Abstract
Differentiation of pluripotent embryonic stem (ES) cells through multipotent neural stem (NS) cells into differentiated neurons is accompanied by wholesale changes in transcriptional programs. One factor that is present at all three stages and a key to neuronal differentiation is the RE1-silencing transcription factor (REST/NRSF). Here, we have used a novel chromatin immunoprecipitation-based cloning strategy (SACHI) to identify 89 REST target genes in ES cells, embryonic hippocampal NS cells and mature hippocampus. The gene products are involved in all aspects of neuronal function, especially neuronal differentiation, axonal growth, vesicular transport and release, and ionic conductance. Most target genes are silent or expressed at low levels in ES and NS cells, but are expressed at much higher levels in hippocampus. These data indicate that the REST regulon is specific to each developmental stage and support the notion that REST plays distinct roles in regulating gene expression in pluripotent ES cells, multipotent NS cells, and mature neurons.
INTRODUCTION
Neural development proceeds via a series of commitments whereby pluripotent embryonic stem (ES) cells give rise to more restricted multipotent neural stem (NS) cells, which in turn generate differentiated neurons. This is orchestrated and accompanied by wholesale changes in transcriptional programs and patterns of gene expression. One factor that has emerged as a key player is a Krüppel-type zinc-finger transcription factor, RE1-silencing transcription factor (REST; also known as the neuron-restrictive silencing factor, NRSF). Understanding the molecular mechanism underlying the REST regulatory roles during neuronal differentiation will shed light on the process of neuronal development. The first step in decoding the REST-directed mechanism is to profile its immediate target genes, which elucidate REST's possible biological functions.
REST interacts with RE1 sites and recruits corepressors including mSin3A (Huang et al., 1999, Roopra et al., 2000) and CoREST (Andrés et al., 1999, Ballas et al., 2001), which in turn, recruit multiple cofactors and transcriptional regulators including histone deacetylases 1 and 2, G9a histone methyltransferase (Roopra et al., 2004), H3 lysine 4 demethylase LSD1 (Shi et al., 2004), and chromatin remodeling machinery, including BRG1, BAF57, and BAF170 (Battaglioli et al., 2002) and BRAF35 (Hakimi et al., 2002). There are over 1300 RE1 sites in the human and murine genomes (Bruce et al., 2004; www.bioinformatics.leeds.ac.uk/group/online/RE1db/re1db_home.htm) and the majority encode ion channels, neurotransmitters, growth factors and hormones, and factors involved in axonal guidance and vesicle trafficking, and molecules involved in maintenance of the cytoskeleton and extracellular matrix (Schoenherr et al., 1996; Bruce et al., 2004). Because many of these target genes are expressed by postmitotic neurons, the role of REST was originally perceived as that of silencer of neuron-specific genes in nonneuronal cells (Chong et al., 1995; Schoenherr and Anderson, 1995). However, several observations have tempered this view. First, REST is expressed in many adult postmitotic neurons, most notably those of the hippocampus, where both REST and its target genes are modulated in the adult hippocampus in response to ischemic or epileptic insults (Palm et al., 1998; Calderone et al., 2003). Second, we have shown that, in most neural and nonneural cell lines, REST principally acts as a repressor of active gene transcription rather than as a silencer (Wood et al., 2003; Belyaev et al., 2004; Bruce et al., 2004). Third, REST has been shown to be sufficient to drive neuronal differentiation of adult neural stem cells (Kuwabara et al., 2004; Su et al., 2004) and even myoblasts (Watanabe et al., 2004). Fourth, REST is present at all stages of neural development from the blastocyst, through embryonic development in most tissues including the neuroepithelium. Finally, REST–/– mice are embryonic lethal around E10 and do not show widespread precocious expression of REST target genes and introduction of a dominant-negative REST construct into chick myotome caused up-regulation of some, but not all, REST target genes (Chen et al., 1998), whereas constitutive expression of REST down-regulated some target genes and caused axon pathfinding errors (Paquette et al., 2000). Taken together, these observations suggest that REST plays distinct roles in embryonic and adult cells and that this complexity might be reflected in the range of target genes that REST interacts with at different developmental stages.
In this study, we directly investigate this hypothesis by using a novel chromatin immunoprecipitation (ChIP) cloning strategy based on selective amplification of ChIP (SACHI) to identify REST target gene interactions in ES cells, hippocampal NS cell line, MHP36 (Kershaw et al., 1994) and mature hippocampus, all of which represent distinct stages of neuronal development. Using this technique, we identify 81 novel REST targets and further, we show that REST occupies a unique subset of target genes in each cell type.
MATERIALS AND METHODS
Preparation of Cells, Tissues, and cDNA
Embryonic stem cells, HM-1 (a gift from Dr. J. McWhir, the Roslin Institute), were cultured in Glasgow minimum essential medium (Sigma, St. Louis, MO) supplemented with 5% fetal calf serum, 5% new born calf serum, 1000 U/ml leukemia inhibitory factor (Chemicon, Temecula, CA), and the following reagents: 2 mM l-glutamine, 1% nonessential amino acids, 1 mM sodium pyruvate and 1% β-mercaptoethanol (Invitrogen, Carlsbad, CA). Neural stem cells, MHP36 (a gift from Prof. J. Price, King's College London), were plated out in 10-cm dishes precoated with 10 μl/ml fibronectin and cultured in DMEM/F12 medium in the presence of 47 μg/ml gentamicin and 200 μg/ml geneticin (Invitrogen). The medium was supplemented with 0.03% bovine serum albumin (BSA) Pathocyte (ICN, Costa Mesa, CA), 94 μg/ml human apo-transferrin, 15 μg/ml human putrescine dihydrochloride, 4.7 μg/ml bovine pancreatic insulin, 380 ng/ml l-thyroxine, 317 ng/ml tri-iodo-l-thyronine, 58 ng/ml progesterone, 1.9 mM l-glutamine, 38 ng/ml sodium selenite, 9.4 U/ml heparin sodium salt (all from Sigma), 10 ng/ml human bFGF, and 10 ng/ml murine interferon (PeproTech, Rocky Hill, NJ). Hippocampal tissues were collected by gross dissection from F1 mice (FVB/N × C57BL6) aged between 16 and 20 d. RNA was extracted using Tri reagent (Sigma) from cells or tissues. RNA was treated with DNAse (Ambion, Austin, TX) and cDNA was made from 2 μg RNA using MMLV (H-point mutant) reverse transcriptase and oligo dT and random hexamers as primers. cDNA was then purified through spin columns (Qiagen, Santa Clarita, CA) using ultra centrifugation and eluted in 50 μl of elution buffer.
