Abstract
Tctex1 and Tctex2 were originally described as potential distorters/sterility factors in the non-Mendelian transmission of t-haplotypes in mice. These proteins have since been identified as subunits of cytoplasmic and/or axonemal dyneins. Within the Chlamydomonas flagellum, Tctex1 is a subunit of inner arm I1. We have now identified a second Tctex1-related protein (here termed LC9) in Chlamydomonas. LC9 copurifies with outer arm dynein in sucrose density gradients and is missing only in those strains completely lacking this motor. Zero-length cross-linking of purified outer arm dynein indicates that LC9 interacts directly with both the IC1 and IC2 intermediate chains. Immunoblot analysis revealed that LC2, LC6, and LC9 are missing in an IC2 mutant strain (oda6-r88) that can assemble outer arms but exhibits significantly reduced flagellar beat frequency. This defect is unlikely to be due to lack of LC6, because an LC6 null mutant (oda13) exhibits only a minor swimming abnormality. Using an LC2 null mutant (oda12-1), we find that although some outer arm dynein components assemble in the absence of LC2, they are nonfunctional. In contrast, dyneins from oda6-r88, which also lack LC2, retain some activity. Furthermore, we observed a synthetic assembly defect in an oda6-r88 oda12-1 double mutant. These data suggest that LC2, LC6, and LC9 have different roles in outer arm assembly and are required for wild-type motor function in the Chlamydomonas flagellum.
INTRODUCTION
Dyneins act as minus end-directed microtubule motors and are required for a variety of fundamental cellular activities. There are two general designs for these enzymes built around either a single or several heavy chain motor units (for recent review, see Sakato and King, 2004). The major cytoplasmic dynein isoform and both axonemal outer arms and inner arms of the I1 type1 contain two, or, in some outer arms, three heavy chains. In all cases, these are associated with two or more intermediate chains (ICs) (usually WD-repeat proteins) and dimers of three different classes of light chains (LCs) (the LC8, Tctex1/Tctex2, and LC7/Roadblock families). These distinct dyneins also contain additional components specific to a particular enzyme class, such as the light intermediate chains of cytoplasmic dynein and the regulatory light chains directly associated with the motor units of the Chlamydomonas outer arm.
The Tctex1 and Tctex2 proteins were first identified as candidates for distorter/sterility factors involved in the non-Mendelian transmission of mouse t-haplotypes (variant forms of a 30- to 40-Mb region of chromosome 17) (Lader et al., 1989; Huw et al., 1995). Subsequently, these proteins were found to be integral components of cytoplasmic (King et al., 1996; King et al., 1998) and/or axonemal (Patel-King et al., 1997; Harrison et al., 1998; Kagami et al., 1998) dyneins and to associate directly with the intermediate chains (Mok et al., 2001). In addition, phosphorylation of Tctex2-related proteins in salmonid fish and sea urchin dyneins has been correlated with the activation of sperm motility (Inaba et al., 1999). These observations led to the hypothesis that defects in axonemal dynein function within sperm flagella provide the underlying molecular basis for this chromosome segregation abnormality (Patel-King et al., 1997; Harrison et al., 1998; Pazour et al., 1999). The subsequent identification of an axonemal outer arm dynein heavy chain as an excellent candidate for the strongest distorter (Fossella et al., 2000) and of a sperm motility kinase as the interacting responder (Herrmann et al., 1999) has provided additional support for this model.
Sequence analysis (DiBella et al., 2001) combined with NMR (Mok et al., 2001; Wu et al., 2001, 2005) and x-ray (Williams et al., 2005) structural studies suggest that both Tctex1 and Tctex2 adopt a similar overall conformation. In cytoplasm, Tctex1 interacts with a variety of proteins in addition to dynein (e.g., Doc-2 [Nagano et al., 1998], rhodopsin [Tai et al., 1999], polio virus receptor [Mueller et al., 2002], mitochondrial voltage-dependent anion channel [Schwarzer et al., 2002], neuronal voltage-gated Ca2+ channels [Lai et al., 2005], Fyn kinase [Kai et al., 1997; Mou et al., 1998], and the Herpes capsid protein VP26 [Douglas et al., 2004]) and has been proposed to act as an adaptor to attach specific cargoes to this motor (Tai et al., 1999). In Chlamydomonas, one Tctex2 homologue (termed LC2) is required for assembly of functional outer arms (Patel-King et al., 1997; Pazour et al., 1999), whereas a related protein imparts stability to inner arm I1 and is necessary for wild-type motor activity (DiBella et al., 2004b). Intriguingly, Tctex1 null alleles in Drosophila are viable, although these mutants are male sterile. Phenotypically, these strains exhibit defects in spermiogenesis where a “nuclear cap” of cytoplasmic dynein is incorrectly localized and a connection between the nucleus and basal body is not formed appropriately; they also have immotile sperm possibly due to the lack of Tctex1 in axonemal dyneins (Caggese et al., 2001; Li et al., 2004).
Although nine light chain components, including the essential Tctex2-related protein LC2, have so far been identified within the Chlamydomonas outer dynein arm (Piperno and Luck, 1979; Pfister et al., 1982; Patel-King et al., 1997; Pazour et al., 1999), a protein clearly similar to Tctex1 has not previously been described in this motor enzyme. This had been somewhat surprising because obvious Tctex1 homologues are present in all other well-characterized dyneins containing multiple heavy chains, e.g., outer arms from sea urchin sperm flagella (Kagami et al., 1998), mammalian cytoplasmic dynein (King et al., 1996), and Chlamydomonas inner dynein arm I1 (Harrison et al., 1998). Here, we identify a second Tctex1-like protein (termed LC9) encoded within the Chlamydomonas genome and demonstrate that it is an integral component of the outer dynein arm associated with both intermediate chains. Thus, in this organism distinct Tctex1-related proteins are used within the flagellar inner and outer arm systems. Furthermore, we find that this LC9 protein together with LC2 (a Tctex2) and LC6 (related to the highly conserved LC8 protein; King and Patel-King, 1995) are missing in a mutant strain (oda6-r88) expressing a modified form of IC2 that allows for outer arm assembly but exhibits a reduced flagellar beat frequency close to that of cells lacking this motor (Mitchell and Kang, 1993). Our results indicate that in the presence of this IC2 mutation, these LCs are not required for assembly of the outer dynein arm but may play a role in the control of wild-type motor function. Analysis of additional mutants defective for LC2 (oda12-1) and LC6 (oda13) as well as an oda6-r88 oda12-1 double mutant provides further support for this model and yields intriguing insight into the mechanisms of outer arm dynein assembly and the molecular architecture of the IC/LC complex.
MATERIALS AND METHODS
Chlamydomonas Strains
The Chlamydomonas reinhardtii strains used in this study are indicated in Table 1. The oda6-r75 and oda6-r88 pseudorevertants were generously provided by Dr. David Mitchell (SUNY Upstate Medical Center, Syracuse, NY). The oda6-r88 oda12-1 double mutant was constructed using standard methods (Harris, 1989). The genotype of the tetrad progeny from this cross was determined using the PCR to obtain appropriate regions from both genes. The oda6-r88 frame-shift mutation forms a novel BamHI restriction site that was used to differentiate between wild-type and mutant versions; the oda12-1 allele is a null and thus only the wild-type LC2 gene was detected by the PCR.
Table 1.
Strains used in this study
| Strain | Description | Reference |
|---|---|---|
| cc124 | Wild type | |
| g1 | Used as parent for generation of null mutants; derived from a nit1–305 × cc124 cross | Pazour et al. (1995) |
| oda6-95 | IC2 frame-shift resulting in a premature stop; cannot assemble outer arms; slow swimming | Kamiya (1988); Mitchell and Kang (1993) |
| oda6-r75 | oda6-95 intragenic pseudorevertant; assembles outer arms and restores wild-type beat frequency | Mitchell and Kang (1993) |
| oda6-r88 | oda6-95 intragenic pseudorevertant; assembles outer arms, but exhibits reduced beat frequency | Mitchell and Kang (1993) |
| ida1 | Defective for 1α HC; lacks inner arm I1 | Kamiya et al. (1991); Porter et al. (1992) |
| ida4 | Defective for p28; lacks a subset of inner arms | Kamiya et al. (1991); LeDizet and Piperno (1995) |
| oda3 | Defective for DC1; lacks outer arms and the outer arm docking complex | Kamiya (1988); Koutoulis et al. (1997) |
| oda9 | Defective for IC1; lacks outer arms, but assembles the docking complex | Kamiya (1988); Wilkerson et al. (1995) |
| oda12-1 | Null mutant for LC2; lacks functional outer arms | Pazour et al. (1999) |
| oda12-2 | Lacks 3′ end of LC2 gene; defective outer arm assembly | Pazour et al. (1999) |
| oda12-1 + pBD14 | Null mutant for LC2, rescued with a 3.1-kb genomic region containing the wild-type LC2 gene | Pazour et al. (1999) |
| oda13 | Null mutant for LC6; assembles outer arms and exhibits a minor swimming defect | Pazour and Witman (2000) |
| oda15 | Null mutant for LC7a; assembles reduced levels of outer arms and inner arm I1 | Pazour and Witman, (2000); DiBella et al. (2004a) |
| oda6-r88 oda12-1 | Double mutant with altered IC2 and null for LC2 | This study; Mitchell and Kang (1993); Pazour et al. (1999) |
| pf14 | Defective for RSP3; lacks radial spokes; paralyzed flagella | Huang et al. (1981) |
| pf18 | Lacks central pair microtubule complex; paralyzed flagella | Adams et al. (1981) |
| pf28pf30 | W4 isolate; lacks outer arms and inner arm I1 | Piperno et al. (1990) |
All strains were grown in R medium (medium I; Sager and Granick, 1953) containing 2.20 mM KH2PO4, 1.71 mM K2HPO4, and 7.5 mM sodium acetate). Flagella beat frequency was determined using the fast Fourier transform (FFT) method of Kamiya (2000).
