Abstract
Actin-related proteins (ARPs) are key players in cytoskeleton activities and nuclear functions. Two complexes, ARP2/3 and ARP1/11, also known as dynactin, are implicated in actin dynamics and in microtubule-based trafficking, respectively. ARP4 to ARP9 are components of many chromatin-modulating complexes. Conventional actins and ARPs codefine a large family of homologous proteins, the actin superfamily, with a tertiary structure known as the actin fold. Because ARPs and actin share high sequence conservation, clear family definition requires distinct features to easily and systematically identify each subfamily. In this study we performed an in depth sequence and comparative genomic analysis of ARP subfamilies. A high-quality multiple alignment of ∼700 complete protein sequences homologous to actin, including 148 ARP sequences, allowed us to extend the ARP classification to new organisms. Sequence alignments revealed conserved residues, motifs, and inserted sequence signatures to define each ARP subfamily. These discriminative characteristics allowed us to develop ARPAnno (http://bips.u-strasbg.fr/ARPAnno), a new web server dedicated to the annotation of ARP sequences. Analyses of sequence conservation among actins and ARPs highlight part of the actin fold and suggest interactions between ARPs and actin-binding proteins. Finally, analysis of ARP distribution across eukaryotic phyla emphasizes the central importance of nuclear ARPs, particularly the multifunctional ARP4.
INTRODUCTION
Since the discovery in the early 1990s of the first genes coding for actin-related proteins (ARPs) called ACT2, now known as ARP2 (Schwob and Martin, 1992) in Saccharomyces cerevisiae and ARP3 (also called ACT2; Lees-Miller et al., 1992b) and ARP1 (called actin-RPV; Lees-Miller et al., 1992a) in Schizosaccharomyces pombe, many new ARPs have been described from unicellular organisms to plants and humans. Sustained investigation of ARP function(s) in yeast, plant, and animal cells has demonstrated that different ARPs, in combination with actin, are required for cytoplasmic or nuclear cellular functions.
The unified classification of ARPs, initially proposed in 1994 (Schroer et al., 1994) was extended in 1997 (Poch and Winsor, 1997). The second study led to the definition of 10 distinct ARP subfamilies according to their relative identity and similarity to conventional actin sequences, where ARP1 is the most similar and ARP10 the least similar. In contrast to the ARP1 to ARP3 subfamily classifications, which were based on multiple sequences from diverse organisms, the ARP4-ARP10 subfamilies were proposed on the basis of only 1 or 2 sequences, in particular from the complete genome of S. cerevisiae (Goffeau et al., 1996). Since then, only one new subfamily, ARP11, has been described (Eckley et al., 1999). This suggested nomenclature has been assessed for major model organisms (Eckley et al., 1999; Harata et al., 2001; Goodson and Hawse, 2002), and a certain number of organisms possess additional “orphan” ARPs that do not group into any of the known subfamilies (Goodson and Hawse, 2002). In fact, had the classification been based on a different organism, ARP7 and ARP9, the yeast specific subfamilies (Blessing et al., 2004), would have been considered as orphans. In this classification, ARP1-ARP3 (and more recently ARP10 and ARP11) are localized in the cytoplasm and perform key functions in the spatiotemporal control of actin assembly and movement of vesicles along microtubules in the cytoplasm (Schafer and Schroer, 1999; Machesky and May, 2001; McKinney et al., 2002). In addition to these well-documented functions, a growing body of evidence supports nuclear functions for ARP4-ARP9 participating in processes like chromatin modulation, regulation of transcription, and DNA repair (Weber et al., 1995; Grava et al., 2000; Harata et al., 2000; Olave et al., 2002; Blessing et al., 2004). This has expanded the palette of actin function and kept ARPs in the limelight of investigative biology.
Together with conventional actins, ARPs define a large family of homologous proteins, the actin superfamily, which share the same structural architecture, known as the “actin fold” (Bork et al., 1992; Holmes et al., 1993; Kabsch and Holmes, 1995). This architecture is also found in heat-shock protein Hsc70, sugar kinases, and bacterial proteins (Bork et al., 1992; Holmes et al., 1993). Although some of these bacterial proteins have recently been shown to retain some actinlike functions (Amos et al., 2004), they show more extreme sequence divergence to actin than ARPs. The actin fold is functionally characterized as an ATPase domain with ATP-binding capacity in the presence of Mg2+ or Ca2+. It is organized in two symmetrical α/β domains I and II, which are connected by a hinge region. Each domain is composed of two subdomains 1 (Ia), 2 (Ib) and 3 (IIa), 4 (IIb). The subdomains 1 and 3 define the “barbed end,” where capping proteins bind actin as opposed to the “pointed end,” composed of subdomains 2 and 4. Each of the two largest subdomains (1, 3) comprises five-stranded β-sheets that are connected by two α-helices. This part of the molecule also forms the hydrophobic cleft that mediates major interactions for actin and actin-binding proteins (ABPs; Dominguez, 2004).
Interestingly, each ARP subfamily has been characterized as part of one or more multisubunit complexes, many of which also contain at least one actin molecule. ARP1, the only ARP known to form a filament (Bingham and Schroer, 1999), is an essential part of the 11-subunit dynactin complex that functions in transport of cargoes and organelles on microtubules. In human cells, this complex also contains the distantly related ARP11 as well as globular actin and the ABP, CapZ (Eckley et al., 1999; Eckley and Schroer, 2003). The ARP2 and ARP3 dimer is part of a seven-subunit complex that nucleates polymerization of de novo actin filaments and branched networks beneath the plasma membrane (reviewed in Pollard et al., 2000). The 3D structure of the ARP2/3 complex has recently been solved in different states (Robinson et al., 2001; Volkmann et al., 2001; Nolen et al., 2004; Rodal et al., 2005).
Nuclear ARPs and actin have been isolated from many nuclear complexes (see Supplementary Data 1). In chromatin-remodeling complexes, ARP4 is generally present with conventional actin in SWI2/SNF2 complexes, with ARP5 and ARP8 in INO80 complexes, and with ARP6 in SWR1 complexes (reviewed in Mohrmann et al., 2004). ARP4 and actin are also components of histone acetyltransferase (HAT) complexes (Doyon et al., 2004). In most cases, the ARP subunits are important for the enzymatic activity of these complexes (Galarneau et al., 2000; Gorzer et al., 2003; Shen et al., 2003). In contrast, ARP7 and ARP9 are not essential for RSC chromatin remodeling complex activity (Szerlong et al., 2003).
With the availability of numerous new ARP sequences in protein databases and recently sequenced genomes, we propose an in-depth comparative genomic analysis of ARP members. We built a new high-quality Multiple Alignment of Complete Sequences (MACS; Lecompte et al., 2001) of ARPs available at http://bips.u-strasbg.fr/ARPAnno/ARPMACS.html. This alignment provides the basis for the extension of the characterization of ARP subfamilies and the classification of ARPs from new organisms. Our sequence analysis differentiates ARPs by determining conserved Family features such as discriminating residues and insertion or deletion (INDEL) sequence signatures. On the basis of our multiple alignment and new discriminating characteristics, we implemented ARPAnno, a freely available web server to annotate ARP sequences http://bips.u-strasbg.fr/ARPAnno. Analysis of the conservation of residues involved in actin ATP-binding capacity predicts weak (or no) interaction for nuclear ARP4-9, ARP10, and ARP11. Furthermore, sequence conservation in the actin fold highlights the importance of the hydrophobic cleft in ARPs, opening new perspectives for interactions between ARPs and ABPs. Finally, the availability of complete genome sequences allowed us to define the distribution of ARPs among eukaryotic phyla and reveals the central importance of the nuclear ARPs, especially ARP4.
MATERIALS AND METHODS
Sequence Searches and Alignment
To cover the maximum diversity of ARPs, sequence searches were performed using as sequence queries: three actin sequences, representative sequences from each ARP subfamily selected from three distantly related organisms (S. cerevisiae, Drosophila melanogaster, and Homo sapiens when available), and five orphan sequences. The sequences were retrieved from Uniprot using SRS (Sequence Retrieval System; Etzold et al., 1996) with their identification (ID) or accession (AC) numbers. This resulted in an initial set of 37 reference proteins shown in Supplementary Data 2.
For each reference protein, a BlastP (Altschul et al., 1990, 1997) search of the Uniprot database (Bairoch et al., 2005; July 2004) was performed and the sequences detected with E < 10-2 were multiply aligned using the PipeAlign program without the clustering step (Plewniak et al., 2003). The PipeAlign web server is available at http://bips.u-strasbg.fr/PipeAlign. These 37 MACS were then merged into a single multiple alignment. Unrelated sequences were removed from the final alignment with LEON (multiple alignment evaluation of neighbors; Thompson et al., 2004). This composite alignment was then refined using RASCAL (rapid scanning and correction of multiple sequence alignment; Thompson et al., 2003) to automatically correct local alignment errors. Finally, manual verification and correction, paying attention to secondary structures, was performed using the seqlab alignment viewer and editor (GCG, 2001). The quality of the final alignment was objectively evaluated using NorMD (normalized mean distance; Thompson et al., 2001). Subfamilies were defined automatically using DPC (density of point clustering; Wicker et al., 2002) and validated by human expertise. Furthermore, a phylogenetic tree based on the final alignment was built with the neighbor joining method (see Supplementary Figure 1). The analysis of this tree confirmed the defined subfamilies.