Neuronal Differentiation
MH-1 ES cells were differentiated to NS cells and subsequently to mature neurons using monolayer culture in N2B27 medium (Ying et al., 2003). ES cells were plated onto 0.1% gelatin-coated cover slips in 24-well plates at a density of 1 × 104/cm2 in ES cell medium. The next day, the ES cell medium was replaced with N2B27 medium, which is a 1:1 mixture of DMEM/F12 medium supplemented with N2 and Neurobasal medium supplemented with B27 (all from Invitrogen). N2 consisted of 25 μg/ml insulin, 100 μg/ml apotransferrin, 6 ng/ml progesterone, 16 μg/ml putrescine, 30 nM sodium selenite, and 50 μg/ml BSA fraction V. Medium was changed every 2 d. Cells were processed for immunocytochemistry at day 0 (prior differentiation) and days 6, 9, and 18 after differentiation.
Immunostaining
Mice (C57BL6) were anesthetized with intraperitoneal Sagatal (60 mg/kg) and transcardially perfused with 4% paraformaldehyde (in 0.1 M phosphate buffer PB; pH 7.4). The brains were removed and postfixed in the same solution for 2 h at 4°C. Transverse sections (50 μm) were then cut on a vibrating microtome (Leica, Milton Keynes, United Kingdom) and collected into phosphate-buffered saline (PBS; pH 7.2). After 10-min washes three times in PBS, the sections were incubated with rabbit anti-REST primary antibody P18 (Santa Cruz Biotechnology, Santa Cruz, CA) for 12–18 h at 4°C. After 10-min washes three times in PBS, sections were incubated with a Cy3-conjugated anti-rabbit secondary antibody (1:1000 in PBS; Jackson ImmunoResearch, Luton, United Kingdom) for ∼ 4 h at room temperature. These sections were then washed 10 min three times in PBS, air-dried onto glass slides, and mounted under a coverslip using VectaMount mounting medium (Vector Laboratories, Peterborough, United Kingdom). The sections were viewed under a Nikon E600 microscope (Melville, NY) equipped with epifluorescence and images obtained via an integrating analogue CCD camera (JVC KYF 55B) attached to an Acquis image capture system (Synoptics Ltd, Cambridge, United Kingdom). The fluorescent images were converted to greyscale, inverted and adjusted for brightness/contrast/intensity and color balance using CorelDraw 11 (Corel Systems, Ottawa, Ontario, Canada).
ES cells, derived NS cells and neurons, and MHP36 cells were fixed in 3.5% paraformaldehyde for 20 min and stained with REST antibody (1: 300; Santa Cruz Biotechnology) alone or in combination with Sox1 (1:1000; Chemicon), Tuj1 (1:200; COVANCE, Princeton, NJ) and NeuN (1:500; Chemicon) antibodies overnight at 4°C.
Chromatin Immunoprecipitation
Cells (1 × 108 cells) or tissues (minced, 100 mg wet weight) were fixed for 10 min in 0.37% or 15 min in 1% formaldehyde, respectively. The fixation reaction was quenched with 0.125 M glycine. Nuclei were isolated by incubation on ice for 10 min in cell lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 0.2% NP40, 10 mM sodium butyrate, and protease inhibitors) and lysed in nuclei lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% SDS, 10 mM sodium butyrate, and protease inhibitors) for 10 min on ice. The lysate was then sonicated to shear the chromatin, cell debris was removed by centrifugation, and IP dilution buffer was added (20 mM Tris-HCl, pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton-X-100, 0.01% SDS, 10 mM sodium butyrate, and proteinase inhibitors). Chromatin was precleared for 3 h at 4°C with 200 μl of protein G-Sepharose blocked with 5% BSA. Whole cell extract was divided into aliquots: 270 μl kept as input chromatin and equal aliquots were incubated separately with anti-REST (P-18) and anti-HA (as a negative control) antibodies (Santa Cruz Biotechnology). Immune complexes were collected by rotation at 4°C for 3 h with 100 μl blocked protein G-Sepharose. Beads were washed twice with wash buffer 1 (20 mM Tris-HCl, pH 8.1, 50 mM NaCl, 2 mM EDTA, 1% Triton-X-100, and 0.1% SDS), once with wash buffer 2 (10 mM Tris-HCl, pH 8.1, 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% deoxycholic acid), and twice with TE buffer. Immune complexes were eluted with elution buffer (100 mM NaHCO3, 1% SDS) and cross-links were reversed by incubation at 65°C for 6 h in the presence of 5 μg RNase A and 0.3 M NaCl. Proteinase K, 90 μg, was added and samples were incubated overnight at 45°C. DNA was purified using phenol:chloroform extraction and ethanol precipitation. DNA from input sample was resuspended in 200 μl water, and DNA was pulled down by REST and HA antibodies in 100 μl water. Two-microliter aliquots were analyzed in real time PCR using primers designed adjacent to the RE1 regions.
ChIP Cloning
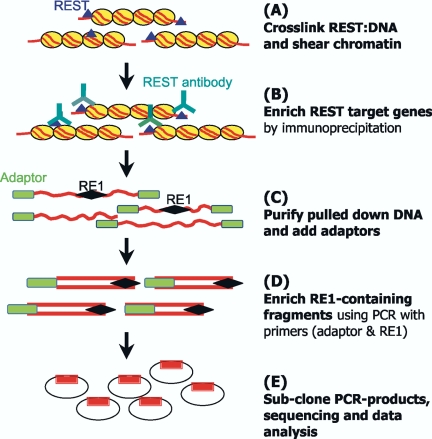
A cloning protocol was devised to specifically amplify RE1 containing sequences from the DNA output of anti-REST immunoprecipitated DNA. The principal of this procedure was to enrich target sequences by a nested PCR strategy using primers corresponding to the RE1 sequence and to end-ligated adaptors. The detailed procedure is summarized in Figure 1. After immunoprecipitation of fixed, sonicated chromatin but before elution of DNA from protein G-Sepharose, the sheared DNA ends were blunt-ended using mung bean nuclease, followed by adding phosphate to the 5′-ends of DNA using T4 polynucleotide kinase (BioLabs, St. Paul, MN). ChIP DNA was ligated with a specific adaptor [5′-CTAATACGACTCACTATAGGGCTCGAGCGGCCGCCCGGGCAGGT-3′]. The RE1-containing fragments were amplified from the enriched ChIP DNA by two rounds of PCR with primer pairs. First-round PCR used an RE1 degenerate primer 5′-[TT(C)AGA(C)G(A)CCA(T)A(T)GGACAG-3′] and adaptor primer A [CTAATACGACTCACTATAGGGC-3′]. PCR cycling parameters were 95°C × 5 min; 94°C × 10 s, 52°C × 20 min, 72°C × 2 min, for 10 cycles; 94°C × 10 s, 56°C × 20 min, 72°C ×x 2 min, for 25 cycles. Second-round PCR used the RE1 degenerate primer and a nested adaptor primer B [5′-TCGAGCGGCCGCCCGGGCAGGT-3′]. Cycling conditions were 95°C × 5 min; 94°C × 10 s, 58°C × 20 min, 72°C × 2 min, for 35 cycles. Second-round PCR-products were size selected at 200–800 base pairs and cloned into the pGEM-easy vector. Seven hundred clones from each cell line were isolated and sequenced. We found that the crucial factor for designing an effective RE1 degenerate primer was to contain no degeneracy within the first six base pairs of the 3′-end.
