Abstract
Kinetochores mediate chromosome attachment to the mitotic spindle to ensure accurate chromosome segregation. Budding yeast is an excellent organism for kinetochore assembly studies because it has a simple defined centromere sequence responsible for the localization of >65 proteins. In addition, yeast is the only organism where a conditional centromere is available to allow studies of de novo kinetochore assembly. Using a conditional centromere, we found that yeast kinetochore assembly is not temporally restricted and can occur in both G1 phase and prometaphase. We performed the first investigation of kinetochore assembly in the absence of the centromeric histone H3 variant Cse4 and found that all proteins tested depend on Cse4 to localize. Consistent with this observation, Cse4-depleted cells had severe chromosome segregation defects. We therefore propose that yeast kinetochore assembly requires both centromeric DNA specificity and centromeric chromatin.
INTRODUCTION
Accurate chromosome segregation in mitosis and meiosis is essential for the maintenance of genomic stability. Chromosomes attach to the mitotic spindle at the kinetochore, the protein complex that assembles onto centromeric DNA. Although kinetochore function is conserved, the underlying centromeric DNA is highly variable. Budding yeast contain a 125-base pair sequence-specific centromere that is sufficient for kinetochore formation (Fitzgerald-Hayes et al., 1982). In contrast, centromeres in multicellular eukaryotes are composed of megabases of highly repetitive DNA that lack sequence specificity (for review, see Sullivan et al., 2001). In these organisms, kinetochore assembly seems to be propagated by unidentified epigenetic component(s) (Karpen and Allshire, 1997; Sullivan et al., 2001).
The best-characterized kinetochore is in budding yeast where >65 components have been identified that constitutively localize to the kinetochore (for reviews, see Biggins and Walczak, 2003; McAinsh et al., 2003). Most of the yeast kinetochore proteins are found in biochemically distinct complexes known as the CBF3, CTF19/COMA, MTW1, NDC80, and DAM1 complexes that seem to assemble on a single centromeric nucleosome (Meluh et al., 1998). Although the exact architecture of the kinetochore is not known, dependency relationships subdivide the kinetochore into inner, central, and outer domains. The inner kinetochore contains the CBF3 complex (Ndc10, Cep3, Skp1, and Ctf13) as well as the DNA binding proteins Mif2, Cbf1, and the yeast centromeric histone H3 variant (CenH3) Cse4. CBF3 binds directly to the centromeric DNA and is thought to nucleate kinetochore assembly because all kinetochore proteins require it for localization (Russell et al., 1999; Goshima and Yanagida, 2000; He et al., 2001; Janke et al., 2001, 2002). The central kinetochore contains the MTW1 (Mtw1, Dsn1, Nnf1, and Nsl1) and CTF19/COMA (Ctf19, Mcm16, Mcm19, Mcm21, Mcm22, Ctf3, Chl4, Okp1, Ame1, Iml3, Nkp1, and Nkp2) complexes. CTF19/COMA can be further divided into two subcomplexes, with Ame1 and Okp1 in one subcomplex (C2.105) and Mcm21 and Ctf19 in another subcomplex (C2.100) (De Wulf et al., 2003). Outer kinetochore complexes include the conserved NDC80 (Ndc80, Spc24, Spc25, and Nuf2) and DAM1 (Dam1, Ask1, Duo1, Dad1, Dad2, Dad3, Dad4, Spc19, Spc34, and Hsk3) complexes. DAM1 is considered to be the outermost complex because it requires microtubules and all other complexes for kinetochore localization (Enquist-Newman et al., 2001; Janke et al., 2002; Li et al., 2002).
One hallmark of all kinetochores is the essential CenH3 (Palmer et al., 1987; Stoler et al., 1995; Buchwitz et al., 1999; Henikoff et al., 2000; Takahashi et al., 2000; Sanyal and Carbon, 2002; Talbert et al., 2002; Zhong et al., 2002; Edwards and Murray, 2005). CenH3s contain a unique N terminus and a well-conserved C terminus that is highly homologous to histone H3 (Malik and Henikoff, 2003). Because all active centromeres contain CenH3, it may be the epigenetic factor that specifies the site of kinetochore formation. Consistent with this hypothesis, RNA interference (RNAi) studies in worm, fly, and human cells have demonstrated that CenH3 is required for the localization of many kinetochore proteins (Blower and Karpen, 2001; Oegema et al., 2001; Goshima et al., 2003). However, several kinetochore proteins do not seem to require CenH3 for localization, suggesting that CenH3-independent assembly pathways also exist (Goshima et al., 2003; Hayashi et al., 2004; Regnier et al., 2005). In addition, the overexpression of human CenH3 fails to nucleate complete kinetochore assembly, despite driving CenH3 localization to euchromatin (Van Hooser et al., 2001). The precise function of CenH3 remains unclear for several reasons. Because RNAi-mediated CenH3 depletion occurs over many cell cycles, it is not known what phenotypes are direct consequences or secondary effects of the loss of CenH3. In addition, these studies cannot distinguish between maintenance and assembly of the kinetochore in the absence of CenH3. Finally, it is difficult to determine whether the CenH3 depletion is complete in multicellular eukaryotes that contain a large number of centromeric nucleosomes.
Little is known about kinetochore assembly in any organism. Budding yeast provide an excellent system to investigate kinetochore assembly due to the large number of identified kinetochore proteins and the defined centromeric DNA sequence. Here, we use a conditional centromere (cCEN), a system unique to budding yeast, to investigate kinetochore assembly in vivo (Hill and Bloom, 1987, 1989).
MATERIALS AND METHODS
Microbial Techniques
Media and microbial techniques were essentially as described previously (Sherman et al., 1974; Rose et al., 1990). All experiments in which cells were released from a G1 arrest were carried out by αF arrest and release, by using αF at 1 μg/ml. Nocodazole was used at 10 μg/ml. Doxycycline (dox) was used at 25 μg/ml, and dox treatment was maintained throughout all Degron-CSE4 experiments. Strains containing the cCEN were maintained in media containing 2% galactose. The cCEN was activated by growth in media containing 2% glucose. Yeast strains used in this study are listed in Table 1 and are derived from the W303 background. cCEN yeast strains were generated by integration of pR285#7 (pGAL-CEN3-URA3; Hill and Bloom, 1989) that was digested with XhoI to direct integration to the HIS4 locus. Strains containing pGAL-UBR1-myc were made by transforming PmeI-digested pLK54#300 (Labib et al., 2000) to direct integration to the UBR1 locus. All strains made by transformation of pLK54#300 were screened by immunoblot to ensure equal expression of UBR1-myc protein. All kinetochore proteins were epitope tagged by the PCR integration technique and generated fusions that are functional at the permissive temperature (Longtine et al., 1998). Primer sequences are available upon request.
