Abstract
Early cellular events associated with tumorigenesis often include loss of cell cycle checkpoints or alteration in growth signaling pathways. Identification of novel genes involved in cellular proliferation may lead to new classes of cancer therapeutics. By screening a tetracycline-inducible cDNA library in A549 cells for genes that interfere with proliferation, we have identified a fragment of UHRF1 (ubiquitin-like protein containing PHD and RING domains 1), a nuclear RING finger protein, that acts as a dominant negative effector of cell growth. Reduction of UHRF1 levels using an UHRF1-specific shRNA decreased growth rates in several tumor cell lines. In addition, treatment of A549 cells with agents that activated different cell cycle checkpoints resulted in down-regulation of UHRF1. The primary sequence of UHRF1 contains a PHD and a RING motif, both of which are structural hallmarks of ubiquitin E3 ligases. We have confirmed using an in vitro autoubiquitination assay that UHRF1 displays RING-dependent E3 ligase activity. Overexpression of a GFP-fused UHRF1 RING mutant that lacks ligase activity sensitizes cells to treatment with various chemotherapeutics. Taken together, our results suggest a general requirement for UHRF1 in tumor cell proliferation and implicate the RING domain of UHRF1 as a functional determinant of growth regulation.
INTRODUCTION
The uncontrolled proliferation that is a hallmark of tumorigenesis is often a result of cellular events such as loss of cell cycle checkpoints or changes in growth signaling pathways. Such cellular processes are tightly regulated through the activation and inactivation of protein function, events that are mediated by protein-protein interactions, various posttranslational modifications, and regulation of protein stability. Deciphering the complexity of these regulatory systems should lead to a better understanding of protein function. These unique functions can be perturbed by overexpression of wild-type proteins, dominant negative mutants, isolated protein domains, or small peptides. Identification of such genetic effectors involved in cellular proliferation can provide significant insight into protein function, and consequently cellular physiology. This information can potentially provide novel avenues for new therapeutic classes in the treatment of cancer.
Retroviral-mediated functional screening is a powerful approach for revealing physiologically relevant gene function. The unique properties of retroviruses make them ideal tools for the introduction of large and diverse libraries of potential genetic effectors to a variety of cell types. Recently, we reported the successful development of a functional approach to identify cell proliferation regulators in human cells utilizing inducible retroviral expression libraries (Hitoshi et al., 2003). Using this strategy, we screened a GFP-fused cDNA library to identify genes that cause cell cycle arrest in A549 lung tumor cells. This screen identified a fragment of UHRF1 (ubiquitin-like protein containing PHD and RING finger domains 1, also known as ICBP90 in humans), a nuclear ring finger protein, which serves as a dominant negative effector of cell growth.
UHRF1 was previously isolated in a screen for proteins that bound to a CCAAT box in the promoter region of the topoisomerase IIα gene. Its overexpression resulted in enhanced expression of topoisomerase IIα protein (Hopfner et al., 2000, 2002). It is a gene whose expression has been positively correlated with cellular proliferation. A murine homolog of UHRF1 (mUHRF1), previously referred to as NP95, has been identified and appears to be subject to similar regulatory mechanisms (Fujimori et al., 1998; Mori et al., 2002). Expression of either UHRF1 or mUHRF1 mRNA and protein is low in quiescent cells and high in proliferating cells and tissues (Fujimori et al., 1998; Hopfner et al., 2000) including breast carcinomas (Mousli et al., 2003), suggesting a role for this family of proteins in proliferation. Indeed, mUHRF1 was isolated in a genetic screen designed to find genes that are induced when terminally differentiated cells are forced into S phase by adenovirus E1A protein (Bonapace et al., 2002).
The primary sequence of UHRF1 contains a PHD and a RING finger motif, domains that have both been linked to E3 ubiquitin ligase activity (Jackson et al., 2000; Coscoy and Ganem, 2003). Recently it was reported that mUHRF1 displayed RING-finger dependent ubiquitination of histones in vitro (Citterio et al., 2004; Mori et al., 2004), although the functional significance of this biochemical activity is still unclear. Because a fragment of UHRF1 was isolated from our proliferation-based functional screen, we decided to further characterize the role of human UHRF1 and its RING finger domain in tumor cell growth.
Here we identify and characterize UHRF1 as a key cellular growth regulator whose E3 ubiquitin ligase activity is critical for cellular recovery from various environmental insults. Abrogation of UHRF1 expression using an UHRF1-specific shRNA resulted in markedly enhanced growth defects in A549, HeLa, and H1299 cells. Using an in vitro autoubiquitination assay, we have confirmed that UHRF1 possesses RING-dependent E3 ubiquitin ligase activity. A549 cells expressing a GFP-tagged UHRF1 RING finger mutant lacking ligase activity display enhanced sensitivity to a wide variety of chemotherapeutics. Our results suggest that the RING finger domain of UHRF1 plays an important role in cellular proliferation through mediating ubiquitination.
MATERIALS AND METHODS
Reagents
The full-length human UHRF1 cDNA was cloned into a tetracycline-regulatable retroviral vector in frame with the upstream GFP to create the expression construct, GFP-UHRF1, used in the chemosensitization experiments. Constructs used for the in vitro autoubiquitination assays contained a myc epitope tag upstream of the GFP. UHRF1 point and deletion mutants were created using PCR-directed mutagenesis (Stratagene QuikChange site-directed mutagenesis kit and Stratagene Ex-site mutagenesis kit, La Jolla, CA). All constructs were verified by sequencing. A rabbit polyclonal antibody was raised against full-length human His6-UHRF1 protein expressed in insect cells (Zymed, South San Francisco, CA) and affinity-purified. Other antibodies used include mouse α-Myc (Covance, Princeton, NJ), mouse α-flag (Sigma, St. Louis, MO), mouse α-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; US Biologicals, Swampscott, MA), rabbit α-actin (Cytoskeleton, Denver, CO) and sheep α-lactate dehydrogenase-1 (LDH; Cortex Biochem, San Leandro, CA)
GFP Fusion Protein Library
A retroviral cDNA library in which cDNA is expressed as a protein with a linker (EEAAKAGGSGGSVVES/R) fused at the C-terminus of GFP, was constructed in a tetracycline (tet)-regulatable (tet-off system) retroviral vector, TRA (Lorens et al., 2000). cDNA encoding GFP and the linker followed by a BstXI cloning site and three stop codons in all three open reading frames was inserted into the TRA vector. Poly A RNA was isolated from A549 lung cancer cells. Double-stranded cDNA was generated from the poly A RNA using the Invitrogen Superscript kit (Carlsbad, CA) with the reverse-transcription primer. The cDNA was sonicated, blunted by exonucleases, ligated to the BstXI adaptor, and cloned into the vector.