Overall, the BlastP similarity searches yielded 73,340 proteins, representing 4200 nonredundant and “nonfragment” proteins. Sequences with <15% amino acid identity, notably some bacterial actinlike proteins, were not included in the final alignment. To obtain an objective evaluation of the true number of ARP sequences in Uniprot, we removed database redundancy by counting only nonidentical sequences for each different organism. The final version of the complete multiple alignment of ARPs (ARP-MACS) contains more than 700 proteins (sequence list included in Supplementary Data 3) clustered into one actin subfamily, 11 ARP subfamilies, and orphans. ARPMACS is available at http://bips.u-strasbg.fr/ARPAnno/ARPMACS.html.
Sequence Analysis
Two statistics were used to characterize each ARP subfamily, RefID and FamID. First, the RefID is defined to compare the current subfamily classification with the one used in 1997 (Poch and Winsor, 1997; called IniID here). It is computed for each ARP subfamily as the mean pairwise percent identity of each sequence in the subset against a reference sequence. Positions in the alignment of these sequences corresponding to gaps were excluded from the calculation. The reference actin sequence used is the human actin gene annotated as “Actin, alpha skeletal muscle (Alpha-actin 1)” Uniprot ID ACTS_HUMAN and AC P02568, that is strictly identical to the actins from Bos taurus, Gallus gallus, Mus musculus, Sus scrofa, Oryctolagus cuniculus, Rattus norvegicus. All indications of amino acid positions used in the following analyses refer to this reference sequence (Supplementary Data 4).
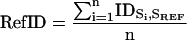 |
where: n is total number of sequence tested, Si and SREF are, respectively, the ith and reference actin sequence, and IDSi,SREF is the pairwise percent identity between the ith and the reference actin sequence, excluding gap regions.
Second, the FamID describes the conservation within each subfamily. The FamID of each ARP subfamily was calculated as the mean pairwise percent identity of each sequence against each other sequence in a given subset. As above, positions in the alignment corresponding to gaps within the subset were excluded from the calculation.
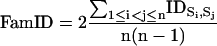 |
where: n is the total number of sequence tested, Si and Sj are the ith and jth sequence, and IDSi,Sj are pairwise percent identity between the ith and jth sequence, excluding gap regions.
Limitations of the RefID and FamID calculations are the absence of certain subfamilies in different organisms and the incomplete representation of ARPs in protein databases. Taking this into account, the two statistics were calculated on a subset of sequences encompassing at least one sequence from mammals, insects, worms, fungi, and plants for each subfamily, except ARP7, ARP9, and ARP10, restricted to yeasts (Accession numbers of sequences used are available upon request).
Discriminating Residues and Insertions/Deletions
The discriminating residues were identified as amino acids strictly present within a particular ARP subfamily and strictly absent in all other sequences. We also identified motifs (2-7 residues) that could distinguish an ARP subfamily (see Figure 2 legend). Insertion and deletions in ARPs were defined as a minimum of 10 residues added to or deleted from the reference actin sequence (Supplementary Data 5). To characterize the INDELs, the entry point was defined as a single position where at least one ARP has an INDEL and the “hot spot” as a short sequence stretch in which many different ARPs have INDELs. The discriminating residues, motifs, and INDELs constitute a knowledge filter used to characterize the ARP subfamilies.
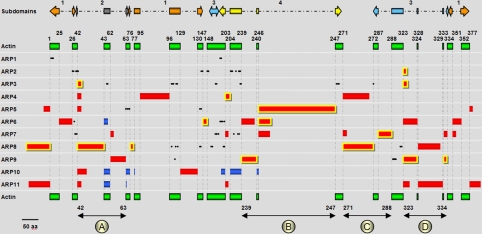
Figure 2.
Schema representing residues, motifs, insertions, and deletions in ARP subfamilies. The reference human α-actin sequence (377 aa) is represented as green rectangles and positions are given according to this reference. The upper arrows illustrate the four subdomains of actin colored in orange (Ia or 1), dark gray (Ib or 2), light blue (IIa or 3), and yellow (IIb or 4). Insertions are represented as red rectangles and deletions as dark blue rectangles (sizes correspond to the mean length). The insertions conserved in all members of a subfamily are in red highlighted with yellow. Hotspots of insertions (circled letters and black double arrow) are indicated at the bottom of the figure (also reported on Figure 4A). Discriminating residues and motifs, indicated as black dots, are as follows: ARP1, N7[A/Q/S]8P9, R196[R/K/H]197; ARP2, N27F28, P40[I/L/M/V]41[I/L/M/V]42R43, Y145Q147G148L149, A206D207F208, Y223, [E/D]234T235T236; ARP3, L52D53, [H/T]103[F/Y/H/T]104F105, P115E116, [G/C]174S175, [R/K]237[F/Y/W]238, D251[G/A]253[Y/F]254, [F/Y/W]272; ARP5, R66[K/R]70F73[D/E]74, [D/R]139[Y/F]142; ARP6, N30, [S/T]196[Y/F/V/L]197[R/V/K]198[H/N/D]201; ARP7, S35Y37[I/L/M/V]38, [K/R]181G184F186[D/N]188[Q/H]190S216[F/Y]229K230; ARP8, [Y/F/H]103[D/E]111[V/L/I]118, [Q/E]138[E/D]139, C166, [E/D]168, [F/W]200P201, [D/E/Q/H]317[R/K]318; ARP9, W113, [R/K]291[W/Y]296[D/E/N]297N298[I/L/V]299
ARPAnno Web Server
To make our results easily available to the scientific community, a web server ARPAnno (actin-related proteins annotation server) has been developed to allow reliable classification and annotation of newly sequenced potential actinlike proteins. ARPAnno is written in Tcl/Tk script or in ANSI C for some functions. ARPAnno also requires the Blast and ClustalW programs. The strategy of ARPAnno is based on a three-step process:
First ARPAnno aligns the query sequence with BlastP against dedicated databases of each subfamily contained in ARP-MACS (actin, 11 ARP subfamilies and orphans). Eligible subfamilies which are the most suitable for further investigation are then determined by the calculation of two cutoffs. First, a global percent identity (GID) is defined as the ratio of the number of identical residues to the total number of residues in all HSPs (high scoring pairs) of the query. Second, a percent coverage (pCover) is defined as the ratio of the number of identical residues to the number of residues that could be aligned between the two sequences.
The query is then aligned against the eligible subfamilies in the ARPMACS using the ClustalW global multiple alignment program (Thompson et al., 1994) in profile mode and filtered according to the knowledge-based criteria (residues and INDELs signatures) defined above. For each eligible subfamily, two scores are calculated: the number of discriminating insertions (pDI) detected as a percentage of the total number of discriminating insertions characterized for the considered subfamily, and the number of discriminating residues (pDR) detected as a percentage of the total number of residues described for the subfamily.
A final score on a scale of 0-100 is computed for each subfamily based on the local and global alignment and the knowledge-based filter.
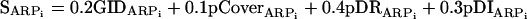 |
where SARPi is the final score, pDRARPi is the percentage of detected residues, and pDIARPi is the percentage of detected insertions for the ith ARP subfamily.
The relative weights of each score were determined experimentally to best separate the subfamilies previously established by ARP-MACS. The alignments of the query with each eligible subfamily are displayed with Family features highlighted in different colors and are available for download in XML or MSF format. ARPAnno is available at http://bips.u-strasbg.fr/ARPAnno and mirrored at www.bioinfomatics.lu.
ATP Binding
To explore the potentiality of ARPs for ATP binding, we calculated the conservation of the 17 key reference actin amino acids for nucleotide binding (D13, S16, G17, L18, K20, Q139, D156, D159, G160, V161, K215, E216, G304, T305, M307, Y308, K338) for all ARP subfamilies as described previously (Kabsch et al., 1990; Lees-Miller et al., 1992a). The mean percent of conserved identical residues and similar residues were computed for each ARP subfamily.
Structural Studies
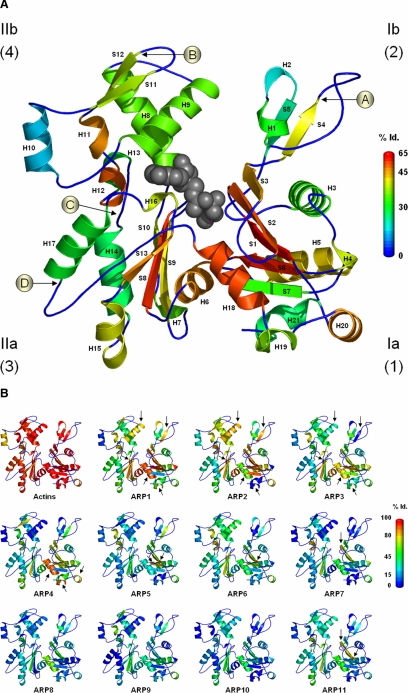
The actin molecule is represented by the 3D structure of yeast actin (Uniprot ID ACT_YEAST, PDB 1YAG; Vorobiev et al., 2003) and secondary structures are named according to the PDB data (see Figure 4). The actin fold is mainly defined by subdomains 1 and 3 excluding helices H15, H19, and H20, as well as helix H11 and the bottom part of helices H8 and H9 in subdomain 4, with no contribution from subdomain 2 (Kabsch and Holmes, 1995). One major actin-binding interface of actin, known as the “hydrophobic cleft” is defined essentially by residues in three helices (H18, H19, and H20) in subdomain 1 (Dominguez, 2004). The mean percent identity to the reference actin in ARPMACS was calculated using a sliding-window corresponding to each secondary structure. This statistic was used to replace the temperature factor field in the PDB file. Figure 4A represents the mean percent identity of all ARP subfamilies and Figure 4B that of each ARP subfamily individually. The sequence conservations are mapped onto the structure with colors ranging from dark blue to red, corresponding to 0-65% identity (Id.; loops excluded) in Figure 4A and to 0-100% Id. in Figure 4B.