Figure 1.
Schematic diagram of ChIP cloning (SACHI) procedure. The technology consists of five steps (A–E). Details are given in Materials and Methods. Briefly, chromatin is cross-linked (A) and immunoprecipitated (B) to enrich for chromatin fragments containing RE1 binding sites. DNA is then purified and polished, and adaptors are ligated (C). RE1 containing fragments are further enriched by nested PCR (D) before subcloning and analysis (E).
Electrophoretic Mobility Shift Assay
The ability of predicted RE1s obtained from the ChIP cloning protocol to bind REST was assayed by electrophoretic mobility shift assay (EMSA), performed as previously described (Bruce et al., 2004). Nuclear protein extract from U373 expressing the REST DNA-binding domain was mixed with a 32P-labeled NaV1.2 consensus RE1 probe and 100-fold molar excess unlabeled 24–28-base pair oligonucleotides, derived from cloned REST target genes, were used to compete the gel shift.
Quantitative Real-time PCR
Primers were designed using the primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_slow.cgi) and “mfold” (http://www.bioinfo.rpi.edu/applications/mfold/old/dna) programs to obtain amplicons using the following parameters: Tm between 58 and 62°C, devoid of secondary structure at Tm 60°C and amplicon size of 50–150 base pairs. ChIP primers were derived from sequence within 500 base pairs of the RE1. Real-time PCR was carried out using SYBR green containing Supermix (Bio-Rad, Hercules, CA) on a Bio-Rad iCycler. All reactions were performed in duplicate. PCR cycle parameters were 95°C for 3 min, followed by 45 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. At the end of the program, the temperature was reduced to 50°C and then gradually increased by 1°C for 10 s up to 96°C, to produce a melt curve. Gene expression was normalized to cyclophilin using the formula 1/2ΔCt, where ΔCt = average unknown gene threshold cycle-average cyclophilin threshold cycle. The expression levels of cyclophilin in ES, MHP cells, and hippocampus showed no significant differences (Supplementary Figure 1).
Data Analysis
The sequences of clones obtained from ChIP cloning were blast-searched against Ensembl mouse database (http://www.ensembl.org/Multi/blastview?Species=Mus_musculus). Homologous sequences were investigated to identify the RE1 consensus: 5′-TY(TC)AGM(AC)R(AG)CCN(ACGT)NRGMCAG-3′ (Bruce et al., 2004). Sequences were classified according to 0, 1, 2, 3, or 4 base pairs mismatch to the consensus RE1. All fragments showing no deviation from the consensus RE1 were then annotated according to the closest genes.
RESULTS
REST Expression Profile during Neuronal Development In Vitro and In Vivo
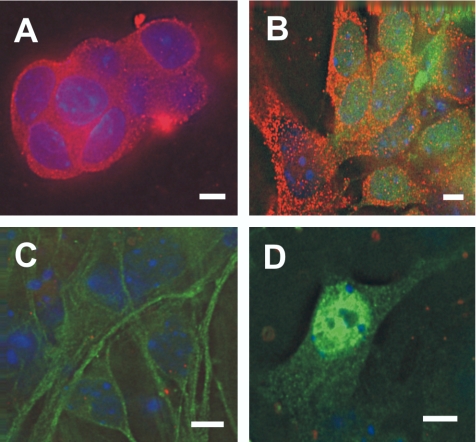
To establish the involvement of REST in neuronal development, we investigated REST expression profiles in neuronal differentiation in vitro and in vivo. The results showed that REST was expressed in both ES cells (Figure 2A) and its derived NS cells, identified by coexpression of the NS marker Sox1 (Figure 2B). Interestingly, no evident REST staining was seen in differentiated neurons either Tuj1+ or NeuN+ cells (Figure 2, C and D). REST was down-regulated in Tuj1+ cells (Figure 2C). We considered these cells as early neurons because some Tuj1+ cells were also nestin+ (unpublished data). The down-regulation of REST persisted in NeuN+ cells (Figure 2D). These observations contrast with the previous report that REST is initially down-regulated in neural stem cell, but are consistent with the observation that REST is down-regulated in early and mature neurons (Ballas et al., 2005). Our results suggest that, during ES cell differentiation, the expression profile of REST declines as neuronal differentiation proceeds.
Figure 2.
REST expression during embryonic stem (ES) cell differentiation into neurons. (A) REST (red) is expressed in both nuclei and cytoplasm of ES cells before differentiation. (B) REST (red) continues to be expressed in ES-derived neural stem cells identified by Sox1+ immunoreactivity (green). REST is down-regulated in differentiated early and mature neurons identified by Tuj1+ (C) and NeuN+ immunoreactivity (D; green). Nuclei are stained with DAPI (blue). Scale bar, 10 μm.
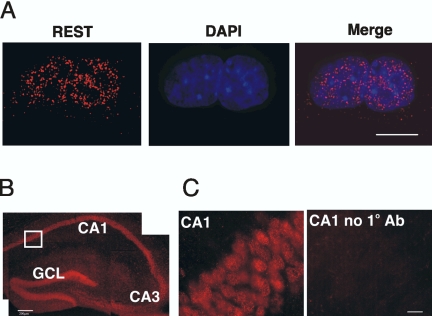
However, REST mRNA and protein have been reported to be expressed in both hippocampal neurons and Tuj1+ cells derived from adult hippocampal NS cells, respectively (Kuwabara et al., 2004). Accordingly we examined REST expression in an embryonic hippocampal NS cell line (MHP36 cells) and in hippocampal neurons. In MHP36 cells, nuclear REST was evident (Figure 3A). MHP36 cells are homogenous with regard to both the expression of REST and the progenitor marker, nestin (unpublished data). Furthermore, in hippocampus, REST was restricted to neuronal cell bodies throughout the pyramidal cell layer and the granular layer of the dentate gyrus (Figure 3B), a distribution that is consistent with the REST mRNA expression profile (Kuwabara et al., 2004). These results are different from those reported in early cortical neurons (Ballas et al., 2005). This is a key difference because it illustrates heterogeneity of REST expression within both progenitors and within differentiated postmitotic neurons. Accordingly, we used immunocytochemistry to demonstrate that REST was expressed in HM-1 ES cells, ES-derived NS cells, MHP36 NS cells, and in neurons of the mature hippocampus.
Figure 3.
REST immunoreactivity in embryonic hippocampal neural stem (NS) cells and in hippocampus (red). (A) REST is expressed in MHP36 NS cells. Nuclei are stained with DAPI in blue. Scale bar, 10 μm. (B) REST is expressed in CA1, CA2, and CA3 regions and the granule cell layer of the dentate gyrus (GCL) of hippocampus (Scale bar, 250 μm). (C) Higher magnification view of the boxed region in B. A negative control of the CA1 region is included (CA1 no 1° antibody). Scale bar, 25 μm.