Fractionation of Flagella and Dynein Purification
Flagella were detached using the dibucaine method and purified by standard procedures (Witman, 1986). After demembranation with 1% IGEPAL CA-630 (replaces Nonidet P-40), dynein was extracted from the resulting axonemes by treatment with 0.6 M NaCl. The outer arm and associated docking complex were then purified by sucrose density gradient centrifugation in the presence of Mg2+ at low hydrostatic pressure (Takada et al., 1992; DiBella et al., 2004a). Samples were routinely electrophoresed in 5–15% acrylamide SDS gradient gels or 12.5% acrylamide slab gels and either stained with Coomassie blue or transferred to nitrocellulose for immunoblotting.
Preparation of Fusion Proteins and Antibodies
The LC9 coding sequence was obtained from first strand cDNA using the PCR, and residues 1–49 were subcloned into the pMAL-c2 vector across the XmnI/XbaI restriction sites. This resulted in the N-terminal region of LC9 attached to the C terminus of maltose-binding protein (MBP) via a short hydrophilic linker containing a factor Xa proteolytic cleavage site. Full-length LC9- and Chlamydomonas Tctex1-MBP fusion proteins were also prepared. The LC9(1-49) N-terminal fusion protein was used as the immunogen for antibody production in rabbit CT231, and the resulting serum was blot-purified against full-length recombinant LC9 after separation from MBP (Olmsted, 1986).
Other antibodies used in this study were blot-purified rabbit polyclonal antibodies R5932 (vs. LC1; Benashski et al., 1999), R5391 (vs. LC2; Patel-King et al., 1997), R4930 (vs. LC3; Patel-King et al., 1996), CT61 (vs. LC4; M. Sakato and King, unpublished data), R4929 (vs. LC5; Patel-King et al., 1996), R4928 (vs. LC6; King and Patel-King, 1995), R7178 (vs. LC7a; Bowman et al., 1999), CT116 (vs. LC7b; DiBella et al., 2004a), R4058 (vs. LC8; King and Patel-King, 1995), and anti-DC 2 (Wakabayashi et al., 2001), and mouse monoclonal antibodies 12γB (vs. γ HC; King et al., 1985), 1878A (vs. IC1; King et al., 1986), and 1869A (vs. IC2; King et al., 1985). Antibody reactivity was detected using peroxidase-conjugated secondary antibodies and an enhanced chemiluminescent substrate (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Zero-Length Cross-linking with 1-Ethyl-3-(3-dimethylaminopropyl)-Carbodiimide (EDC)
Cross-linking with EDC was performed essentially as described previously (King et al., 1991; DiBella et al., 2004a). Isolated axonemes and purified dynein samples in 30 mM HEPES, pH 7.4, 5 mM MgSO4, 0.5 mM EDTA, and 25 mM KCl (HMEK) buffer were treated with either 0 or 20 mM EDC in 100 mM HEPES, pH 7.4, for 60 min at room temperature. The cross-linking reaction was terminated by addition of β-mercaptoethanol in HMEK to a final concentration of 0.1 M before preparation for electrophoresis and immunoblotting.
Multi-Angle Laser Light Scattering
To assess the oligomeric state of LC9 and other related dynein components, MBP fusion proteins in 20 mM Tris-Cl, pH 7.4, 200 mM NaCl, and 1 mM EDTA were fractionated by gel filtration chromatography using a Superdex 200 column, and the peak fractions were analyzed by multi-angle laser light scattering using a Wyatt miniDAWN laser photometer (Wyatt Technology, Santa Barbara, CA). This allowed the native molecular mass of each protein peak to be determined directly and also provided a measure of the poly-dispersity of the protein sample.
Northern and Southern Blot Analysis
Northern and Southern blot analysis was performed as described previously (King and Patel-King, 1995). RNA was prepared both from nondeflagellated cells, and from cells that had been deflagellated and allowed to regenerate flagella for 30 min. For Southern analysis, wild-type genomic DNA was restricted with BamHI, PstI, PvuII, and SmaI. The entire LC9 coding region was used to generate a 32P-labeled probe.
Electron Microscopy
Wild-type and oda6-r88 oda12-1 mutant axonemes were fixed with 2.5% glutaraldehyde, 1% tannic acid in 100 mM sodium cacodylate, pH 7.4, and postfixed with 1% osmium tetroxide, 0.8% potassium ferricyanide. Samples were stained en bloc with 1% tannic acid and 0.5% uranyl acetate, dehydrated in ethanol and propylene oxide, and embedded in Polybed (Polysciences, Warrington, PA). Sections were poststained with uranyl acetate and lead citrate and examined in a Philips CM-10 electron microscope at an accelerating voltage of 60 kV.
Computational Methods
Searches of the Chlamydomonas genome were performed using the BLAST interface at the Joint Genome Institute (Walnut Creek, CA) (http://genome.jgi-psf.org/chlre2/chlre2.home.html). The Chlamydomonas expressed sequence tag (EST) databases were searched using BLAST at the National Center for Biotechnology Information and at the Kazusa DNA Research Center (Chiba, Japan). Sequence assembly was performed with the GCG suite of programs. Alignments were generated using ClustalW and processed with BOXSHADE. Residues within LC9 that are not conserved in Chlamydomonas inner arm Tctex1 were mapped onto the molecular surface of the Tctex1 NMR solution structure (PDB accession 1XDX; Wu et al., 2005) and displayed using MOLMOL (Koradi et al., 1996).
RESULTS
Identification of a Second Tctex1-like Protein (LC9) in Chlamydomonas
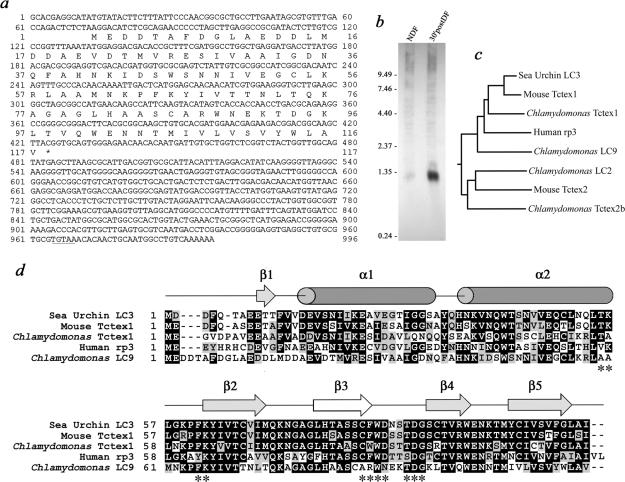
Previously, we reported the identification of a Tctex1-related LC within inner arm I1 from Chlamydomonas flagella (Harrison et al., 1998). To assess whether additional members of this LC family exist in Chlamydomonas, we searched the genome sequence (version 1.0) for related molecules using inner arm Tctex1 (AAC18035) as the initial query sequence. We obtained a single high-scoring hit that was then used to examine the EST database. The entire coding sequence was initially assembled from a series of overlapping EST clones (BG849022, BG847628, BI721765, BI530285, BG847627, and BG849020). Subsequently, we used specific oligonucleotides to obtain this region from first strand cDNA using the PCR. This product was sequenced to confirm the assembly and used to generate fusion proteins (see below). The complete cDNA sequence is 996 base pairs in length and contains a single open reading frame that encodes a protein of 117 residues (here termed LC9) with a mass of 12,981 Da and a predicted pI of 4.37 (Figure 1a). The assembled sequence also contains two in-frame stop codons in the 5′-untranslated region (UTR) and a perfect copy of the Chlamydomonas polyadenylation signal in the 3′-UTR. Search of the genome sequence (version 2.0) and Southern blotting (our unpublished data) indicates that there is a single intron-less gene for LC9 in Chlamydomonas located on linkage group X between the KAT gene (encodes p60 katanin) and the GP441 marker. Northern analysis demonstrates that LC9 mRNA is of low abundance in nondeflagellated cells, but it is highly up-regulated after flagellar excision as is characteristic of integral flagellar components (Figure 1b).
Figure 1.