Figure 4.
Actin amino acid conservation in secondary structure throughout ARP subfamilies. Actin 3D structure is drawn from the yeast PDB data 1YAG in standard orientation with secondary structures labeled H for helices and S for strands, numbered in order of appearance from N- to C-terminus. The secondary structures were colored according to percent identity (Id.) with the reference human α-actin (Uniprot ID ACTS_HUMAN and AC P02568) by replacing the temperature factor field in the PDB file. The figures were made with PyMol (Delano, 2002). (A) Mean ARP subfamilies' global conservation. The conservation scale 0-65% Id. is colored from dark blue to red. An ATP molecule is represented in dark gray. The four circled letters indicate with an arrowhead the four hot spot positions of insertions. The four subdomains of actin are indicated as Ia (1), Ib (2), IIa (3), and IIb (4). (B) Individual ARP subfamily conservation structures. Arrows mark specific details for each subfamily as described in results. The conservation scale 0-100% Id. is colored from dark blue to red.
Phylogenetic Distribution of ARPs in Complete Genomes
The ARP distribution was examined in 20 eukaryotic organisms for which the complete genome sequences are available. The presence/absence of each ARP was cross-validated at both the proteomic and genomic levels. Inspection of recently reported genomic sequences identified potential new ARP genes missed during the gene prediction process. A table summarizing proteomic and genomic searches is included in Supplementary Data 6. Where available, the nucleotide sequence was retrieved from the NCBI nucleotide sequence database known as GenBank (Benson et al., 2005) and RefSeq (Pruitt et al., 2005) and queried with the 37 reference sequences using the TBlastN program.
The 20 complete eukaryotic genomes used are: Oryza sativa (Goff et al., 2002), Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000), Plasmodium falciparum (Gardner et al., 2002), Encephalitozoon cuniculi (Katinka et al., 2001), Neurospora crassa (Galagan et al., 2003), S. cerevisiae (Goffeau et al., 1996), Candida glabrata, Yarrowia lipolytica (Dujon et al., 2004), S. pombe (Wood et al., 2002), Anopheles gambiae (Holt et al., 2002), D. melanogaster (Adams et al., 2000), Caenorhabditis elegans (Chervitz et al., 1998; C. elegans Sequencing Consortium, 1998), Ciona intestinalis (Dehal et al., 2002), Tetraodon nigroviridis (Jaillon et al., 2004), M. musculus (Waterston et al., 2002), H. sapiens (Lander et al., 2001; Venter et al., 2001). We also used dedicated websites in order to retrieve the latest sequence version for Thalassiosira pseudonana (http://genome.jgipsf.org/thaps1; Armbrust et al., 2004), Dictyostelium discoideum (http://dictybase.org; Kreppel et al., 2004; Eichinger et al., 2005), and other sites for additional Blast searches for Cryptosporidium parvum (http://cryptodb.org/CryptoDB.shtml; Abrahamsen et al., 2004; Puiu et al., 2004), and Cyanidioschyzon merolae (http://merolae.biol.s.u-tokyo.ac.jp; Matsuzaki et al., 2004).
An extended exploration of all complete and incomplete Fungi proteomes and genomes reported and existing at the NCBI Blast server was made. We used 31 Fungi, divided into Ascomycota Saccharomycotina (C. albicans, C. glabrata, D. hansenii, E. gossypii, K. lactis, K waltii, N. castellii, S. bayanus 623-6C, S. bayanus MCYC 623, S. cerevisiae, S. kluyveri, S. kudriavzevii, S. mikatae, S. paradoxus and Y. lipolytica), Ascomycota Pezizomycotina (A. fumigatus, A. nidulans, A. terreus, C. immitis, C. posadasii, G. zeae, M. grisea, N. crassa), Ascomycota Schizosaccharomycetes (S. pombe), Basidiomycota (C. cinerea okayama, C. neoformans var. grubii H99, C. neoformans var. neoformans B-3501A, C. neoformans var. neoformans JEC21, P. chrysosporium, U. maydis) and Microsporidia (E. cuniculi).
RESULTS
Sequence Analysis and Subfamily Definition
We built ARP-MACS, a new high quality multiple alignment of complete sequences of all ARPs and actins available in Uniprot (July 2004) as the basis for an extended characterization of ARP subfamilies. In our earlier study, the previously defined ARP1-ARP3 subfamilies (Schroer et al., 1994) were confirmed on the basis of 5-8 sequences, and the remaining ARP subfamilies were proposed essentially on the basis of S. cerevisiae sequences (Poch and Winsor, 1997). Later these subfamilies were established by phylogenetic analyses (Eckley et al., 1999; Harata et al., 2001; Goodson and Hawse, 2002). Since 1997, including this analysis, the only new major ARP that was identified is ARP11 (Eckley et al., 1999). The growing number of ARP proteins available in protein databases and classified in ARP-MACS (Table 1) consolidates the ARP4-ARP11 subfamilies classification. In agreement with previous studies, the major ARP subfamilies are ARP1-6, ARP8, and ARP11, whereas fewer sequences are available for subfamilies ARP7, ARP9, and ARP10. As illustrated below, these subfamilies are restricted to certain phyla. Twenty-seven orphan protein sequences were found in Metazoa (H. sapiens, M. fascicularis, M. musculus, and C. elegans), plants (O. sativa and A. thaliana) and in the parasites E. cuniculi, P. falciparum, and P. yoelii. These sequences range in size from 328 to 1207 amino acids, and share from 21 to 49% Id. with the reference actin. They have been included in the overall alignment but are not considered as a subfamily because they lack common defining characteristics.
Table 1.
Evolution of the number of actin and ARP sequences since 1997
| ARP-MACS Uniprot (2004)
|
||||
|---|---|---|---|---|
| Subfamilies | Poch and Winsor (1997) | No redundancy | Total | 20 complete genomes |
| Actin | 29a | – | 546 | 20 |
| ARP1 | 8 | 20 | 26 | 16 |
| ARP2 | 5 | 19 | 30 | 15 |
| ARP3 | 7 | 23 | 34 | 15 |
| ARP4 | 2 | 21 | 39 | 21b |
| ARP5 | 1 | 12 | 19 | 14 |
| ARP6 | 2 | 16 | 21 | 19 |
| ARP7 | 1 | 5 | 5 | 2 |
| ARP8 | 1 | 11 | 13 | 13 |
| ARP9 | 1 | 7 | 7 | 4 |
| ARP10 | 1 | 6 | 6 | 3 |
| ARP11 | 0 | 8 | 10 | 10 |
| Total | 29 | – | 756 | 152 |
| Total ARP | 29 | 148 | 210 | 132 |
Sequences were collected and analyzed as described in Materials and Methods.
Twenty-nine actin sequences out of 194 available (Poch and Winsor, 1997)
Second ARP4 in Y. lypolytica and S. pombe
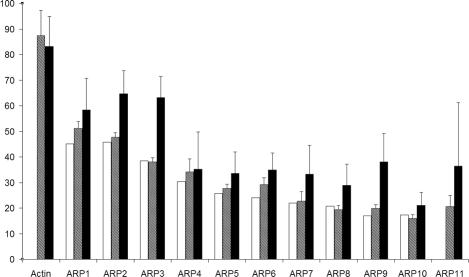
To validate the reliability of the ARP classification based on the mean percent identity between ARP sequences and the reference actin (RefID), we compared the ranking obtained here with the ARP ranking based on initial percent identities (IniID) deduced from the data available in 1997 (Poch and Winsor, 1997). IniID and RefID are highly correlated (Figure 1); ARP1 is the closest to actin and ARP10 and ARP11 are the most distant in both cases, reinforcing the universal classification. Nevertheless, with our recent refinements and definitions, the relative order of some subfamilies could have been exchanged, e.g., ARP5 with ARP6. In spite of these small variations, to avoid confusion in naming genes and proteins, we do not recommend changing the existing nomenclature. The growing number of sequences per ARP subfamily allows an evaluation of the intrasubfamily conservation (FamID). Three groups of proteins were distinguished (Figure 1). As expected, the conventional actins (546 sequences) are the most conserved subfamily (FamID > 80%). The second group is composed of cytoplasmic ARPs (ARP1-ARP3), and shares significantly more intrasubfamily conservation (50% < FamID < 80%) than the last group including all nuclear ARPs and ARP10 and ARP11 (FamID < 40%).