ChIP Cloning
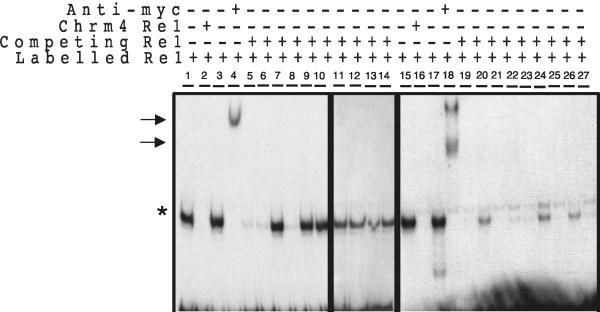
ChIP has become increasingly the method of choice to identify specific targets of transcription factors. However, the relatively poor enrichment afforded by ChIP means that the vast majority of isolated DNA is not specifically bound to the transcription factor. This massive background precludes any direct cloning approach aimed at identifying novel targets. Here, we have used a selection strategy based on the use of nested PCR to specifically amplify bona fide targets. By using a degenerate RE1 sequence as one PCR primer and two nested linker primers, we were able to amplify those genomic fragments that contained RE1 consensus sequences. Next, we distinguished those sequences that represented bona fide REST targets with EMSAs using unlabeled candidate RE1s to compete the gel shift produced by incubation of the NaV1.2 consensus RE1 with U373 nuclear extract. This analysis showed that all consensus RE1s inhibited the NaV1.2 gel shift with varying affinities, whereas only 30% of those sequences containing more than 1-base pair mismatches were bound by REST (Figure 4). Accordingly, in this study, we have focused only on those REST target genes adjacent to consensus RE1s.
Figure 4.
In vitro binding of predicted RE1s using EMSA. The ability of putative RE1 sequences to bind REST was judged by their ability to compete the shift produced by incubation of a radiolabeled rat NaV1.2 promoter RE1 with a myc-tagged REST DNA binding domain construct (REST DBD), when used at 100-fold molar excess. Lanes 5–14 represent RE1s diverging by more than 1 base pairs mismatch, while lanes 19–27 represent perfect-match consensus RE1s. Specific REST DBD:RE1 complexes are indicated by an asterisk (*). Arrows indicate complexes supershifted by an anti-c-Myc antibody. Lanes 2 and 16 represent the EMSA conducted in the presence of excess Chrm4 RE1, used as a positive control. Lanes 3 and 17 are EMSAs conducted in the presence of excess nonspecific ds oligonucleotide as a negative control. Lanes 4 and 18 are EMSAs conducted in the presence of an anti-myc antibody.
Identification of REST Target Genes in Embryonic Stem Cells, Neural Stem Cells, and Hippocampus
Ninety-three independent clones containing consensus RE1s were isolated from ES, NS, and hippocampus using our ChIP cloning procedure. Eighty-nine of these sequences were proximal to a known or predicted transcriptional unit. Sixty RE1 sites were located within intragenic regions, of which 50% were located in the first intron. Another 25 (28%) and 7 (7.8%) RE1s were located in the 5′flanking and 3′flanking regions, respectively (Tables 1 and 2). Four genes comprising L1cam, Syt2, GEF1, and ENSMUSG00000041339 were adjacent to, or contained, two RE1 sites. Thirty-eight genes (42%) were annotated in the Ensembl database (Supplementary Table 1) and were classified into six groups according to their biological functions (Table 1), whereas the remaining 51 genes (58%) corresponded to novel, predicted transcriptional units (Table 2). Of the annotated genes, 24 (65%) are expressed specifically or selectively in the nervous system (Table 1), including those concerned with neurite outgrowth, axonal guidance, vesicle trafficking, signaling, and transcription, consonant with the role of REST in regulating neuronal gene expression. The fact that more than 85% of these genes have not previously been shown to be direct targets of REST is testament to the power of this ChIP cloning protocol to reveal novel target genes.
Table 1.
Categorization of REST target genes from embryonic stem cells, embryonic hippocampal neural stem cells, and hippocampus
| Gene (Protein) | RE1 location | Biological process | Expression |
|---|---|---|---|
| Syt7 (Synaptotagmin-7) | (Intron 1) | Vesicular transport/release | Neuronal |
| Syt2 (Synaptotagmin-2) | (Intron 1 and 42 kb 5′) | Vesicular transport | Neuronal |
| Rab4a (Ras-related protein Rab-4A) | (3.2 kb 3′) | Vesicular transport | ? |
| Apba2 (Amyloid β A4 precursor protein-binding family A member 2) | (Intron 4) | Vesicular release | Neuronal |
| Foxp4 (Foxp4 protein) | (53 kb 5′) | Transcription | Neuronal |
| Fev (ETS-domain transcription factor) | (6.4 kb 5′) | Transcription | Neuronal |
| CREBBP/EP300 inhibitory protein 2 (E1A-like inhibition of differentiation 2) | (3.9 kb 5′) | Transcription | Widely expressed |
| Zfm1 (Nuclear protein, NP220) | (Intron I) | Transcription | ? |
| ENSMUSG00000058422 (human homolog LMTK2 kinase) | (1.7 kb 5′) | Signalling | ? |
| Resp18 (Regulated endocrine-specific protein 18 precursor) | (Intron 1) | Signalling | Neuronal |
| NM_172994 (human homolog PP2A, subunit B, B-γ isoform, Ppp2r2c) | (Intron 1) | Signal transduction | Neuronal |
| Cdk5r2 (Cyclin-dependent kinase 5 activator 2 precursor) | (2.9 kb 3′) | Signal transduction | Neuronal |
| Q8BY52 (BFA-resistant GEF1) | (0.7 kb 5′ and ex6/in5) | Signal transduction | Ubiquitous |
| Vrk3 (Serine/threonine-protein kinase VRK3) | (Intron 12) | Signal transduction | Non-neuronal |
| Pdgfra (α platelet-derived growth factor receptor precursor) | (40 kb 5′) | Organogenesis | Widely expressed |
| Drd2 (D(2) dopamine receptor) | (Intron 1) | Neurotransmission | Neuronal |
| Olfr1336 (Olfactory receptor MOR103-7) | (4.5 kb 5′) | Neurotransmission | Neuronal |
| Ntrk3 (Neurotrophin-3 receptor non-catalytic isoform 2) | (Intron 2) | Neuronal differentiation | Neuronal |
| Catnd2 (catenin δ-2, neurojungin) | (Intron 13) | Neurite outgrowth/cell adhesion | Neuronal |
| Cdh4 (Cadherin-4 precursor) | (Intron 1) | Neurite outgrowth/cell adhesion | Neuronal |
| Shank2 (SH3 and multiple ankyrin repeat domains protein 2) | (Intron 7) | Neurite outgrowth | Neuronal |
| L1cam (Neural cell adhesion molecule L1 precursor) | (15 kb 5′ and intron 1) | Neurite outgrowth | Neuronal |
| Extl3 (Exostosin-like 3) | (Exon 4) | Neurite outgrowth | Ubiquitous |
| Arc (Growth factor ARC) | (15 kb 5′) | Neurite outgrowth/cell-matrix | Neuronal |
| Cspg3 (Neurocan core protein precursor) | (Intron 1) | Neurite outgrowth/cell adhesion | Neuronal |
| Unc5d (Netrin receptor UNC5D precursor) | (Intron 1) | Neurite outgrowth/cell adhesion | Mesenchymal |
| Mbp (Myelin basic protein) | (63 kb 5′) | Myelination | Glial |
| Pkd1l3 (Polycystic kidney disease 1-like 3) | (Intron 14) | Ion channel | ? |
| Accn1 (Amiloride-sensitive cation channel 1, neuronal) | (Intron 1) | Ion channel | Neuronal |
| Cacna2d3 (Calcium channel α-2-δ-C subunit) | (Intron 1) | Ion channel | Neuronal |
| Kcnh1 (Potassium voltage-gated channel subfamily H member 1) | (Intron 10) | ion channel | Neuronal+muscle |
| Atp2b2 (Plasma membrane calcium-transporting ATPase 2) | (Intron 2) | Ion channel | Neuronal |
| Hcn3 (K/Na hyperpolarization-activated cyclic nucleotide-gated channel 3) | (Intron 1) | Ion channel | Neuronal |
| Kirrel3 (Kin of IRRE-like protein 3 precursor) | (74 kb 5′) | Hemopoeisis | Neuronal+stromal |
| Adam23 (ADAM 23 precursor) | (22 kb 3′) | Cell-matrix interaction | Neuronal |
| Crhr2 (Corticotropin-releasing factor receptor 2 precursor) | (Intron 3) | Angiogenesis | Widely expressed |
| ENSMUSG00000028804 (human homolog cub and sushi multiple domain 2) | (Intron 1) | ? | ? |
| Fndc1 (fibronectin type III domain containing 1) | (Intron 1) | ? | ? |
Table 2.
