Abstract
Autophagy is a catabolic multitask transport route that takes place in all eukaryotic cells. During starvation, cytoplasmic components are randomly sequestered into huge double-membrane vesicles called autophagosomes and delivered into the lysosome/vacuole where they are destroyed. Cells are able to modulate autophagy in response to their needs, and under certain circumstances, cargoes such as aberrant protein aggregates, organelles and bacteria can be selectively and exclusively incorporated into autophagosomes. In the yeast Saccharomyces cerevisiae, for example, double-membrane vesicles are used to transport the Ape1 protease into the vacuole, or for the elimination of superfluous peroxisomes. In the present study we reveal that in this organism, actin plays a role in these two types of selective autophagy but not in the nonselective, bulk process. In particular, we show that precursor Ape1 is not correctly recruited to the PAS, the putative site of double-membrane vesicle biogenesis, and superfluous peroxisomes are not degraded in a conditional actin mutant. These phenomena correlate with a defect in Atg9 trafficking from the mitochondria to the PAS.
INTRODUCTION
Eukaryotic cells are often confronted with dramatic intracellular changes arising from developmental remodeling, pathological anomalies, and pathogen invasion. Autophagy provides one effective way to adapt and cope with these various extreme situations by rapidly delivering large fractions of the cytosol, aberrant protein aggregates, organelles and pathogens into the lysosome/vacuole interior where they are destroyed by resident hydrolases (Kirkegaard et al., 2004; Levine and Klionsky, 2004; Nakagawa et al., 2004; Shintani and Klionsky, 2004a; Rubinsztein et al., 2005). The hallmark of this catabolic pathway is the sequestration of cargoes by large cytosolic double-membrane vesicles called autophagosomes that successively dock and fuse with the lysosome/vacuole releasing the inner vesicles into the lumen of this organelle (Reggiori and Klionsky, 2002, 2005). Despite a series of recent studies that have revealed the versatility of this transport route, it remains largely unknown how autophagy is induced in these various situations and how the different cargoes are specifically recognized. Currently, autophagy is divided into selective and nonselective types (Reggiori and Klionsky, 2005). This process is considered selective when a precise cargo is specifically and exclusively incorporated into autophagosomes. In contrast, cytoplasmic structures are randomly enwrapped into double-membrane vesicles during nonselective autophagy.
The yeast Saccharomyces cerevisiae provides an excellent example to illustrate how cells adapt autophagy to their needs by changing its specificity. Yeast use this pathway to generate an internal pool of nutrients during starvation and to participate in the cellular reorganization induced by sporulation (Tsukada and Ohsumi, 1993; Schlumpberger et al., 1997). In addition, this organism possesses at least two other transport routes that are morphologically and mechanistically similar to autophagy but are used for different purposes. First, the precursor form of the oligomeric protease aminopeptidase I (prApe1) is unconventionally delivered from the cytoplasm directly to the vacuole lumen through a process termed the cytoplasm to vacuole targeting (Cvt) pathway. The Cvt pathway is an example of specific autophagy that is biosynthetic, and prApe1 is packaged into double-membrane vesicles, called Cvt vesicles, that are substantially smaller than autophagosomes (Baba et al., 1997; Scott et al., 1997). As in all eukaryotic organisms, starvation conditions trigger autophagy, which is characterized by the expansion of these structures and a concomitant change in their specificity that allows, in addition to prApe1, the sequestration of large portions of the cytoplasm including organelles such as mitochondria (Baba et al., 1997; Scott et al., 1997). Second, when yeast cells are grown in the presence of oleic acid as a sole carbon source, peroxisomes are proliferated because they contain the enzymes required to metabolize this substrate. Once the cells are shifted to a different carbon source such as glucose, the superfluous peroxisomes are selectively degraded by a specific mechanism, termed pexophagy, which also uses double-membrane vesicles (Hutchins et al., 1999).
Genetic screens designed for the selection of yeast mutant strains blocked in one of these pathways have been the most successful approaches in the discovery of proteins mediating autophagy (summarized in Klionsky et al., 2003). Approximately 27 proteins named Atg that are shared among these various transport routes have been characterized and most of them appear to be primarily restricted to the pre-autophagosomal structure (PAS), the potential site of organization for Cvt vesicle/autophagosome formation (Suzuki et al., 2001; Kim et al., 2002; Klionsky et al., 2003). Many of the Atg components have been shown to have orthologues among all eukaryotic organisms (Reggiori and Klionsky, 2002; Levine and Klionsky, 2004).
Despite the identification of the Atg proteins and the uncovering of several interactions among these factors, it remains unclear how autophagosomes are created. Analyses of autophagy in mammalian cells have provided substantial pharmacological information concerning the effects of a range of inhibitors. These data have provided insight into the nature of the autophagic mechanism. For example, treatment of normal rat kidney epithelial cells with microfilament depolymerizing agents such as cytochalasins blocks formation of the autophagosome (Aplin et al., 1992). However, it still remains completely unclear if actin plays a direct role in the biogenesis of these large vesicles. In contrast, microtubule inhibitors interfere with fusion of the autophagosome with the lysosome in both rat hepatocytes and normal rat kidney epithelial cells (Aplin et al., 1992; Blankson et al., 1995; Fengsrud et al., 1995; Seglen et al., 1996). At present no studies have indicated a role for the yeast cytoskeleton in autophagy. Rather, the only published data suggest that microtubules do not play a role in the process (Kirisako et al., 1999). In the present study we show that the actin cytoskeleton is required for the Cvt pathway and pexophagy, two types of yeast-selective autophagy, but is dispensable for the nonselective process.