Isolation of cDNAs That Encode Antiproliferative Effectors
A549 lung tumor cells expressing the tetracycline regulatable transactivator (tTA; A549.tTA cells) were infected with a tet-inducible (tet-off) retroviral GFP fusion protein library (TRA_GFP-cDNA containing 2.0 × 107 independent inserts) and selected according to both high fluorescent intensity of the lipophilic cell membrane staining dye, DiI (D-282, Molecular Probes, Eugene, OR) with a fluorescence-activated cell sorter, MoFlo (Dako-Cytomation, Fort Collins, CO) and resistance to infection with retrovirally encoded diphtheria toxin alpha chain (DT-α virus) as described previously (Hitoshi et al., 2003). After two rounds of DT-α virus selection and DiI-based cell sorting, cells were plated for single-cell cloning in the presence of the tetracycline analog, doxycycline (Dox) to suppress GFP-cDNA expression. After careful observation, 134 colonies were picked and analyzed for their effect on growth in both Dox-containing (expression off) and Dox-free media (expression on). The cDNA inserts from these clones were amplified by PCR, sequenced, recloned, and transduced into naïve A549.tTA cells to evaluate their effect on cell proliferation with the DiI staining intensity assay.
Cell Tracker Assay
A549.tTA cells were stained with the cell tracker dye, DiI, 1 d before infection, infected with tet-regulatable retroviruses expressing GFP fusion proteins, and incubated at 37°C for 5 d. After the incubation, fluorescent intensity of GFP and DiI was analyzed with MoFlo. The effect of GFP fusion proteins on proliferation was assessed with comparison of the DiI intensity distribution between the GFP-positive population (gating on subpopulations consisting of the high expressers) and the GFP-negative population in the same sample. Nondividing cells or slow growing cells remain DiI bright as a result of the decreased rate of cell division.
Quantitative Analysis of UHRF1 Transcripts
The real-time PCR method was used for quantification of UHRF1 mRNA. Expression of UHRF1 mRNA was normalized to that of 18S rRNA. The primer sets and the probe for UHRF1 are as follows: the UHRF1 forward primer 5′-CCAGCAGAGCAGCCTCATC-3′, the UHRF1 reverse primer 5′-TCCTTGAGTGACGCCAGGA-3′ and the UHRF1 probe 5′-FAM-CAAGAGCAACGCCAAGCTGTGGAAT-TAMRA-3′. Primers for quantification of 18S rRNA were purchased from Applied Biosystems (Foster City, CA). Total RNAs from human mammary gland epithelial cells (HMECs), human prostate epithelial cells (PrEC), human cervical carcinoma cells (HeLa), lung carcinoma cells (A549 and H1299), colon carcinoma cells (HCT116), and breast tumor cells (MCF7, MBA-MD231, and HCC1937) were isolated using RNA easy kit (Qiagen, Valencia, CA). Total RNA isolated from primary tumor tissues and associated normal tissues were purchased from Ardais (Lexington, MA). The real-time PCR was performed using a Taqman 8000 instrument.
siRNA Studies
The sequences used for the transient transfections were purchased from Dharmacon (Lafayette, CO) and are as follows: UHRF1-1 sense strand (AAACAGAUGGAGGACGGCCAdTdT), UHRF1–2 sense strand (AAUUGGGGCUGUACAAGGUCAdTdT), and luciferase control sense strand (CUUACGCUGAGUACUUCGAdTdT). The duplex 1 of UHRF1-1 was also introduced into cells as a short hairpin RNA (shRNA) under the regulation of a U6 promoter using a retroviral construct that also expressed GFP as a marker (Holland et al., 2005). Cells were seeded at 1 × 105 per well into a six-well dish 1 d before transfection. The next day, cells were transfected with 100 nM siRNA duplexes using Oligofectamine (Invitrogen) according to reagent technical literature. Twenty-four hours after transfection, siRNA-containing medium was removed and cells were transferred to a fresh six-well dish for Western or cell cycle analysis. For cell cycle analysis, cells pulsed for 4 h with 10 μM bromodeoxyuridine (BrdU) were trypsinized and fixed with cold 70% ethanol. The nuclei were isolated by digesting the fixed cells with 0.08% pepsin in 0.1 N HCl for 20 min at 37°C followed by neutralization with two volumes of 0.1 M sodium borate. The nuclei were vortexed, spun down, and resuspended in buffer containing 10 mM HEPES, pH 7.4, 150 mM NaCl, 4% fetal calf serum, 0.1% sodium azide, and 0.5% Tween-20. The resulting nuclear suspension was incubated overnight with FITC-conjugated α-BrdU followed by brief propidium iodide treatment before flow cytometric analysis.
GFP Positivity and Chemosensitization Assays
A549.tTA cells were infected with tet-regulatable retroviruses encoding either GFP or GFP-fused UHRF1 proteins. FACS analysis was performed on the total infected population at set intervals after infection to determine the percentage of GFP-positive cells. For chemosensitization experiments, retrovirus-infected cells were divided into two aliquots 3 d after infection. One aliquot was treated with drug, and the other aliquot was treated with dimethyl sulfoxide (DMSO) at the same dilution. Cells were treated with etoposide or cis-platinum for 3 h at 37°C, and washed with phosphate-buffered saline (PBS) and then fresh medium was added. Cells treated with either taxol or hydroxyurea contained those drugs in the medium for the duration of the experiment. All drugs were purchased from Sigma. At indicated time points after drug treatment, FACS analysis was performed on both the DMSO-treated and drug-treated populations to determine the percentage of GFP-positive cells.
In Vitro Autoubiquitination Assays
293T cells were transfected with myc-GFP or myc-GFP-UHRF1 protein expression constructs using Fugene (Roche Applied Science, Indianapolis, IN). Forty-eight hours after transfection, cells were washed with PBS and lysed in 20 mM HEPES, 300 mM NaCl, 1 mM EDTA, 0.1% NP-40. After clarification of lysates, myc-tagged proteins were immunoprecipitated using α-myc agarose conjugate (Santa Cruz Biotechnology, Santa Cruz, CA). After washing beads twice with lysis buffer, beads were washed twice with ubiquitination assay buffer (62.5 mM Tris, pH 7.5, 6.25 mM MgCl2, 50 μM ATP, 1.0 mM dithiothreitol [DTT]). Ubiquitination reaction was initiated by adding 5 ng human E1, 20 ng UbcH5, and 100 ng of flag-ubiquitin (J. Huang, unpublished results) to beads in a total reaction volume of 30 μl. Reactions were performed for 1 h at room temperature and then stopped by adding 30 μl 2× SDS-PAGE gel loading buffer.
For the plate-based ubiquitination assay, 300 ng of recombinant UHRF1 protein or 75 ng recombinant APC2/APC11 was incubated with 5 ng E1, 20 ng UbcH5, and 100 ng flag-ubiquitin in 62.5 mM Tris, pH 7.5, 6.25 mM MgCl2, 1.0 mM DTT, 0.05 mM ATP in 100 μl total volume in a 96-well Ni(II) coated plate (Pierce Biotechnology, Rockford, IL) for 1 h at room temperature. After washing with PBS, incorporated flag-ubiquitin was detected using mouse anti-flag antibody (Sigma), anti-mouse IgG-horseradish peroxidase (HRP; Jackson ImmunoResearch Laboratories, West Grove, PA), and SuperSignal chemiluminescent reagent (Pierce). Reactions were performed in quadruplicate.
