J. Physiol Lung Cell Mol. Physiol 284, L533–L540. [DOI] [PubMed] [Google Scholar]
- Goetzl, E. J., An, S., and Smith, W. L. (1995). Specificity of expression and effects of eicosanoid mediators in normal physiology and human diseases. FASEB J. 9, 1051–1058. [DOI] [PubMed] [Google Scholar]
- Grunstein, M. (1997). Histone acetylation in chromatin structure and transcription. Nature 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Hideshima, T., Richardson, P., Chauhan, D., Palombella, V. J., Elliott, P. J., Adams, J., and Anderson, K. C. (2001). The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 61, 3071–3076. [PubMed] [Google Scholar]
- Hla, T. and Neilson, K. (1992). Human cyclooxygenase-2 cDNA. Proc. Natl. Acad. Sci. USA 89, 7384–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. M., Tian, Q., Baer, M., Spooner, C. J., Williams, S. C., Johnson, P. F., and Schwartz, R. C. (2000). The C/EBP bZIP domain can mediate lipopolysaccharide induction of the proinflammatory cytokines interleukin-6 and monocyte chemoattractant protein-1. J. Biol. Chem. 275, 16373–16381. [DOI] [PubMed] [Google Scholar]
- Huang, W. C. and Chen, C. C. (2005). Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional;Activity. Mol. Cell. Biol. 25, 6592–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof, A., Yang, X. J., Ogryzko, V. V., Nakatani, Y., Wolffe, A. P., and Ge, H. (1997). Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7, 689–692. [DOI] [PubMed] [Google Scholar]
- Inoue, H., Yokoyama, C., Hara, S., Tone, Y., and Tanabe, T. (1995). Transcriptional regulation of human prostaglandin-endoperoxide synthase-2 gene by lipopolysaccharide and phorbol ester in vascular endothelial cells. Involvement of both nuclear factor for interleukin-6 expression site and cAMP response element. J. Biol. Chem. 270, 24965–24971. [DOI] [PubMed] [Google Scholar]
- Joshi-Barve, S., Barve, S. S., Butt, W., Klein, J., and McClain, C. J. (2003). Inhibition of proteasome function leads to NF-kappaB-independent IL-8 expression in human hepatocytes. Hepatology 38, 1178–1187. [DOI] [PubMed] [Google Scholar]
- Kosaka, T., Miyata, A., Ihara, H., Hara, S., Sugimoto, T., Takeda, O., Takahashi, E., and Tanabe, T. (1994). Characterization of the human gene (PTGS2) encoding prostaglandin-endoperoxide synthase 2. Eur. J. Biochem. 221, 889–897. [DOI] [PubMed] [Google Scholar]
- Kovacs, K. A., Steinmann, M., Magistretti, P. J., Halfon, O., and Cardinaux, J. R. (2003). CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J. Biol. Chem. 278, 36959–36965. [DOI] [PubMed] [Google Scholar]
- Kraemer, S. A., Meade, E. A., and DeWitt, D. L. (1992). Prostaglandin endoperoxide synthase gene structure: identification of the transcriptional start site and 5′-flanking regulatory sequences. Arch. Biochem. Biophys. 293, 391–400. [DOI] [PubMed] [Google Scholar]
- Lee, J. Y., Kim, N. A., Sanford, A., and Sullivan, K. E. (2003). Histone acetylation and chromatin conformation are regulated separately at the TNF-alpha promoter in monocytes and macrophages. J. Leukoc. Biol. 73, 862–871. [DOI] [PubMed] [Google Scholar]
- Liao, J., Piwien-Pilipuk, G., Ross, S. E., Hodge, C. L., Sealy, L., MacDougald, O. A., and Schwartz, J. (1999). CCAAT/enhancer-binding protein beta (C/EBPbeta) and C/EBPdelta contribute to growth hormone-regulated transcription of c-fos. J. Biol. Chem. 274, 31597–31604. [DOI] [PubMed] [Google Scholar]
- Ling, Y. H., Liebes, L., Zou, Y., and Perez-Soler, R. (2003). Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to Bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. J. Biol. Chem. 278, 33714–33723. [DOI] [PubMed] [Google Scholar]
- Marmorstein, R. (2001). Structure and function of histone acetyltransferases. Cell Mol. Life Sci. 58, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin, A. B., Gabai, V. L., Yaglom, J., Shifrin, V. I., and Sherman, M. Y. (1998). Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J. Biol. Chem. 273, 6373–6379. [DOI] [PubMed] [Google Scholar]
- Milosavljevic, T. S., Petrovic, M. V., Cvetkovic, I. D., and Grigorov, I. I. (2002). DNA binding activity of C/EBPbeta and C/EBPdelta for the rat alpha2-macroglobulin gene promoter is regulated in an acute-phase dependent manner. Biochemistry (Mosc.) 67, 918–926. [DOI] [PubMed] [Google Scholar]
- Nakajima, T., Kinoshita, S., Sasagawa, T., Sasaki, K., Naruto, M., Kishimoto, T., and Akira, S. (1993). Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 90, 2207–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, K., Furusu, A., Xu, Q., Konta, T., and Kitamura, M. (2001). Unexpected transcriptional induction of monocyte chemoattractant protein 1 by proteasome inhibition: involvement of the c-Jun N-terminal kinase-activator protein 1 pathway. J. Immunol. 167, 1145–1150. [DOI] [PubMed] [Google Scholar]
- Nie, M., Pang, L., Inoue, H., and Knox, A. J. (2003). Transcriptional regulation of cyclooxygenase 2 by bradykinin and interleukin-1beta in human airway smooth muscle cells: involvement of different promoter elements, transcription factors, and histone h4 acetylation. Mol. Cell. Biol. 23, 9233–9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli, V. (1998). The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273, 29279–29282. [DOI] [PubMed] [Google Scholar]