MATERIALS AND METHODS
Strains, Media, and Plasmids
The S. cerevisiae strains used in this study are listed in Table 1. For gene disruptions, the entire ATG1 coding region was replaced with the Escherichia coli kanr gene using PCR primers containing ∼40 bases of identity to the regions flanking the open reading frame. PCR-based integration of YFP, GFP, and RFP at the 3′ end of ATG9, PEX14, and ABP140 was used to generate strains expressing fusion proteins under the control of their native promoters. The templates for integration were pDH3, pFA6a-GFP(S65T)-His3MX6, pFA6a-mRFP-His3MX6, and pFA6a-mRFP-KanMX6 (Longtine et al., 1998; Drees et al., 2001). The pFA6a-RFP-His3MX6 and pFA6a-RFP-KanMX6 vectors were created by replacing GFP with mRFP (PacI-AscI digestions) in the pFA6a-GFP(S65T)-His3MX6 and pFA6a-RFP-KanMX6 plasmids, respectively (Longtine et al., 1998; Campbell et al., 2002). PCR verification and prApe1 processing were used to confirm the functionality of all genomic fusions. The RFP-Vam3 (pRFPVAM3416) fluorescent chimera was generated by excising GFP with HindIII and EcoRI from the pVAM3416 vector and replacing it with RFP (Reggiori et al., 2000).
Table 1.
Strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| AHY001 | SEY6210 atg11Δ::HIS3 | Kim et al. (2001b) |
| atg1Δ | BY4742 atg1Δ::KAN | ResGen |
| BY4742 | MATα ura3Δ leu2Δ his3Δ lys2Δ | ResGen |
| D3Y103 | TN124 atg13Δ::URA3 | Scott et al. (2000) |
| DDY336 | MATa ura3-52 leu2-3,112 his3-Δ200 can1-1 tub2–201 cry1 act1–133::HIS3 | Drubin et al. (1993) |
| DDY1493 | MATα ura3-52 leu2-3,112 his3-Δ200 tub2-201 act1-159::HIS3 | Drubin et al. (1993) |
| FRY138 | SEY6210 ATG9-YFP::HIS3 S.k. atg1Δ::URA3 | Reggiori et al. (2004a) |
| FRY162 | SEY6210 ATG9-GFP::HIS3 S.k. | Reggiori et al. (2005) |
| FRY239 | SEY6210 ATG9-YFP::KAN atg1Δ::HIS3 | This study |
| FRY245 | SEY6210 ATG9-RFP::HIS3 S.k. | This study |
| IRA001 | BY4742 PEX14-GFP::HIS3 S.k. | This study |
| IRA002 | BY4742 PEX14-GFP::HIS3 S.k. atg1Δ::KAN | This study |
| IRA003 | DDY1493 pep4Δ::LEU2 | This study |
| IRA004 | DDY1493 ATG9-RFP::KAN | This study |
| IRA005 | DDY1493 atg1Δ::URA3 ATG9-RFP::KAN | This study |
| IRA006 | SEY6210 ATG9-RFP::HIS3 S.k. atg1Δ::URA3 | This study |
| IRA010 | LWY1412 ABP140-RFP::KAN | This study |
| JKY007 | SEY6210 atg9Δ::HIS3 | Noda et al. (1995) |
| LWY1412 | MATa ura3-52 leu2-3,112 | Drubin et al. (1993) |
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel GAL | Robinson et al. (1988) |
| TN124 | MATa leu2-3,112 ura3-52 trp1 pho8::pho8Δ60 pho13Δ::LEU2 | Noda et al. (1995) |
| WPHYD2 | SEY6210 atg4Δ::LEU2 | Kim et al. (2001a) |
Yeast cells were grown in rich (YPD; 1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal media (SMD; 0.67% yeast nitrogen base, 2% glucose, amino acids, and vitamins as needed). Starvation experiments were conducted in synthetic media lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids, 2% glucose).
Plasmids expressing GFP-Atg8 (pRS316GFP-Aut7), YFP-Atg8 (pRS414EYFP-Aut7), CFP-Ape1 (pTS470), GFP-Ape1 (pTS466), pCu416GFPSKL, and Atg1ts have been described elsewhere (Guan et al., 2001; Suzuki et al., 2001; Shintani et al., 2002).
Fluorescence Microscopy
Yeast cells were grown or starved in the appropriate medium before imaging. For S. cerevisiae mitochondrial fluorescent labeling, 1 ml of growing culture was incubated for 30 min in the presence of 1 μM Mitofluor Red 589 (Molecular Probes, Eugene, OR). Cells were then washed with the same culture medium before imaging in order to remove the excess dye (Reggiori et al., 2005).