Production of Recombinant UHRF1 Proteins
UHRF1 wild-type and mutant protein sequences were cloned into the pDEST10 baculovirus transfer vector using Gateway technology (Invitrogen) followed by transposition into DH10bac Escherichia coli (Invitrogen) to produce the recombinant baculoviruses. After a 48-h infection, SF9 cells were lysed in 25 mM Tris, pH 8.5, 0.5 M NaCl, 0.2% NP-40, 10% glycerol, and 1 mM Tris(2-carboxyethyl) phosphine (TCEP; Pierce). Cell lysates were clarified by centrifugation and then the supernatants were incubated with ProBond Ni(II) resin (Invitrogen). Bound protein was washed with 20 mM imidazole in lysis buffer and eluted with 250 mM imidazole. D-salt columns (Pierce) were used to transfer the protein into 25 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP-40, 10% glycerol, and 1 mM TCEP.
Fluorescence Microscopy
A549 cells expressing either GFP-UHRF1 wild-type or GFP-UHRF1 ΔRING proteins were fixed in 3.7% paraformaldehyde in PBS and then stained with Hoechst 33342. Cells were visualized using an Axiovert S100 fluorescent microscope (Carl Zeiss MicroImaging, Thornwood, NY) and imaged using a CCD camera (Hamamatsu Photonics K.K., Hamamatsu, Japan).
RESULTS
Isolation of an Antiproliferative Fragment of UHRF1
A dual selection screen for the isolation of antiproliferative molecules was performed using a GFP fusion protein library in a tetracycline (tet) regulatable retroviral vector using the protocol described (Hitoshi et al., 2003). The screen entailed both 1) positive selection for cells with high staining of a lipophilic cell membrane dye, DiI, indicative of reduced cell division; and 2) negative selection of cycling cells by infection with retrovirally encoded diphtheria toxin alpha chain (DT-α virus). In this screen, retrovirally delivered DT-α virus preferentially kills cycling cells due to the requirement of nuclear envelope breakdown for efficient retroviral integration (Roe et al., 1993).
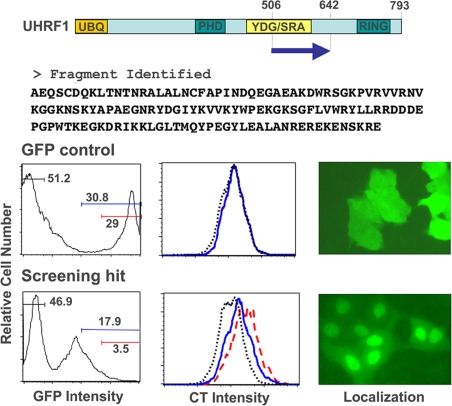
Of the hits identified, one clone encoded a fragment of UHRF1 containing amino acid positions 506–642 that is a part of the YDG/SRA domain. This domain in UHRF1 has been shown recently to be sufficient for binding to methylated CpG sites and for forming complexes with HDAC1 and DNA (Unoki et al., 2004). The isolated fragment of UHRF1, which was expressed as a GFP fusion protein, showed both reduction of cell growth in the absence of doxycycline, a condition where GFP-UHRF1 transcription occurs, and normal cell growth in the presence of doxycycline, where GFP-UHRF1 transcription is turned off. Because hits were isolated from the screen based on the fluorescence intensity of the cell tracker dye, DiI, we utilized the same dye-based proliferation assay to confirm the effect of both the GFP control and the GFP-fused UHRF1 hit on cell proliferation (Figure 1). The effects of these constructs on proliferation were assessed by comparing the DiI intensity distributions between the GFP-positive population (gating on either the entire positive population or on subpopulations consisting of the high GFP expressers) and the GFP-negative population within the same sample. The negative control, GFP, showed no significant difference in the DiI fluorescence intensity distribution between the GFP-positive and -negative populations. Cells infected with retroviruses encoding the fragment of UHRF1 exhibited higher DiI fluorescence in the GFP-positive population, indicative of their inhibitory effect on cell proliferation. The antiproliferative effect (maintenance of DiI staining intensity) was more pronounced in cells demonstrating higher GFP expression, indicating that the antiproliferative activity is dose-dependent. Because the hit identified in the functional screen was fused to GFP, we examined the localization of the GFP fusion in A549 cells expressing the tetracycline transactivator, A549.tTA cells (Figure 1). Although GFP alone was distributed uniformly throughout both the cytoplasm and the nucleus, the hit was concentrated in the nucleus.
Figure 1.
Identification of an antiproliferative fragment of UHRF1. The fragment (amino acid positions 506–642) was identified as a hit that showed both reduction of cell growth in the absence of doxycycline (Dox, a tetracycline analog) and normal cell growth in the presence of Dox. The position of the fragment in the full-length UHRF1 protein is indicated as an arrow and the amino acid sequence of the fragment is shown. The primary sequence of UHRF1 contains a UBQ domain (ubiquitin like, AA 14–89), a PHD domain (AA 330–379), a YDG/SRA domain (AA 427–599), and a RING finger domain (AA 724–762). A549.tTA cells were stained with the cell tracker dye, DiI, infected with tet-regulatable retroviruses expressing GFP or the GFP fused UHRF1 fragment, and incubated at 37°C for 5 d. The effect of the fragment on proliferation was assessed with comparison of the DiI intensity distribution between the GFP-positive population (gating on subpopulations consisting of the GFP-high expressers; blue lines and red lines in the figure) and the GFP-negative population (black lines) in the same sample. The localization of the GFP fused UHRF1 fragment and GFP alone in A549.tTA cells is shown in the right panels.
UHRF1 Is Required for Proliferation in Human Cells
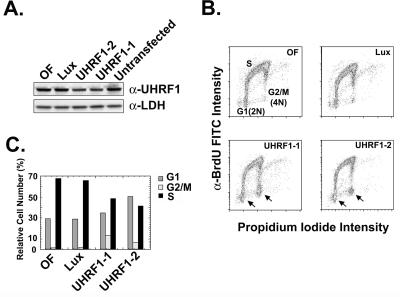
Our dye-based proliferation assay established that the 506–642 fragment of UHRF1 fused to GFP had antiproliferative effects in A549 cells. Previous functional studies using antisense oligonucleotides against mUHRF1 suggested that mUHRF1 may play a role in the G1 to S transition (Bonapace et al., 2002). To determine whether UHRF1 played a similar role in cell cycle progression as mUHRF1, H1299 lung tumor cells were transfected using several siRNAs targeted against UHRF1 (Elbashir et al., 2001). Western blot analysis of UHRF1 protein levels in H1299 cells harvested 24 h posttransfection (Figure 2A) demonstrated at least a twofold knockdown in UHRF1 protein levels with either UHRF1-specific siRNA 1 or 2.
Figure 2.
Down-regulation of UHRF1 in H1299 cells results in G1 and G2/M cell cycle arrest. (A) Reduction of UHRF1 protein in H1299 cells treated with oligofectamine or transfected with siRNAs against either luciferase or UHRF1. Measurement was performed 24 h after transfection. Lactate dehydrogenase was used as the loading control. (B) Flow cytometric analysis of siRNA transfected H1299 cells pulsed with BrdU 72 h after transfection and then stained with FITC-conjugated α-BrdU and propidium iodide. DNA content is indicated on the x-axis and extent of BrdU incorporation is indicated on the y-axis. Arrows indicate increased G1 and G2/M phase cell populations for the UHRF1-transfected cells compared with the controls. Color coding in the plots goes in the order from blue to green to red, with red representing the highest cell density. (C) Graphical representation of flow cytometric data from B.