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| SBY1524 | Mata CTF13-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY1525 | Mata MIF2-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY1526 | Mata CTF3-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY1823 | Mata DAM1-myc9:TRP1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY2061 | Mata MTW1-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY2770 | Mata NUF2-myc13:KanMX ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY3456 | Mata DAM1-myc9:TRP1 ura3-1 leu2-3112 his3-11 trp1-1 lys2Δ can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX HIS4:pGAL-CEN3-URA3 |
| SBY3910 | Mata ASK1-myc13:HIS3 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY3912 | Mata ASK1-myc13:HIS3 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX HIS4:pGAL-CEN3-URA3 |
| SBY3665 | Mata DAM1-myc9:TRP1 ura3-1 leu2-3112 his3-11 trp1-1 lys2Δ can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY3920 | Mata OKP1-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 HIS4:pGAL-CEN3-URA3 |
| SBY3933 | Mata bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 mad2ΔURA3 |
| SBY4355 | Mata CTF-myc13:KanMX ura3-1 leu2-3112 his3-11 trp1-1 lys2Δ can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4356 | Matα ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4391 | Mata bar1 ura3-1 leu2-3112 his3-11 trp1-1:SPC42-GFP:TRP1 lys2Δ can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 mad2ΔURA3 |
| SBY4452 | Mata MIF2-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 lys2Δ can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4453 | Mata MIF2-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4456 | Mata MTW1-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4457 | Mata MTW1-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4458 | Mata OKP1-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4459 | Mata OKP1-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 HIS4 pGAL-CEN3-URA3 |
| SBY4460 | Mata NDC80-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4461 | Mata NDC80-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4547 | Mata bar1 ura3-1 leu2-3112 his3-11 trp1-1:SPC42-GFP:TRP1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 mad2ΔURA3 |
| SBY4708 | Mata CTF19-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4711 | Mata CTF19-myc13:KanMX bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4712 | Mata STU2-myc13:HIS3 bar1 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4713 | Mata NDC10-myc13:KanMX ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1 pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4805 | Mata STU2-myc13:HIS3 ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc: HIS3 HIS4:pGAL-CEN3-URA3 |
| SBY4806 | Mata NDC10-myc13:KanMX ura3-1 leu2-3112 his3-11 trp1-1 can1-100 ade2-1:7tetop-UB-R-CSE4:ADE cse4ΔKanMX pGAL-UBR1-myc:HIS3 HIS4:pGAL-CEN3-URA3 |
All strains were generated for this study and are isogenic with the W303 background.
Strains Used in Figures
The following strains were used in the figures. For Figure 1, Ctf13-myc13 (SBY1524) (B), Mif2-myc13 (SBY1525) (C), Ctf3-myc13 (SBY1526) (D), Okp1-myc13 (SBY3920) (E), Mtw1-myc13 (SBY2061) (F), and Nuf2-myc13 (SBY2770) (G). For Figure 2, Dam1-myc9 (SBY3665). For Figure 3, Degron-CSE4, pGAL-UBR1-myc (SBY4355). For Figure 4, Ndc10-myc13 (CSE4+, SBY4713) and (Degron-CSE4, SBY4806) (A and E), Mif2-myc13 (CSE4+, SBY4453) and (Degron-CSE4, SBY4452) (B), Mtw1-myc13 (CSE4+, SBY4457) and (Degron-CSE4, SBY4456) (C and F), and Okp1-myc13 (CSE4+, SBY4459) and (Degron-CSE4, SBY4458) (D). For Figure 5, Ctf19-myc13 (CSE4+, SBY4711) and (Degron-CSE4, SBY4708) (A), Ndc80-myc13 (CSE4+, SBY4461) and (Degron-CSE4, SBY4460) (B), Dam1-myc9 (CSE4+, SBY1823) and (Degron-CSE4, SBY3456) (C), and Ask1-myc13 (CSE4+, SBY3910) and (Degron-CSE4, SBY3912) (D). For Figure 6, Degron-CSE4, pGAL-UBR1-myc, cse4Δ, mad2Δ (SBY3933). For Figure 7, Degron-CSE4, mad2Δ, Spc42-GFP, cse4Δ, pGAL-UBR1-myc (SBY4391) and mad2Δ, Spc42-GFP, pGAL-UBR1-myc (SBY4547). For Supplemental Figure, pGAL-UBR1-myc (SBY4356) (A), Okp1-myc13 (CSE4+, SBY4459) and (Degron-CSE4, SBY4458) (B), Mif2-myc13 (CSE4+, SBY4453) and (Degron-CSE4, SBY4452) (C), and Stu2-myc13 (CSE4+, SBY4712) and (Degron-CSE4, SBY4805) (D).
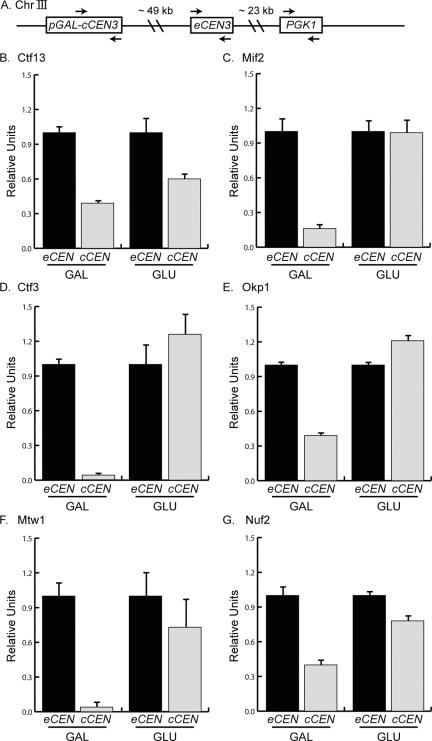
Figure 1.
Kinetochore proteins from each subcomplex can assemble in G1. (A). Schematic of chromosome III (Chr III) indicates the position of the cCEN (pGAL-cCEN3) and the negative control locus (PGK1) with respect to the eCEN (eCEN3). Arrows indicate the PCR primer sets used to detect these loci in ChIP experiments shown in the subsequent figures. (B–G). Strains expressing Ctf13-myc13, Mif2-myc13, Ctf3-myc13, Okp1-myc13, Mtw1-myc13, and Nuf2-myc13 were arrested in G1 with αF and then transferred to media containing GAL or GLU. ChIP was performed with anti-myc antibody. Relative units indicate the eCEN:PGK1 ratio or the cCEN:PGK1 ratio and are normalized to an eCEN:PGK1 value of 1.0 for each experiment. All kinetochore proteins tested localize to the active cCEN in G1.
Figure 2.
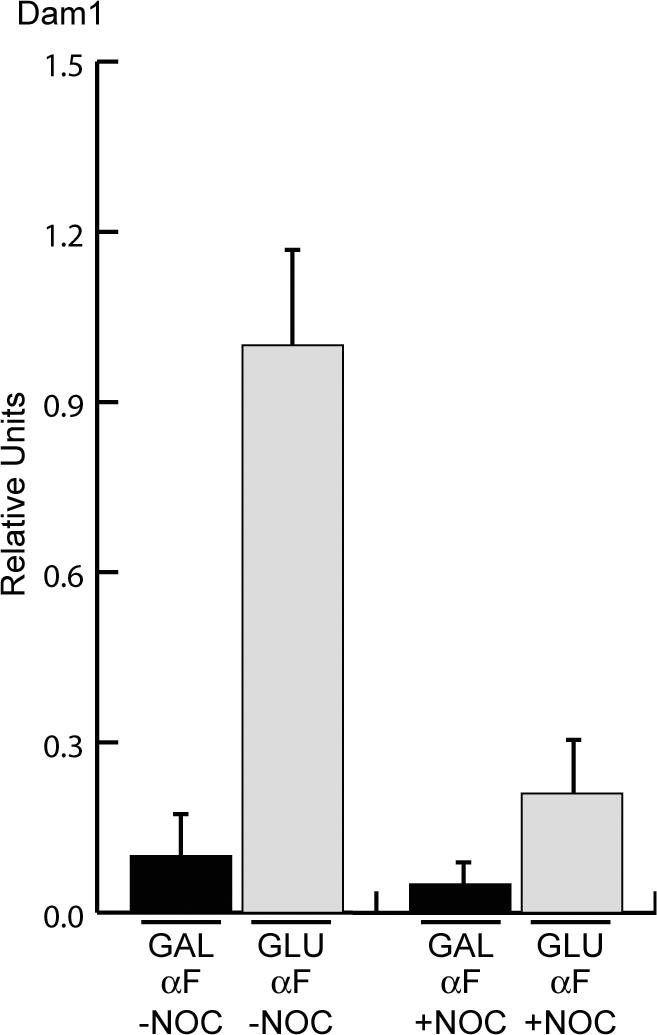
The G1 cCEN requires microtubules for kinetochore assembly. Cells expressing Dam1-myc9 were arrested in G1 with αFin galactose and then treated with (+NOC) or without (–NOC) the microtubule-depolymerizing drug nocodazole. Subsequently, the cultures were split into media containing GAL or GLU while maintaining the nocodazole treatment in αF. ChIP was performed with anti-myc antibody. Relative units indicate the cCEN:PGK1 ratio and the amount of Dam1-myc9 in GLU-NOC is normalized to a value of 1.0. Dam1 localization to the cCEN in G1 depends on microtubules.