To stain the actin cytoskeleton with Texas Red-X phalloidin (TR phalloidin; Molecular Probes), 10 ml of cells at a very early logarithmic (log) phase (OD600 = 0.2-0.4) were harvested, resuspended in 1 ml of phosphate-buffered saline (PBS) buffer and fixed by adding 130 μl of 37% formaldehyde for 20 min at room temperature. Cells were then washed twice with 1 ml PBS buffer and permeabilized in 1 ml of the same solution containing 1% TX-100 for 4 min. After centrifugation at 8000 rpm 1 min, cells were washed twice with PBS and resuspended in 20 μl of the same buffer. This cell suspension was mixed with 10 μl of 6.6 μM TR phalloidin in methanol and incubated for 2 h with occasional shaking at room temperature in the dark. Cells were finally washed three times with 1 ml of ice-cold PBS buffer, resuspended in the mounting solution (1 ml/ml p-phenylenediamine, 90% glycerol in PBS buffer), and viewed immediately.
Fluorescence signals were visualized with the use of an Axiovert S1002TV fluorescence microscope (Carl Zeiss, Thornwood, NY). The images were captured with a Photometrix camera and deconvolved using DeltaVision software (Applied Precision, Issaquah, WA).
Miscellaneous Procedures
Protein extraction, pulse-chase radiolabeling experiments, the protease protection assay, subcellular fractionation, monitoring of pexophagy, and the Pho8Δ60 activity assays were conducted as described previously (Hutchins et al., 1999; Kim et al., 2001b; Scott et al., 2001; Reggiori et al., 2003, 2004a).
Reagents
Proteinase K was from Roche Molecular Biochemicals (Indianapolis, IN). Anti-Pho8 antibodies were from Molecular Probes. Supersignal West Pico Chemiluminescent Substrate was from Pierce Chemical Co. (Rockford, IL). Antiserum to Pgk1 was generously provided by Dr. Jeremy Thorner (University of California, Berkeley). Antiserum to Ape1 was described previously (Klionsky et al., 1992). All other reagents were from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific Company (Pittsburgh, PA) unless specified otherwise.
RESULTS
The Actin Cytoskeleton Plays an Essential Role in Precursor Ape1 Transport During Vegetative Growing Conditions and in Pexophagy
In addition to nonselective autophagy, the yeast S. cerevisiae possesses at least two types of selective autophagy: The cytoplasm to vacuole targeting (Cvt) pathway and pexophagy (Klionsky et al., 1992; Hutchins et al., 1999). The first transport route assures the delivery of two resident vacuolar hydrolases, aminopeptidase I and α-mannosidase, whereas the second specifically disposes of superfluous peroxisomes. To assess the role of the cytoskeleton, and in particular actin, in these three pathways it is necessary to utilize methods that allow an assessment of the direct effects of any perturbations. That is, secondary defects due to interference with cytoskeletal function can be quite problematic. Accordingly, we decided to begin by analyzing the effects on specific and nonspecific autophagy of latrunculin A (LatA), a toxin that inhibits actin polymerization and therefore disrupts the cytoskeleton (Spector et al., 1983; Ayscough et al., 1997). This type of pharmacological treatment allows an assessment of acute effects following the inhibition of actin function.
Aminopeptidase I is synthesized in the cytoplasm as a zymogen (prApe1) and then delivered into the vacuole where it is processed to its active, lower molecular weight form (Ape1) by resident proteases (Klionsky et al., 1992). Precursor Ape1 transport is mediated by the Cvt pathway during vegetative growth conditions and by autophagy when cells are starved (Klionsky et al., 1992; Scott et al., 1997, 2000). We analyzed the effects of cytoskeleton disruption on prApe1 maturation in these two different physiological situations with pulse-chase radiolabeling experiments in the presence or absence of 200 μg/ml LatA, a concentration that has been shown to block actin function in yeast cells (Ayscough et al., 1997). As expected, maturation of prApe1 was complete in mock samples after the 2 h chase in both growing and autophagy-inducing (treatment with rapamycin) conditions (Figure 1A). In contrast, the processing of this hydrolase was severely impaired (average of 50%) in cells kept in rich medium in the presence of LatA for 20 min but not in those where autophagy was induced. Longer preincubation periods with LatA (40-60 min) caused the almost complete block (90%) of prApe1 maturation (our unpublished observations). To verify that the LatA effects were due to the dissolution of actin structures, we examined a wild-type strain expressing Abp140-RFP under the same conditions. Abp140 is a protein that binds actin cables and therefore can be used to specifically stain these structures (Fehrenbacher et al., 2004). The wild-type strain was incubated in the presence or absence of 200 μg/ml LatA for 1 h before imaging. As shown in Figure 1B, actin cables were clearly visible in untreated cells but were disrupted and clumped when LatA was added to the culture medium. On the basis of these results we concluded that cytoskeleton integrity was necessary for efficient prApe1 transport by the Cvt pathway.
Figure 1.

The cytoskeleton is essential for Cvt pathway progression. (A) LatA blocks prApe1 maturation. The wild-type (SEY6210) strain grown at 30°C was incubated in the presence or absence of 200 μg/ml LatA for 20 min. Each sample was split in two, pulse-radiolabeled with [35S]methionine/cysteine for 12 min and subjected to a nonradioactive chase for 2 h. Rapamycin (rap; 0.2 μg/ml) was added to one of the two samples at the end of the pulse to induce autophagy as indicated. Ape1 was immunoprecipitated before and after the chase and resolved by SDS-PAGE. (B) LatA disrupts actin cables. A wild-type (IRA010) strain expressing Abp140-RFP grown at 30°C was incubated in the presence or absence of 200 μg/ml LatA for 1 h before imaging. (C) Actin depolymerization does not affect autophagy. Wild-type (TN124) and atg13Δ (D3Y103) cells expressing Pho8Δ60 were incubated with or without 200 μg/ml LatA for 20 min before inducing autophagy with 0.2 μg/ml rapamycin for 4 h. Pho8 activity was determined for each sample before (black bars) and after (white bars) rapamycin addition. Results are expressed as a percentage of the activity measured for the wild-type strain (set to 100%) treated with rapamycin only and they represent the average of two experiments.