To measure changes in the cell cycle distribution in UHRF1 siRNA-transfected cells, flow cytometric analysis was performed 72 h after transfection on cells pulsed with BrdU. Reduction of UHRF1 mRNA levels in H1299 cells led to reproducible increases in the G1 and G2/M phase cell populations, as shown by the increase in the number of cells with either a 2N or 4N DNA content and low α-BrdU FITC intensity (Figure 2B, depicted graphically in Figure 2C) for UHRF1-specific siRNAs 1 and 2 compared with the controls. Accordingly, we also observe a concomitant decrease in the number of cells in S phase for the UHRF1 siRNA-treated cells. These data suggest that UHRF1 is required for progression from G1 to S phase and also for G2/M phase progression.
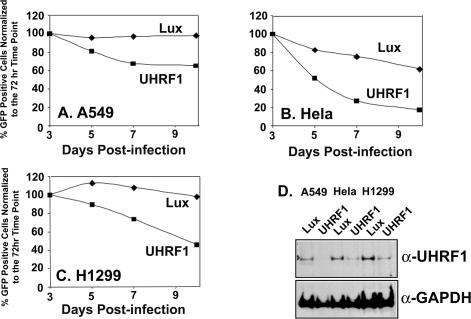
Reduction of UHRF1 message levels as measured by quantitative PCR analysis was most efficient in siRNA-transfected H1299 cells compared with other cell lines such as A549 and HeLa (unpublished data). Depending on the turnover rate of UHRF1 mRNA and protein levels in different cell types, the ability to detect siRNA-dependent phenotypes is likely increased in cell lines with the most efficient message knockdown due to the transient nature of siRNA effects. To circumvent this experimental limitation, UHRF1 siRNA sequence number 1 was incorporated into a retroviral vector for sustained delivery of the UHRF1 shRNA to target cells. In addition to the UHRF1 shRNA, the retroviral targeting vector also expressed GFP as a marker for cellular infection. A549, HeLa, and H1299 cells were infected with the UHRF1 shRNA retroviral targeting vector. As a control, the three cell lines were also infected separately with an shRNA targeting vector expressing an shRNA against luciferase. Three days after infection, UHRF1 shRNA-infected cells were separated from noninfected cells using flow cytometry and then analyzed for UHRF1 protein levels using an UHRF1-specific antibody. Potent down-regulation of UHRF1 protein by the UHRF1-specific shRNA but not by the shRNA against luciferase was observed in all three cell lines (Figure 3D). The effect of the UHRF1 shRNA on cellular proliferation was measured by analyzing the percentage of GFP-positive cells in the total cell population as a function of time. Although the luciferase shRNA vector had either no effect (A549 or H1299) or minimal antiproliferative effects (HeLa), the UHRF1 shRNA caused a dramatic reduction in the number of GFP-positive cells, consistent with its suggested role from our functional screen (Figure 3, A–C).
Figure 3.
Retroviral mediated delivery of UHRF1-specific shRNA is antiproliferative in A549, HeLa, and H1299 cells. (A–C) GFP positivity profiles for A549 (A), HeLa (B), and H1299 (C) cells infected with retrovirus expression either UHRF1-specific shRNA or control luciferase (Lux) shRNA. The number of GFP-positive cells at each time point is plotted relative to the %GFP-positive cells at day 3. (D) Reduction of UHRF1 protein levels in cells infected with either retrovirus expressing UHRF1-specific shRNA or retrovirus-encoding control luciferase shRNA. Three days after infection, cells were sorted using the GFP marker present in the retroviral vector and lysed in gel running buffer, and then Western blot analysis was performed for the presence of UHRF1 protein and the loading control, GAPDH.
Primary Tumors Display Elevated Expression of UHRF1
In primary lung fibroblasts, UHRF1 expression is positively correlated with cellular proliferation status, whereas in tumor cells, its expression is uncoupled from cell cycle status and, moreover, elevated compared with levels in nontumor cells (Mousli et al., 2003). Elevated UHRF1 expression has also been shown in breast carcinomas (Hopfner et al., 2000). These expression analyses, along with our data showing that UHRF1 is involved in cell cycle progression, suggests that UHRF1 may be an attractive cancer target. Therefore we analyzed the expression of UHRF1 mRNA across several tumor and primary cell lines, as well as in different primary tumors using quantitative PCR analysis. No striking differences were observed when UHRF1 message levels were compared across a panel of tumor cell lines with the primary cell lines, HMECs and PrECs (Figure 4A). This is in contrast with the results obtained from quantitative PCR analysis of primary tumors. We analyzed UHRF1 mRNA levels in total RNA extracted from primary tumors from patients with lung, breast, or prostate carcinomas and compared it with UHRF1 mRNA levels in corresponding normal tissue. In both breast and lung samples, UHRF1 message levels consistently showed elevated expression in the tumor tissue versus the corresponding normal tissue across the entire population tested (Figure 4, B and C). When matched samples from individual patients were compared (unpublished data), the same trend was seen in prostate tumors, although the magnitude of the effect was not as dramatic as that observed in the breast and lung tumor samples (Figure 4B).
Figure 4.
Quantitative analysis of UHRF1 mRNA in several tumor cell lines (A) and primary tumors (B and C). The real-time PCR method was used for quantification of UHRF1 mRNA, as described in Materials and Methods. Data are shown as a mean (indicated as bars or circles) ± SD of triplicate reactions. Each circle represents a RNA sample from an individual patient. Expression levels of RNA from tumor tissues and tumor-associated normal tissues are shown as closed circles and open circles, respectively.
UHRF1 Displays RING-dependent Ubiquitin E3 Ligase Activity
UHRF1 contains a RING finger domain at its C-terminus. RING fingers are small zinc-binding domains with the consensus sequence C-X2-C-X9-39-C-X1-3-H-X2-3-C/H-X2-C-X9-39-C-X2-C, where X is any amino acid C is cysteine, and H, is histidine, and is a domain often associated with ubiquitin ligase activity (Freemont, 2000; Pickart, 2001). E3 ubiquitin ligases, in conjunction with E1 and E2 proteins, are involved in conjugation of ubiquitin to a protein substrate (Hershko and Ciechanover, 1998). The functional consequence of ubiquitin modification varies depending on the valency of ubiquitin addition (mono vs. polyubiquitination) or the site of cross-linking on the ubiquitin molecule in polyubiquitination (Zhang, 2003; Herrera and Triezenberg, 2004; Hochstrasser, 2004; Marmor and Yarden, 2004; Sun and Chen, 2004). The most familiar ubiquitin modification and outcome is polyubiquitination at lysine 48 leading to substrate degradation by the 26S proteasome. Murine UHRF1 has been shown to polyubiquitinate histones in vitro although the functional consequence of this specific modification is unclear (Citterio et al., 2004).