Hypothetical genes isolated by ChIP cloning using REST antibody
| NM_177374 (intron 1)a | GENSCAN00000142787 (intron 4) |
| NM_175461 (intron 2) | GENSCAN00000090077 (intron 1) |
| NM_198617 (17.6 kb 5′) | GENSCAN00000117131 (8.8 kb 5′) |
| NM_153584 (intron 1) | GENSCAN00000131824 (intron 4) |
| NM_173446 (intron 1) | GENSCAN00000072415 (intron 8) |
| Q8BS13 (8.9 kb 3′) | GENSCAN00000099949 (intron 2) |
| Q8BMM4 (54 kb 3′) | GENSCAN00000005818 (intron 1) |
| Q8BPU5 (intron 2) | GENSCAN00000001446 (intron 4) |
| Q8C817 (3.2 kb 5′) (human KIAA1913) | GENSCAN00000133834 (intron 2) |
| Q80T91 (intron 4) | GENSCAN00000123398 (intron 4) |
| ENSMUSG00000039098 (3 kb 5′) | GENSCAN00000164262 (2 kb 3′) |
| ENSMUSG00000034324 (intron 3) | GENSCAN00000161449 (0.6 kb 5′) |
| ENSMUSG00000041544 (5.6 kb 5′) | GENSCAN00000165253 (7 kb 5′) |
| ENSMUSG00000064130 (38.1 kb 5′) | GENSCAN00000119112 (intron 1) |
| ENSMUSG00000046182 (intron 2) | GENSCAN000000152268 (intron 1) |
| ENSMUSG00000061706 (17 kb 5′) | GENSCAN00000017182 (intron 1) |
| ENSMUSG00000028804 (intron 1) | GENSCAN00000180246 (intron 1) |
| ENSMUSG00000051719 (intron 1) | GENSCAN00000023281 (intron 3) |
| ENSMUSG00000038457 (intron 5) | GENSCAN00000123398 (intron 4) |
| ENSMUSG00000041339 (12 kb and 17 kb 5′) | GENSCAN00000149179 (5 kb 5′) |
| ENSMUSG00000051978 (intron 1) | GENSCAN00000134958 (intron 1) |
| ENSMUSESTG00000031508 (intron 1) | GENSCAN00000137792 (12 kb 5′) |
| ENSMUSESTG00000025135 (intron 2) | GENSCAN00000126950 (intron 9) |
| ENSMUSESTG00000018966 (intron 1) | GENSCAN00000129663 (intron 1) |
| ENSMUSESTG00000020906 (10 kb 5′) | GENSCAN00000057808 (intron 3) |
| GENSCAN00000103896 (17 kb 3′) |
The locations of RE1 sites are indicated in parentheses.
Validation of Bona Fide REST Target Genes
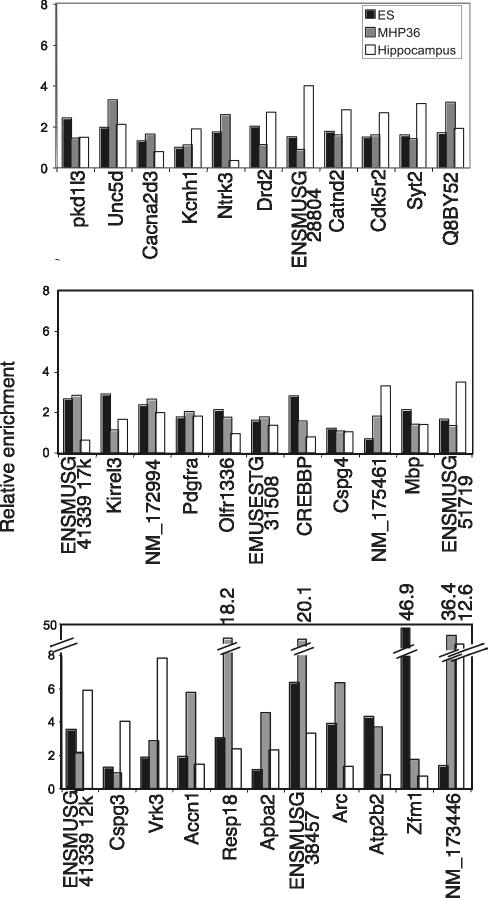
Validation of REST target genes isolated from ChIP cloning was carried out using conventional ChIP analysis. Thirty-three genes were randomly selected for testing from the isolated 89 genes and included 23 annotated genes and 10 predicted genes. Thirty-two genes (97%) were occupied by REST and showed greater than 1.5-fold enrichments in ChIP assays, in at least one of the three cell types (Figure 5). Four genes comprising Resp18 and ENSMUSG00000038457 in NS cells, Zfm1 in ES cells, and NM_173446 in NS cells and hippocampus exhibited markedly high enrichments of 18-, 20-, 46-, 36-, and 12.6-fold, respectively.
Figure 5.
Validation of the isolated RE1 sites from ChIP cloning using in vivo ChIP assay. Thirty-three RE1 containing regions were tested in conventional ChIP for REST occupancy in ES cells (black bars), NS cells (gray bars), and hippocampus (white bars). Only the RE1 associated with Cspg4 showed no enrichment for REST over background in any cell types, demonstrating the high efficiency of SAHI to identify bona fide REST targets.