The processing of prApe1, however, is not by itself an adequate marker to monitor autophagy. For example, in the absence of Atg17 the delivery of this protease is normal, whereas autophagy is almost completely blocked (Cheong et al., 2005). To directly quantify autophagy, we decided to measure the vacuolar processing of the cytosolic marker protein Pho8Δ60. This truncated form of the vacuolar alkaline phosphatase (ALP; Pho8) lacks the transmembrane domain and consequently localizes to the cytosol (Noda et al., 1995). This factor is delivered to the vacuole exclusively by autophagy. Proteolytic cleavage of the Pho8Δ60 propeptide in the vacuole lumen generates the active form of the enzyme, which can be detected by an activity assay (Noda et al., 1995). A strain where the chromosomal PHO8 gene is replaced with pho8Δ60 was incubated in the presence or absence of 200 μg/ml LatA, and phosphatase activity was determined either before or after induction of autophagy with rapamycin. In wild-type cells, there was low ALP activity when cells were grown in rich medium (Figure 1C). There was an approximately eightfold increase in activity after the induction of autophagy after treatment with rapamycin for 4 h. As a control, we examined Pho8Δ60 activity in an atg13Δ strain; Atg13 is essential for autophagy (Funakoshi et al., 1997; Scott et al., 2000). In contrast to wild-type cells, there was no increase in ALP activity from Pho8Δ60 when atg13Δ cells were shifted to starvation conditions. As shown in Figure 1C, LatA did not impair autophagy in wild-type cells, indicating that the actin cytoskeleton was unnecessary for this nonselective catabolic process.
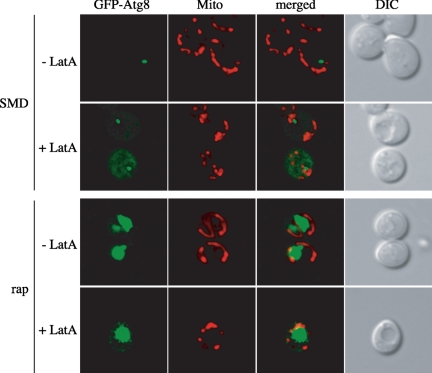
During the course of these experiments we noticed that the wild-type strain treated with LatA displayed a consistently higher level of Pho8Δ60 activity than the untreated cells (Figure 1C). One possibility was that a prolonged incubation with LatA leads to a damaged vacuole and causes the partial release of proteases into the cytoplasm with consequent processing of inactive Pho8Δ60. Accordingly, we decided to simultaneously examine the intactness of this organelle and the LatA effects on autophagy by an assay that did not rely on proteolytic processing. As a result, we chose to follow the transport of the GFP-Atg8 chimera. Part of Atg8 remains associated with autophagosomal structures from the initial formation stage until their complete breakdown in the vacuole; therefore GFP-Atg8 is an optimal marker to follow the itinerary of double-membrane vesicles (Kirisako et al., 1999; Huang et al., 2000). We also used Mitofluor Red, a dye that permits visualization of mitochondria (Reggiori et al., 2005), to allow a simultaneous assessment of the efficacy of the LatA treatment; LatA disrupts normal mitochondrial morphology (Boldogh et al., 1998).
A strain carrying a plasmid expressing the GFP-Atg8 chimera under the control of its authentic promoter was grown at 30°C and labeled with Mitofluor Red, in the presence or absence of 200 μg/ml LatA for 30 min before the addition of 0.2 μg/ml rapamycin for 2 h. As previously shown, the GFP-Atg8 chimera was concentrated at the PAS in untreated growing cells, whereas it was mostly localized in the vacuole interior when rapamycin was added to the same culture to trigger autophagy (Kirisako et al., 1999; Suzuki et al., 2001; Kim et al., 2002; Figure 2). In both situations, mitochondria were organized in their typical tubular network. As reported, LatA severely altered the morphology of this organelle confirming its activity in the imaged cells (Boldogh et al., 1998; Figure 2). Nonetheless, this toxin did not affect GFP-Atg8 localization to a single punctate structure in rich medium, suggesting that assembly of the PAS is normal and does not account for the defect in prApe1 maturation (Figure 1A). More to the point, this fluorescent fusion protein was normally transported into the vacuole lumen in conditions where autophagy was active in the presence of LatA. This observation supports our Pho8Δ60 assay data and leads to the conclusion that actin filaments do not play a relevant role during autophagy in yeast. It should be noted that after a 2-h incubation in the presence of both LatA and rapamycin, several cells were dead and their intracellular compartments lysed (our unpublished observations). This lysis may account for the higher Pho8Δ60 activity measured in LatA-treated cells.
Figure 2.
LatA causes mitochondrial reticulum fragmentation. Wild-type (SEY6210) cells carrying a plasmid expressing the GFP-Atg8 fusion under the control of the authentic promoter were incubated with Mitofluor Red in the presence or absence of 200 μg/ml LatA for 30 min before inducing autophagy by the addition of 0.2 μg/ml rapamycin for 3 h. Mitochondria (Mito) and GFP-Atg8 were imaged with a fluorescence microscope before and after rapamycin (rap) treatment. DIC, differential interference contrast.