Molecular analysis of LC9. (a) Nucleotide and predicted protein sequence of LC9 originally derived from the overlapping ESTs BG849022, BG847628, BI721765, BI530285, BG847627, and BG849020 and confirmed by analysis of a full-length cDNA. The 5′-UTR contains two in-frame stop codons, and a perfect copy of the Chlamydomonas polyadenylation signal (underline) is present in the 3′-UTR. This sequence is available under GenBank accession no. DQ114947. (b) RNA samples from nondeflagellated cells (NDF) and from cells that had been deflagellated by pH shock and allowed to regenerate flagella for 30 min (30′postDF) were probed with the LC9 coding region. A single message of ∼1.3 kb is greatly up-regulated after flagellar excision. (c) Rooted neighbor-joining phylogenetic tree illustrating the relationship between LC9, Chlamydomonas inner arm Tctex1 (AF039437), sea urchin outer arm LC3 (JC6573), murine Tctex1 (A32995), and human rp3 (U02556). Also included are three members of the Tctex2 light chain subclass, which form a distinct grouping: murine Tctex2 (U21673), Chlamydomonas outer arm LC2 (U89649) and Chlamydomonas inner arm Tctex2b (BK004867). (d) Sequence alignment of the Tctex1-related proteins shown in c was performed using ClustalW. The secondary structure of Chlamydomonas Tctex1 determined by NMR spectroscopy (Wu et al., 2001, 2005) is shown above the alignment. The strand-switched dimer interface involves the β3 strand (white), which hydrogen bonds to the β2 strand from the other monomer. Residues in mammalian Tctex1 that interact with the cytoplasmic dynein IC (Mok et al., 2001) are indicated by *.
Phylogenetic analysis and sequence comparisons with other members of this protein family are shown in Figure 1, c and d, respectively. The proteins used in the comparison include Chlamydomonas LC9, Chlamydomonas inner arm Tctex1, sea urchin outer arm LC3, murine Tctex1, human rp3, and three members of the Tctex2 LC subclass. Chlamydomonas LC9 clearly groups with other members of the Tctex1 subclass. Overall, LC9 shares 40% sequence identity (52% similarity) with Chlamydomonas inner arm Tctex1. The most highly conserved regions of LC9 include the C-terminal portion of helix α2 and the β2, β3, and β4 strands and their connecting loops (Figure 1d). Even so, mapping the amino acid differences between LC9 and Chlamydomonas Tctex1 onto the NMR solution structure of the latter (PDB accession 1XDX) revealed considerable variation over the entire molecular surface (our unpublished data).
Solution Properties of an MBP-LC9 Fusion Protein
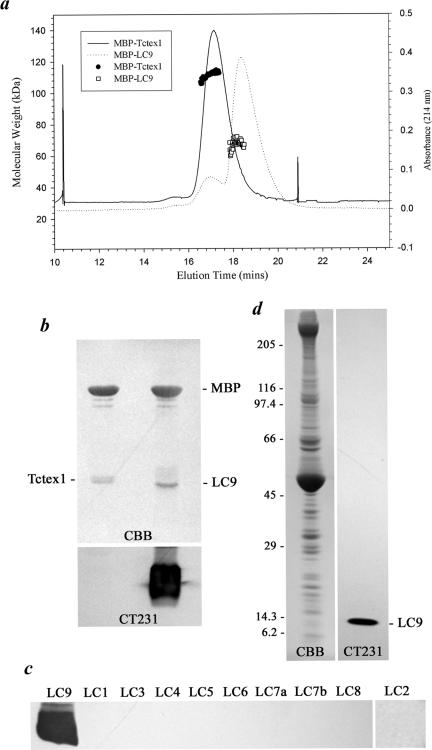
Canonical Tctex1 is a dimeric protein as determined by native gel electrophoresis (DiBella et al., 2001) and high-resolution structural studies (Williams et al., 2005; Wu et al., 2005). To assess whether LC9 can also form dimers, the coding region was subcloned into the pMAL-c2 expression vector, and the protein was expressed as a fusion with MBP. We then directly determined the molecular mass of the native MBP fusion protein using multi-angle laser light scattering (Figure 2a). An MBP fusion with Tctex1 from Chlamydomonas inner arm I1 is dimeric in this assay as expected from our structural studies (native molecular weight [mol. wt.]= 111,300 Da; calculated [calc.] monomer mol. wt. = 55,274 Da). The MBP-LC2 protein is monomeric as observed previously (native mol. wt. = 56,310 Da; calc. monomer mol. wt. = 58,351 Da) (DiBella et al., 2001), whereas the MBP-LC9 fusion protein behaved as a mixture of both monomer and dimer under the same conditions (native mol. wt. = 72,150/126,200 Da; calc. monomer mol. wt. = 55,450 Da) (Figure 2a).
Figure 2.
Oligomeric state of LC9 and specificity of the CT231 antibody. (a) The oligomeric state of fusion proteins containing LC9 and Chlamydomonas inner arm Tctex1 attached to MBP was determined by multi-angle laser light scattering. The absorbance at 214 nm of the gel filtration column elution profile for MBP-Tctex1 (solid line) and MBP-LC9 (dotted line) is shown. Individual molecular weight determinations are indicated by • (MBP-Tctex1) and □ (MBP-LC9), respectively. MBP-Tctex1 is dimeric, whereas MBP-LC9 is mostly monomeric under the same solution conditions; a small peak of MBP-LC9 dimer is evident coeluting with MBP-Tctex1. (b) The Tctex1 and LC9 fusion proteins were digested with factor Xa, electrophoresed in a 15% acrylamide gel and either stained with Coomassie blue (top) or blotted to nitrocellulose and probed with CT231 (bottom). The antibody recognizes LC9 but not Tctex1 with which it shares 40% sequence identity. (c) Recombinant outer arm dynein LCs were probed with CT231 raised against LC9. No other outer arm LC component is recognized by this antibody. (d) Approximately 120 μgof wild-type axonemes were electrophoresed in a 5–15% acrylamide gradient gel and stained with Coomassie blue (left) or probed with CT231 (right). Only a single axonemal protein of the appropriate Mr is recognized.
LC9 is Specifically Associated with the Outer Dynein Arm
To generate an antibody that specifically recognizes LC9, the MBP-LC9(1-49) protein was used as the immunogen and the resulting CT231 rabbit antiserum was blot-purified against full-length recombinant LC9 following digestion of the fusion protein with factor Xa. To initially test the specificity of the CT231 antibody, Chlamydomonas Tctex1 and LC9 MBP fusion proteins were digested with factor Xa and electrophoresed to separate the LC moieties (Figure 2b). Immunoblotting revealed that LC9 was detected by CT231 but that no signal was obtained for the inner arm Tctex1 homologue with which it shares 40% sequence identity. The antibody was also tested against all recombinant outer arm dynein LCs and again was found to react solely with LC9 (Figure 2c). When used to probe axoneme samples, the CT231 antibody detected a single band of Mr ∼13,000 corresponding to the predicted size of LC9 (Figure 2d).
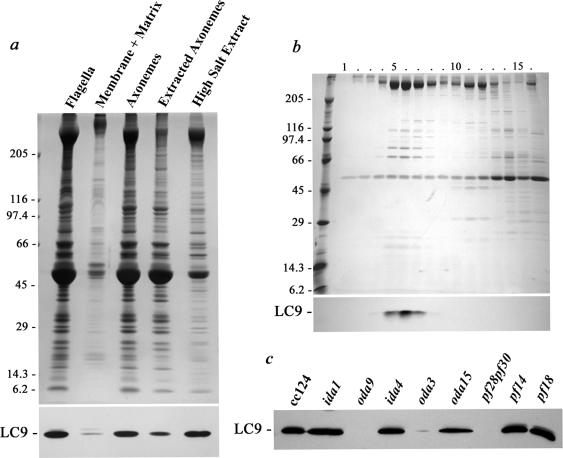
To define the flagellar location of LC9, isolated flagella were first fractionated (Figure 3a) by detergent extraction to remove the membrane and matrix components. Nearly all LC9 protein was associated with the resulting demembranated axonemes. Treatment with 0.6 M NaCl solubilized ∼60–70% of LC9. Subsequent fractionation of the high salt extract by sucrose density gradient centrifugation in the presence of Mg2+ and at low hydrostatic pressure revealed that LC9 copurified with the intact outer dynein arm (Figure 3b, fractions 5–7). To further assess this observation, axonemes were prepared from a variety of mutants lacking specific axonemal structures including inner and outer arms, radial spokes and the central pair microtubule complex. LC9 was greatly reduced or missing only in those strains (oda3, oda9, and pf28pf30) that fail to assemble outer dynein arms (Figure 3c); the oda15 strain assembles ∼30% of outer arms (Pazour and Witman, 2000; DiBella et al., 2004a) and therefore retains considerable amounts of LC9.
Figure 3.