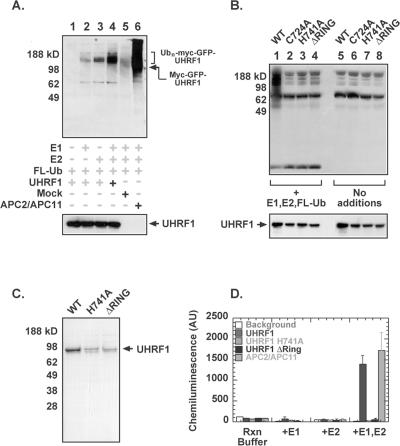
To determine whether UHRF1 has ubiquitin ligase activity in vitro, 293T cells were transfected with a myc-GFP-tagged UHRF1 construct. UHRF1 was immunoprecipitated using an anti-myc antibody and divided equally into four samples. Ubiquitination reactions were performed by addition of the following recombinant proteins: flag-tagged ubiquitin, human E1, and the E2 enzyme UbcH5. The UHRF1 ubiquitination reactions were performed in parallel with a complex composed of recombinant APC2/APC11 as a positive control. APC2 and APC11 are the cullin and the RING protein subunits of the Anaphase Promoting Complex, a RING domain containing multiprotein complex that is involved in the transition from metaphase to anaphase. In the presence of Ubc4, APC11 alone is sufficient for the formation of multiubiquitin chains (Gmachl et al., 2000; Leverson et al., 2000). Untransfected 293T lysates treated with anti-myc antibody were included as a negative control. Figure 5 shows the appearance of high-molecular-weight flag-tagged ubiquitin-protein conjugates in the presence of UHRF1, E1, and E2 (Figure 5A, lane 4). Some residual ubiquitination (albeit not as efficient) is seen when either E1 (lane 3) or E2 (lane 2) are omitted, which could be a result of coimmunoprecipitation of endogenous E1 or E2 with UHRF1.
Figure 5.
UHRF1 displays RING-dependent autoubiquitination activity in vitro. (A) 293T cells were transfected using a myc-GFP-UHRF1 expression construct. Forty-eight hours after transfection, cells were harvested and treated as described in Materials and Methods. The UHRF1-transfected lysates were split into four aliquots that were subjected to different combinations of enzymes required for the ubiquitination reaction. Recombinant APC2/APC11 was used as a positive control. Gel was probed using α-Flag antibody to look at extent of ubiquitination and also with α-Myc to look at protein expression levels. (B) 293T cells were transfected using myc-GFP-UHRF1 wild-type and RING finger mutant protein expression constructs and then treated as described in Materials and Methods. Gel was probed with α-Flag antibody to look at polyubiquitination (B, top panel) and also with α-Myc to look at protein expression levels (B, bottom panel). (C) Purified recombinant His6-UHRF1 wild-type, H741A, and ΔRING proteins expressed in SF9 cells. (D) Recombinant His6-UHRF1 proteins or APC2/APC11 were incubated with recombinant E1, UbcH5, and flag-ubiquitin in a 96-well Ni(II) coated plate. After washing to remove unbound proteins, incorporated flag-ubiquitin was detected using an anti-flag primary antibody and an HRP-conjugated secondary antibody.
Zinc coordination is required to maintain the structural integrity of the RING domain and hence has been shown to be absolutely required for E3 ligase activity. According to the consensus sequence of the RING domain, cysteine 724 and histidine 741 are residues in the UHRF1 RING motif that appear to be important for zinc binding and for ligase activity (Freemont, 2000). The point mutants UHRF1 C724A and UHRF1 H741A, as well as an UHRF1 RING finger deletion mutant (Δ724–762), were constructed in the myc-GFP tagged expression vector and transfected into 293T cells alongside the myc-GFP-UHRF1 wild-type construct. UHRF1 wild-type and RING mutant proteins were immunoprecipitated using an anti-myc antibody, and ubiquitination reactions were performed as previously described by adding recombinant flag-tagged ubiquitin, human E1, and E2. Figure 5B shows that disruption or deletion of the RING domain of UHRF1 abrogates its ubiquitination activity (compare lanes 2–4 with lane 1), demonstrating that the ubiquitination activity of UHRF1 requires an intact RING finger domain.
To confirm that the polyubiquitinated products were indeed due to the ligase activity of UHRF1 and not of a coimmunoprecipitated protein, we expressed and purified UHRF1 wild-type and mutant proteins from SF9 cells and tested them for their ligase activity using a ubiquitination assay in which recombinant E1, UbcH5, and flag-ubiquitin are added to His6-UHRF1 immobilized on a plate. Incorporation of the flag-ubiquitin is detected using a primary antibody against the flag epitope and a HRP-conjugated secondary antibody. Figure 5C is a Coomassie blue-stained gel of purified UHRF1 wild-type, H741A, and ΔRING proteins. The activity of these UHRF1 recombinant proteins in the plate-based ubiquitination assay is shown in Figure 5D. The results obtained with the recombinant UHRF1 proteins parallel the results obtained with transfected UHRF1 immunoprecipitated from 293T cells. The ubiquitination activity is dependent on the presence of both the E1 and E2 enzymes and requires an intact RING finger domain. The positive signal observed in the plate-based assay for UHRF1 wild-type protein along with the ubiquitination defects obtained for the RING finger mutants also shows that the polyubiquitinated protein in the immunoprecipitated UHRF1 experiment is autoubiquitinated UHRF1 and not polyubiquitination of a coimmunoprecipitated substrate.
The UHRF1 RING Domain Is Required for Survival in the Presence of Cytotoxic and Genotoxic Agents
Because UHRF1 appears to be critical for cellular growth, we wanted to determine whether the UHRF1 RING finger and its associated E3 ligase activity contributed to this function. To analyze the role of the UHRF1 RING finger on proliferation, we infected A549.tTA cells with retroviruses expressing either GFP alone, GFP fused to wild-type UHRF1, or GFP fused to a UHRF1 RING finger deletion mutant (Δ724–762), previously shown to lack ligase activity. We then monitored the change in the percentage of cells in the total population expressing either GFP or GFP-UHRF1 fusion proteins over a period of 10 d after infection by flow cytometry. The results of the analysis are plotted as the change in the GFP-positive population relative to the percentage GFP-positive cells recorded at the first time point monitored. Figure 6A shows that no significant changes in the percent GFP-positive cells are observed for all three constructs during the period monitored, indicating that the RING finger of UHRF1 does not appear to contribute to general cellular proliferation in A549.tTA cells.
Figure 6.
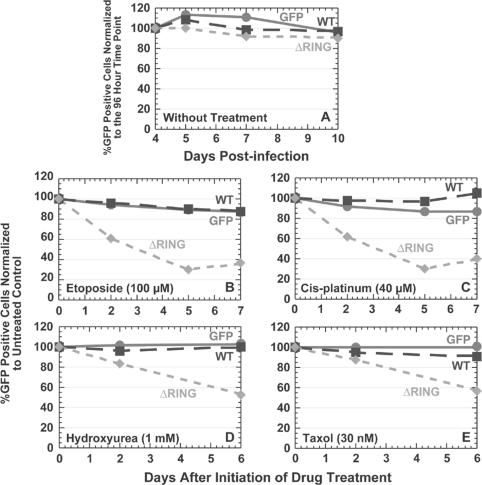
Overexpression of GFP-UHRF1 ΔRING mutant in A549 cells sensitizes cells to the effects of different chemotherapeutics. (A) GFP positivity profile for A549.tTA cells infected with retroviruses expressing GFP, GFP-UHRF1 wild-type, or GFP-UHRF1 ΔRING mutant proteins. The number of GFP-positive cells at each time point is plotted relative to the %GFP-positive cells at day 4. (B–E) GFP positivity profiles for A549.tTA cells infected with the same retroviruses and then treated with different drugs as described in Materials and Methods. The number of GFP-positive cells at each time point is plotted relative to the %GFP-positive cells in an untreated sample on the same day.