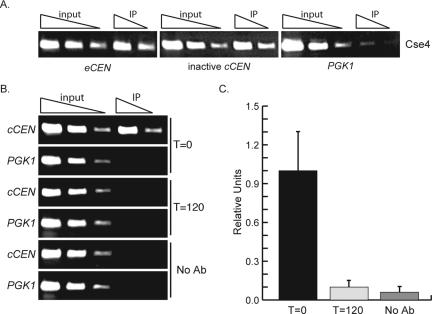
Figure 3.
Cse4 localizes to the inactive cCEN and can be depleted using a Degron-Cse4 protein. (A) Degron-CSE4 pGAL-UBR1-myc cells were grown in galactose and arrested with nocodazole. ChIP was performed with anti-Cse4 antibody. Serial dilutions of lysates (input) and immunoprecipitated DNA (IP) are shown. Cse4 localizes to the inactive cCEN at levels similar to the eCEN. (B) Cells were grown as described above (A), and dox was added to repress transcription of Degron-CSE4. ChIP was performed using either anti-Cse4 antibody or no antibody at the indicated times after dox addition. Cse4 is depleted from the cCEN within 120 min. (C). Quantification of B. Relative units indicate the cCEN: PGK1 ratio.
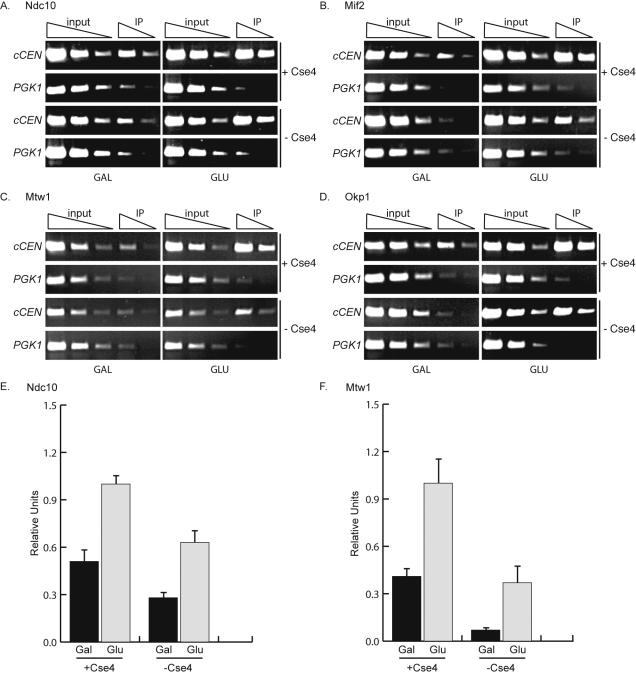
Figure 4.
Cse4 is required for the full occupancy of inner and some central kinetochore components. (A–F). pGAL-UBR1-myc cCEN cells expressing the indicated myc-epitope tagged kinetochore protein and either wild-type CSE4 (+Cse4) or Degron-CSE4 (–Cse4) were grown in media containing galactose and arrested with nocodazole. Dox was added to repress transcription of Degron-CSE4, and cells were transferred into media containing GAL (inactive cCEN) or GLU (active cCEN) and analyzed by ChIP with anti-myc antibody. (E and F) Quantification of A and C. Relative units indicate the cCEN:PGK1 ratio. The cCEN:PGK1 value in GLU +Cse4 is normalized to a value of 1.0. Cse4 is required for the complete occupancy of Ndc10-myc13, Mif2-myc13, Mtw1-myc13, and Okp1-myc13 (also see Supplemental Figure 1, B and C).
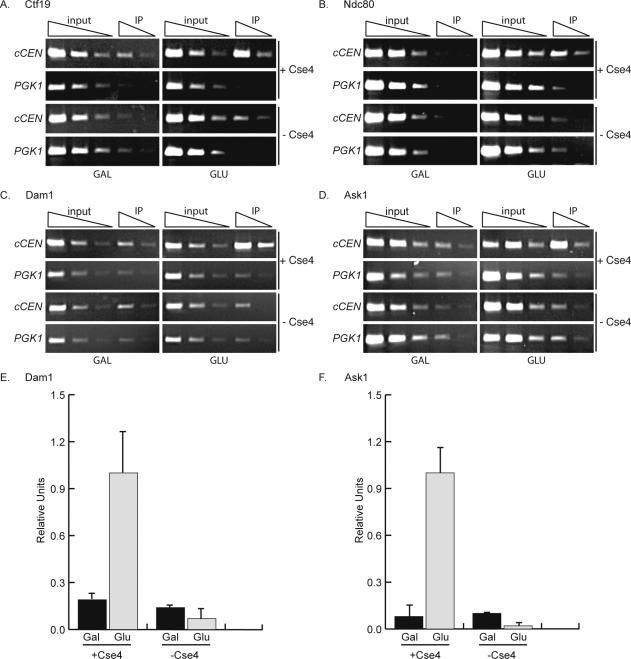
Figure 5.
Cse4 is required to localize some central and outer kinetochore components. (A and B). pGAL-UBR1-myc cCEN cells and the indicated myc-epitope tagged kinetochore protein with either wild-type CSE4 (+Cse4) or Degron-CSE4 (–Cse4) were grown in media containing galactose (cCEN inactive) and arrested in prometaphase with nocodazole. Dox was added to repress transcription of Degron-CSE4, and cells were transferred into media containing GAL (inactive cCEN) or GLU, active cCEN) and analyzed by ChIP with anti-myc antibody. Cse4 is required to localize Ctf19-myc13 and Ndc80-myc13 to the cCEN. (C and D). Cse4 was depleted from the cCEN in cells expressing Dam1-myc9 or Ask1-myc13 by incubation with dox for 4 h. Cells were transferred to media containing GAL (inactive cCEN) or GLU (active cCEN) in the presence of dox and then harvested for ChIP with anti-myc antibody. Cse4 is required to localize Dam1-myc9 and Ask1-myc13. (E and F). Quantification of C and D. Relative units indicate the cCEN:PGK1 ratio. The cCEN:PGK1 value in GLU +Cse4 is normalized to a value of 1.0.
Figure 6.
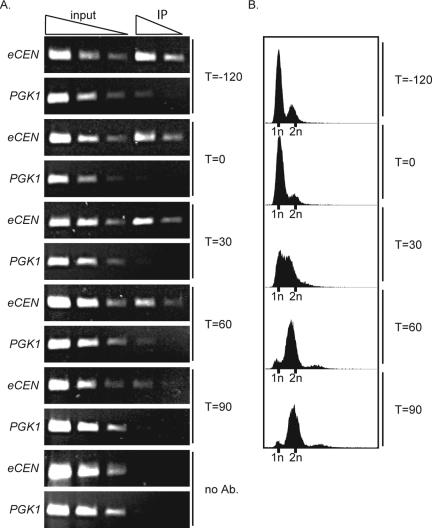
Cse4 can be depleted from the eCEN. Cells expressing Degron-CSE4 (Degron-CSE4 pGAL-UBR1-myc cse4Δ mad2Δ) were arrested in αF in media containing galactose. Dox was added to repress transcription of Degron-CSE4, and then cells were released into the cell cycle in the absence of Cse4. Cells were harvested for Cse4 ChIP (A) and FACS (B) at –120 min (when dox was added) and 0, 30, 60, and 90 min after αF release. Cse4 is completely depleted from the eCEN within 90 min.