LC9 is a component of the outer dynein arm. (a) Approximately 150 μg of flagella were treated with detergent to yield the microtubular axoneme and solubilize the membrane and flagellar matrix components. Subsequently, the axonemes were extracted with 0.6 M NaCl to remove proteins associated via ionic interactions. Equivalent amounts of each sample were separated in a 5–15% acrylamide gradient gel and stained with Coomassie blue (top) or blotted and probed with CT231 (bottom). LC9 is specifically associated with the axonemes and most is solubilized by high salt treatment. The Mr markers are indicated at left. (b) The 0.6 M NaCl extract was sedimented through a 5–20% sucrose density gradient in the presence of Mg2+, and equivalent volumes of each fraction were electrophoresed in a 5–15% acrylamide gradient gel (top, Coomassie blue; bottom, CT231 blot); the outer dynein arm sediments at ∼23 S in fractions 5–7, as does LC9. The Mr markers are indicated at left. (c) Axonemes from wild-type Chlamydomonas (strain cc124) and from mutants lacking inner arm I1 (ida1), a subset of inner arms I2/3 (ida4), the outer arm (oda9), the outer arm and docking complex (oda3), both outer arms and inner arm I1 (pf28pf30), radial spokes (pf14), and central pair microtubule complex (pf18) were electrophoresed and probed with CT231. Also included is the strain oda15, which lacks LC7a and only assembles ∼30% of outer arms (DiBella et al., 2004a). The LC9 protein is specifically missing only in those strains that do not assemble outer dynein arms.
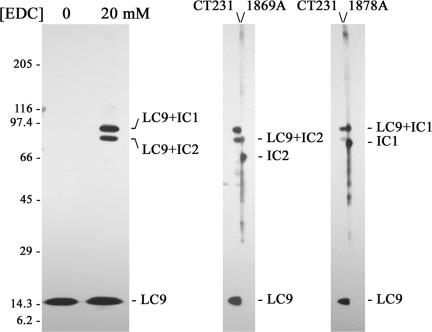
LC9 Interacts Directly with IC1 and IC2
To gain insight into the outer arm proteins with which LC9 associates, we used chemical cross-linking with the zero-length carbodiimide reagent EDC. Cross-links generated with EDC contain no cross-linker components in the final product because the carbodiimide is converted to a urea derivative, and thus two proteins must be in direct contact to be covalently joined. Treatment of purified outer arm dynein with 20 mM EDC generated two prominent bands (Mr ∼80,000 and 90,000) containing LC9 (Figure 4, left); these same bands were observed after EDC treatment of isolated axonemes (our unpublished data). Previously, we observed that both IC1 and IC2 yielded products of Mr 82,000 and 91,000, respectively, after EDC treatment (King et al., 1991; DiBella et al., 2004a), and the Mr of these LC9-containing products suggested that they may derive from attachment of LC9 to both IC1 (76,623 Da) and IC2 (63,519 Da). To test whether these bands indeed represent LC9-IC complexes, blots of dynein samples treated with 20 mM EDC were cut lengthwise and one-half probed with CT231 and the other with either 1878A (vs. IC1; King et al., 1986) or 1869A (vs. IC2; King et al., 1985) (Figure 4, middle and right). After reassembly of the blots, we found that the lower and upper LC9-containing bands precisely comigrated with bands detected by 1869A and 1878A, respectively. These results indicate that LC9 is in direct contact with both ICs within the purified dynein particle and the axoneme.
Figure 4.
LC9 interacts with both outer arm dynein intermediate chains. Purified outer arm dynein from the ida1 strain that lacks inner arm I1 was treated with 0 or 20 mM EDC, electrophoresed, and probed with the CT231 antibody (left). In the absence of EDC, only the LC9 band was observed. However, after EDC treatment two additional prominent bands of Mr ∼80,000–90,000 were obtained. In the center and right panels, single lanes of 20 mM EDC-treated dynein were cut lengthwise, and half were probed for LC9 and the other half for either IC2 (monoclonal antibody [mAb] 1869A) or IC1 (mAb 1878A). Reassembly of the blots revealed that the two LC9 cross-linked products also contain IC2 (Mr ∼80,000 band) and IC1 (Mr ∼90,000 band).
Previously, a Tctex1-binding motif (K/R)(K/R)XX(K/R) has been identified in the mammalian cytoplasmic dynein IC (Mok et al., 2001). In addition, a second binding region VS(K/H)(T/S)X(V/T)(T/S)(N/Q)V has been proposed to exist in certain Tctex1-associated proteins (Sugai et al., 2003), although this motif is either truncated or does not occur in some known binding partners. Examination of the Chlamydomonas IC1 and IC2 sequences identified perfect copies of the Lys/Arg-rich motif in the N-terminal regions of both proteins (18KKTRK22 in IC1 and 98KKVEK102 in IC2) that may represent LC9 interaction sites. No complete correlate of the second motif is discernible in either IC, although the IC2 region (residues 31–54) altered in oda6-r88 (see below) does exhibit some weak similarity to the C-terminal part of this motif.
Incomplete Assembly of the IC/LC Complex in an oda6 Pseudorevertant
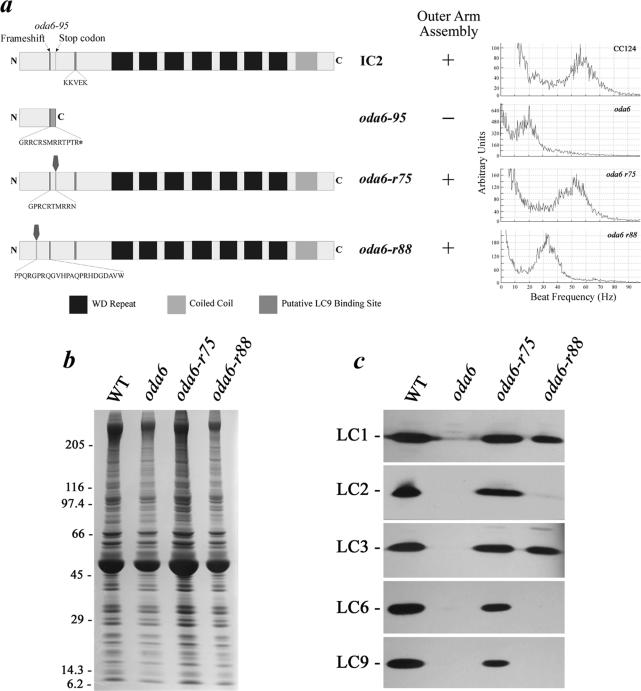
Strains defective at the oda6 locus that encodes IC2 are unable to assemble outer dynein arms (Kamiya, 1988; Mitchell and Kang, 1991). The original oda6-95 mutation removes a single base pair and results in a frame-shift at residue 54 that inserts 13 novel residues before a premature stop codon (Figure 5a). Because the oda6-95 strain does not assemble outer arms, it consequently exhibits a reduced flagellar beat frequency of ∼20–25 Hz rather than 50–60 Hz observed for wild-type Chlamydomonas. Mitchell and Kang (1993) isolated two classes of oda6 intragenic pseudorevertants exemplified by strains oda6-r75 and oda6-r88. Both strains contain additional frame-shifts that, when combined with the original oda6-95 alteration, restore the IC2 reading frame (Figure 5a). However, although both pseudorevertant strains assemble outer dynein arms, only the oda6-r75 mutation also results in restoration of wild-type flagellar beat frequency; in oda6-r88 beat frequency increases to only 30–35 Hz (Figure 5a) (Mitchell and Kang, 1993). Because the putative LC9 binding site in IC2 is located near the region disrupted by the oda6-95 mutation, we reasoned that the different phenotypes of the pseudorevertants might reflect differential LC assembly. To test this possibility, axonemes were prepared from wild-type, oda6, oda6-r75, and oda6-r88 strains (Figure 5b) and probed with antibodies against all outer arm dynein LCs. Axonemes from oda6 lack most outer arm LCs with the exception of LC7a, LC7b, and LC8, which are present because they are also components of other axonemal structures (King, 2002; DiBella et al., 2004a). Both pseudorevertant classes contain LC1, LC3, LC4, LC5, LC7a, LC7b, and LC8 (the data for LC1 and LC3 are shown in Figure 5c). However, although LC2, LC6, and LC9 are present in oda6-r75, they are completely absent from both intact flagella (our unpublished data) and demembranated axonemes (Figure 5c) prepared from oda6-r88.
Figure 5.
An oda6 pseudorevertant lacks LC2, LC6, and LC9. (a) Diagram of the IC2 protein indicating the location of WD-repeats, the C-terminal coiled coil domain, a putative LC9 interaction site, and the original oda6-95 frame-shift that results in a premature stop codon is shown at top. Maps of IC2 from oda6-95 and the two pseudorevertants (oda6-r75 and oda6-r88) are shown below the wild type (data are from Mitchell and Kang, 1993). In these diagrams, residues that are altered from the original and their location are indicated. Whether the strain is capable of outer arm dynein assembly and flagellar beat frequency analysis is indicated at right. (b) Axonemes from wild-type, oda6, oda6-r75, and oda6-r88 strains were electrophoresed in a 5–15% acrylamide gradient gel and stained with Coomassie blue. The Mr markers are at left. (c) Identical samples to those shown in b were blotted and probed with antibodies R5932, R5391, R4930, R4928, and CT231 to detect LC1, LC2, LC3, LC6, and LC9, respectively. All these outer arm proteins are missing in oda6 but are restored in oda6-r75. In contrast, LC2, LC6, and LC9 are not present in axonemes from the oda6-r88 pseudorevertant, which assembles outer arms but does not rescue the beat frequency defect.