Murine UHRF1 knockout studies using murine embryonic stem cells showed no change in the proliferation rate in the absence of mUHRF1 but an increased sensitivity to the effects of chemotherapeutics, suggesting that mUHRF1 may play a role in the maintenance of genomic stability rather than general proliferation in this cell type (Muto et al., 2002). Because we did not see any proliferation defects upon overexpression of the GFP-UHRF1 ΔRING mutant, we decided to check if perhaps the UHRF1 RING finger played a role similar to that of mUHRF1 in the cellular response to either cytotoxic or genotoxic stress. We used a similar flow cytometric-based experiment where GFP-positive cells are counted over time except that 3 d after infection of A549.tTA cells, the samples were split, and half of the infected population treated with either genotoxic or cytotoxic agents. Instead of reporting the results as percent GFP-positive cells normalized to day 1, the results were plotted as percent GFP-positive cells in the drug-treated population normalized to the GFP-positive cells in the untreated population on the same day of data collection. Although no difference between the drug-treated and the control populations were found for cells overexpressing either GFP or GFP-UHRF1 wild-type proteins, a dramatic loss in GFP-positive cells was observed in cells overexpressing the GFP-UHRF1 ΔRING mutant (Figure 6, B–E) with etoposide, cis-platinum, hydroxyurea, and taxol treatment. The same results were obtained in both A549 and HeLa cells overexpressing GFP-UHRF1 C724A (Supplementary Material), a RING finger point mutant defective in E3 ligase activity (Figure 5B).
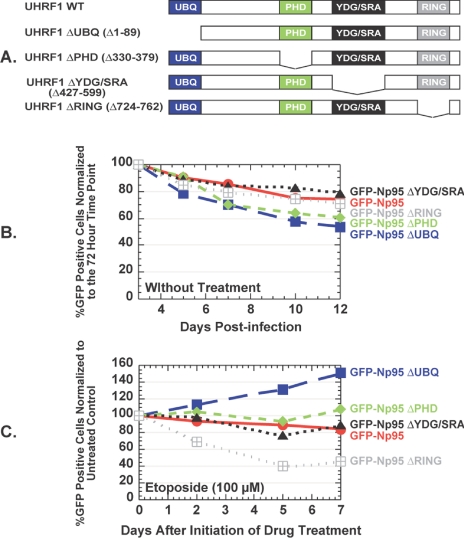
To determine whether the UHRF1 RING domain alone conferred this protective effect, we made GFP-UHRF1 deletion mutants (Figure 7A) lacking the ubiquitin-like domain (ΔUBQ), the PHD domain (ΔPHD), or the YDG/SRA domain (ΔYDG/SRA), all domains known to mediate protein-protein interactions. None of the deletion mutants displayed strong antiproliferative effects compared with wild-type UHRF1 in untreated A549 cells (Figure 7B). Although no effect was observed with the PHD and YDG/SRA deletion mutants, a profound loss of GFP-positive cells upon etoposide treatment occurred only with cells overexpressing the GFP-UHRF1 ΔRING mutant (Figure 7C). Surprisingly, the loss of cells containing GFP-UHRF1 ΔUBL is dramatically attenuated in the presence of drug.
Figure 7.
Cellular survival upon etoposide treatment requires the RING domain of UHRF1. (A) Schematic diagram of deletion mutants used in domain analysis. (B) GFP positivity profile for A549.tTA cells infected with retroviruses expressing different GFP-fused deletion mutants of UHRF1. The number of GFP-positive cells at each time point is plotted relative to the %GFP-positive cells at day 3. (C) GFP positivity profiles for A549.tTA cells infected with the same retroviruses and then treated with etoposide as described in Materials and Methods. The number of GFP-positive cells at each time point is plotted relative to the %GFP-positive cells in an untreated sample on the same day.
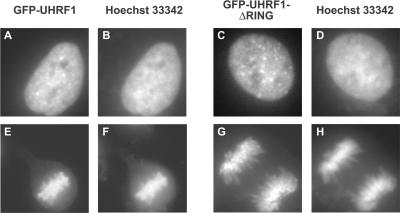
To rule out the possibility that the striking dominant negative effects observed for the RING mutant were due to cellular mislocalization caused by the GFP fusion, we looked at the localization of both GFP-UHRF1 wild-type and GFP-UHRF1 ΔRING proteins in A549 cells using the fluorescence of the GFP label. Both GFP-UHRF1 and GFP-UHRF1 ΔRING proteins are localized to the nucleus during interphase and remain chromatin associated during mitosis (Figure 8), localization that is consistent with the reported subcellular localization of endogenous mUHRF1 (Citterio et al., 2004). Thus, cellular survival appears to depend up an intact RING finger with presumably intact ubiquitin ligase activity when cells are challenged with these agents and disruption of the RING motif results in heightened sensitivity of cells to these agents.
Figure 8.
Both GFP-UHRF1 and GFP-UHRF1 ΔRING localize to the nucleus. A549.tTA cells were infected with either GFP-UHRF1 wild-type or GFP-UHRF1 ΔRING retrovirus. Three days postinfection, cells were fixed using 3.7% paraformaldehyde in PBS, stained with Hoechst 33342, washed with PBS, and imaged in a six-well dish using an inverted fluorescence microscope equipped with a CCD camera. Localization of GFP-UHRF1 is shown in A and E, localization of GFP-UHRF1 ΔRING is shown in C and G, and DNA staining is shown in B, D, F, and H.
UHRF1 mRNA and Protein Levels Are Down-regulated in Response to Cellular Damage
HCT116 cells treated with the DNA-damaging agent adriamycin have been shown to display a dramatic p53-dependent reduction in UHRF1 mRNA and protein levels (Arima et al., 2004). Reduction of UHRF1 protein under conditions of cell cycle arrest supports the requirement of cells for UHRF1 for growth (Figure 3). Our data suggests that under these conditions, UHRF1 must still be present because cells require UHRF1 RING-finger dependent ligase activity for their survival (Figure 6). We wondered if cells down-regulated UHRF1 levels in response to 1) DNA damaging agents other than adriamycin and 2) agents that activate other cell cycle checkpoints. A549 cells were treated with various genotoxic and cytotoxic agents, and then poly A RNA was isolated and analyzed for UHRF1 mRNA using quantitative PCR and cell extracts were analyzed by Western blotting for UHRF1 protein. We observed that all treatments used in our experiment had the effect of decreasing both the UHRF1 mRNA and protein levels (Figure 9, A and B). The extent of UHRF1 down-regulation was not similar for all treatments. Treatment with agents that cause DNA damage (cisplatinum, etoposide, and bleomycin) resulted in greater UHRF1 down-regulation than agents that either disrupted mitotic spindle formation (taxol and nocodazole) or inhibited DNA synthesis (hydroxyurea). We cannot at this time distinguish whether this dramatic decrease in UHRF1 levels observed with genotoxic agents is a direct effect caused by the damage itself or an indirect effect caused by activation of the G2 checkpoint. Regardless, cells that are induced to stop cycling by activation of different cell cycle checkpoints down-regulate UHRF1 and the extent of UHRF1 reduction appears to be treatment-dependent. At this time, we do not know if this dramatic down-regulation of UHRF1 occurs solely at the transcriptional level or is a result of transcriptional and translational effects. Addition of proteasome inhibitors to cells treated with genotoxic agents 8 h before harvesting had no significant effect on UHRF1 protein levels (unpublished data), suggesting that the loss of UHRF1 protein that is observed in this experiment is due to rapid transcriptional down-regulation coupled with slow protein turnover.