In the yeast S. cerevisiae, pexophagy is induced when cells are shifted from conditions that necessitate peroxisome function (e.g., oleic acid) to glucose, or media lacking nitrogen (Evers et al., 1991; Hutchins et al., 1999). To monitor visually the degradation of excess peroxisomes during this process, we transformed the wild-type and atg1Δ strains with a plasmid expressing GFP C-terminally tagged with the peroxisome targeting tripeptide SKL. Transformants were grown in YPD and then shifted for 12 h to a medium containing glycerol, a suboptimal carbon source for yeast, and subsequently transferred for 19 h into a medium with oleic acid as the sole carbon source. As shown in Figure 3A and quantified in Figure 3B, this procedure induces peroxisome proliferation. These cultures were then harvested, resuspended in SD-N medium with or without 200 μg/ml LatA and incubated for an additional 6 h before being imaged. As expected, most of the peroxisomes were eliminated in wild-type cells under nitrogen starvation, resulting in approximately one peroxisome per cell (Guan et al., 2001; Wang et al., 2001; Figure 3, A and B). In contrast, the presence of LatA in the medium prevented the degradation of these organelles. The effects of this toxin were comparable to those caused by the deletion of ATG1, a gene essential for pexophagy (Figure 3, A and B). It should be noted that incubation of cells with LatA alone for 6 h did not result in a substantial loss of viability (our unpublished observations), in contrast to the results we observed in the presence of LatA and rapamycin.
Figure 3.
LatA blocks pexophagy. (A) Actin filaments are required for peroxisome elimination by pexophagy. The wild-type (BY4742) and atg1Δ (atg1Δ) strains transformed with a plasmid expressing GFP-SKL were grown in conditions that induce peroxisomes, washed, and resuspended in SD-N medium for 6 h in the presence or absence of 200 μg/ml LatA. Cells were imaged before and after starvation. (B) Quantification of peroxisome accumulation in LatA-treated cells. Peroxisomes were counted in ∼30 cells prepared for the imaging shown in A. The experiment was repeated twice, and error bars represent the peroxisome number deviation between cells. (C) LatA inhibits the proteolytic degradation of peroxisomal proteins during pexophagy. The Pex14-GFP (IRA001) and Pex14-GFP atg1Δ (IRA002) strains were grown and treated as in A. Protein extracts were prepared from cells before and after nitrogen starvation and resolved by SDS-PAGE, and after Western blot, membranes were probed with anti-GFP monoclonal antibody (Covance, Princeton, NJ). The positions of the full-length Pex14-GFP fusion protein and free GFP are indicated.
To confirm our microscopy data, we decided to analyze the fate of Pex14, a peroxisome integral membrane protein (Albertini et al., 1997), through a biochemical approach. This protein was C-terminally fused with GFP, which would be exposed on the cytosolic face of the peroxisome; delivery of the tagged peroxisome to the vacuole would allow cleavage of the GFP moiety, allowing us to monitor autophagy through the appearance of free GFP. Peroxisome proliferation was induced in wild-type and atg1Δ cells expressing this chimera as described above. No free GFP was observed at that stage in any of the strains (Figure 3C). Free GFP was detected in the wild-type strain, however, after transfer into SD-N medium for 6 h. In contrast, free GFP was not present in the atg1Δ protein extracts obtained from cells starved in the same way, indicating that pexophagy was involved in the transport and processing of the Pex14-GFP fusion in the vacuole. Treatment of the wild-type strain with LatA also inhibited cleavage of the Pex14-GFP chimera. Taken together with the microscopy analysis, these data indicate that actin filaments are essential for the normal progression of pexophagy.
Actin Filaments are Involved in Atg9 Cycling
We have recently shown that Atg9 cycles between the mitochondria and the PAS where it participates in the formation of double-membrane vesicles (Noda et al., 2000; Reggiori et al., 2004a, 2005). Once the vesicle is completed, Atg9 is retrieved; it is not found associated with the final Cvt vesicle or autophagosome (Noda et al., 2000). As noted above, actin filaments are essential to maintain the mitochondrial reticulum morphology (Drubin et al., 1993; Boldogh et al., 1998; Figure 2). We decided to determine if the defect in prApe1 transport through the Cvt pathway observed when actin filaments are disrupted correlated with an alteration of the Atg9 trafficking. To do so, we decided to utilize the Transport of Atg9 after Knocking-out Atg1 (TAKA) assay (Shintani and Klionsky, 2004b; Cheong et al., 2005) that allows us to analyze the transport of this protein from mitochondria to the PAS. In brief, this is an epistasis assay based on the observation that atg1Δ cells accumulate Atg9 almost exclusively at the PAS (Reggiori et al., 2004a). The atg1Δ ATG9-YFP strain carrying the Atg1ts-expressing plasmid was grown at permissive temperature and then incubated in the presence or absence of LatA for 1 h before transferring the cells to 35°C for 1 h. As reported, Atg9-YFP was dispersed in several punctate structures when the mock-treated strain was grown at 24°C (Figure 4A). The temperature shift caused the Atg9-YFP to accumulate at the PAS as expected. The presence of LatA in the medium affected Atg9-YFP localization even at the permissive temperature. The fluorescent fusion protein was still in part concentrated to punctate structures, but in addition part of the population was homogeneously distributed in tubular conformations (Figure 4A). Only a fraction of Atg9-YFP was able to accumulate at the PAS when the same cells were transferred to 35°C for 1 h. This result indicates that a possible explanation for the prApe1 maturation defect observed in LatA-treated cells is the failure of Atg9 to reach the PAS.