LC6 Is Not Required for Assembly of LC2 and LC9
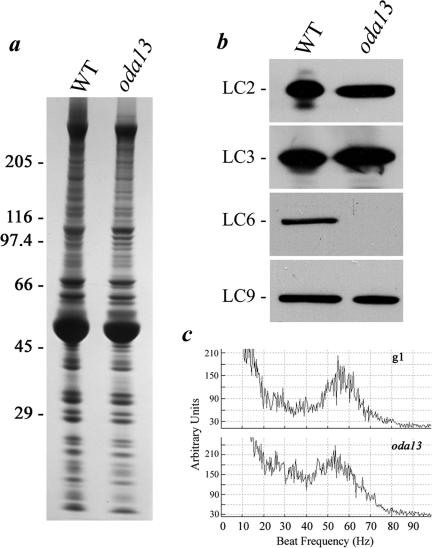
No mutants defective for LC9 have been identified so far. However, the oda13 strain is an LC6 insertional null allele that assembles outer dynein arms and exhibits a minor swimming defect (Pazour and Witman, 2000). Therefore, to test whether the failure of LC9 assembly in oda6-r88 is a consequence of the lack of LC6 or whether the oda6-r88 alteration affects LC9 association more directly, we prepared axonemes from oda13 and examined the LC content (Figure 6, a and b). We observed that the oda13 mutant completely lacks LC6 as reported previously (Pazour and Witman, 2000), but it contains wild-type amounts of LC2, LC3, and LC9. Thus, LC9 assembly onto the ICs is not contingent on LC6. Although oda13 exhibits a clear alteration in flagellar beat frequency compared with the parental strain (the peak in the FFT spectrum becomes much broader although the maximal value remains close to the wild-type level; Figure 6c, bottom), the oda6-r88 defect is much more severe (Figure 5a). This suggests that lack of LC6 alone is not responsible for the oda6-r88 phenotype.
Figure 6.
LC9 assembly is not dependent on LC6. (a) Flagellar axonemes were isolated from wild-type and the oda13 strain, and ∼100 μg of each was electrophoresed in a 5–15% acrylamide gradient gel and stained with Coomassie blue. The Mr markers are at left. (b) Similar samples were blotted to nitrocellulose and probed with antibodies R5391, R4930, R4928, and CT231 to detect LC2, LC3, LC6, and LC9, respectively (right). Only LC6 is missing in the oda13 mutant axonemes, indicating that this protein is not required for the assembly of either LC2 or LC9. (c) FFT analysis of the oda13 mutant and the parental wild-type strain (g1) reveals only a subtle reduction in the peak flagellar beat frequency for the mutant.
The oda6-r88 Mutation Is Not an Extragenic Suppressor of oda12-1
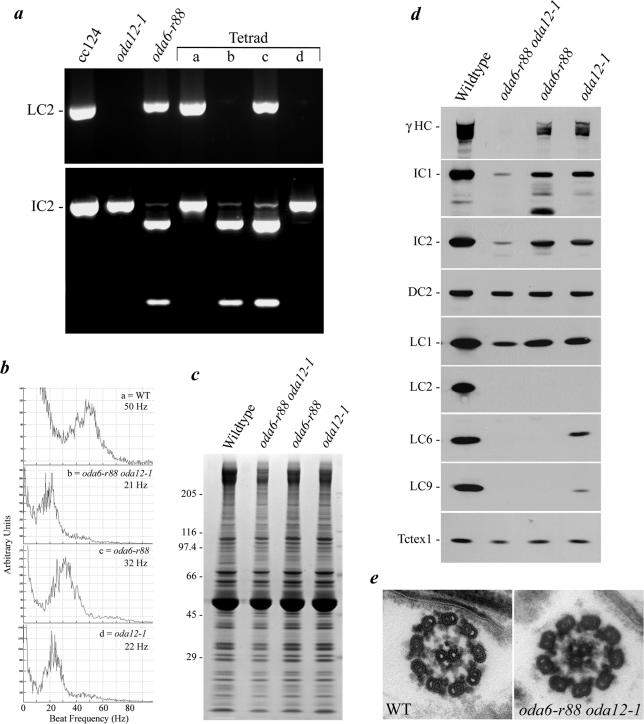
The absence of LC2 in outer arms of the oda6-r88 strain is intriguing, because we observed previously that an LC2-null mutant (oda12-1) does not assemble functional outer arms and that this phenotype may be rescued by transformation with a genomic fragment containing only the wild-type LC2 gene (Pazour et al., 1999). To test whether the oda6-r88 mutation might act as an extragenic suppressor of oda12-1 and bypass the requirement for LC2 in outer arm assembly, we constructed an oda6-r88 oda12-1 double mutant. The genotype of the tetrad progeny from the cross was determined using the PCR to assay for the presence of the LC2 gene and to assess whether the IC2 gene contained the novel BamHI restriction site introduced by the oda6-r88 frame-shift mutation (Figure 7a). FFT analysis of the tetrad progeny revealed that the oda6-r88 oda12-1 strain had the characteristic beat frequency of oda mutants (∼20 Hz) and was clearly distinguishable from the oda6-r88 revertant (∼30–35 Hz) (Figure 7b). Thus, oda6-r88 does not functionally suppress the oda12-1 motility phenotype.
Figure 7.
The oda6-r88 allele is not an extragenic suppressor of oda12-1. (a) PCR analysis of genomic DNA from the progeny of an oda6-r88× oda12-1 cross and both wild-type and parental mutant strains. Top, a 585-base pair segment of the LC2 gene that is completely missing in the oda12-1 null allele. Bottom, a BamHI digest of a 495-base pair region of the IC2 gene that includes the novel restriction site introduced by the oda6-r88 reversion. In this tetratype tetrad, progeny “b” represents the double mutant. (b) Flagellar beat frequency analysis of the four tetrad products identified in a. The double mutant exhibits a beat frequency similar to oda12-1 and is clearly reduced from that observed for oda6-r88. (c) Axoneme samples from each tetrad product were electrophoresed in a 5–15% acrylamide gradient gel and stained with Coomassie blue. The Mr markers are indicated at left. (d) Samples identical to those shown in c were blotted to nitrocellulose and probed to detect various components of the outer dynein arm. The Tctex1 protein from inner arm I1 was used as a loading control. (e) Cross-section electron micrographs of wild-type and oda6-r88 oda12-1 mutant axonemes. No outer arms are detectable in the double mutant.
The oda6-r88 oda12-1 Double Mutant Exhibits a Synthetic Assembly Defect
Previously, we demonstrated that no morphologically distinguishable outer arms were identified in oda12-1 axonemes by thin-section electron microscopy (Pazour et al., 1999). In an attempt to further define the differences between oda6-r88 and oda12-1, which both completely lack LC2, we tested oda12-1 axonemes for the presence of several other outer arm components (Figure 7, c and d). Immunoblot analysis revealed that this mutant retains considerable amounts of several outer arm-specific proteins including the γ HC, IC1, IC2, and LC1; a reduced amount of LC6 and very minor amounts of LC9 were also detectable. We also examined axonemes from the oda12-2 allele, which lacks only the 3′ end of the LC2 gene and found that they incorporate similar levels of LC9 to those observed in oda12-1. Rescue of the oda12-1 strain with a 3.1-kb HindIII fragment (pBD14) containing the LC2 gene (Pazour et al., 1999) restored LC9 assembly to wild-type levels (our unpublished data).
Assembly of the outer arm docking complex was unaffected by either the oda6-r88 or oda12-1 mutations. However, we did observe a synthetic interaction between these alleles as the double mutant contained essentially undetectable amounts of the γ HC and greatly reduced quantities of both IC1 and IC2 (Figure 7d); as expected, essentially no outer arms were detected by EM (Figure 7e). Intriguingly however, LC1 was assembled in this strain even in the absence of its target heavy chain (HC), suggesting that this LC can be incorporated independently of the remainder of the arm.
DISCUSSION
In this report, we have identified an additional member (LC9) of the Tctex1 family as an integral component of Chlamydomonas outer arm dynein. Mutant analysis has revealed that LC2, LC6, and LC9 play an important and inter-connected role in generating a functional dynein particle. Furthermore, we have made several intriguing observations concerning the interplay of light chain incorporation and outer arm dynein assembly that yield insight into the mechanisms by which these motor enzymes are constructed.
Distinct Members of the Tctex1 Family are Present in Inner and Outer Arm Dyneins
Genomic analysis revealed that Chlamydomonas contains two members of the Tctex1 family of dynein LCs. Previously, we found that Chlamydomonas Tctex1 is a component of inner arm I1 (Harrison et al., 1998) and demonstrate here that the second member of this family (LC9) is also an integral axonemal component. Sequence analysis indicates that the LC9 protein is a rather divergent member of this LC class as is the alternate mammalian cytoplasmic dynein LC (currently termed rp3) (King et al., 1998). LC9 copurifies with outer arm dynein in sucrose density gradients and is missing only in those mutant strains that are devoid of outer arms. Thus, the presence of a Tctex1-like protein seems to be a general feature of the design of dyneins containing two or more HCs [see Sakato and King (2004) for further discussion].