Figure 9.
UHRF1 mRNA (A) and protein (B) is down-regulated in response to genotoxic and cytotoxic agents in A549 cells. A549 cells were treated with different agents and then 24 h after initiation of treatment, cells were harvested for either quantitative PCR analysis of UHRF1 mRNA or Western blot analysis of UHRF1 protein and the loading control, actin. Data for PCR analysis are shown as a mean ± SD of triplicate reactions. Cells were treated with 100 μM etoposide or 40 μM cis-platinum for 3 h at 37°C and washed with PBS, and then fresh medium was added. Other drugs were left in the medium until cells were harvested (20 nM taxol, 1 mM hydroxyurea, 330 nM nocodazole, and 3 mU/ml bleomycin).
DISCUSSION
In this report, we show results characterizing UHRF1, a gene isolated from a proliferation-based functional genetic screen, as a critical regulator of human tumor cell proliferation. We also show that UHRF1 has RING-dependent E3 ubiquitin ligase activity and that cells overexpressing a GFP-fused UHRF1 RING mutant are rendered sensitive to the effects of chemotherapeutics, suggesting a role for UHRF1 ligase activity in the cellular response to cytotoxic damage.
UHRF1 Is Essential for Proliferation in Human Cells
Previous studies performed on mUHRF1 have collectively suggested a role for this UHRF1 homolog in the G1/S phase transition in mouse cells (Bonapace et al., 2002). Our results obtained in human tumor cells confirm and extend these results, suggesting that human UHRF1 is not only important but required for proliferation. We isolated a fragment encoding amino acids 506–642 from the UHRF1 gene from our proliferation-based functional screen. This fragment partially overlaps with the YDG/SRA domain (amino acids 414–586) that was shown to be sufficient for binding to methylated CpG sites and also for recruitment of HDAC1 complexes. This same study found that UHRF1 binds to methylated regions within the promoters of several tumor suppressor genes, including p16INK4A and p14ARF, suggesting that UHRF1 may regulate the expression of these genes through the recruitment of HDAC activity (Unoki et al., 2004). The homologous region in the mUHRF1 gene was identified as a histone-binding domain. Accordingly, mUHRF1 localizes to chromatin (Citterio et al., 2004). Although we have also observed UHRF1 localization to chromatin (Figure 6), the GFP-fused fragment is nuclear but does not appear to localize to chromatin (Figure 1). Thus, the dominant negative effect exerted by the UHRF1 fragment could be due to titration of binding factors critical for UHRF1 function, such as HDAC complexes. If UHRF1 did regulate transcription of tumor suppressors through recruitment of HDAC activity, then inhibition of this function by the GFP-fused UHRF1 fragment would activate tumor suppressor expression, resulting in an antiproliferative phenotype.
To further probe the requirement of UHRF1 for cellular proliferation, we used siRNA-mediated mRNA reduction to knock down UHRF1 levels. Cell cycle analysis of transiently transfected H1299 cells showed an increase of cells in G1 and a decrease of cells in S phase (Figure 2), similar to results obtained for mUHRF1. Additionally, we also observed for both of the UHRF1-specific siRNAs a reproducible increase in the number of cells in G2/M, indicating that UHRF1 function is critical during multiple phases of the cell cycle. Surprisingly, we see significant changes in the cell cycle distribution of UHRF1-transfected H1299 cells resulting from a reduction of UHRF1 protein levels of only about twofold. This result suggests that a threshold level may exist for UHRF1 protein levels required to support cell cycle progression.
When UHRF1 siRNA is delivered using a retroviral vector, significant growth defects are observed in A549, HeLa, and H1299 cells for the specific shRNA-infected population relative to the control luciferase shRNA-infected population (Figure 3). The extent of the antiproliferative effect is variable among the three cell lines and is not directly proportional to the amount of UHRF1 protein present after UHRF1 shRNA treatment. The results of this experiment would be consistent with HeLa and H1299 cells requiring a higher threshold level of UHRF1 for proliferation than A549 cells. Presently, we have not been able to determine for technical reasons whether the growth defect occurs through a cytotoxic activation of apoptosis or a cytostatic arrest caused by lack of UHRF1.
The striking difference in growth rates that we see with reduction of UHRF1 protein is both similar and in contrast to the findings obtained in murine cells. Although mUHRF1–/– ES cells exhibit no differences in general proliferation rate compared with wild-type ES cells (Muto et al., 2002), NIH 3T3 cells treated with antisense oligonucleotides to mUHRF1 do not enter S phase (Bonapace et al., 2002). Although mUHRF1 protein was not detectable in either experiment after gene targeting, as is suggested by the results of our shRNA experiment, differences may exist among cell types in the threshold level of UHRF1 protein required to support proliferation.
The UHRF1 RING Finger Is Critical for Ubiquitin Ligase Activity In Vitro and Plays a Role in Cellular Sensitization to Cytotoxic Agents
We tested and found that UHRF1 possesses RING-dependent ubiquitin ligase activity in vitro (Figure 5). We wanted to know what the functional consequence of deleting the RING finger-dependent ligase activity was on cellular proliferation. No obvious growth defects in A549 cells expressing the GFP-UHRF1 ΔRING mutant were apparent. However, treatment of these same cells with a cytotoxic agent revealed a striking difference in the proliferation rate. Although no change in the growth rate of cells infected with wild-type UHRF1 is observed with treatment, the percentage of cells harboring the UHRF1 ΔRING mutant decreases dramatically (Figure 6), implicating a role for this domain in cellular survival under these conditions. This is reminiscent of results obtained with mUHRF1–/– ES cells. No change in growth rate occurs in the mUHRF1–/– cells, but the viability of the cells becomes compromised when they are treated with genotoxic agents such as x-rays, UV light, and alkylating agents.
In human tumor lines, we find that knock down of the UHRF1 gene using siRNA induces a general proliferation defect and deletion of the UHRF1 RING finger causes a proliferation defect that is uncovered only when cells are damaged, suggesting that some ubiquitin ligase-independent activity of UHRF1 may be critical for its role in general proliferation and that the ligase activity of UHRF1 is important only in cells challenged by different environmental stresses. UHRF1 may play a key role in assembly of a protein complex that is required for cellular proliferation, a function supported by the presence of several domains such as a ubiquitin-like domain, a PHD domain, and a YDG/SRA domain, which have all been shown to be important for mediating protein-protein interactions.