Figure 7.
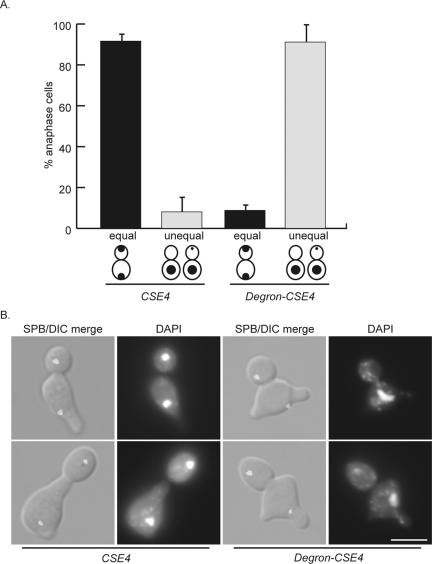
Cse4-depleted cells are defective in chromosome segregation. Cells expressing Degron-CSE4 (mad2Δ Spc42-GFP cse4Δ Degron-CSE4 pGAL-UBR1-myc) or wild-type Cse4 (mad2Δ Spc42-GFP pGAL-UBR1-myc) were grown in galactose and arrested in αF. Dox was added, and cells were released into the cell cycle in the absence of Cse4. After 150 min, cells were fixed and stained with DAPI. (A). At 150 min, cells that had segregated spindle pole bodies (anaphase cells) were scored as having equal DAPI masses (equal) or unequal DAPI masses (unequal). (B). Representative cells at the 150-min time point. The top images of the Degron-CSE4 show a cell with one, decondensed DAPI mass, and the bottom images represent a cell that has undergone unequal segregation where a small amount of DNA is in the bud. Bar, 5 μm.
Protein and Immunological Techniques
Protein extracts were made and immunoblotted as described previously (Minshull et al., 1996). 9E10 antibodies (Covance, Princeton, NJ) that recognize the Myc tag were used at a 1:10,000 dilution.
Chromatin Immunoprecipitation (ChIP) and Quantification
ChIP analysis was performed as described with the following modifications (Strahl-Bolsinger et al., 1997). Samples were fixed for 15 min, except Dam1 and Ask1 epitope-tagged cells, which required a 2-h fixation. Samples were washed once in 25 ml of Tris-buffered saline. Cells were lysed in 500 μl of ice-cold lysis buffer with glass beads by beating in the cold room (mini bead-beater; Biospec Products, Bartlesville, OK) for 2 × 30 s (1-min rest). Chromatin was sheared by sonicating 5 × 10 s using a Sonifier cell disruptor (model W185; Misonix, Farmingdale, NY) on setting 3 (average fragment size 500 base pairs) and clarified for 10 min in microfuge. Rabbit polyclonal anti-myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or Cse4 antibodies (Pinsky et al., 2003) were used for immunoprecipitations with protein G-conjugated Dyna beads (Dynal Biotech, Lake Success, NY). Washes were 1 min each. Primers used to amplify the eCEN3 locus (202 bp) were SB773 and SB774, the cCEN locus (389 bp) were SB712 and SB717, and PGK1 (242bp) were SB775 and SB776. Specific sequences are available upon request. Taq polymerase was used for all PCR amplifications (New England Biolabs, Beverly, MA). For all nonquantitative analyses, input and immunoprecipitated (IP) template concentrations were titrated into the linear range. PCR volumes were 25 μl, and primers were added to each reaction at a final concentration of 1 μM. Fivefold serial dilutions of the crude lysates (input) and immunoprecipitated DNA (IP) are shown in all figures. All IPs were confirmed to be equal by immunoblotting.
For quantitative analyses, input and IP template concentrations were diluted to give linear PCR amplification with 24 cycles. Only samples that fell in the linear range were included in the analysis, and all experiments shown are averages of at least two independent experiments with error bars representing 1 SD. PCR reagent concentrations were the same as described above. PCR products were resolved on 6% nondenaturing polyacrylamide gels for SYBR Green analysis and on 1.4% agarose gels for Vistra Green analysis. SYBR Green and Vistra Green were used at 1:10,000 in 1× Tris-Borate-EDTA for 10 and 60 min, respectively. SYBR Green was used in Figures 1, 2, 3 and 5, and Vistra Green was used in Figure 4. Relative amplification of each PCR product was determined using the Typhoon PhosphorImager and ImageQuant software (GE Healthcare, Little Chalfont, England). The efficiency with which each PCR product was amplified in the input was used to normalize the corresponding IP sample PCR products.
Flow Cytometry
Flow cytometry was performed as described previously (Hutter and Eipel, 1979) with propidium iodide (Sigma-Aldrich, St. Louis, MO). A BD Biosciences FACScan flow cytometer was used and analyzed on CellQuest software (BD Biosciences, San Jose, CA).
Microscopy
Microscopy was performed as described previously (Biggins et al., 1999) on a Nikon Eclipse E600 microscope with a 60× A/1.40 oil immersion lens. A CoolSNAPfx camera (Photometrics, Tucson, AZ) was used to acquire images. 4,6-Diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, CA) was used at 1 μg/ml final concentration. At least 100 cells were analyzed for all reported experiments.
RESULTS
Yeast Kinetochores Can Assemble in G1 Phase
It is not known what cell cycle stages are permissive for kinetochore assembly in budding yeast. One possibility is that kinetochore assembly is temporally restricted to occur after centromere replication. We therefore set out to address the cell cycle requirements for kinetochore assembly by using a cCEN (Figure 1A). The cCEN is integrated 49 kb to the left of endogenous centromere 3 (eCEN3) and is controlled by the galactose promoter (pGAL). When cells are grown in galactose media, transcription through the cCEN keeps it inactive. The addition of glucose represses transcription at the cCEN, allowing a functional kinetochore to form within 20 min (Hill and Bloom, 1989; Neff and Burke, 1992; Dewar et al., 2004; Tanaka et al., 2005).
To begin analyzing the cell cycle restrictions over kinetochore assembly, we first determined whether kinetochores could assemble in G1 cells. Kinetochore assembly at the cCEN was analyzed using ChIP. cCEN cells containing an epitope-tagged kinetochore protein were arrested in G1 by using the mating pheromone α factor (αF) and then shifted for 30 min into either galactose to keep the cCEN inactive or glucose to activate the cCEN. ChIP was performed to compare the localization of each protein at the endogenous centromere (eCEN) relative to the cCEN and a control locus, PGK1. We tested a representative subset of the yeast kinetochore complexes, including the inner kinetochore proteins Mif2-myc13 and Ctf13-myc13 (CBF3 complex), the central kinetochore proteins Ctf3-myc13 (CTF19/COMA complex), Okp1-myc13 (CTF19/COMA complex), and Mtw1-myc13 (MTW1 complex), and the outer kinetochore protein Nuf2-myc13 (NDC80 complex). As expected, all of the proteins were enriched to similar levels at the eCEN relative to PGK1 in both glucose and galactose, indicating that none of the proteins are limiting for assembly (our unpublished data). To determine the occupancy of proteins at the cCEN, the eCEN and cCEN localization were normalized to the control locus PGK1. We then set the eCEN enrichment to 1.0 and calculated the relative amount of protein at the cCEN. When the cCEN was inactive (GAL), the relative protein occupancy at the cCEN relative to the eCEN varied (Figure 1, B–G). Ctf13, Mif2, Okp1, and Nuf2 were at 10–30% occupancy relative to the eCEN, whereas Ctf3 and Mtw1 were not detected at the cCEN (Figure 1B). Although it is unclear whether partial occupancy reflects a population of cells with reduced protein binding or a small population of cells with fully occupied kinetochores, none of the proteins that we tested fully localized to the inactive cCEN during G1 phase.