Lack of LC2, LC6, and LC9 Correlates with Defective Outer Arm Motor Function
Zero-length cross-linking with EDC revealed that LC9 interacts directly with both IC1 and IC2. The oda6-95 frame-shift mutant cannot synthesize full-length IC2 and consequently fails to assemble outer arms (Kamiya, 1988; Mitchell and Kang, 1993). Two classes of intragenic pseudorevertants have been identified that insert additional frame-shifts and allow for dynein assembly (Mitchell and Kang, 1993). Importantly, the beat frequency of oda6-r75 reverts almost to wild-type (>50 Hz), whereas that of oda6-r88 remains relatively low (∼30–35 Hz), although it is still greater than mutants lacking all outer arms (e.g., oda1 ∼20–25 Hz). Based on these data, Mitchell and Kang (1993) suggested that the region defective in IC2 from oda6-r88 may in some way regulate motor activity or be required for assembly of a LC. We have now found that oda6-r88 outer arm dynein completely lacks three LC components, which likely all interact independently with this region (discussed below). Tctex1 is a structural homologue of the highly conserved LC8 protein (Mok et al., 2001; Wu et al., 2001, 2005; Williams et al., 2005), and structural studies of LC8 interactions with its target peptides (Liang et al., 1999) indicate that each LC-IC interface involves at least seven residues. Because the IC segment defective in oda6-r88 is only 23 residues in length, the independent binding of these three LCs would require this entire region. Thus, it becomes highly probable that it is the LC deficiency in this strain, rather than the IC alteration per se, that results in nonfunctional dynein.
Analysis of the oda13 null allele indicates that lack of LC6 has only a minor effect on flagellar beat frequency and does not result in a severe swimming defect as seen in oda6-r88. This suggests that it is defective assembly of LC2 and/or LC9 in oda6-r88 that causes outer arm dysfunction. Previously, we demonstrated that strains lacking LC2 did not assemble outer arms that could be identified by electron microscopy nor was any IC1 observed (Pazour et al., 1999). We have now found that oda12-1 does contain some outer arm proteins, including IC1. The reason for this apparent discrepancy remains uncertain, although we have observed that both ICs are much more sensitive to proteolysis in strains lacking LC2 (oda12-1 and oda6-r88), presumably because the N-terminal region is more exposed. Even so, the outer arm dyneins assembled in oda12-1 are completely nonfunctional as flagellar beat frequency of this strain is indistinguishable from those mutants (e.g., oda1) that completely lack this motor. These observations present an intriguing conundrum as they indicate that outer arms assembled in an LC2-null background are not functionally equivalent to assembled arms that lack this component.
A number of possibilities might account for these observations. First, these LCs may actually assemble in oda6-r88 flagella but be so weakly bound to the dynein arm that they dissociate after demembranation. This scenario does not seem to be correct because none of these LCs could be detected in intact flagella from this strain. Second, the oda12 insertional alleles may be missing a second gene close to LC2 that is actually responsible for the assembly/functional defects. Our examination of the Chlamydomonas genome revealed that another apparent dynein LC related to LC8 is encoded close to the ODA12 locus. This gene is missing in the oda12-1 null allele but is present in oda12-2, which lacks only the 3′ end of the LC2 gene. Furthermore, this LC8-related gene is not restored in oda12-1 strains transformed with a wild-type LC2 gene construct (pBD14), which rescues both outer arm assembly and the motility defect (Patel-King., Pazour, and King, unpublished observations) (Pazour et al., 1999). Thus, although an additional potential dynein gene is missing in oda12-1, it is not responsible for the differential outer arm function observed in the oda6-r88 and oda12-1 strains.
A third possibility is that the oda6-r88 mutation acts as an extragenic suppressor of oda12-1 and removes the requirement for LC2 in outer arm assembly. If correct, the oda6-r88 oda12-1 double mutant should have had a flagellar beat frequency similar to oda6-r88 rather than oda12-1; however, we observed the converse. Moreover, we found that the double mutant had a synthetic assembly defect in that several outer arm-specific proteins including the γ HC and both ICs were dramatically reduced compared with either parent. One clear compositional difference between oda6-r88 and oda12-1 is the presence of LC6 and a small amount of LC9 in axonemes of the latter strain. Thus, it is feasible that assembly of LC6 in the absence of LC2 results in defective motility, but that the absence of both proteins is less detrimental.
Finally, it is also possible that LC2 is not essential for dynein assembly/function per se, but rather that it interacts in the cytoplasm with another protein that is itself required for assembly or activation of a functional motor. If this second protein were unstable in the absence of LC2, then the oda12 mutant would contain nonfunctional dyneins as we have observed. In contrast, although oda6-r88 does not incorporate LC2 into the flagellum, that protein is likely still present in the cell body and thus could interact with and stabilize the hypothetical assembly/activating factor. This could allow for the production of at least some active dynein motors.
Implications for Intermediate Chain/Light Chain Complex Architecture
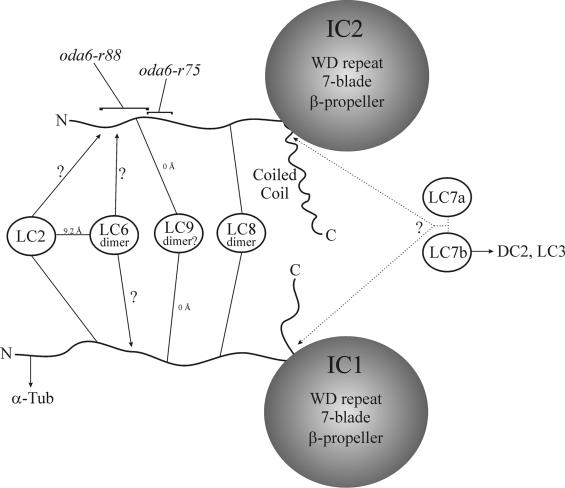
The IC/LC complex of the Chlamydomonas outer arm consists of two ICs and LC2, LC6, LC7a, LC7b, LC8, and LC9. We demonstrated previously using dimethylpimelimidate (DMP) cross-linking that LC2 and LC6 are within 9.2 Å of each other in situ and potentially interact directly (DiBella et al., 2001). Analysis of the LC6-null mutant oda13 revealed that both LC2 and LC9 assemble at wild-type levels in the absence of LC6. We also found in the oda12-1 mutant that reduced amounts of LC6 and low levels of LC9 could assemble in the total absence of LC2, whereas in oda6-r88 all three LCs were completely missing. We have obtained no evidence to suggest that either LC2 or LC6 associates directly with LC9, although lack of LC2 dramatically reduces the amount of LC9 that is incorporated. Thus, it seems most likely that each of these LCs interacts directly with one or both ICs and that disruption of the interface with IC2 is sufficient to disallow LC assembly. A model depicting IC/LC interactions is shown in Figure 8.
Figure 8.
Model for IC/LC interactions within the outer dynein arm. A model for protein-protein associations within the IC/LC complex of outer arm dynein is shown. Both ICs consist of an N-terminal region (in IC1 this interacts directly with α-tubulin), seven WD-repeats that form a β-propeller, and a C-terminal segment (in IC2 this is predicted to form a coiled-coil). LC2, LC6, and LC9 likely interact with IC2 via the region defective in oda6-r88; the region altered in oda6-r75 is not necessary for their assembly. Both LC2 (Mitchell and Rosenbaum, 1986) and LC9 (this study) have been shown to interact directly with IC1. Putative LC8 binding sites within both ICs are located relatively close to the WD-repeat region. The precise order of association sites on IC1 and within the oda6-r88-defective region of IC2 is speculative. Analysis of other dyneins has suggested that members of the LC7/Roadblock family interact with IC regions either N-terminal to the first WD-repeat (Susalka et al., 2002) or C-terminal of the last repeat (Hendrickson et al., 2004); due to the interlocking design of the WD-repeat β-propeller, these regions are spatially close together. Maximal distances (in angstroms) between components are based on DMP and EDC cross-linking (King et al., 1991; DiBella et al., 2001, 2004a).
Although the regions of IC1 and IC2 N-terminal of the WD-repeats are almost completely distinct, there are at least four short motifs that are apparently conserved between the two proteins. One of these occurs within the 23-residue region of IC2 that is altered in oda6-r88 and contains both basic and acidic residues. Because we found that LC9 could be directly cross-linked to both ICs using EDC, this motif [(K/Q)(D/E)YIPxxP] represents a potential interaction site. Both axonemal ICs also contain a KKxxK motif suggested to be involved in Tctex1 binding to the cytoplasmic dynein IC (Mok et al., 2001). In IC2, this motif is located C-terminal of the regions defective in oda6-r88 and oda6-r75, whereas in IC1 it is immediately N-terminal to the (K/Q)(D/E)YIPxxP motif.