Figure 1.
Actin and ARP conservation. The initial percent identity (IniID) used in 1997 (Poch and Winsor, 1997) to classify the ARPs is represented as open bars. A new percent identity (RefID) is shown as hatched bars. The closed bars are the mean percent identity inside a given subfamily (FamID). Error bars, SDs.
ARP Subfamily Characterization
Because of high sequence identity and similarity between ARPs and actin sequences, it is frequently difficult to unambiguously detect and classify an ARP sequence from BlastP database searches. Indeed the Blast score and ranking of ARP homologous sequences is perturbed by the presence of insertions and deletions and the existence of a very limited number of discriminating residues (see Materials and Methods). As an example, the search of homologues (BlastP) for the human ARP1 in Uniprot leads to 1653 protein “hits” exhibiting a significant E-value (E ≥ 10-2). Among these, ARP1 sequences are dispersed among conventional actin and other ARPs. The last ARP1 detected was the yeast ARP1 at rank 769, lower than many non-ARP1 sequences. This prompted us to define discriminating criteria, i.e., sequence features conserved in a given subfamily and strictly absent in any other, for each ARP subfamily using specific residues, motifs or INDELs as shown in Figure 2 and Supplementary Data 5.
The analysis of the regions aligned in all ARP and actin sequences, revealed single discriminating residues and motifs for 8 ARP subfamilies, that is, 6 single residues and 23 motifs covering 76 residues. ARP2 and ARP3 subfamilies have the highest number of motifs (6 and 7, respectively), whereas no specific residues or motifs were found for the ARP4, ARP10, and ARP11 subfamilies. We next built a map of INDELs relative to the reference actin for each subfamily sequence (Figure 2). Strikingly, ARPs show a high number of different insertions (41) of different sizes at various positions along the total 377 amino acid length of actin, but only four deletions. Excluding the N- and C-terminal extensions, a total of 16 entry points was observed. Four hot spots (named A, B, C, and D in Figure 2) are found at positions 42-63, 239-247, 271-288, and 323-334. INDELS have a maximum size of ∼330 amino acids and occur mainly in loops. No large insertions were identified in the core structure of the actin fold. In terms of domain distribution, the discontinuous subdomain 1 is the least susceptible to INDELs with only ARP4, ARP6, and ARP10 having intrasubdomain insertions. In contrast, the smallest subdomain 2 is the most sensitive to adaptation, with the main hot spot (A) comprising 11 INDELs distributed in ARP3-ARP6 and ARP8-ARP11. Hot spot (A) includes one deletion from each of ARP6, ARP10, and ARP11 but none of these are characteristic of all subfamily members. The largest deletion is observed in subdomain 4 of S. cerevisiae ARP10 and results in the complete loss of almost all the subdomain. Another remarkable feature, if we consider the ARPs cellular localization, is the paucity of insertions larger than 10 residues in cytoplasmic ARP1, ARP2, and ARP3, in contrast to the nuclear ARPs, which contain from 1 to 5 insertions.
Although many entry points are common to different ARP subfamilies, it is noteworthy that no sequence similarity was found between the insertions from different ARP subfamilies. Thus, ARP characterization can be completed by describing the presence of an insertion common to all members of a given ARP subfamily (Family Insertion, highlighted in yellow in Figure 2). However, ARP1 has no insertion ≥10 aa, and ARP10 and ARP11 have many different INDELs but none are conserved in all members of the subfamily. We also found that the N-terminal motif MS[G/A][G/A][V/L]YGG in ARP4 (Choi et al., 2001), previously described as characteristic, is absent from 6 ARP4 sequences from different organisms (plasmodia and yeast). Two other Family Insertions are of particular interest. The largest insertion in ARP5 (position 246) is rich in charged residues, and the ARP9 Family Insertion at position 333 contains a pattern rich in rare aromatic amino acids [P/S][D/E]YF[P/S][E/S]WK. Taken together, the specific residues, motifs, and Family Insertions constitute a knowledge filter that defines at least one discriminative feature for each ARP subfamily except for ARP10 and ARP11, which are defined only by sequence similarity.
ARPAnno Web Server
Our approach, based on ARP-MACS, combines three complementary strategies with local and global sequence information and a knowledge filter (see Materials and Methods). Based on this, we implemented a web server to annotate ARP sequences. The web server, called ARPAnno, is available at http://bips.u-strasbg.fr/ARPAnno and allows the user to submit a sequence in FASTA format. The analysis of actin and ARP conservation (Figure 1) shows that a query is identified as an actin if it has a GID >80% and a pCover >80% compared with any conventional actin sequence (see Materials and Methods). To estimate the accuracy and reliability of the ARPAnno annotations, we submitted each of the ∼700 previously identified actin and ARP proteins in ARPMACS for automatic classification. In this large-scale test, all proteins were assigned to the correct subfamilies. To evaluate the predictive strength of our server, we performed a second test involving the newly detected proteins from a later version of Uniprot (January 2005). The second set was composed of 68 sequences that were classified by the program with best SARPi ranging from 36.9 to 99.0 as follows: 36 conventional actins, 3 Orphans, 6 ARP1, 7 ARP2, 6 ARP3, 8 ARP4, 1 ARP9, and 1 ARP10 from diverse organisms such as Y. lipolytica, D. hansenii, Caenorhabditis briggsae, Paramecium tetraurelia, Xenopus tropicalis or Gallus gallus. For complete sequences, an SARPi > 55 was highly reliable to assign a subfamily. Further validation by visual inspection suggested that the only 2 sequences with SARPi < 55 corresponds to 2 proteins from P. tetraurelia, classified by ARPAnno as an actin and annotated as putative actin in Uniprot.
ATP Binding
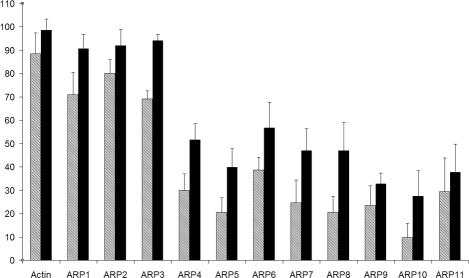
In addition to actin, ARP1, ARP2, and ARP3 bind an ATP molecule, for which the hydrolysis is proposed to induce a conformational change required for their biological function (Otterbein et al., 2001; Nolen et al., 2004; Martin et al., 2005). The mean conservation of 17 key reference residues involved in nucleotide binding was computed for each ARP subfamily and is illustrated in Figure 3. The same analysis was carried out using a slightly different set of nucleotide binding residues (Beltzner and Pollard, 2004) and gave similar results (our unpublished results). We observed two groups. The first group, composed of conventional actin and cytoplasmic ARPs (ARP1-ARP3), has >60% identical and >90% similar residues, whereas the second group (ARP4-ARP11) has <35% identical and <46% similar residues. Thus, this predicts that the first group is able to bind ATP (Kd ≤ μM), which has been shown to be the case (Belmont et al., 1999; Bingham and Schroer, 1999; Sablin et al., 2002). In contrast, this analysis of key binding residues predicts that, the second group, mainly composed of nuclear ARPs, might not bind ATP or might bind with significantly less affinity and/or through other residues.
Figure 3.
Conservation pattern of the 17 residues (D13, S16, G17, L18, K20, Q139, D156, D159, G160, V161, K215, E216, G304, T305, M307, Y308, and K338) known to participate in nucleotide binding to actin. For the 11 ARP subfamilies and actin, percent identity is represented as hatched bars and percent similarity as closed bars. Error bars, SDs.
From Sequence to Structure
To visualize the sequence to structure conservation of ARP subfamilies, we mapped the degree of amino acid conservation relative to the actin reference sequence onto the secondary structures using yeast actin as a model structure. The mean conservation for all ARP sequences in ARP-MACS is represented in Figure 4A, whereas the detail for individual ARP subfamilies is represented in Figure 4B. The secondary structures defining the actin fold are widely conserved in all ARP subfamilies as seen by the abundant green color corresponding to 25-35% Id. (Figure 4A). Some specific regions are highly conserved (> 45% Id. from orange to red) corresponding to secondary structures in three subdomains: in subdomain 1 a complete beta sheet composed of 4 strands (S1, S2, S3, and S6) and two alpha-helices (H18 and H20), in subdomain 3, two beta strands (S8 and S13), and one helix (H6), and in subdomain 4, the two helices (H11 and H12). All these secondary structures, with the exception of H20, are part of the previously defined actin fold (Kabsch and Holmes, 1995). The observed conservation points are localized in the bottom half of the actin fold and more precisely, in the hydrophobic cleft (Dominguez, 2004), a key region for actin dimerization and for interaction with ABPs.
The analyses of individual ARP subfamily conservation highlight specific patterns. As expected in view of the FamID, the main cytoplasmic ARP1-ARP3 share more conserved elements than nuclear ARPs. Surprisingly, ARP2 is less conserved in the helix H18 and H19 involved in the hydrophobic cleft than in either ARP1 or ARP3. Additional features can be observed in subdomains 2 and 4 for ARP1 and ARP2. We noticed that ARP1 and ARP2 reveal better sequence conservation in helix H9 and in strand S4 and S10 than ARP3. Within the nuclear ARPs, ARP4 unexpectedly maintains high conservation in the lower part of subdomain 1 (H18, H19, and H20). This observation underlines functional perspectives for ARP4 through its hydrophobic cleft. Finally, with regard to other secondary structures that are part of subdomain 1, S1 is highly conserved in ARP5, ARP6, and ARP11; S6 in ARP5; and S2 in ARP7 and ARP11.