We next tested whether the proteins could achieve full occupancy at the cCEN when it was activated (GLU). All proteins tested assembled to levels comparable with the eCEN (Figure 1), suggesting that the G1 phase of the cell cycle is permissive for kinetochore assembly. In addition, because the levels of protein assembled at the cCEN are comparable with the eCEN, the cCEN is a valid system for kinetochore assembly studies.
Although all of the kinetochore proteins tested assembled at the cCEN during a G1 arrest, these experiments did not address whether the newly assembled kinetochore is functional to bind microtubules. Because kinetochore proteins localized to the cCEN cannot be distinguished from the eCEN by microscopy, we could not perform a functional assay for microtubule binding to the cCEN (our unpublished data). We therefore analyzed the localization of the Dam1 protein to the cCEN as an indirect test for microtubule binding because Dam1 requires microtubules for its assembly at the kinetochore. Cells expressing Dam1-myc9 were arrested in G1 and then transferred into either galactose (GAL) or glucose (GLU) media and harvested for ChIP analysis. Similar to the other kinetochore proteins we assayed, Dam1-myc9 localized to the active cCEN in G1-arrested cells (Figure 2). To confirm that Dam1 localization to the cCEN is microtubule dependent, cells expressing Dam1-myc9 were arrested in G1 and then treated with or without the microtubule-destabilizing drug nocodazole (NOC) for 60 min. The cultures were then split into GAL or GLU media. As expected, ChIP analysis confirmed the previously reported observation that Dam1 localization to the eCEN depends on microtubules (our unpublished data) (Enquist-Newman et al., 2001; Li et al., 2002). To determine Dam1 occupancy at the cCEN, Dam1 localization to the active cCEN relative to PGK1 in the absence of nocodazole was set to 1.0 and then compared with the cCEN:PGK1 ratio in the other conditions. We found that Dam1 association with the cCEN also depends on microtubules (Figure 2). Thus, a kinetochore assembled on the cCEN in G1 can bind microtubules, suggesting that a functional kinetochore can assemble in G1 cells.
Cse4 Localizes to the Inactive cCEN and Can Be Removed by Using a Degradable Cse4 Protein
Because all of the kinetochore proteins tested localize to the cCEN in a glucose-dependent manner, the cCEN is an excellent system to further study kinetochore assembly. We were particularly interested in understanding the role of the budding yeast CenH3 in de novo kinetochore assembly. We first investigated whether Cse4 localizes to the inactive cCEN by performing ChIP with Cse4 antibodies in asynchronously growing cells, G1-arrested cells and prometaphase-arrested cells. Unlike the other kinetochore proteins analyzed above, Cse4 localized to the inactive cCEN at levels comparable with the eCEN (Figure 3A; our unpublished data).
To analyze kinetochore assembly in the absence of Cse4, it was first critical to deplete Cse4 from the inactive cCEN. Cse4 was therefore fused to an N-degron sequence (Degron-CSE4) to conditionally target it for destruction (Collins et al., 2004). Transcription of Degron-CSE4 is repressed by the addition of dox, and the protein is targeted for ubiquitin-mediated proteolysis by the N-end rule (Dohmen et al., 1994; Turner and Varshavsky, 2000). To enhance the kinetics of Cse4 depletion, the ubiquitin-protein ligase UBR1 was overexpressed from the inducible galactose promoter (pGAL) (Labib et al., 2000). Strains expressing Degron-CSE4 as the sole copy of Cse4 had similar occupancy at the inactive cCEN as the endogenous Cse4 protein (our unpublished data), consistent with our observations that Degron-Cse4 can complement a cse4 deletion (Collins et al., 2004). To determine whether Cse4 could be depleted from the inactive cCEN, pGAL-UBR1-myc cCEN Degron-CSE4 cells were grown in galactose and arrested in prometaphase with nocodazole to eliminate cell cycle variation and to prevent cells from attempting mitosis with dicentric chromosomes. Dox was added to repress transcription of Degron-CSE4 and ChIP analysis revealed that the Degron-Cse4 protein is depleted from the inactive cCEN by120 min (Figure 3B). To confirm that Cse4 was completely depleted, we performed quantitative PCR and found that the amount of Degron-Cse4 that remained at the inactive cCEN after 120 min is comparable with the control ChIP where antibody was not added (Figure 3C). In contrast, a significant amount of Degron-Cse4 protein remained at the eCEN 120 min after dox addition (Collins et al., 2004). The difference in the amount of Cse4 that remains at the eCEN compared with the cCEN may be due to the kinetochore structure protecting Cse4 from destruction at the eCEN and/or transcription aiding depletion at the cCEN.
Cse4 Is Required for Kinetochore Assembly
Having established a way to deplete Degron-Cse4 from the inactive cCEN, we next investigated the assembly of representative members of the kinetochore complexes in the absence of Cse4. pGAL-UBR1-myc cCEN cells containing an epitope-tagged kinetochore protein and either Degron-CSE4 or wild-type CSE4 were grown in galactose and arrested in prometaphase with nocodazole (cCEN inactive). Dox was added to repress transcription of Degron-CSE4, and then cells were shifted into either galactose (GAL, inactive cCEN) or glucose (GLU, active cCEN) and harvested for ChIP analysis. Quantitative PCR confirmed that Cse4 remained depleted when the cells were harvested (our unpublished data). In addition, a cCEN strain that expressed pGAL-UBR1-myc as the only myc-tagged protein was used in control ChIP experiments. As expected, Ubr1-myc does not localize to the cCEN, or the negative control locus, PGK1 (Supplemental Figure 1A). We also confirmed that pGAL-UBR1-myc expression does not alter the levels of any of the kinetochore proteins tested in Figures 4 and 5 (our unpublished data).
We first assayed kinetochore assembly in the absence of Cse4 by analyzing the localization of the inner kinetochore proteins Ndc10-myc13 (CBF3 complex) and Mif2-myc13 as well as the central kinetochore proteins Mtw1-myc13 (MTW1 complex) and Okp1-myc13 (CTF19/COMA complex). All of these proteins were enriched at the cCEN when it was activated in either the presence or absence of Cse4 (Figure 4, A–D). However, because the proteins did not seem to be enriched to the same extent, we performed quantitative PCR to determine whether the occupancy of the proteins at the cCEN was the same in the presence and absence of Cse4. The amount of protein that localized to the active cCEN relative to PGK1 in the presence of Cse4 was set to 1.0 and compared with the cCEN:PGK1 ratio for the other conditions. Because Cse4 at the eCEN was partially reduced under these conditions (Collins et al., 2004), a comparison of the cCEN to the eCEN was not possible. Ndc10 and Mtw1 both partially localized to the inactive cCEN in the presence of Cse4 (Figure 4, E and F). The occupancy of these proteins at the inactive cCEN was further reduced in the absence of Cse4, suggesting that Cse4 may have a role in localizing these proteins. Consistent with this observation, Ndc10 and Mtw1 did not achieve the same level of occupancy at the active cCEN in the absence of Cse4 compared with the presence of Cse4. Similar results were obtained for Mif2 and Okp1 (Figure 4, B and D, and Supplemental Figure 1, B and C), indicating that Cse4 is required to fully localize these proteins to the cCEN.