The results showed that the equivalent RE1 sites were differentially bound by REST in ES, NS cells, and hippocampus. RE1 sites associated with Ntrk3, ENMUSG000000 41339/17k, Olfr1336, ENSMUSESTG00000031508, CREBBP/EP300 inhibitory protein 2, Accn1, Arc, Atp2b2, and Zfm1 genes were bound by REST in ES and NS cells, but not in hippocampus, whereas the RE1 sites associated with Drd2, ENSMUSG00000028804, Syt2, ENSMUSG00000015719, and Kirrel3 genes were bound by REST in ES and hippocampus, but not in NS cells. Conversely, RE1 sites associated with NM_175461, Apba2, and NM_173446 genes were bound by REST in NS cells and hippocampus, but not in ES cells. Furthermore, some RE1 sites were bound by REST in only one cell type including Pkd1l3 and Mbp genes in ES cells, the Cacna2d3 gene in NS cells, and Kcnh1 and Cspg3 genes in hippocampus. Only one RE1 site, which was located 30 kb upstream from the coding region of Cspg4 gene, failed to show any occupancy by REST in any cell type. Collectively, these data show that each cell type bears a unique REST occupancy signature.
Gene Expression Profile of the REST Differentially Targeted Genes
To examine the relationship between REST occupancy and expression of the corresponding gene, we carried out real-time quantitative PCR to determine the expression profile of REST target genes in each cell type. Of the 33 verified REST target genes, 20 were randomly selected to determine their expression pattern. One striking difference was evident in the contrast of the expression profile seen in ES and NS cells compared with that seen in the hippocampus. Although most genes were either silent or expressed at relatively low level in both ES and NS cells, with one exception, all genes were expressed at relatively high levels in hippocampus (Table 3). In ES cells, only the Olfr1336 was silent. In NS cells, 10 of the 20 genes were silent, whereas CREBBP/EP300 inhibitory protein 2, Kirrel3, and ENSMUSG00000051719 genes were expressed at relatively high levels. In hippocampus, only the olfactory receptor gene, Olfr1336, was silent. All other genes, including the five unannotated predicted genes were expressed at high levels relative to ES and NS cells.
Table 3.
Quantitative PCR analysis of REST target gene expression levels in embryonic stem (ES) cells, embryonic hippocampal neural stem (NS) cells, and hippocampus
| REST target genes | ES | NS | Hippocampus |
|---|---|---|---|
| Cspg3 | 0.3 | 0 | 230.8 |
| Arc | 82.0 | 19.0 | 276.0 |
| Syt2 | 4.0 | 0.4 | 115.0 |
| Apba2 | 48.0 | 0.7 | 6684.0 |
| Kcnh1 | 0.1 | 0 | 22.0 |
| Atp2b2 | 48.0 | 0 | 2179.0 |
| Accn1 | 12.0 | 0 | 1858.0 |
| Cacna2d3 | 14.0 | 10.0 | 468.0 |
| Drd2 | 5.0 | 0 | 101.0 |
| Olfr1336 | 0 | 2.0 | 0 |
| Ntrk3 | 0.6 | 0 | 2330.0 |
| Mbp | 7.0 | 0 | 39473.0 |
| Zfm1 | 727.6 | 5.5 | 5048.0 |
| CREBBP/EP300 inhibitory protein 2 | 239.1 | 214.5 | 1120.1 |
| Kirrel3 | 0.1 | 231.0 | 434.0 |
| NM_175461 | 9.0 | 6.0 | 1385.0 |
| NM_173446 | 0.3 | 0 | 2700.0 |
| ENSMUSG00000051719 | 4.0 | 688 | 21234.0 |
| ENSMUSG00000028804 | 1.0 | 0 | 741.0 |
| ENSMUSESTG00000031508 | 2.0 | 0 | 1773.0 |
Expression levels are shown relative to cyclophilin (×10-5).
Most of these target genes are reported to be specifically or selectively expressed in neurons. Exceptions include CREBBP/EP300 inhibitory protein 2 and Zfm1, which are widely expressed and Kirrel3, which is expressed in both neural and stromal cells. The fact that REST and so many target genes are coexpressed in the hippocampus, further underscores the principle role of REST in the adult CNS as a regulator, but not a silencer of neuronal gene expression.
DISCUSSION
Transcription factors play crucial roles in all aspects of cellular function including cell proliferation, differentiation, and survival. Yet, a key issue remains identification of their target genes. It is anticipated that the repertoire of target genes operated on by any given transcription factor will vary according to cell type and developmental stage. Despite this assumption, we have very little knowledge of the cell-specific regulon for any transcription factor in the genome of higher eukaryotes. Several strategies have evolved to identify transcription factor targets including ChIP-chip (Lee et al., 2002), ChIP Display (Barski and Frenkel, 2004), SACO (Impey et al., 2004), GMAT (Roh et al., 2005), and STAGE (Kim et al., 2005). All of these methodologies combine ChIP with SAGE, differential display or microarray to reduce the overwhelming contribution of nonspecific products to ChIP pulldown. Their success rate in identifying validated target genes ranged from 60 to 85%. In this study, we have developed a novel strategy that uses a selective amplification of ChIP (SACHI) to identify cell and stage specific interactions of REST with its target genes with a success rate up to 97%. Among the isolated REST target genes, Crhr2, Cdk5r2, Syt2, Syt7, Kcnh1, Cspg3, L1cam, and Ntrk3 have been identified in our previous study (Bruce et al., 2004). We have used this new strategy to identify 81 additional REST target genes from ES, NS, and hippocampus. This new protocol could also be used to isolate the target genes of other transcription factors as long as the length of their DNA binding sites exceeds five base pairs.
REST Target Genes and Neuronal Function
Many REST target genes involved in regulating core aspects of neuronal phenotype such as vesicular transport and release, signaling, and neurite outgrowth. Several known REST target genes are involved in neurite outgrowth and/or axonal guidance through regulation of intercellular and cell matrix interactions and of the intracellular cytoskeleton and include L1cam, Cdh4, Adam23, Catnd2, Ppp2r2c, and Unc5d (Gerhardt et al., 2000; Kim et al., 2002, Strack, 2002; Nishimura et al., 2003; Frappé et al., 2004; Goldsmith et al., 2004; Liu et al., 2004). This list can now be expanded by the addition of a further four REST target genes, Cspg3, Shank2, Extl3, and Arc, all of which are involved in regulation of cell adhesion or modulation of the cytoskeleton (Fujimoto et al., 2004; Osman et al., 2004; Qualmann et al., 2004). This cohort of target genes lends mechanistic insight into the axon pathfinding errors resulting from constitutive expression of REST (Paquette et al., 2000). Two new targets regulating vesicular trafficking and release include Rab4a and Apba2. These add to the known REST targets, Snap25, Syt2, Syt7, synapsin 1, and synaptophysin and illustrate the ability of REST to regulate every step of neurosecretion from trafficking, through docking to fusion and release.