Phylogenetic Distribution
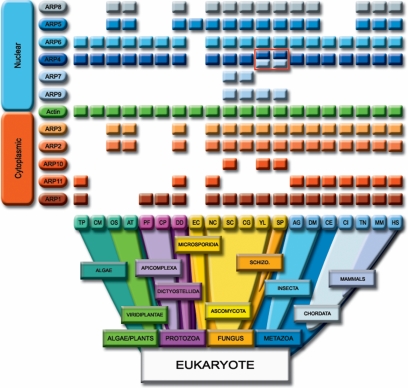
The growing number of completely sequenced genomes available allows us to define the edges of the distribution of eukaryotic ARPs by in depth analysis of the proteomes and genomes of 20 organisms ranging across eukaryotic phyla. As observed in many organisms (T. pseudonana, D. discoideum...; Supplementary Data 6), the genomic validation is essential to assess the presence of a given ARP, considering that a certain number of genes present have not been annotated as proteins. The phylogenetic distribution of ARP subfamilies and conventional actin is represented in Figure 5. According to defined ARP signatures, we detected 132 ARP proteins in 11 subfamilies from algae to mammals, and at least 1 actin and 1 ARP in each organism analyzed. It is noteworthy that the organisms with limited numbers of ARP (E. cuniculi, C. merolae) have no detectable cytoplasmic ARPs but include at least one nuclear ARP. In all other organisms, both nuclear and cytoplasmic ARPs are present. Remarkably, the examination of the presence/absence profiles led to the definition of pairs of copresent/coabsent ARPs such as ARP2 with ARP3, ARP4 with ARP6, ARP5 with ARP8, ARP7, with ARP9, and to lesser extent ARP1 with ARP10 or ARP11. Surprisingly, the most widely distributed ARPs in evolution, copresent in all organisms studied with the exception of the small obligate parasite E. cuniculi, are the nuclear ARPs, ARP4, and ARP6. This result was unexpected and leads to the conclusion that ARP4 and ARP6 represent the most universal ARPs conserved throughout the eukaryotic phyla. The second most widely distributed pair of proteins is ARP2 and ARP3, well studied components of the actin nucleation complex. They are copresent in plants, fungi, and Metazoa but are coabsent in algae and in Apicomplexa.
Figure 5.
Schematic representation of ARP distribution among the eukaryotic phyla. The columns represent different organisms with a completely sequenced genome: T. pseudonana (TP), C. merolae (CM), O. sativa (OS), A. thaliana (AT), P. falciparum (PF), C. parvum (CP), D. discoideum (DD), E. cuniculi (EC), N. crassa (NC), S. cerevisiae (SC), C. glabrata (CG), Y. lipolytica (YL), S. pombe (SP), A. gambiae (AG), D. melanogaster (DM), C. elegans (CE), C. intestinalis (CI), T. nigroviridis (TN), M. musculus (MM), and H. sapiens (HS). The existence of a colored rectangle indicates the presence of the protein in the organism considered. Conventional actin is represented as green rectangles in between the two groups of ARPs, the cytoplasmic ARPs (ARP1-ARP3, ARP10, and ARP11) as indicated by orange rectangles and the nuclear ARPs (ARP4-ARP9) indicated by blue rectangles. The four rectangles outlined in red highlight the presence of a second distinct ARP4, named ARP4* in Y. lipolytica (YL) and in S. pombe (SP).
ARP1, the closest ARP to conventional actin, is individually more widely distributed than ARP2 and ARP3. However, when one considers the functional complex dynactin where the ARP1 filament is capped by ARP11 (Eckley et al., 1999), the pattern of presence/absence appears more complex than other pairs. In fact, although ARP11 is not present without ARP1, it is not found in every organism bearing ARP1. It is interesting to notice that ARP10, restricted to fungi, only partially complements the ARP11 pattern. Furthermore, our extended exploration of fungi (see Materials and Methods) confirms the presence of ARP1 in 30 out of 31 organisms (except E. cuniculi) and restricts ARP10 to only 5 Ascomycota Saccharomycotina (D hansenii, E. gossypii, K lactis, S. cerevisiae, and Y. lipolytica) and 1 Ascomycota Schizosaccharomycetes (S. pombe). One ARP11 was found in Ascomycota Pezizomycotina (N. crassa).
The coabsence profile of ARP5 and ARP8 is puzzling since they are missing in a number of different phyla such as the algae, the Apicomplexa, and two Metazoan phyla, C. elegans and C. intestinalis. Our results also confirm that the functionally obligate heterodimeric partners, ARP7 and ARP9 (Szerlong et al., 2003), were restricted to fungi as previously suggested (Goodson and Hawse, 2002; Blessing et al., 2004). The presence of ARP7 and ARP9 has been assessed in the 31 fungi genomes available at NCBI and we could clearly restrict ARP7 and ARP9 to Ascomycota Saccharomycotina and Ascomycota Schizosaccharomycetes. Neither ARP7 nor ARP9 were found in the Ascomycota Pezizomycotina, Basidiomycota, or Microsporidia. Surprisingly, the copresence of ARP7 and ARP9 is not observed in two completely sequenced organisms of Ascomycota, Y. lipolytica, and S. pombe, where ARP9 is present but ARP7 is absent. In this context it is noteworthy that these two organisms are the only fungi that encode an additional and distinct ARP4 (red box in Figure 5, Uniprot accession numbers Q6C0A9 and Q09849; annotated here as ARP4*). This strongly suggests that ARP4* may complement the lack of ARP7 in these yeasts.
DISCUSSION
The alignment of all available actin and ARP sequences in Uniprot combined with a detailed comparative analysis of 20 completely sequenced eukaryotic genomes reinforces the existing ARP subfamilies and finds them present in more organisms. Our calculation of conservation of ARPs to actin led to a classification in strong agreement with previous studies (Poch and Winsor, 1997; Eckley et al., 1999; Harata et al., 2001; Goodson and Hawse, 2002). It has been proposed that yeast ARP10 and metazoan ARP11 subfamilies might form only one highly divergent ARP family, based on the presence of an ARP11 in an Ascomycota (N. crassa) dynactin complex (Lee et al., 2001). However, based on functional and genomic analyses (Eckley and Schroer, 2003; Borkovich et al., 2004) and our sequence analysis, there was no compelling evidence to force this grouping. The question remains open and a decisive argument would be to group them if yeast ARP10 is found in the dynactin complex or to separate them if both an ARP10 and an ARP11 are found in a single organism. No new, more distantly related ARP subfamilies were created but 27 orphans with relatively high identity to actin have been noted and aligned. However, because no characteristic features could be defined for these proteins, they have been left as orphans. In light of the status of ARP7 and ARP9 which are restricted to fungi, some orphans, such as the Actinlike 7A and 7B found in human (Q9Y615 and Q9Y614) and mouse (Q9QY84 and Q9QY83), could be considered as new ARP subfamilies dedicated to specific tissues or functions. It will be of interest to know whether they carry out new functions or overlap with those of the defined ARP subfamilies. In particular, one plant specific orphan from A. thaliana called AtARP7 (although it does not belong to the ARP7 subfamily!) is localized in the nucleus during interphase, suggesting a potential nuclear function (Kandasamy et al., 2003).
Here, we propose that ARPAnno can be used to assign new actinlike genes to an ARP subfamily before assigning a name. A protein sequence can be submitted either as an existing entry in a protein database or as a translated open reading frame (ORF) prediction. In this latter case caution should be used because some mispredicted ORFs (wrong insertions or deletions) will give incorrect scores. According to our analysis an SARPi > 55 is highly reliable for complete sequences. Ambiguous situations can often be clarified by direct comparison to the MACS of the closest subfamilies. Our strategy of combining local and global approaches, together with a knowledge-based filter is essential to the study of this family of proteins. The ARP-MACS will be updated and the discriminating criteria revised regularly. It would be now of interest to build an automatic procedure to mine the ARPs in new genomes and combine prediction, extraction, and annotation.
The ARP-MACS has allowed us to better characterize ARP subfamilies with specific residues, motifs, and/or INDELs found in all subfamilies except ARP10 and ARP11. Considering the high number of insertions, the strength and plasticity of the actin fold is remarkable, further illustrated by the peripheral positions of the hot spots of insertion (see Figure 4A). A restricted part of the fold is highlighted by the average structural conservation of all ARP subfamilies (Figure 4A). Furthermore, high conservation in the hydrophobic cleft (helices H18, H19, and H20), which forms an actin-binding interface (Dominguez, 2004), opens new perspectives for possible interactions between ARPs and ABPs. In line with this, ARP4 stands out as having high sequence conservation in H18 and H19. In fact, recent exploration across ARP subfamilies of 27 actin residues involved in gelsolin binding showed the best conservation in ARP4 and ARP1 (Archer et al., 2004). Analysis of a larger pool of ARPs from the ARP-MACS (148 vs. 63) confirmed this result (our unpublished results). Indeed, it has been reported that the gelsolinlike domain in the ABP, Fli-I, binds to ARP4 in the SWI/SNF complex, and contributes to transcriptional activation (Lee et al., 2004).