We extended our analysis of kinetochore assembly in the absence of Cse4 to the central kinetochore component Ctf19 (CTF19/COMA complex) and the outer kinetochore components Ndc80 (NDC80 complex), Dam1, and Ask1 (DAM1 complex). Cells containing Ctf19-myc13 and Ndc80-myc13 were grown as described above for the inner kinetochore proteins and harvested for ChIP. Ctf19-myc13 (Figure 5A) and Ndc80-myc13 (Figure 5B) were not enriched at the inactive cCEN and showed little enrichment at the active cCEN in the absence of Cse4. Immunoblot analysis confirmed that equal amounts of Ctf19-myc13 and Ndc80-myc13 were immunoprecipitated (our unpublished data).
We also analyzed Dam1 and Ask1, members of the outer kinetochore DAM1 complex. However, because this complex requires microtubules for assembly, the experiment was performed using a four-hour asynchronous Cse4 depletion instead of arresting cells in prometaphase with nocodazole. After 4 h of Cse4 depletion, 87% of the cells were arrested in metaphase. Therefore, although nocodazole was not added, the cells were arrested at the same cell cycle stage as the other experiments. In addition, quantitative PCR analysis determined that Cse4 depletion at the cCEN was complete under these conditions (our unpublished data). We found that Cse4 is required for the localization of the outer kinetochore proteins Dam1-myc9 and Ask1-myc13 (Figure 5, C–F). Consistent with the mislocalization of the outer kinetochore components, the microtubule associated protein Stu2-myc13 also failed to localize to the active cCEN in the absence of Cse4 (Supplemental Figure 1D). Immunoblot analysis confirmed that equal amounts of Dam1-myc9, Ask1-myc13, and Stu2-myc13 were immunoprecipitated (our unpublished data). In conclusion, inner kinetochore proteins and some central components show a partial dependency on Cse4 for localization, whereas other central and all outer kinetochore components completely require Cse4 for kinetochore localization. Together, these data indicate that Cse4 is essential for kinetochore assembly.
Cse4 Can Be Depleted from the Endogenous Centromere
To determine how the defects in kinetochore assembly that occur in the absence of Cse4 alter chromosome segregation, we analyzed the phenotype of Cse4-depleted cells. To do these experiments, we first investigated whether cells could be depleted of Cse4 at the eCEN. We hypothesized that cells would need to pass through S phase in the absence of Cse4 to achieve depletion at the eCEN because fluorescence recovery after photobleaching experiments demonstrated that Cse4 is replaced at the centromere during S phase (Pearson et al., 2004). To facilitate G1 arrest and allow the analysis of cells at anaphase, Degron-CSE4 pGAL-UBR1-myc cells were also deleted for the MAD2 spindle checkpoint gene, which halts the cell cycle when there is a defect in proper spindle assembly (for review, see Lew and Burke, 2003). Although Degron-CSE4 cells do not respond to αF, Degron-CSE4 mad2Δ cells arrest in αF for reasons that are unclear (Biggins et al., 2001). To analyze Cse4 depletion at the eCEN, Degron-CSE4 pGAL-UBR1-myc mad2Δ cells were arrested with αF, and dox was added to repress transcription of Degron-CSE4. Cells were then released into the cell cycle and monitored by ChIP and fluorescence-activated cell sorting (FACS) for Cse4 localization and DNA content, respectively. Consistent with our hypothesis, Cse4 was completely depleted from the eCEN after cells had completed replication at 90 min, although some Cse4 remained if cells did not pass synchronously through S phase (Figure 6; our unpublished data). Quantitative PCR was used to confirm that Cse4 was completely depleted at the eCEN (our unpublished data).
Cells Depleted of Cse4 Exhibit Severe Chromosome Segregation Defects
The depletion of Cse4 at the eCEN allowed us to characterize the phenotype of cells completely lacking Cse4. Although phenotypic analyses of temperature sensitive cse4 alleles have been carried out (Stoler et al., 1995; Keith et al., 1999; Chen et al., 2000; Glowczewski et al., 2000; Keith and Fitzgerald-Hayes, 2000; Biggins et al., 2001), the consequences of depleting the centromeric nucleosome have not been analyzed previously. Mad2Δ pGAL-UBR1-myc cells containing Degron-CSE4 or wild-type CSE4 were arrested in G1 with αF, treated with dox to repress transcription of Degron-CSE4, and then released into the cell cycle. Cells also contained a fluorescent spindle pole body (SPB) component, Spc42-GFP, to allow determination of the cell cycle stage. Cells were analyzed for chromosome segregation when the majority of cells were in anaphase, as defined by having fully separated SPBs. Chromosome segregation was scored as equal when similar amounts of DNA were at each pole (equal). Chromosome missegregation fell into two categories (unequal): one completely unsegregated DAPI mass, or unequal segregation where a small amount of DNA was in the daughter cell and the majority of the DNA was in the mother cell. Most control cells underwent normal DNA segregation (92%). However, the majority of Cse4-depleted cells (91%) underwent massive chromosome missegregation and had either completely unsegregated DNA (50%), or a very small amount of DNA in the daughter cell (41%) (Figure 7). It is important to note that even the cells with unequal segregation had very little DNA that had actually segregated to the daughter cell (Figure 7B). Thus, Cse4-depleted cells have severe defects in chromosome segregation, consistent with the defects in kinetochore assembly observed at the cCEN in the absence of Cse4.
DISCUSSION
Using a cCEN, we found that yeast cells do not restrict kinetochore assembly to a specific cell cycle phase. In addition, every kinetochore protein tested exhibited some dependence on Cse4 for assembly, consistent with our observation that Cse4-depleted cells cannot segregate chromosomes. We therefore propose that yeast use a combination of centromeric chromatin and centromeric DNA specificity to mediate kinetochore assembly.
A Conditional Centromere Provides an Assay to Study Yeast Kinetochore Assembly
We used a cCEN that is controlled by transcription to analyze kinetochore assembly. Because kinetochore proteins at the cCEN cannot be distinguished from the eCEN by microscopy (our unpublished data), we analyzed the assembly of representative proteins from each kinetochore complex using ChIP. There was variation in the occupancy of proteins at the inactive cCEN depending on the subcomplex analyzed. Cse4 was the only protein that fully localized to the inactive cCEN, suggesting that Cse4 may have sequence-specific centromere binding like other inner centromere proteins. Although our results contrast with a previous study that did not detect Cse4 at the inactive cCEN (Mythreye and Bloom, 2003), our results are consistent with the maintenance of the centromere-specific chromatin structure at the inactive cCEN (Hill and Bloom, 1987). Although no other protein fully occupied the inactive cCEN, several inner (Ctf13, Ndc10, and Mif2) and central kinetochore proteins (Okp1, Nuf2, and Mtw1) showed enrichment that is likely because of their transient association with the inactive cCEN. Because the inner proteins are required for outer kinetochore assembly, their transient association should be longer than the outer proteins and therefore easier to detect. Consistent with this idea, we did not detect outer kinetochore proteins at the inactive cCEN. Despite the variation at the inactive cCEN, all of the proteins tested localized to the active cCEN at levels comparable with the eCEN.
Although we detected full occupancy of Cse4 at the inactive cCEN, it is not clear why we detected only partial localization of Ndc10, because Ndc10 is required for the localization of Cse4 (Ortiz et al., 1999). One possibility is that Cse4 and Ndc10 transiently bind to the inactive cCEN, but Cse4 is easier to detect because it has a stronger binding affinity. Another possibility is that the partial localization of Ndc10 is sufficient for Cse4 localization. Further studies will be required to understand the dependency of Cse4 localization on Ndc10.