REST Interacts with the Chromatin of Transcriptionally Active Genes in Differentiated Neurons
Our results show that REST occupies distinct subsets of target genes in ES cells, NS cells and hippocampus. A total of 89 genes were identified in this study, 38 of which are annotated, whereas 51 have no annotation and represent hypothetical genes. Of the 38 annotated REST target genes 24 (65%) encode gene products that are specifically or selectively expressed in neural tissue. Most target genes are transcribed at high levels in the hippocampus compared with ES and NS cells.
All target genes selected for expression analysis were transcribed in hippocampus, with the exception of Olfr1336. Most of the RE1 sites of these genes are occupied by REST. How does REST act principally as a repressor and become an activator in hippocampus? A previous study showed that NRSE dsRNA (RE1 dsRNA) appeared in the early stage of neurogenesis and in hippocampus. During the differentiation of NS cell to neurons, NRSE dsRNA triggered gene expression of neuron-specific genes through interaction with REST transcriptional machinery, by which REST was converted from a repressor to an activator (Kuwabara et al., 2004). If this is the case, why are not all the REST target genes occupied by REST? For example, five genes comprising Atp2b2, Cacna2d3, Ntrk3, Zfm1, and CREBBP/EP300 inhibitory protein 2 are actively transcribed but not occupied by REST in hippocampus. This indicates that existence of NRSE dsRNA cannot explain all facets of REST interaction with its target genes. Equally clear, is that REST cannot operate purely as an “on/off” switch whereby REST acts as a silencer.
REST Interaction with Target Genes in Stem Cells
In contrast to the hippocampus, the majority of target genes are silent in MHP36 neural stem cells and most are occupied by REST, irrespective of their transcriptional status. The only unoccupied sites lie within the Cspg3, Kcnh1, Drd2, and Kirrel 3 genes. The profile of REST occupancy in ES cells is very similar to that of MHP36 cells but unlike MHP36 cells, most REST target genes are transcribed in ES cells, albeit at low levels compared with the hippocampus. As with MHP36 cells, Cspg3, and Kcnh1 are both silent and occupied by REST. This low level of transcription likely reflects a transcriptionally poised state whereby the chromatin is an open configuration before full transcriptional activation upon differentiation (Schubeler et al., 2000; Ragoczy et al., 2003). In general, this study shows that many REST target genes are either silent or weakly expressed in ES and MHP36 cells but are strongly expressed in hippocampus. This may imply that REST is acting to silence or strongly repress these genes in both multipotent and pluripotent cells and then switches to become a regulator in postmitotic differentiated neurons.
A recent study reported that REST was present in ES cells but upon retinoic acid differentiation into neurons, REST was rapidly down-regulated in NS cells and was absent from both cultured cortical progenitors and cortical neurons (Ballas et al., 2005). However, in our study, REST continues to be expressed in NS cells, embryonic hippocampal stem cells, and ES cell-derived NS cells. In vivo, REST is expressed widely throughout the ventricular neuroepithelium (Schoenherr et al., 1995) and is present in many adult cortical neurons (Palm et al., 1998). In the hippocampus, most neurons of the pyramidal and granular layers express REST. In fact, REST levels and REST target gene levels are dynamically regulated in response to ischemic or epileptic insults (Palm et al., 1998; Calderone et al., 2003). Here, we have used the MHP36 neural stem cell line, derived from embryonic mouse hippocampus and which is capable of differentiating into neurons and glia both in vitro and in vivo (Mellodew et al., 2004; Hugnot et al., 2001). As such it represents a homogenous, tractable system to examine REST interactions with its target genes at the neural stem cell stage. In contrast to the study of Ballas et al. (2005) on cortical progenitors and neurons, we find REST is expressed in both hippocampal stem cells and neurons. This difference probably indicates diversity of REST actions in different populations of CNS progenitors and neurons but may also reflect differences in developmental stage.
In summary, we have used a novel ChIP cloning protocol (SACHI) to identify 89 REST target genes. We show that the REST occupies a distinct set of target genes in ES cells, NS cells and hippocampal neurons and that occupancy is not simply related to expression levels. The results suggest that the REST-directed mechanism in the gene expression of its target genes extends beyond the simple “on/off” model and the mechanism may be compounded by combinatorial regulation from other transcription factors or corepressors.
Supplementary Material
Acknowledgments
This work was funded by the Wellcome Trust.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–07–0687) on September 29, 2005.
Abbreviations used: RE1, repressor element 1; NRSE, neuron-restrictive silencer element; REST, repressor element 1 silencing transcription factor; NRSF, neuron-restrictive silencing factor; ES, embryonic stem cell; NS, neural stem cell; EMSA, electrophoretic mobility shift assay.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Andrés, M. E., Burger, C., Peral-Rubio, M. J., Battaglioli, E., Anderson, M. E., Grimes, J., Dallman, J., Balls, N., and Mandel, G. (1999). CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 96, 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas, N. et al. (2001). Regulation of neuronal traits by a novel transcriptional complex. Neuron 31, 353–365. [DOI] [PubMed] [Google Scholar]
- Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C., and Mandel, G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657. [DOI] [PubMed] [Google Scholar]
- Barski, A., and Frenkel, B. (2004). ChIP Display: novel method for identification of genomic targets of transcription factors. Nucleic Acids Res. 32, e104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglioli, E., Andres, M. E., Rose, D. W., Chenoweth, J. G., Rosenfeld, M. G., Anderson, M. E., and Mandel, G. (2002). REST repression of neuronal genes requires components of the hSWI.SNF complex. J. Biol. Chem. 277, 41038–41045. [DOI] [PubMed] [Google Scholar]
- Belyaev, N. D., Wood, I. C., Bruce, A. W., Street, M., Trinh, J-B., and Buckley, N. J. (2004). Distinct RE-1 silencing transcription factor-containing complexes interact with different target genes. J. Biol. Chem. 279, 556–561. [DOI] [PubMed] [Google Scholar]
- Bruce, A. W., Donaldson, I. J., Wood, I. C., Yerbury, S. A., Sadowski, M. I., Chapman, M., Göttgens, B., and Buckley, N. J. (2004). Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA 101, 10458–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone, A., Jover, T., Noh, K. M., Tanaka, H., Yokota, H., Lin, Y., Grooms, S. Y., Regis, R., Bennett, M. V., and Zukin, R. S. (2003). Ischemic insults derepress the gene silencer REST in neurons destined to die. J. Neurosci. 23, 2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z-F., Paquette, A. J., and Anderson, D. J. (1998). NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20, 136–142. [DOI] [PubMed] [Google Scholar]