Figure 4.
Actin filament disruption affects Atg9 sorting from mitochondria in growing conditions. (A) LatA blocks Atg9 transport to the PAS. The Atg9-YFP atg1Δ (FRY138) strain transformed with the plasmid carrying the atg1ts allele was grown at permissive temperature, incubated with or without 200 μg/ml LatA for 1 h, and imaged. The same cells were then shifted to 35°C for 1 h before again visualizing the fluorescent signals. (B) Atg9 cycling is normal in LatA-treated cells when nitrogen-starved. The experiment described in A was repeated but incubation with LatA, and the temperature shift was done in SD-N medium. (C) LatA interferes with Cvt complex targeting to the PAS. Wild-type (SEY6210) cells cotransformed with plasmids expressing YFP-Atg8 and CFP-Ape1 were grown to an early log phase and then incubated in the presence of LatA for 90 min. Cells were imaged before and after the addition of toxin. A white arrow shows the position of CFP-Ape1. DIC, differential interference contrast.
Treatment with LatA, however, did not inhibit autophagy (Figures 1C and 2) despite the fact that this process requires Atg9 (Noda et al., 2000). Consequently, we decided to determine if Atg9 transport to the PAS was normal in the presence of LatA in conditions where autophagy is active. The strain analyzed above was nitrogen-starved at permissive temperature for 1 h in the presence or absence of 200 μg/ml LatA before being incubated for an additional hour at 35°C. In mock-treated cells, Atg9-YFP was normally distributed at 24°C and accumulated at the PAS at restrictive temperature as expected (Figure 4B). Similar to growing cells, the same chimera was relocated in tubular structures adjacent to the plasma membrane when LatA was added to starvation medium at permissive temperature. Surprisingly, the TAKA assay revealed that Atg9-YFP trafficked normally to the PAS under starvation conditions at the nonpermissive temperature in the presence of LatA, despite its abnormal localization in permissive conditions (Figure 4B). We concluded that LatA interfered with the proper Atg9 localization in growing and starved cells, but that it blocked Atg9 transport to the PAS only in growing cells.
Part of the yeast Atg9 population is found in very close proximity to, or on the surface of, mitochondria (Reggiori et al., 2005). In addition, certain mutants that disrupt the morphology of this organelle interfere with Atg9 sorting from this location and therefore double-membrane vesicle formation (F. Reggiori and D. J. Klionsky, unpublished results). Accordingly, we decided to explore if LatA was having a similar effect. We stained an Atg9-GFP expressing strain with Mitofluor Red in both growing and nitrogen starvation conditions in the presence or absence of LatA. Indeed, we found that this toxin caused the partial dissolution of Atg9 punctate structures and resulted in its homogeneous redistribution on the mitochondrial reticulum surface (our unpublished results).
It has previously been shown that certain mutants that affect proper PAS biogenesis interfere with recruitment of the Cvt complex (prApe1 oligomer-Atg19 receptor) to this location (Reggiori et al., 2003, 2004a). It seemed possible that could also be the case in LatA-treated cells because Atg9 failed to be efficiently transported to the PAS and consequently this structure was not properly organized (Figure 4A). To test this hypothesis, the wild-type strain was cotransformed with plasmids expressing a fluorescent PAS protein marker, YFP-Atg8, and a Cvt complex component, CFP-Ape1 (Shintani et al., 2002; Reggiori et al., 2003, 2004b). Transformants were grown to an early log phase then incubated in the presence of 200 μg/ml LatA for 90 min. Cells were imaged at the beginning and end of this incubation. As reported, prApe1 and therefore the Cvt complex colocalized with the PAS identified with the YFP-Atg8 chimera in almost all the untreated cells (Figure 4C). In contrast, the two fluorescent signals were separated in 45-50% of the cells when LatA was added to the growing culture (50 cells with clear expression of both YFP and CFP chimera were scored; data are the average of two experiments), indicating that the recruitment of the Cvt complex to the PAS was significantly compromised.
Taken together, these results suggest that the cytoskeleton plays an essential role in Atg9 trafficking during vegetative growth conditions by guaranteeing at least the proper organization of the mitochondrial reticulum. Conversely, actin filament disruption severely affects the Cvt pathway and pexophagy.