The IC location at which the LC7a and LC7b components (axonemal homologues of the roadblock cytoplasmic dynein LCs) bind remains uncertain. Analysis of cytoplasmic dynein has suggested that an IC region immediately N-terminal of the first WD-repeat (or possibly including the repeat) is necessary (Susalka et al., 2002), whereas study of the bop5 mutant version of IC138 within Chlamydomonas inner arm I1 has implicated the seventh WD-repeat and/or the C-terminal region in the interaction of LC7b with that dynein (Hendrickson et al., 2004). Due to the interlocking assembly of the WD-repeat β-propeller (the first β strand of repeat #1 hydrogen bonds to the last strand of repeat #7; see, e.g., Sondek et al., 1996), these regions are likely in proximity at the tertiary structure level and may both contribute to LC7/roadblock LC binding (Hendrickson et al., 2004).
Heavy Chain-independent Assembly of the Motor Domain-associated Light Chain
Previously, we demonstrated that the leucine-rich repeat protein LC1 associates directly with the motor domain of the γ HC via a strong hydrophobic association (Benashski et al., 1999; Wu et al., 2000). Furthermore, we determined by EDC cross-linking that LC1 interacts directly with an additional as yet unknown axonemal protein (p45) that does not copurify with the outer dynein arm. The interaction between LC1 and p45 is ionic and is disrupted by the high salt treatment required to extract the outer arm from the axoneme (Benashski et al., 1999; Wu et al., 2000). Analysis of axonemes from the oda6-r88 oda12-1 double mutant revealed that considerable amounts of LC1 assemble even in the complete absence of the γ HC. This implies an alternative, dynein HC-independent, assembly pathway for LC1, and further suggests that this protein mediates a direct linkage between the γ HC motor domain and other axonemal components.
In conclusion, identification of a Tctex1-like protein within Chlamydomonas outer arm dynein and analysis of mutants defective for components of the IC/LC complex, has revealed a functional contribution of LC binding to dynein motor activity and provided insight into the molecular architecture of this portion of the dynein particle.
Acknowledgments
We thank Dr. David Mitchell (SUNY Upstate Medical Center) for providing the oda6 pseudorevertant strains and for helpful discussions. We are also grateful to Nancy Haas and Dr. Carolyn Silflow (University of Minnesota, St. Paul, MN) for locating the LC9 gene on the Chlamydomonas genetic map. This study was supported by Grants GM-51293 (to S.M.K.), GM-60992 (to G.J.P.), and GM-30626 (to G.B.W.) from the National Institutes of Health. S.M.K. is an investigator of the Patrick and Catherine Weldon Donaghue Medical Research Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0732) on September 29, 2005.
Abbreviations used: DMP, dimethylpimelimidate; EDC, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide; HC, heavy chain; IC, intermediate chain; LC, light chain; MBP, maltose-binding protein; UTR, untranslated region.
Footnotes
This inner arm dynein is also known as subspecies f (Kagami and Kamiya, 1992).
References
- Adams, G., Huang, B., Piperno, G., and Luck, D. (d1981). Central pair microtubule complex of Chlamydomonas flagella: polypeptide composition as revealed by analysis of mutants. J. Cell Biol. 91, 69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benashski, S. E., Patel-King, R. S., and King, S. M. (1999). Light chain 1 from the Chlamydomonas outer dynein arm is a leucine-rich repeat protein associated with the motor domain of the γ heavy chain. Biochemistry 38, 7253-7264. [DOI] [PubMed] [Google Scholar]
- Bowman, A. B., Patel-King, R. S., Benashski, S. E., McCaffery, J. M., Goldstein, L. S., and King, S. M. (1999). Drosophila roadblock and Chlamydomonas LC 7, a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J. Cell Biol. 146, 165-180. [PMC free article] [PubMed] [Google Scholar]
- Caggese, C., Moschetti, R., Ragone, G., Barsanti, P., and Caizzi, R. (2001). dtctex-1, the Drosophila melanogaster homolog of a putative murine t-complex distorter encoding a dynein light chain, is required for production of functional sperm. Mol Genet Genomics 265, 436-444. [DOI] [PubMed] [Google Scholar]
- DiBella, L. M., Benashski, S. E., Tedford, H. W., Harrison, A., Patel-King, R. S., and King, S. M. (2001). The Tctex1/Tctex2 class of dynein light chains. Dimerization, differential expression, and interaction with the LC8 protein family. J. Biol. Chem. 276, 14366-14373. [DOI] [PubMed] [Google Scholar]
- DiBella, L. M., Sakato, M., Patel-King, R. S., Pazour, G. J., and King, S. M. (2004a). The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol. Biol. Cell 15, 4633-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella, L. M., Smith, E. F., Patel-King, R. S., Wakabayashi, K., and King, S. M. (2004b). A novel Tctex2-related light chain is required for stability of inner dynein arm I1 and motor function in the Chlamydomonas flagellum. J. Biol. Chem. 279, 21666-21676. [DOI] [PubMed] [Google Scholar]
- Douglas, M., Diefenbach, R., Homa, F., Miranda-Saksena, M., Rixon, F., Vittone, V., Byth, K., and Cunningham, A. (2004). Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains rp3 and Tctex1 and plays a role in retrograde intracellular transport. J. Biol. Chem. 279, 28522-28530. [DOI] [PubMed] [Google Scholar]
- Fossella, J., Samant, S. A., Silver, L. M., King, S. M., Vaughan, K. T., Olds-Clarke, P., Johnson, K. A., Mikami, A., Vallee, R. B., and Pilder, S. H. (2000). An axonemal dynein at the Hybrid Sterility 6 locus: implications for t haplotype-specific male sterility and the evolution of species barriers. Mamm. Genome 11, 8-15. [DOI] [PubMed] [Google Scholar]
- Harris, E. H. (1989). The Chlamydomonas Sourcebook, San Diego: Academic Press.
- Harrison, A., Olds-Clarke, P., and King, S. M. (1998). Identification of the t complex-encoded cytoplasmic dynein light chain Tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J. Cell Biol. 140, 1137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, T. W., Perrone, C. A., Griffin, P., Wuichet, K., Mueller, J., Yang, P., Porter, M. E., and Sale, W. S. (2004). IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol. Biol. Cell 15, 5431-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, B. G., Koschorz, B., Wertz, K., McLaughlin, J., and Kispert, A. (1999). A protein kinase encoded by the t complex responder gene causes non-Mendelian inheritance. Nature 402, 141-146. [DOI] [PubMed] [Google Scholar]
- Huang, B., Piperno, G., Ramanis, Z., and Luck, D. (1981). Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88, 80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huw, L. Y., Goldsborough, A. S., Willison, K., and Artzt, K. (1995). Tctex 2, a sperm tail surface protein mapping to the t-complex. Dev. Biol. 170, 183-194. [DOI] [PubMed] [Google Scholar]
- Inaba, K., Kagami, O., and Ogawa, K. (1999). Tctex2-related outer arm dynein light chain is phosphorylated at activation of sperm motility. Biochem. Biophys. Res. Commun. 256, 177-183. [DOI] [PubMed] [Google Scholar]
- Kagami, O., Gotoh, M., Makino, Y., Mohri, H., Kamiya, R., and Ogawa, K. (1998). A dynein light chain of sea urchin sperm flagella is a homolog of mouse Tctex1, which is encoded by a gene of the t complex sterility locus. Gene 211, 383-386. [DOI] [PubMed] [Google Scholar]
- Kagami, O., and Kamiya, R. (1992). Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J. Cell Sci. 103, 653-664. [Google Scholar]
- Kai, N., Mishina, M., and Yagi, T. (1997). Molecular cloning of Fyn-associated molecules in the mouse central nervous system. J. Neurosci. Res 48, 407-424. [PubMed] [Google Scholar]
- Kamiya, R. (1988). Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J. Cell Biol. 107, 2253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, R. (2000). Analysis of cell vibration for assessing axonemal motility in Chlamydomonas. Methods 22, 383-387. [DOI] [PubMed] [Google Scholar]
- Kamiya, R., Kurimoto, E., and Muto, E. (1991). Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell Biol. 112, 441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S. M. (2002). Dynein Motors: structure, mechanochemistry and regulation. In: Molecular Motors, ed. M. Schliwa, Weinheim: Wiley-VCH Verlag, 45-78.