- Chong, J. A., Tapia-Ramirez, J., Kim, S., Toledo-Aral, J. J., Zheng, Y., Boutros, M. C., Altshuller, Y. M., Frohman, M. A., Kraner, S. D., and Mandel, G. (1995). REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957. [DOI] [PubMed] [Google Scholar]
- Frappé, I., Wang, C., Caines, G., Rideout-Gros, S., and Aubert, I. (2004). Cell adhesion molecule L1 promotes neurite outgrowth of septal neurons. J. Neurosci. Res. 75, 667–677. [DOI] [PubMed] [Google Scholar]
- Fujimoto, T., Tanaka, H., Kumamaru, E., Okamura, K., and Miki, N. (2004). Arc interacts with microtubules/microtubule-associated protein 2 and attenuates microtubule-associated protein 2 immunoreactivity in the dendrites. J. Neurosci. Res. 76, 51–63. [DOI] [PubMed] [Google Scholar]
- Gerhardt, H., Rascher, G., Schuck, J., Weigold, U., Reidies, C., and Wolburg, H. (2000). R- and B-cadherin expression defines subpopulations of glial cells involved in axonal guidance in the optic nerve head of the chicken. Glia 31, 131–143. [PubMed] [Google Scholar]
- Goldsmith, A. P., Gossage, S. J., and ffrench-Constant, C. (2004). ADAM23 is a cell-surface glycoprotein expressed by central nervous system neurons. J. Neurosci. Res. 78, 647–658. [DOI] [PubMed] [Google Scholar]
- Hakimi, M. A., Bochar, D. A., Chenoweth, J., Lane, W. S., Mandel, G., and Shiekhattar, R. (2002). A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA 99, 7420–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Myers, S. J., and Dingledine, R. (1999). Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2, 867–872. [DOI] [PubMed] [Google Scholar]
- Hugnot, J. P., Pilcher, H., Rashid-Doubell, F., Sinden, J., and Price, J. (2001). Regulation of glial differentiation of MHP36 neural multipotent cell line. Neuroreport 12, 2237–2241. [DOI] [PubMed] [Google Scholar]
- Impey, S., McCorkle, S. R., Cha-Molstad, H., Dwyer, J. M., Yochum, G. S., Boss, J. M., McWeeney, S., Dunn, J. J., Mandel, G., and Goodman, R. H. (2004). Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119, 1041–1054. [DOI] [PubMed] [Google Scholar]
- Kershaw, T.R., Rashid-Doubell, F., and Sinden, J. D. (1994). Immunocharacterization of H-2Kb-tsA58 transgenic mouse hippocampal neuroepithelial cells. Neuroreport 5, 2197–2200. [DOI] [PubMed] [Google Scholar]
- Kim, K., Sirota, A., Chen, Y-H., Jones, S. B., Dudek, R., Lanford, G. W., Thakore, C., and Lu, Q. (2002). Dendrite-like process formation and cytoskeletal remodeling regulated by delta-catenin expression. Exp. Cell Res. 275, 171–184. [DOI] [PubMed] [Google Scholar]
- Kim, J., Bhinge, A. A., Morgan, X. C., and Iyer, V. R. (2005). Mapping DNA-protein interactions in large genomes by sequence tag analysis of genomic enrichment. Nat. Methods 2, 47–53. [DOI] [PubMed] [Google Scholar]
- Kuwabara, T., Hsieh, J., Nakashima, K., Taira, K., and Gage, F. H. (2004). A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116, 779–793. [DOI] [PubMed] [Google Scholar]
- Lee, T. I. et al. (2002). Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- Liu, G., Beggs, H., Jürgensen, C., Park, H-T., Gorski, J., Jones, K., Reichardt, L., Wu, J., and Rao, Y. (2004). Netrin requires focal adhesion kinase and Src family kinases for axon outgrowth and attraction. Nat. Neurosci. 7, 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellodew, K., Suhr, R., Uwanogho, D. A., Reuter, I., Lendahl, U., Hodges, H., and Price, J. (2004). Nestin expression is lost in a neural stem cell line through a mechanism involving the proteasome and Notch signalling. Brain Res. Dev. Brain Res. 151, 13–23. [DOI] [PubMed] [Google Scholar]
- Nishimura, K., Yoshihara, F., Tojima, T., Ooashi, N., Yoon, W., Mikoshiba, K., Bennett, V., and Kamiguchi, H. (2003). L1-dependent neuritogenesis involves ankyrinB that mediates L1-CAM coupling with retrograde actin flow. J. Cell Biol. 163, 1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, N.M.S., Naora, H., and Otani, H. (2004). Glycosyltransferase encoding gene EXTL3 is differentially expressed in the developing and adult mouse cerebral cortex. Brain Res. Dev. Brain Res. 151, 111–117. [DOI] [PubMed] [Google Scholar]
- Ragoczy, T., Telling, A., Sawado, T., Groudine, M., and Kosak, S. T. (2003). A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 11, 513–525. [DOI] [PubMed] [Google Scholar]
- Roh, T. Y., Cuddapah, S., and Zhao, K. (2005). Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19, 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra, A., Sharling, L., Wood, I. C., Briggs, T., Bachfischer, U., Paquette, A. J., and Buckley, N. J. (2000). Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell. Biol. 20, 2147–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra, A., Qazi, R., Schoenike, B., Daley, T. J., and Morrison, J. F. (2004). Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 14, 727–738. [DOI] [PubMed] [Google Scholar]
- Palm, K., Belluardo, N., Metsis, M., and Timmusk, T. (1998). Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J. Neurosci. 18, 1280–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette, A. J., Perez, S. E., and Anderson, D. J. (2000). Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc. Natl. Acad. Sci. USA 97, 12318–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann, B., Boeckers, T. M., Jeromin, M., Gundelfinger, E. D., and Kessels, M. M. (2004). Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J. Neurosci. 24, 2481–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr, C. J., and Anderson, D. J. (1995). The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Schoenherr, C. J., Paquette, A. J., and Anderson, D. J. (1996). Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. USA 93, 9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler, D., Francastel, C., Cimbora, D. M., Reik, A., Martin, D. I., and Groudine, M. (2000). Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P.A., Casero, R. A., and Shi, Y. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. [DOI] [PubMed] [Google Scholar]
- Strack, S. (2002). Overexpression of the protein phosphatase 2A regulatory subunit Bgamma promotes neuronal differentiation by activating the MAP kinase (MAPK) cascade. J. Biol. Chem. 277, 41525–41532. [DOI] [PubMed] [Google Scholar]
- Su, X., Kameoka, S., Lentz, S., and Majumder, S. (2004). Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol. Cell. Biol. 24, 8018–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Kameoka, S., Gopalakrishnan, V., Aldape, K. D., Pan, A. A., Lang, F. F., and Majumder, S. (2004). Conversion of myoblasts to physiologically active neuronal phenotype. Genes Dev. 18, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, I. C., Belyaev, N. D., Bruce, A. W., Jones, C., Mistry, M., Roopra, A., and Buckley, N. J. (2003). Interaction of the repressor element 1-silencing transcription factor (REST) with target genes. J. Mol. Biol. 334, 863–874. [DOI] [PubMed] [Google Scholar]
- Ying, Q. L., Stavridis, M., Griffiths, D., Li, M., and Smith, A. (2003). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.