The most prominent property of actin is to self-assemble into a filamentous structure that implicates actin-actin interfaces distributed over the surfaces of the four subdomains. Presumed actin-actin interfaces in ARP1-ARP3 harbor only a few short insertions proposed to be compatible with their role in actin assembly (Robinson et al., 2001). However, ARP1 is the sole ARP able to form a homopolymer filament in vivo. In vitro, mammalian ARP1 has been shown to polymerize with actin (Eckley and Schroer, 2003) and D. dictyostelium ARP1 was found with ARP2 and an orphan ARP (Uniprot ID Q54HE9_DICDI) in a filament (Gomez-Garcia and Kornberg, 2004). In the dynactin complex, ARP11 is found at the pointed end of the ARP1 filament, but in certain organisms (P. falciparum, C. parvum, and most of the fungi), genomic inspection revealed no ARP11 (or ARP10), suggesting that ARP11 is not an obligate partner for ARP1. This is consistent with weak conservation in the ARP11 subfamily. Like the situation in green plants (Kandasamy et al., 2004), we found that the C. merolae genome lacks ARP1 and ARP11 (Figure 5). In keeping with this, we did not detect any sequences similar to any other subunits of the human dynactin complex.
The specificity of the ARP2 and ARP3 function relies on the fact that neither is able to homopolymerize. Indeed, these ARPs heterodimerize to bind the first actin monomer of a new filament. ARP2 and ARP3 compared to actin high-light secondary structures that are differentially conserved; ARP2 is more conserved at the pointed end in subdomains 2 and 4 (H9, S4, and S10), whereas ARP3 is better conserved at the barbed end in subdomain 1 (H18 and H19; Figure 4B). This differential conservation is in agreement with their inability to self-polymerize and with their role in the ARP2/3 complex. According to the recent analysis of the ARP2/3 complex (Beltzner and Pollard, 2004), ARP2 interacts through its pointed end with ARP3, and ARP3 is in contact through its barbed end with the first actin monomer of the nascent filament. Our comparative analyses show that ARP2 and ARP3 are absent from the genomes of Apicomplexa, Algae and from Macrosporidia. Thus, it is tempting to speculate that other nucleators might replace these ARPs. For example, formins nucleate ARP2/3-independent actin polymerization (Pruyne et al., 2002; Sagot et al., 2002; Moseley et al., 2004). Inspection of the five genomes lacking ARP2 and ARP3 revealed one or more genes coding for forminlike proteins (Higgs and Peterson, 2005 and our unpublished results).
Nuclear ARP4-ARP9 have been isolated in many complexes involved in chromatin modulating functions and localized predominantly in the nucleus (reviewed in Olave et al., 2002; Blessing et al., 2004). The nuclear ARPs show less intrasubfamily conservation (FamID) than actin and major cytoplasmic ARPs. Additionally, ARP-MACS revealed that nuclear ARPs have many insertions, which when conserved characterize the subfamily (Family Insertion). For example, the sole ARP5 large Family Insertion contains several bipartite NLS sequences and shows an overall negative charge. Thus, it might interact with positively charged molecules such as histones. For ARP4, inspection of the ARP-MACS shows that the Family Insertion at position 203 (Insertion I in yeast; Harata et al., 1999) also contains putative NLS sequences (Weber et al., 1995). As an example of a function in a limited number of organisms, S. cerevisiae ARP4 contains a nonconserved insertion (known as Insertion II, position 271) shown to bind core histones (Harata et al., 1999). We found this insertion in only four organisms, S. cerevisiae, S. pombe, P. falciparum, and P. yoelii. The absence of this functionally characterized insertion in the majority of organisms opens the question of whether other ARP4 proteins bind histones. Recent studies in A. thaliana found no evidence of AtARP4 binding putative histone H2B (Kandasamy et al., 2003). Thus, characterization of insertions might serve as a guide for future in vivo studies to determine whether conserved insertions bear subfamily functions.
The decreasing sequence conservation in ARPs relative to actin raises the important question of nucleotide binding capacity. Indeed, for actin and ARP1-ARP3 the binding and hydrolysis of ATP (Bingham and Schroer, 1999; Dayel et al., 2001; Le Clainche et al., 2001) is important for their functions. Although the measured binding affinities differ between different reports, the fact that ARP2 has greater ATP binding affinity than ARP3 is consistent with their percent identity difference of nucleotide binding residues (Figure 3). According to the average conservation of nucleotide contacts, our analysis predicts that nuclear ARPs, if able to bind ATP, would do so with much less affinity than cytoplasmic ARPs. Indeed, a recent report suggests that yeast ARP4 binds weakly to ATP because 5 mM ATP precluded the binding of a competitive inhibitor. Mutation in the ATP-binding pocket appeared to increase ARP4 occupancy in complexes, whereas excess ATP released it from the wild-type NuA4 complex, suggesting a role in modulating complex dynamics (Sunada et al., 2005). To date, no direct ATP binding data have been reported for nuclear ARPs other than ARP4.
In apparent contrast, some nuclear ARPs may not bind ATP for their most important functions because they remain functional when mutated in nucleotide contact residues. Mutations in D13 and/or S16 in ARP5 and ARP8 did not change either inositol growth rate or transcriptional activations (Shen et al., 2003). Similarly, in yeast ARP7 and ARP9, mutations (D13, S16, Q139, D156, D159, G304) did not alter the activity of the RSC complex (Cairns et al., 1998; Szerlong et al., 2003). These results correspond to our predictions. Given that yeast ARP5, ARP7, and ARP9 are essential in certain strains, the lack of ATPase phenotypes implies roles other than direct implication in remodeling. Notably, our results predict that the best conservation of nucleotide contact residues should be in ARP6. Other open questions are whether a restricted number of conserved residues might be sufficient for weak binding and whether other residues present in the nuclear ARPs could contribute to an ATP-binding site. These predictions await validation with biological tests on individual proteins.
Presence/absence patterns of proteins over a wide range of phyla are a potent tool to predict the partners involved in complexes. ARP subfamilies represent a textbook case where identical patterns correspond to their copresence in functional complexes: ARP2 and ARP3 in ARP2/3 complex, ARP4 and ARP6 in SWR1 complex, and ARP5 and ARP8 in INO80 complex (Supplementary Data 1). Thus, ARP distribution should predict the presence of the corresponding complexes in different organisms. Effectively, the lack of ARP5 and ARP8 is correlated with the lack of the catalytic subunit INO80 in the corresponding genomes (Bakshi et al., 2004 and our unpublished results). Although their absence from the lower phyla is understandable during evolution, the mystery of their disappearance from certain Metazoa remains. As observed recently (Goodson and Hawse, 2002), ARP7 and ARP9 are only present in fungi. We further restricted them to Saccharomycotina and Schizosaccharomyceta subphyla. Surprisingly, the absence of ARP7 and the unearthing of a second ARP4 (ARP4*) in Y. lipolytica and S. pombe make ARP4* a good candidate for functional replacement of ARP7. In light of this hypothesis, it is noteworthy that the SWI/SNF complexes of vertebrates and Drosophila contain actin and ARP4 (Papoulas et al., 1998; Zhao et al., 1998), whereas the SWI/SNF and RSC complexes of S. cerevisiae contain ARP7 and an ARP9 (Cairns et al., 1998; Peterson et al., 1998). Do the complexes from Y. lipolytica and S. pombe contain the newly described ARP4*?
Perhaps the most unexpected result of our analyses is the revelation of the omnipresence of the nuclear ARPs, in particular ARP4 and ARP6. ARP4 stands out as the family cornerstone, because it is present in all phyla, except the parasite E. cuniculi, and in almost all complexes that contain nuclear ARPs, both ATP-dependent chromatin remodeling complexes, and in HAT complexes (Supplementary Data 1). As such, ARP4 is implicated in multiple functions such as chromatin remodeling (reviewed in (Olave et al., 2002), transcriptional activation (Harata et al., 2002; Percipalle et al., 2003), DNA double-stranded break repair (van Attikum et al., 2004), apoptosis (Ikura et al., 2000), tumor suppression (Medjkane et al., 2004), histone acetylation (Galarneau et al., 2000), histone chaperone activity (Shen et al., 2003), kinetochore-spindle attachment, and gene silencing at centromeres (Minoda et al., 2005). Because ARP4 is a primary subunit with actin, with ARP6 or with ARP5 and ARP8 in different complexes, ARP4 may be an ancestor of nuclear ARPs. In support of this hypothesis, our results suggest that ARP4* may replace ARP7 in Y. lipolytica and S. pombe. When other ARP-containing complexes are isolated, it would not be surprising to find an ARP4.
In conclusion, comparative genomics revealing the copresence or coabsence of ARP subfamilies among eukaryotic phyla largely confirms the biological data that ARPs are associated in multiprotein complexes. A major and unexpected finding of our study is that the major ARPs and the minimum package for eukaryotic organisms are the nuclear ARPs, ARP4 and ARP6.