Specific Actin Point Mutants Block the Cvt Pathway and Pexophagy without Affecting the Mitochondrial Morphology
Actin filaments are a key structural element of the cytoskeleton. Their dissolution affects multiple cellular pathways and the morphology of several organelles, including that of the mitochondrial reticulum (Drubin et al., 1993; Boldogh et al., 1998; Eitzen et al., 2002; Winder and Ayscough, 2005; Figures 2 and 4C). Atg9 shuttles between this location and the PAS (Reggiori et al., 2005). It is possible that an alteration of mitochondrial structure and function interferes with Atg9 transport indirectly. This idea is supported by the observation that LatA affects normal Atg9 localization (Figure 4, A and B). In addition, we have recently identified mutants that block the trafficking of this factor by disrupting the mitochondrial morphology (F. Reggiori and D. J. Klionsky, unpublished results). To examine if actin filaments have additional roles in the Cvt pathway, beside that of maintaining mitochondrial structure, we decided to screen a collection of 10 actin point mutants (Drubin et al., 1993; Belmont and Drubin, 1998). We thought that the Cvt pathway might be affected in some of the strains with a normal mitochondrial reticulum. Accordingly, prApe1 maturation was analyzed in these mutants by pulse-chase radiolabeling followed by immunoprecipitation and, when not described in the literature, mitochondrial morphology was evaluated by staining the cells with Mitofluor Red (Table 2). Experiments were conducted at both the semipermissive and nonpermissive temperatures of 24 and 37°C, respectively.
Table 2.
Actin mutant phenotypes
| Mutant | Cvt pathway (24°C) | Mitochondrial morphology (24°C) | Cvt pathway (37°C) | Mitochondrial morphology (37°C) | Actin dynamic defect |
|---|---|---|---|---|---|
| Wild type | + | + | + | + | None |
| act1-101 (D363A, E364A) | + | + | – | – | Cable formation, delocalized patches |
| act1-102 (K359A, E361A) | + | + | + | + | None |
| act1-105 (E311A, R312A) | + | + | + | + | ? |
| act1-111 (D222A, E224A, E226A) | + | + | – | – | Cable formation, delocalized patches |
| act1-121 (E83A, K84A) | +/- | + | – | + | Delocalized patches |
| act1-122 (D80A, D81A) | + | + | + | + | ? |
| act1-133 (D24A, D25A) | – | – | – | – | Cable formation |
| act1-135 (E4A) | + | + | + | + | None |
| act1-157 (D157E) | +/- | ND | ND | ND | None |
| act1-159 (V159N) | – | + | – | + | Disassembly |
Precursor Ape1 transport was determined with pulse-chase radiolabeling experiments followed by immunoprecipitation, whereas the mitochondrial reticulum structure was analyzed by staining cells with Mitofluor Red. The plus sign indicates the complete processing of prApe1 or normal mitochondrial morphology. The minus sign describes the aberrant organization of this organelle or prApe1 accumulation ≥ 70%. The plus/minus sign specifies that a 50% block of prApe1 transport was observed. The mutant phenotypes with regard to actin dynamics that have been reported in the literature are also indicated (Wertman et al., 1992; Drubin et al., 1993; Smith et al., 1995; Belmont and Drubin, 1998; Wolven et al., 2000). The strains that were further analyzed are highlighted in bold. ND, not determined.
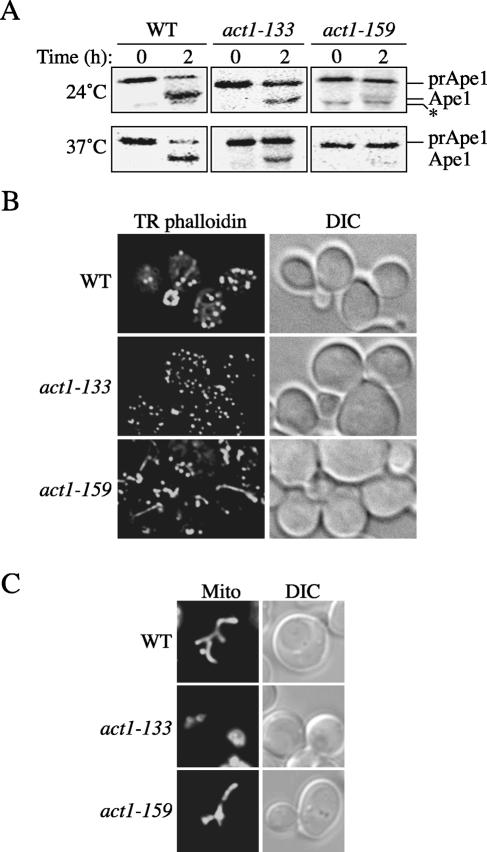
Wild-type cells and 4 mutants (act1-102, act1-105, act1-122, and act1-135) showed no prApe1 maturation block at both temperatures, whereas two others (act1-101 and act1-111) revealed a defect only at 37°C (Table 2). Two strains (act1-121 and act1-157) had a partial block at 24°C that became exacerbated when cells were analyzed at the higher temperature (Table 2). Finally, prApe1 transport was almost completely inhibited in the act1-133 and act1-159 strains at both temperatures (Figure 5A). The mutants with an impaired Cvt pathway could be divided in two groups. The first one includes cells that have both a prApe1 maturation defect and an aberrant mitochondrial reticulum (act1-101, act1-111, and act1-133). It should be noted that all the mutants with altered mitochondrial morphology showed a block in the Cvt pathway. Interestingly, these strains are not able to form actin cables (Table 2) and therefore have phenotypes similar to LatA-treated cells. The second group comprises strains with a normal mitochondrial organization but a severe prApe1 processing impairment (act1-121 and act1-159). This unique phenotype seems to correlate with the fact that in these two mutants only some actin functions are abolished (Table 2). We decided to analyze further one member of each group. We selected the act1-133 and act1-159 mutants because they already displayed clear defects at 24°C and consequently allowed us to avoid working at high temperatures where several cellular adaptation responses are induced. Analysis at the lower temperature should also minimize defects due to indirect effects on other processes.