- King, S. M., Barbarese, E., Dillman, J. F., Benashski, S. E., Do, K. T., Patel-King, R. S., and Pfister, K. K. (1998). Cytoplasmic dynein contains a family of differentially expressed light chains. Biochemistry 37, 15033-15041. [DOI] [PubMed] [Google Scholar]
- King, S. M., Dillman, J. F., Benashski, S. E., Lye, R. J., Patel-King, R. S., and Pfister, K. K. (1996). The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J. Biol. Chem. 271, 32281-32287. [DOI] [PubMed] [Google Scholar]
- King, S. M., Otter, T., and Witman, G. B. (1985). Characterization of monoclonal antibodies against Chlamydomonas flagellar dyneins by high-resolution protein blotting. Proc. Natl. Acad. Sci. USA 82, 4717-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S. M., Otter, T., and Witman, G. B. (1986). Purification and characterization of Chlamydomonas flagellar dyneins. Methods Enzymol. 134, 291-306. [DOI] [PubMed] [Google Scholar]
- King, S. M., and Patel-King, R. S. (1995). The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J. Biol. Chem. 270, 11445-11452. [DOI] [PubMed] [Google Scholar]
- King, S. M., Wilkerson, C. G., and Witman, G. B. (1991). The Mr78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with α-tubulin in situ. J. Biol. Chem. 266, 8401-8407. [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. (1996). MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51-55. [DOI] [PubMed] [Google Scholar]
- Koutoulis, A., Pazour, G. J., Wilkerson, C. G., Inaba, K., Sheng, H., Takada, S., and Witman, G. B. (1997). The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J. Cell Biol. 137, 1069-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lader, E., Ha, H. S., O'Neill, M., Artzt, K., and Bennett, D. (1989). tctex-1: a candidate gene family for a mouse t complex sterility locus. Cell 58, 969-979. [DOI] [PubMed] [Google Scholar]
- Lai, M., Wang, F., Rohan, J., Maeno-Hikichi, Y., Chen, Y., Zhou, Y., Gao, G., Sather, W., and Zhang, J. (2005). A Tctex1-Ca2+ channel complex for selective surface expression of Ca2+ channels in neurons. Nat. Neurosci. 8, 435-442. [DOI] [PubMed] [Google Scholar]
- LeDizet, M., and Piperno, G. (1995). The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol. Biol. Cell 6, 697-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.-G., Serr, M., Newman, E., and Hays, T. (2004). The Drosophila Tctex-1 light chain is dispensable for essential cytoplasmic dynein functions but is required during spermatid differentiation. Mol. Biol. Cell 15, 3005-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, J., Jaffrey, S. R., Guo, W., Snyder, S. H., and Clardy, J. (1999). Structure of the PIN/LC8 dimer with a bound peptide. Nat. Struct. Biol. 6, 735-740. [DOI] [PubMed] [Google Scholar]
- Mitchell, D. R., and Kang, Y. (1991). Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J. Cell Biol. 113, 835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D. R., and Kang, Y. (1993). Reversion analysis of dynein intermediate chain function. J. Cell Sci. 105, 1069-1078. [DOI] [PubMed] [Google Scholar]
- Mitchell, D. R., and Rosenbaum, J. L. (1986). Protein-protein interactions in the 18 S ATPase of Chlamydomonas outer dynein arms. Cell Motil. Cytoskeleton 6, 510-520. [DOI] [PubMed] [Google Scholar]
- Mok, Y.-K., Lo, K.W.-H., and Zhang, M. (2001). Structure of Tctex-1 and its interaction with cytoplasmic dynein intermediate chain. J. Biol. Chem. 276, 14067-14074. [DOI] [PubMed] [Google Scholar]
- Mou, T., Kraas, J. R., Fung, E. T., and Swope, S. L. (1998). Identification of a dynein molecular motor component in Torpedo electroplax; binding and phosphorylation of Tctex-1 by Fyn. FEBS Lett. 435, 275-281. [DOI] [PubMed] [Google Scholar]
- Mueller, S., Cao, X., Welker, R., and Wimmer, E. (2002). Interaction of the poliovirus receptor CD155 with the dynein light chain Tctex-1 and its implication for poliovirus pathogenesis. J. Biol. Chem. 277, 7897-7904. [DOI] [PubMed] [Google Scholar]
- Nagano, F., Orita, S., Sasaki, T., Naito, A., Sakaguchi, G., Maeda, M., Watanabe, T., Kominami, E., Uchiyama, Y., and Takai, Y. (1998). Interaction of Doc2 with Tctex-1, a light chain of cytoplasmic dynein. Implication in dynein-dependent vesicle transport. J. Biol. Chem. 273, 30065-30068. [DOI] [PubMed] [Google Scholar]
- Olmsted, J. B. (1986). Analysis of cytoskeletal structures using blot-purified monospecific antibodies. Methods Enzymol. 134, 467-472. [DOI] [PubMed] [Google Scholar]
- Patel-King, R. S., Benashki, S. E., Harrison, A., and King, S. M. (1996). Two functional thioredoxins containing redox-sensitive vicinal dithiols from the Chlamydomonas outer dynein arm. J. Biol. Chem. 271, 6283-6291. [DOI] [PubMed] [Google Scholar]
- Patel-King, R. S., Benashski, S. E., Harrison, A., and King, S. M. (1997). A Chlamydomonas homologue of the putative murine t complex distorter Tctex-2 is an outer arm dynein light chain. J. Cell Biol. 137, 1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G., Sineschekov, O., and Witman, G. (1995). Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J. Cell Biol. 131, 427-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G. J., Koutoulis, A., Benashski, S. E., Dickert, B. L., Sheng, H., Patel-King, R. S., King, S. M., and Witman, G. B. (1999). LC2, the Chlamydomonas homologue of the t complex-encoded protein Tctex2, is essential for outer dynein arm assembly. Mol. Biol. Cell 10, 3507-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G. J., and Witman, G. B. (2000). Forward and reverse genetic analysis of microtubule motors in Chlamydomonas. Methods 22, 285-298. [DOI] [PubMed] [Google Scholar]
- Pfister, K. K., Fay, R. B., and Witman, G. B. (1982). Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil. 2, 525-547. [DOI] [PubMed] [Google Scholar]
- Piperno, G., and Luck, D. J. (1979). Axonemal adenosine triphosphatases from flagella of Chlamydomonas reinhardtii. Purification of two dyneins. J. Biol. Chem. 254, 3084-3090. [PubMed] [Google Scholar]
- Piperno, G., Ramanis, Z., Smith, E. F., and Sale, W. S. (1990). Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J. Cell Biol. 110, 379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M. E., Power, J., and Dutcher, S. K. (1992). Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J. Cell Biol. 118, 1163-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager, R., and Granick, S. (1953). Nutritional studies with Chlamydomonas reinhardtii. Ann. NY Acad. Sci. 56, 831-838. [DOI] [PubMed] [Google Scholar]
- Sakato, M., and King, S. (2004). Design and regulation of the AAA+ microtubule motor dynein. J. Struct. Biol. 146, 58-71. [DOI] [PubMed] [Google Scholar]
- Schwarzer, C., Barnikol-Watanabe, S., Thinnes, F. P., and Hilschmann, N. (2002). Voltage-dependent anion-selective channel (VDAC) interacts with the dynein light chain Tctex1 and the heat-shock protein PBP74. Int J Biochem. Cell Biol. 34, 1059-1070. [DOI] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D. G., Hamm, H. E., and Sigler, P. B. (1996). Crystal structure of a G-protein β/γ dimer at 2.1 Å resolution. Nature 379, 369-374. [DOI] [PubMed] [Google Scholar]
- Sugai, M., Saito, M., Sukegawa, I., Katsushima, Y., Kinouchi, Y., Nakahata, N., Shimosegawa, T., Yanagisawa, T., and Sukegawa, J. (2003). PTH/PTH-related protein receptor interacts directly with Tctex-1 through its C-terminal domain. Biochem. Biophys. Res. Commun. 311, 24-31. [DOI] [PubMed] [Google Scholar]
- Susalka, S. J., Nikulina, K., Salata, M. W., Vaughan, P. S., King, S. M., Vaughan, K. T., and Pfister, K. K. (2002). The roadblock light chain binds a novel region of the cytoplasmic dynein intermediate chain. J. Biol. Chem. 277, 32939-32946. [DOI] [PubMed] [Google Scholar]
- Tai, A. W., Chuang, J. Z., Bode, C., Wolfrum, U., and Sung, C. H. (1999). Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell 97, 877-887. [DOI] [PubMed] [Google Scholar]
- Takada, S., Sakakibara, H., and Kamiya, R. (1992). Three-headed outer arm dynein from Chlamydomonas that can functionally combine with outer-arm-missing axonemes. J. Biochem. 111, 758-762. [DOI] [PubMed] [Google Scholar]
- Wakabayashi, K., Takada, S., Witman, G. B., and Kamiya, R. (2001). Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil. Cytoskeleton 48, 277-286. [DOI] [PubMed] [Google Scholar]
- Wilkerson, C. G., King, S. M., Koutoulis, A., Pazour, G. J., and Witman, G. B. (1995). The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 129, 169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J., Xie, H., and Hendrickson, W. (2005). Crystal structure of dynein light chain Tctex1. J. Biol. Chem. 280, 21981-21986. [DOI] [PubMed] [Google Scholar]
- Witman, G. B. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280-290. [DOI] [PubMed] [Google Scholar]
- Wu, H., Maciejewski, M. W., Benashski, S. E., Mullen, G. P., and King, S. M. (2001). 1H, 15N and 13C resonance assignments for the Tctex1 dynein light chain from Chlamydomonas flagella. J. Biomol. NMR 20, 89-90. [DOI] [PubMed] [Google Scholar]
- Wu, H., Maciejewski, M. W., Marintchev, A., Benashski, S. E., Mullen, G. P., and King, S. M. (2000). Solution structure of a dynein motor domain associated light chain. Nat. Struct. Biol. 7, 575-579. [DOI] [PubMed] [Google Scholar]
- Wu, H., Maciejewski, M. W., Takebe, S., and King, S. M. (2005). Solution structure of the Tctex1 dimer reveals a mechanism for dynein-cargo interactions. Structure 13, 213-223. [DOI] [PubMed] [Google Scholar]