Supplementary Material
Acknowledgments
We thank Raymond Ripp and Luc Moulinier for stimulating discussions, Julie Thompson and Frederic Chalmel for their useful programs, Arnaud Muller for the mirrored web site, and Agatha Schulter for the representation of Figure 5. We are also grateful to two very careful reviewers and the monitoring editor whose comments significantly helped remodelling of our manuscript. This work was supported by the National Research Fund, the “Fondation Luxembourgeoise Contre le Cancer,” the Fondation “Aide aux Enfants Atteints d'un Cancer,” Luxembourg, the Centre National de Recherche Scientifique and the Direction des Relations Internationales of the CNRS. This work was achieved using the RNG (Réseau National de Génopoles) Strasbourg Bioinformatics Platform infrastructures and competencies. J. Muller is supported by a fellowship from the MCESR, Luxembourg. Y.O. thanks the Japan Society for the Promotion of Science (JSPS) for a Young Scientist Fellowship.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-06-0508) on September 29, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abrahamsen, M. S. et al. (2004). Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441-445. [DOI] [PubMed] [Google Scholar]
- Adams, M. D. et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos, L. A., van den Ent, F., and Lowe, J. (2004). Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr. Opin. Cell Biol. 16, 24-31. [DOI] [PubMed] [Google Scholar]
- Arabidospis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- Archer, S. K., Behm, C. A., Claudianos, C., and Campbell, H. D. (2004). The flightless I protein and the gelsolin family in nuclear hormone receptor-mediated signalling. Biochem. Soc. Trans. 32, 940-942. [DOI] [PubMed] [Google Scholar]
- Armbrust, E. V. et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79-86. [DOI] [PubMed] [Google Scholar]
- Bairoch, A. et al. (2005). The Universal Protein Resource (UniProt). Nucleic Acids Res. 33 Database Issue, D154-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi, R., Prakash, T., Dash, D., and Brahmachari, V. (2004). In silico characterization of the INO80 subfamily of SWI2/SNF2 chromatin remodeling proteins. Biochem. Biophys. Res. Commun. 320, 197-204. [DOI] [PubMed] [Google Scholar]
- Belmont, L. D., Orlova, A., Drubin, D. G., and Egelman, E. H. (1999). A change in actin conformation associated with filament instability after Pi release. Proc. Natl. Acad. Sci. USA 96, 29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltzner, C. C., and Pollard, T. D. (2004). Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J. Mol. Biol. 336, 551-565. [DOI] [PubMed] [Google Scholar]
- Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Wheeler, D. L. (2005). GenBank. Nucleic Acids Res. 33 Database Issue, D34-D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham, J. B., and Schroer, T. A. (1999). Self-regulated polymerization of the actin-related protein Arp1. Curr. Biol. 9, 223-226. [DOI] [PubMed] [Google Scholar]
- Blessing, C. A., Ugrinova, G. T., and Goodson, H. V. (2004). Actin and ARPs: action in the nucleus. Trends Cell Biol. 14, 435-442. [DOI] [PubMed] [Google Scholar]
- Bork, P., Sander, C., and Valencia, A. (1992). An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89, 7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich, K. A. et al. (2004). Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68, 1-108, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, B. R., Erdjument-Bromage, H., Tempst, P., Winston, F., and Kornberg, R. D. (1998). Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 2, 639-651. [DOI] [PubMed] [Google Scholar]
- Chervitz, S. A. et al. (1998). Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science 282, 2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, E. Y., Park, J. A., Sung, Y. H., and Kwon, H. (2001). Generation of the dominant-negative mutant of hArpNbeta: a component of human SWI/SNF chromatin remodeling complex. Exp. Cell Res. 271, 180-188. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. The C. elegans Sequencing Consortium. Science 282, 2012-2018. [DOI] [PubMed] [Google Scholar]
- Dayel, M. J., Holleran, E. A., and Mullins, R. D. (2001). Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc. Natl. Acad Sci. USA 98, 14871-14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal, P. et al. (2002). The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157-2167. [DOI] [PubMed] [Google Scholar]
- Delano, W. L. (2002). The PyMOL User's Manual.
- Dominguez, R. (2004). Actin-binding proteins—a unifying hypothesis. Trends Biochem. Sci. 29, 572-578. [DOI] [PubMed] [Google Scholar]
- Doyon, Y., Selleck, W., Lane, W. S., Tan, S., and Cote, J. (2004). Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell Biol. 24, 1884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B. et al. (2004). Genome evolution in yeasts. Nature 430, 35-44. [DOI] [PubMed] [Google Scholar]
- Eckley, D. M., Gill, S. R., Melkonian, K. A., Bingham, J. B., Goodson, H. V., Heuser, J. E., and Schroer, T. A. (1999). Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J. Cell Biol. 147, 307-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley, D. M., and Schroer, T. A. (2003). Interactions between the evolutionarily conserved, actin-related protein, Arp11, actin, and Arp1. Mol. Biol. Cell 14, 2645-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger, L. et al. (2005). The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold, T., Ulyanov, A., and Argos, P. (1996). SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 266, 114-128. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E. et al. (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859-868. [DOI] [PubMed] [Google Scholar]
- Galarneau, L., Nourani, A., Boudreault, A. A., Zhang, Y., Heliot, L., Allard, S., Savard, J., Lane, W. S., Stillman, D. J., and Cote, J. (2000). Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Cell 5, 927-937. [DOI] [PubMed] [Google Scholar]
- Gardner, M. J. et al. (2002). Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GCG. (2001). Wisconsin Package Version 10.2, Genetics Computer Group (GCG), Madison, WI.
- Goff, S. A. et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92-100. [DOI] [PubMed] [Google Scholar]
- Goffeau, A. et al. (1996). Life with 6000 genes. Science 274, 546, 563-567. [DOI] [PubMed] [Google Scholar]
- Gomez-Garcia, M. R., and Kornberg, A. (2004). Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc. Natl. Acad. Sci. USA 101, 15876-15880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson, H. V., and Hawse, W. F. (2002). Molecular evolution of the actin family. J. Cell Sci. 115, 2619-2622. [DOI] [PubMed] [Google Scholar]
- Gorzer, I., Schuller, C., Heidenreich, E., Krupanska, L., Kuchler, K., and Wintersberger, U. (2003). The nuclear actin-related protein Act3p/Arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Mol. Microbiol. 50, 1155-1171. [DOI] [PubMed] [Google Scholar]
- Grava, S., Dumoulin, P., Madania, A., Tarassov, I., and Winsor, B. (2000). Functional analysis of six genes from chromosomes XIV and XV of Saccharomyces cerevisiae reveals YOR145c as an essential gene and YNL059c/ARP5 as a strain-dependent essential gene encoding nuclear proteins. Yeast 16, 1025-1033. [DOI] [PubMed] [Google Scholar]
- Harata, M., Nishimori, K., and Hatta, S. (2001). Identification of two cDNAs for human actin-related proteins (Arps) that have remarkable similarity to conventional actin. Biochim. Biophys. Acta 1522, 130-133. [DOI] [PubMed] [Google Scholar]
- Harata, M., Oma, Y., Mizuno, S., Jiang, Y. W., Stillman, D. J., and Wintersberger, U. (1999). The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol. Biol. Cell 10, 2595-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harata, M., Oma, Y., Tabuchi, T., Zhang, Y., Stillman, D. J., and Mizuno, S. (2000). Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J. Biochem. (Tokyo) 128, 665-671. [DOI] [PubMed] [Google Scholar]
- Harata, M., Zhang, Y., Stillman, D. J., Matsui, D., Oma, Y., Nishimori, K., and Mochizuki, R. (2002). Correlation between chromatin association and transcriptional regulation for the Act3p/Arp4 nuclear actin-related protein of Saccharomyces cerevisiae. Nucleic Acids Res. 30, 1743-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, H. N., and Peterson, K. J. (2005). Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 16, 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, K. C., Sander, C., and Valencia, A. (1993). A new ATP-binding fold in actin, hexokinase and Hsc70. Trends Cell Biol. 3, 53-59. [DOI] [PubMed] [Google Scholar]
- Holt, R. A. et al. (2002). The genome sequence of the malaria mosquito Anopheles gambiae. Science 298, 129-149. [DOI] [PubMed] [Google Scholar]
- Ikura, T., Ogryzko, V. V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J., and Nakatani, Y. (2000). Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463-473. [DOI] [PubMed] [Google Scholar]
- Jaillon, O. et al. (2004). Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946-957. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., and Holmes, K. C. (1995). The actin fold. FASEB J. 9, 167-174. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., Mannherz, H. G., Suck, D., Pai, E. F., and Holmes, K. C. (1990). Atomic structure of the actin:DNase I complex. Nature 347, 37-44. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M. K., Deal, R. B., McKinney, E. C., and Meagher, R. B. (2004). Plant actin-related proteins. Trends Plant Sci. 9, 196-202. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M. K., McKinney, E. C., and Meagher, R. B. (2003). Cell cycle-dependent association of Arabidopsis actin-related proteins AtARP4 and AtARP7 with the nucleus. Plant J. 33, 939-948. [DOI] [PubMed] [Google Scholar]