Interestingly, putative substrates for mUHRF1 ubiquitination activity have been reported. All four nucleosomal core histones, histone H3, histone H2A, histone H2B, and histone H4 were shown to be ubiquitinated in vitro by mUHRF1 (Citterio et al., 2004). Histone H3 could also be coimmunoprecipitated with mUHRF1, demonstrating association of the two proteins in vivo. In addition, mUHRF1 also localizes to chromatin. That mUHRF1 ubiquitinates histones is a likely possibility, given the in vitro ligase activity and the localization data, but it is not clear what the consequence of histone H3 ubiquitination by mUHRF1 would be for cellular proliferation. Monoubiquitination of histones H2A and H2B has been linked to transcriptional activation but may occur through multiple pathways as an E2/E3 pair, Rad6p/Bre1p, has been shown to mediate monoubiquitination of histone H2B (Hwang et al., 2003; Wood et al., 2003). Additionally, substrates for the ubiquitination activity of mUHRF1 other than histones likely exist to account for the chemosensitization phenotype that we observe with the UHRF1 ΔRING mutant because no strong connection between histone ubiquitination and cellular recovery from different insults currently exists.
We have shown that UHRF1, as an E3 ubiquitin ligase, plays an important role in tumor cell growth. The success of proteasome inhibitors such as Velcade in the treatment of multiple myeloma has made the targeting of intracellular protein degradation pathways an attractive therapeutic strategy. Therefore, E3 ligases currently represent an important class of potential drug targets for anticancer therapy (Sun, 2003). Selective E3 ligase inhibitors should lead to higher specificity and lower toxicity for cancer patients. Taken together, our results suggest that UHRF1 is an attractive cancer target although further validation is needed.
Supplementary Material
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–03–0194) on September 29, 2005.
Abbreviations used: GFP, green fluorescent protein; tTA, tetracycline regulatable transactivator; siRNA, short interfering ribonucleic acid; shRNA, short hairpin ribonucleic acid; BrdU, bromodeoxyuridine; HMECs, human mammary epithelial cells; PrECs, human prostate epithelial cells.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Arima, Y., Hirota, T., Bronner, C., Mousli, M., Fujiwara, T., Niwa, S., Ishikawa, H., and Saya, H. (2004). Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells 9, 131–142. [DOI] [PubMed] [Google Scholar]
- Bonapace, I. M., Latella, L., Papait, R., Nicassio, F., Sacco, A., Muto, M., Crescenzi, M., and Di Fiore, P. P. (2002). Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J. Cell Biol. 157, 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio, E., Papait, R., Nicassio, F., Vecchi, M., Gomiero, P., Mantovani, R., Di Fiore, P. P., and Bonapace, I. M. (2004). Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell Biol. 24, 2526–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy, L., and Ganem, D. (2003). PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 13, 7–12. [DOI] [PubMed] [Google Scholar]
- Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Freemont, P. S. (2000). RING for destruction? Curr. Biol. 10, R84–R87. [DOI] [PubMed] [Google Scholar]
- Fujimori, A., Matsuda, Y., Takemoto, Y., Hashimoto, Y., Kubo, E., Araki, R., Fukumura, R., Mita, K., Tatsumi, K., and Muto, M. (1998). Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm. Genome 9, 1032–1035. [DOI] [PubMed] [Google Scholar]
- Gmachl, M., Gieffers, C., Podtelejnikov, A. V., Mann, M., and Peters, J. M. (2000). The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 97, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera, F. J., and Triezenberg, S. J. (2004). Molecular biology: what ubiquitin can do for transcription. Curr. Biol. 14, R622–R624. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hitoshi, Y. et al. (2003). Cellular localization and antiproliferative effect of peptides discovered from a functional screen of a retrovirally delivered random peptide library. Chem. Biol. 10, 975–987. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2004). Ubiquitin signalling: what's in a chain? Nat. Cell Biol. 6, 571–572. [DOI] [PubMed] [Google Scholar]
- Holland, S. J. et al. (2005). Multiple roles for the receptor tyrosine kinase axl in tumor formation. Cancer Res. 65, 9294–9303. [DOI] [PubMed] [Google Scholar]
- Hopfner, R., Mousli, M., Jeltsch, J., Voulgaris, A., Lutz, Y., Marin, C., Bellocq, J., Oudet, P., and Bronner, C. (2000). ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIa expression. Cancer Res. 60, 121–128. [PubMed] [Google Scholar]
- Hopfner, R., Mousli, M., Oudet, P., and Bronner, C. (2002). Overexpression of ICBP90, a novel CCAAT-binding protein, overcomes cell contact inhibition by forcing topoisomerase II alpha expression. Anticancer Res. 22, 3165–3170. [PubMed] [Google Scholar]
- Hwang, W. W., Venkatasubrahmanyam, S., Ianculescu, A. G., Tong, A., Boone, C., and Madhani, H. D. (2003). A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 11, 261–266. [DOI] [PubMed] [Google Scholar]
- Jackson, P. K., Eldridge, A. G., Freed, E., Furstenthal, L., Hsu, J. Y., Kaiser, B. K., and Reimann, J. D. (2000). The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10, 429–439. [DOI] [PubMed] [Google Scholar]
- Leverson, J. D., Joazeiro, C. A., Page, A. M., Huang, H., Hieter, P., and Hunter, T. (2000). The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol. Biol. Cell 11, 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorens, J. B., Jang, Y., Rossi, A. B., Payan, D. G., and Bogenberger, J. M. (2000). Optimization of regulated LTR-mediated expression. Virology 272, 7–15. [DOI] [PubMed] [Google Scholar]
- Marmor, M. D., and Yarden, Y. (2004). Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 23, 2057–2070. [DOI] [PubMed] [Google Scholar]
- Mori, T., Li, Y., Hata, H., and Kochi, H. (2004). NIRF is a ubiquitin ligase that is capable of ubiquitinating PCNP, a PEST-containing nuclear protein. FEBS Lett. 557, 209–214. [DOI] [PubMed] [Google Scholar]
- Mori, T., Li, Y., Hata, H., Ono, K., and Kochi, H. (2002). NIRF, a novel RING finger protein, is involved in cell-cycle regulation. Biochem. Biophys. Res. Commun. 296, 530–536. [DOI] [PubMed] [Google Scholar]
- Mousli, M., Hopfner, R., Abbady, A. Q., Monte, D., Jeanblanc, M., Oudet, P., Louis, B., and Bronner, C. (2003). ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br. J. Cancer 89, 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto, M., Kanari, Y., Kubo, E., Takabe, T., Kurihara, T., Fujimori, A., and Tatsumi, K. (2002). Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J. Biol. Chem. 277, 34549–34555. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Roe, T., Reynolds, T. C., Yu, G., and Brown, P. O. (1993). Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12, 2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., and Chen, Z. J. (2004). The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16, 119–126. [DOI] [PubMed] [Google Scholar]
- Sun, Y. (2003). Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol. Ther. 2, 623–629. [PubMed] [Google Scholar]
- Unoki, M., Nishidate, T., and Nakamura, Y. (2004). ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 23, 7601–7610. [DOI] [PubMed] [Google Scholar]
- Wood, A. et al. (2003). Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 11, 267–274. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. (2003). Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 17, 2733–2740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.