Yeast Kinetochore Assembly Is Not Cell Cycle Restricted
We found that representative proteins from all kinetochore complexes assembled at the cCEN during G1 phase and prometaphase, suggesting that the entire kinetochore assembles. Due to the rapid assembly of kinetochore proteins (within 10 min), kinetic studies to test the order of assembly were not feasible (our unpublished data). Similar to the eCEN, Dam1 recruitment to the G1 cCEN was microtubule dependent, strongly suggesting that a fully functional kinetochore can assemble in G1. Together, these data suggest that budding yeast kinetochores can assemble and become functional throughout the cell cycle.
This is the first study to demonstrate that yeast cells maintain the ability to assemble kinetochores in multiple cell cycle stages. The lack of temporal restriction on kinetochore assembly may allow the repair of damaged kinetochores. Alternatively, yeast cells may not temporally restrict kinetochore assembly because a specific centromere sequence is required for assembly. The lack of cell cycle control indicates that all posttranslational modifications required for assembly are either maintained from the previous cell cycle or can occur during G1. In addition, none of the kinetochore proteins seem to be limiting for assembly because changes in eCEN localization were not detected when the cCEN was activated.
Although yeast cells maintain the ability to assemble kinetochores at several cell-cycle stages, it remains unclear when kinetochores normally form. It is likely that assembly is coupled to centromere replication because yeast kinetochores quickly achieve bioriented attachments after centromere duplication and Cse4 deposition occurs at this time (Goshima and Yanagida, 2000; He et al., 2000; Pearson et al., 2004).
The Functions of CenH3
We analyzed kinetochore assembly at the cCEN in the absence of the CenH3. We were not able to analyze the maintenance of a previously assembled kinetochore because cells needed to pass through S phase to deplete Cse4 from the eCEN. Because kinetochores likely disassemble during S phase (Tanaka et al., 2005), studies at the eCEN would not distinguish between kinetochore assembly and maintenance. All proteins tested exhibited some dependence on Cse4 for localization to the cCEN. The inner and central kinetochore proteins Ndc10, Mif2, Mtw1, and Okp1 achieved ∼50% occupancy at the active cCEN in the absence of Cse4, whereas the central protein Ctf19 and all outer proteins (Ndc80, Dam1, Ask1, and Stu2) completely failed to localize. Although residual Cse4 may remain, the partial occupancy of the inner kinetochore proteins may instead be due to their Cse4-independent sequence-specific binding (Lechner and Carbon, 1991; Meluh and Koshland, 1995; Stoler et al., 1995). It is likely that central kinetochore proteins achieve partial localization in the absence of Cse4 due to interactions with inner kinetochore proteins. The differences in the dependence of the CTF19/COMA complex proteins Ctf19 and Okp1 may reveal different requirements for Cse4 in localizing the C2.100 (Ctf19 and Mcm21) and C2.105 (Ame1 and Okp1) subcomplexes (De Wulf et al., 2003).
We extended our analyses to examine the phenotype of Cse4-depleted cells in a single cell cycle. In contrast to studies on cse4 temperature-sensitive alleles that exhibited chromosome missegregation (Stoler et al., 1995; Keith et al., 1999; Biggins et al., 2001), the majority of DNA did not segregate in the Cse4-depleted cells. Strikingly, this phenotype most strongly resembles ndc10-1 mutants that completely lack kinetochore assembly and microtubule attachment (Goh and Kilmartin, 1993), suggesting that microtubule–kinetochore attachments are not made in the absence of Cse4. This is consistent with our ChIP experiments that revealed severe defects in kinetochore assembly. Although ndc10-1 cells do not engage the spindle checkpoint (Goh and Kilmartin, 1993), we were unable to determine whether Cse4-depleted cells are also defective in the checkpoint. Degron-CSE4 cells do not respond to αF unless the MAD2 spindle checkpoint gene is mutated, so we used cells defective in the checkpoint to facilitate the cell cycle arrest for the reported experiments.
Our studies of Cse4-depleted cells revealed two additional phenotypes that were not observed in cse4 temperature-sensitive mutants. First, Cse4-depleted cells had a larger and more diffuse DAPI mass than wild-type cells, suggesting a role in condensation. This may be a conserved function because the Caenorhabditis elegans CenH3 is also required for condensation (Chan et al, 2004; Maddox, Portier, Desai, and Oegema, unpublished data) and a Drosophila condensin protein Cap-G interacts with CenH3 (Jager et al., 2005). We also found that Cse4-depleted cells were delayed in cytokinesis (our unpublished data), similar to the delay observed in human cells overexpressing a CenH3 mutant (Zeitlin et al., 2001). Future work will be aimed at determining the role of Cse4 in condensation and cytokinesis.
The ability to deplete Cse4 in a single cell cycle allowed us to perform the first study of de novo kinetochore assembly in the absence of CenH3. In agreement with other studies using RNAi to deplete CenH3 over multiple cell cycles, we found that Cse4 is required for the localization of many kinetochore proteins (Howman et al., 2000; Oegema et al., 2001; Regnier et al., 2005). However, in contrast to studies that suggested the existence of CenH3-independent assembly pathways based on immunofluorescence data (Oegema et al., 2001; Goshima et al., 2003; Hayashi et al., 2004; Regnier et al., 2005), we found that all proteins tested exhibited some dependence on Cse4 when the ChIP technique was used to quantify occupancy. Although the inner kinetochore complex CBF3 was previously proposed to nucleate yeast kinetochore assembly (Russell et al., 1999; Goshima and Yanagida, 2000; He et al., 2001; Janke et al., 2001, 2002), our work shows that the centromeric nucleosome also has an essential role in kinetochore assembly. We therefore favor a model where the centromeric nucleosome creates a chromatin structure that cooperates with the binding of the inner CBF3 complex to initiate yeast kinetochore assembly.
Supplementary Material
Acknowledgments
We thank Kerry Bloom for plasmids and technical advice regarding the cCEN, Toshi Tsukiyama for advice regarding ChIP, and Bodo Stern for the pGAL-UBR1-myc plasmid. We thank Linda Breeden, Suzanne Furuyama, Rich Gardner, Chitra Kotwaliwale, Harmit Malik, Ben Pinsky, Toshi Tsukiyama, and Danielle Vermaak for discussions and helpful comments on the manuscript. A Viral Oncology Training grant to K. C., a grant from the National Institutes of Health to A. C., and a National Institutes of Health grant and a Beckman Young Investigator Award to S. B. supported this work. S. B. is a Leukemia and Lymphoma Society Scholar.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–08–0771) on October 5, 2005.