- Katinka, M. D. et al. (2001). Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450-453 [DOI] [PubMed] [Google Scholar]
- Kreppel, L., Fey, P., Gaudet, P., Just, E., Kibbe, W. A., Chisholm, R. L., and Kimmel, A. R. (2004). dictyBase: a new Dictyostelium discoideum genome database. Nucleic Acids Res. 32 Database issue, D332-D333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S. et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- Le Clainche, C., Didry, D., Carlier, M. F., and Pantaloni, D. (2001). Activation of Arp2/3 complex by Wiskott-Aldrich Syndrome protein is linked to enhanced binding of ATP to Arp2. J. Biol. Chem. 276, 46689-46692. [DOI] [PubMed] [Google Scholar]
- Lecompte, O., Thompson, J. D., Plewniak, F., Thierry, J., and Poch, O. (2001). Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene 270, 17-30. [DOI] [PubMed] [Google Scholar]
- Lee, I. H., Kumar, S., and Plamann, M. (2001). Null mutants of the neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol. Biol. Cell 12, 2195-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. H., Campbell, H. D., and Stallcup, M. R. (2004). Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol. Cell. Biol. 24, 2103-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller, J. P., Helfman, D. M., and Schroer, T. A. (1992a). A vertebrate actin-related protein is a component of a multisubunit complex involved in microtubule-based vesicle motility. Nature 359, 244-246. [DOI] [PubMed] [Google Scholar]
- Lees-Miller, J. P., Henry, G., and Helfman, D. M. (1992b). Identification of act2, an essential gene in the fission yeast Schizosaccharomyces pombe that encodes a protein related to actin. Proc. Natl. Acad. Sci. USA 89, 80-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L. M., and May, R. C. (2001). Arps: actin-related proteins. Results Probl. Cell Differ. 32, 213-229. [DOI] [PubMed] [Google Scholar]
- Martin, A. C., Xu, X. P., Rouiller, I., Kaksonen, M., Sun, Y., Belmont, L., Volkmann, N., Hanein, D., Welch, M., and Drubin, D. G. (2005). Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J. Cell Biol. 168, 315-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, M. et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653-657. [DOI] [PubMed] [Google Scholar]
- McKinney, E. C., Kandasamy, M. K., and Meagher, R. B. (2002). Arabidopsis contains ancient classes of differentially expressed actin-related protein genes. Plant Physiol. 128, 997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medjkane, S., Novikov, E., Versteege, I., and Delattre, O. (2004). The tumor suppressor hSNF5/INI1 modulates cell growth and actin cytoskeleton organization. Cancer Res. 64, 3406-3413. [DOI] [PubMed] [Google Scholar]
- Minoda, A., Saitoh, S., Takahashi, K., and Toda, T. (2005). BAF53/Arp4 homolog Alp5 in fission yeast is required for histone H4 acetylation, kinetochore-spindle attachment, and gene silencing at centromere. Mol. Biol. Cell 16, 316-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann, L., Langenberg, K., Krijgsveld, J., Kal, A. J., Heck, A. J., and Verrijzer, C. P. (2004). Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24, 3077-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, J. B., Sagot, I., Manning, A. L., Xu, Y., Eck, M. J., Pellman, D., and Goode, B. L. (2004). A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 15, 896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen, B. J., Littlefield, R. S., and Pollard, T. D. (2004). Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc. Natl. Acad. Sci. USA 101, 15627-15632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave, I. A., Reck-Peterson, S. L., and Crabtree, G. R. (2002). Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71, 755-781. [DOI] [PubMed] [Google Scholar]
- Otterbein, L. R., Graceffa, P., and Dominguez, R. (2001). The crystal structure of uncomplexed actin in the ADP state. Science 293, 708-711. [DOI] [PubMed] [Google Scholar]
- Papoulas, O., Beek, S. J., Moseley, S. L., McCallum, C. M., Sarte, M., Shearn, A., and Tamkun, J. W. (1998). The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125, 3955-3966. [DOI] [PubMed] [Google Scholar]
- Percipalle, P., Fomproix, N., Kylberg, K., Miralles, F., Bjorkroth, B., Daneholt, B., and Visa, N. (2003). An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA 100, 6475-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C. L., Zhao, Y., and Chait, B. T. (1998). Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J. Biol. Chem. 273, 23641-23644. [DOI] [PubMed] [Google Scholar]
- Plewniak, F. et al. (2003). PipeAlign: A new toolkit for protein family analysis. Nucleic Acids Res. 31, 3829-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch, O., and Winsor, B. (1997). Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast 13, 1053-1058. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., Blanchoin, L., and Mullins, R. D. (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct 29, 545-576. [DOI] [PubMed] [Google Scholar]
- Pruitt, K. D., Tatusova, T., and Maglott, D. R. (2005). NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33 Database Issue, D501-D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A., and Boone, C. (2002). Role of formins in actin assembly: nucleation and barbedend association. Science 297, 612-615. [DOI] [PubMed] [Google Scholar]
- Puiu, D., Enomoto, S., Buck, G. A., Abrahamsen, M. S., and Kissinger, J. C. (2004). CryptoDB: the Cryptosporidium genome resource. Nucleic Acids Res. 32 Database issue, D329-D331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, R. C., Turbedsky, K., Kaiser, D. A., Marchand, J. B., Higgs, H. N., Choe, S., and Pollard, T. D. (2001). Crystal structure of Arp2/3 complex. Science 294, 1679-1684. [DOI] [PubMed] [Google Scholar]
- Rodal, A. A., Sokolova, O., Robins, D. B., Daugherty, K. M., Hippenmeyer, S., Riezman, H., Grigorieff, N., and Goode, B. L. (2005). Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol. 12, 26-31. [DOI] [PubMed] [Google Scholar]
- Sablin, E. P., Dawson, J. F., VanLoock, M. S., Spudich, J. A., Egelman, E. H., and Fletterick, R. J. (2002). How does ATP hydrolysis control actin's associations? Proc. Natl. Acad. Sci. USA 99, 10945-10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot, I., Rodal, A. A., Moseley, J., Goode, B. L., and Pellman, D. (2002). An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4, 626-631. [DOI] [PubMed] [Google Scholar]
- Schafer, D. A., and Schroer, T. A. (1999). Actin-related proteins. Annu. Rev. Cell Dev. Biol. 15, 341-363. [DOI] [PubMed] [Google Scholar]
- Schroer, T. A., Fyrberg, E., Cooper, J. A., Waterston, R. H., Helfman, D., Pollard, T. D., and Meyer, D. I. (1994). Actin-related protein nomenclature and classification. J. Cell Biol. 127, 1777-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob, E., and Martin, R. P. (1992). New yeast actin-like gene required late in the cell cycle. Nature 355, 179-182. [DOI] [PubMed] [Google Scholar]
- Shen, X., Ranallo, R., Choi, E., and Wu, C. (2003). Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12, 147-155. [DOI] [PubMed] [Google Scholar]
- Sunada, R., Görzer, I., Oma, Y., Yoshida, T., Suka, N., Wintersberger, U., and Harata, M. (2005). The nuclear actin-related protein Act3p/Arp4p is involved in the dynamics of chromatin-modulating complexes. Yeast 22, 753-768. [DOI] [PubMed] [Google Scholar]
- Szerlong, H., Saha, A., and Cairns, B. R. (2003). The nuclear actin-related proteins Arp7 and Arp9, a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 22, 3175-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Plewniak, F., Ripp, R., Thierry, J. C., and Poch, O. (2001). Towards a reliable objective function for multiple sequence alignments. J. Mol. Biol. 314, 937-951. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Prigent, V., and Poch, O. (2004). LEON: multiple alignment evaluation of neighbours. Nucleic Acids Res. 32, 1298-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Thierry, J. C., and Poch, O. (2003). RASCAL: rapid scanning and correction of multiple sequence alignments. Bioinformatics 19, 1155-1161. [DOI] [PubMed] [Google Scholar]
- van Attikum, H., Fritsch, O., Hohn, B., and Gasser, S. M. (2004). Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777-788. [DOI] [PubMed] [Google Scholar]
- Venter, J. C. et al. (2001). The sequence of the human genome. Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- Volkmann, N., Amann, K. J., Stoilova-McPhie, S., Egile, C., Winter, D. C., Hazelwood, L., Heuser, J. E., Li, R., Pollard, T. D., and Hanein, D. (2001). Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science 293, 2456-2459. [DOI] [PubMed] [Google Scholar]
- Vorobiev, S., Strokopytov, B., Drubin, D. G., Frieden, C., Ono, S., Condeelis, J., Rubenstein, P. A., and Almo, S. C. (2003). The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. USA 100, 5760-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R. H. et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- Weber, V., Harata, M., Hauser, H., and Wintersberger, U. (1995). The actin-related protein Act3p of Saccharomyces cerevisiae is located in the nucleus. Mol. Biol. Cell 6, 1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, N., Dembele, D., Raffelsberger, W., and Poch, O. (2002). Density of points clustering, application to transcriptomic data analysis. Nucleic Acids Res. 30, 3992-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, V. et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415, 871-880. [DOI] [PubMed] [Google Scholar]
- Zhao, K., Wang, W., Rando, O. J., Xue, Y., Swiderek, K., Kuo, A., and Crabtree, G. R. (1998). Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625-636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.