- Potter, S., Mitchell, M. D., Hansen, W. R., and Marvin, K. W. (2000). NF-IL6 and CRE elements principally account for both basal and interleukin-1 betainduced transcriptional activity of the proximal 528bp of the PGHS-2 promoter in amnion-derived AV3 cells: evidence for involvement of C/EBP beta. Mol. Hum. Reprod. 6, 771–778. [DOI] [PubMed] [Google Scholar]
- Rahman, I. (2002). Oxidative stress and gene transcription in asthma and chronic obstructive pulmonary disease: antioxidant therapeutic targets. Curr. Drug Targets. Inflamm. Allergy 1, 291–315. [DOI] [PubMed] [Google Scholar]
- Ramji, D. P. and Foka, P. (2002). CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramji, D. P., Vitelli, A., Tronche, F., Cortese, R., and Ciliberto, G. (1993). The two C/EBP isoforms, IL-6DBP/NF-IL6 and C/EBP delta/NF-IL6 beta, are induced by IL-6 to promote acute phase gene transcription via different mechanisms. Nucleic Acids Res. 21, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, A. and Ray, B. K. (1994). Serum amyloid A gene expression under acute-phase conditions involves participation of inducible C/EBP-beta and C/EBP-delta and their activation by phosphorylation. Mol. Cell. Biol. 14, 4324–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell, P., Yuan, H., Magnusson, R., and Figueiredo-Pereira, M. E. (2000). Proteasome inhibition in neuronal cells induces a proinflammatory response manifested by upregulation of cyclooxygenase-2, its accumulation as ubiquitin conjugates, and production of the prostaglandin PGE(2). Arch. Biochem. Biophys. 374, 325–333. [DOI] [PubMed] [Google Scholar]
- Schroer, K., Zhu, Y., Saunders, M. A., Deng, W. G., Xu, X. M., MeyerKirchrath, J., and Wu, K. K. (2002). Obligatory role of cyclic adenosine monophosphate response element in cyclooxygenase-2 promoter induction and feedback regulation by inflammatory mediators. Circulation 105, 2760–2765. [DOI] [PubMed] [Google Scholar]
- Schwartz, A. L. and Ciechanover, A. (1999). The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu. Rev. Med. 50, 57–74. [DOI] [PubMed] [Google Scholar]
- Shibata, T., Imaizumi, T., Tamo, W., Matsumiya, T., Kumagai, M., Cui, X. F., Yoshida, H., Takaya, S., Fukuda, I., and Satoh, K. (2002). Proteasome inhibitor MG-132 enhances the expression of interleukin-6 in human umbilical vein endothelial cells: Involvement of MAP/ERK kinase. Immunol. Cell Biol. 80, 226–230. [DOI] [PubMed] [Google Scholar]
- Smith, W. L. (1992). Prostanoid biosynthesis and mechanisms of action. Am. J. Physiol 263, F181–F191. [DOI] [PubMed] [Google Scholar]
- Svotelis, A., Doyon, G., Bernatchez, G., Desilets, A., Rivard, N., and Asselin, C. (2005). IL-1 beta-dependent regulation of C/EBP delta transcriptional activity. Biochem. Biophys. Res. Commun. 328, 461–470. [DOI] [PubMed] [Google Scholar]
- Tamura, M., Sebastian, S., Yang, S., Gurates, B., Fang, Z., Okamura, K., and Bulun, S. E. (2003). Induction of cyclooxygenase-2 in human endometrial stromal cells by malignant endometrial epithelial cells: evidence for the involvement of extracellularly regulated kinases and CCAAT/enhancer binding proteins. J. Mol. Endocrinol. 31, 95–104. [DOI] [PubMed] [Google Scholar]
- Tanabe, T. and Tohnai, N. (2002). Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat. 68–69, 95–114. [DOI] [PubMed] [Google Scholar]
- Tang, Q., Chen, W., Gonzales, M. S., Finch, J., Inoue, H., and Bowden, G. T. (2001). Role of cyclic AMP responsive element in the UVB induction of cyclooxygenase-2 transcription in human keratinocytes. Oncogene 20, 5164–5172. [DOI] [PubMed] [Google Scholar]
- Tazawa, R., Xu, X. M., Wu, K. K., and Wang, L. H. (1994). Characterization of the genomic structure, chromosomal location and promoter of human prostaglandin H synthase-2 gene. Biochem. Biophys. Res. Commun. 203, 190–199. [DOI] [PubMed] [Google Scholar]
- Trautwein, C., Caelles, C., van der Geer, P., Hunter, T., Karin, M., and Chojkier, M. (1993). Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature 364, 544–547. [DOI] [PubMed] [Google Scholar]
- Tsuji, S., Tsujii, M., Kawano, S., and Hori, M. (2001). Cyclooxygenase-2 upregulation as a perigenetic change in carcinogenesis. J. Exp. Clin. Cancer Res. 20, 117–129. [PubMed] [Google Scholar]
- Xie, W., Fletcher, B. S., Andersen, R. D., and Herschman, H. R. (1994). v-src induction of the TIS10/PGS2 prostaglandin synthase gene is mediated by an ATF/CRE transcription response element. Mol. Cell. Biol. 14, 6531–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. G., Wang, J., Yang, X., Hsu, H. C., and Mountz, J. D. (2004). Regulation of apoptosis proteins in cancer cells by ubiquitin. Oncogene 23, 2009–2015. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Saunders, M. A., Yeh, H., Deng, W. G., and Wu, K. K. (2002). Dynamic regulation of cyclooxygenase-2 promoter activity by isoforms of CCAAT/enhancer-binding proteins. J. Biol. Chem. 277, 6923–6928. [DOI] [PubMed] [Google Scholar]