Abbreviations used: αF, α factor; CenH3, centromeric histone H3 variant; ChIP, chromatin immunoprecipitation; cCEN, conditional centromere; dox, doxycycline; eCEN, endogenous centromere; FACS, fluorescence-activated cell sorter; IP, immunoprecipitate; SPB, spindle pole body.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Biggins, S., Bhalla, N., Chang, A., Smith, D. L., and Murray, A. W. (2001). Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics 159, 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., Severin, F. F., Bhalla, N., Sassoon, I., Hyman, A. A., and Murray, A. W. (1999). The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and Walczak, C. E. (2003). Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13, R449–R460. [DOI] [PubMed] [Google Scholar]
- Blower, M. D., and Karpen, G. H. (2001). The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3, 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz, B. J., Ahmad, K., Moore, L. L., Roth, M. B., and Henikoff, S. (1999). A histone-H3-like protein in C. elegans. Nature 401, 547–548. [DOI] [PubMed] [Google Scholar]
- Chan, R. C., Severson, A. F., and Meyer, B. J. (2004). Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 167, 613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Baker, R. E., Keith, K. C., Harris, K., Stoler, S., and Fitzgerald-Hayes, M. (2000). The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20, 7037–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, K. A., Furuyama, S., and Biggins, S. (2004). Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14, 1968–1972. [DOI] [PubMed] [Google Scholar]
- De Wulf, P., McAinsh, A. D., and Sorger, P. K. (2003). Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17, 2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar, H., Tanaka, K., Nasmyth, K., and Tanaka, T. U. (2004). Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 428, 93–97. [DOI] [PubMed] [Google Scholar]
- Dohmen, R. J., Wu, P., and Varshavsky, A. (1994). Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263, 1273–1276. [DOI] [PubMed] [Google Scholar]
- Edwards, N. S., and Murray, A. W. (2005). Identification of Xenopus CENP-A and an associated centromeric DNA repeat. Mol. Biol. Cell 16, 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman, M., Cheeseman, I. M., Van Goor, D., Drubin, D. G., Meluh, P. B., and Barnes, G. (2001). Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., Clarke, L., and Carbon, J. (1982). Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29, 235–244. [DOI] [PubMed] [Google Scholar]
- Glowczewski, L., Yang, P., Kalashnikova, T., Santisteban, M. S., and Smith, M. M. (2000). Histone-histone interactions and centromere function. Mol. Cell. Biol. 20, 5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, P. Y., and Kilmartin, J. V. (1993). NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., Kiyomitsu, T., Yoda, K., and Yanagida, M. (2003). Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Yanagida, M. (2000). Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100, 619–633. [DOI] [PubMed] [Google Scholar]
- Hayashi, T., Fujita, Y., Iwasaki, O., Adachi, Y., Takahashi, K., and Yanagida, M. (2004). Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729. [DOI] [PubMed] [Google Scholar]
- He, X., Asthana, S., and Sorger, P. K. (2000). Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101, 763–775. [DOI] [PubMed] [Google Scholar]
- He, X., Rines, D. R., Espelin, C. W., and Sorger, P. K. (2001). Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106, 195–206. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., Platero, J. S., and van Steensel, B. (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A., and Bloom, K. (1987). Genetic manipulation of centromere function. Mol. Cell. Biol. 7, 2397–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A., and Bloom, K. (1989). Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 1368–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman, E. V., Fowler, K. J., Newson, A. J., Redward, S., MacDonald, A. C., Kalitsis, P., and Choo, K. H. (2000). Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter, K. J., and Eipel, H. E. (1979). Microbial determination by flow cytometry. J. Gen. Microbiol. 113, 369–375. [DOI] [PubMed] [Google Scholar]
- Jager, H., Rauch, M., and Heidmann, S. (2005). The Drosophila melanogaster condensin subunit Cap-G interacts with the centromere-specific histone H3 variant CID. Chromosoma 113, 350–361. [DOI] [PubMed] [Google Scholar]
- Janke, C., Ortiz, J., Lechner, J., Shevchenko, A., Magiera, M. M., Schramm, C., and Schiebel, E. (2001). The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Ortiz, J., Tanaka, T. U., Lechner, J., and Schiebel, E. (2002). Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G. H., and Allshire, R. C. (1997). The case for epigenetic effects on centromere identity and function. Trends Genet. 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Keith, K. C., Baker, R. E., Chen, Y., Harris, K., Stoler, S., and Fitzgerald-Hayes, M. (1999). Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19, 6130–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, K. C., and Fitzgerald-Hayes, M. (2000). CSE4 genetically interacts with the saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a cse4p variant nucleosome. Genetics 156, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib, K., Tercero, J. A., and Diffley, J. F. (2000). Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Lechner, J., and Carbon, J. (1991). A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64, 717–725. [DOI] [PubMed] [Google Scholar]
- Lew, D. J., and Burke, D. J. (2003). The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37, 251–282. [DOI] [PubMed] [Google Scholar]
- Li, Y., Bachant, J., Alcasabas, A. A., Wang, Y., Qin, J., and Elledge, S. J. (2002). The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and Henikoff, S. (2003). Phylogenomics of the nucleosome. Nat. Struct. Biol. 10, 882–891. [DOI] [PubMed] [Google Scholar]
- McAinsh, A. D., Tytell, J. D., and Sorger, P. K. (2003). Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19, 519–539. [DOI] [PubMed] [Google Scholar]
- Meluh, P. B., and Koshland, D. (1995). Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., Yang, P., Glowczewski, L., Koshland, D., and Smith, M. M. (1998). Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94, 607–613. [DOI] [PubMed] [Google Scholar]
- Minshull, J., Straight, A., Rudner, A., Dernburg, A., Belmont, A., and Murray, A. W. (1996). Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6, 1609–1620. [DOI] [PubMed] [Google Scholar]
- Mythreye, K., and Bloom, K. S. (2003). Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 160, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M. W., and Burke, D. J. (1992). A delay in the Saccharomyces cerevisiae cell cycle that is induced by a dicentric chromosome and dependent upon mitotic checkpoints. Mol. Cell. Biol. 12, 3857–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Desai, A., Rybina, S., Kirkham, M., and Hyman, A. A. (2001). Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, J., Stemmann, O., Rank, S., and Lechner, J. (1999). A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13, 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D. K., O'Day, K., Wener, M. H., Andrews, B. S., and Margolis, R. L. (1987). A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C. G., Yeh, E., Gardner, M., Odde, D., Salmon, E. D., and Bloom, K. (2004). Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 14, 1962–1967. [DOI] [PubMed] [Google Scholar]
- Pinsky, B. A., Tatsutani, S. Y., Collins, K. A., and Biggins, S. (2003). An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell 5, 735–745. [DOI] [PubMed] [Google Scholar]
- Regnier, V., Vagnarelli, P., Fukagawa, T., Zerjal, T., Burns, E., Trouche, D., Earnshaw, W., and Brown, W. (2005). CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25, 3967–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., Winston, F., and Heiter, P. (1990). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Russell, I. D., Grancell, A. S., and Sorger, P. K. (1999). The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145, 933–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal, K., and Carbon, J. (2002). The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc. Natl. Acad. Sci. USA 99, 12969–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G., and Lawrence, C. (1974). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Stoler, S., Keith, K. C., Curnick, K. E., and Fitzgerald-Hayes, M. (1995). A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9, 573–586. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., Hecht, A., Luo, K., and Grunstein, M. (1997). SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11, 83–93. [DOI] [PubMed] [Google Scholar]
- Sullivan, B. A., Blower, M. D., and Karpen, G. H. (2001). Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2, 584–596. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., Chen, E. S., and Yanagida, M. (2000). Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288, 2215–2219. [DOI] [PubMed] [Google Scholar]
- Talbert, P. B., Masuelli, R., Tyagi, A. P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Mukae, N., Dewar, H., van Breugel, M., James, E. K., Prescott, A. R., Antony, C., and Tanaka, T. U. (2005). Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434, 987–994. [DOI] [PubMed] [Google Scholar]
- Turner, G. C., and Varshavsky, A. (2000). Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117–2120. [DOI] [PubMed] [Google Scholar]
- Van Hooser, A. A., Ouspenski, I. I., Gregson, H. C., Starr, D. A., Yen, T. J., Goldberg, M. L., Yokomori, K., Earnshaw, W. C., Sullivan, K. F., and Brinkley, B. R. (2001). Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114, 3529–3542. [DOI] [PubMed] [Google Scholar]
- Zeitlin, S. G., Shelby, R. D., and Sullivan, K. F. (2001). CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C. X., Marshall, J. B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J. A., Jiang, J., and Dawe, R. K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.