Figure 5.
Identification of act1 mutants that specifically affect the Cvt pathway without altering mitochondrial structure. (A) Precursor Ape1 processing is blocked in several act1 mutants. The wild-type (LWY1412), act1-133 (DDY336), and act1-159 (DDY1493) strains were pulse-labeled for 10 min and subjected to a nonradioactive chase for 2 h at either 24 or 37°C in SMD medium. Ape1 was immunoprecipitated and resolved by SDS-PAGE. The asterisk marks a cross-reacting band that migrates jut below mature Ape1. (B) Actin cables in the act1-133 and act1-159 mutants. The strains analyzed above were grown to an early log phase, fixed, permeabilized, and incubated with TR phalloidin as described in Materials and Methods before collecting fluorescence images. (C) Morphology of the mitochondrial reticulum in the act1-133 and act1-159 mutants. The wild-type (LWY1412), act1-133 (DDY336), and act1-159 (DDY1493) strains were grown at semipermissive temperature and then stained with Mitofluor Red as described in Materials and Methods.
We first examined the actin cable organization and mitochondrial morphology, and compared it to that described in the literature (Drubin et al., 1993; Belmont and Drubin, 1998). To visualize the actin cables in these mutants, we genomically tagged ABP140 with the RFP gene. Images of the cells expressing this fluorescent chimera revealed the presence of polymeric structures in both wild-type and act1-159 strains but not in act1-133 (our unpublished results). Because we could not exclude that the mutations of the act1-133 allele were compromising the Abp140 binding site, we decided to stain actin cables with an alternate method. The Texas Red-X phalloidin (TR phalloidin) fluorescent conjugate associates with actin filaments and consequently it labels both the actin cables and patches. The wild-type, act1-133, and act1-159 cells were grown to an early log phase at 24°C, fixed, permeabilized and incubated with TR phalloidin dye as described in Materials and Methods. Actin cables were clearly visible in both wild-type and act1-159 strains but not in the act1-133 mutant (Figure 5B). In cells from the latter strain, however, actin patches were stained. This result demonstrates that TR phalloidin did not label actin cables in the act1-133 mutant because they were absent, and not because of an inability to bind the mutant filaments. The same strains were then grown again to an early log phase at 24°C but this time they were incubated with Mitofluor Red for 40 min in order to stain mitochondria. As shown in Figure 5C and in complete agreement with the previous reports, wild-type and act1-159 cells have a normal mitochondrial network, whereas the organization of this organelle is severely compromised in the act1-133 mutant where the mitochondrial reticulum was often observed clumped together. On the basis of the result with the act1-159 allele, we concluded that actin plays a direct role in the Cvt pathway, which is separable from that of maintaining the correct structural architecture of the mitochondrial reticulum.
We next analyzed prApe1 transport in autophagy-inducing conditions in the act1 mutants. Wild-type and mutant cells were grown to an early log phase and then pulse-chase radiolabeled in the presence of rapamycin in order to trigger autophagy. As shown in Figure 6A, prApe1 was essentially completely processed in both act1 mutants similar to wild-type cells. To verify that autophagy was functional in these strains, we then monitored GFP-Atg8 delivery into the vacuole lumen in nitrogen starvation conditions as in Figure 2. In rich medium, this fluorescent chimera was concentrated at the PAS in both wild-type and act1-133 cells but not in the act1-159 mutant, where it was completely diffuse in the cytosol (Figure 6B). Nonetheless, GFP-Atg8 was normally transported to the vacuole in all the strains when nitrogen-starved, indicating that the act1-133 and act1-159 point mutations did not interfere with bulk autophagy. An identical result was also obtained with the rest of the act1 mutants (our unpublished results). It should be noted that the PAS was clearly recognizable in starved act-159 cells.
Figure 6.
Autophagy proceeds normally in act1 mutants. (A) The processing of prApe1 is not perturbed in act1 mutants under starvation conditions. The wild-type (LWY1412), act1-133 (DDY336), and act1-159 (DDY1493) strains were pulse-labeled for 10 min and subjected to a nonradioactive chase for 2 h at 24°C in SMD medium. Rapamycin (rap; 0.2 μg/ml) was added after the pulse-labeling in order to induce autophagy. (B) Autophagy is fully functional in act1 mutants. The wild-type, act1-133, and act1-159 strains carrying a plasmid expressing GFP-Atg8 and grown in rich medium at 24°C were nitrogen-deprived for 4 h in SD-N medium at the same semipermissive temperature. Fluorescence images were collected at the beginning and the end of the starvation period. (C) Atg8 lipidation is normal in the act1-159 strain. Protein extracts were prepared from wild-type, atg4Δ (WPHYD2), and act1-159 cells grown at 24°C and then resolved by SDS-PAGE. After Western blot, membranes were probed with anti-Atg8 antiserum (Huang et al., 2000). The positions of nonlipidated and phosphatidylethanolamine (PE)-conjugated forms of Atg8 are indicated. DIC, differential interference contrast.
Atg8 is not seen at the PAS in deletion strains that are unable to conjugate this protein to phosphatidylethanolamine but also in others where Atg8 is normally lipidated but fails to be transported to this specialized site (Suzuki et al., 2001). To unveil which of these two causes prevented the GFP-Atg8 concentration at the PAS in the act1-159 mutant when grown in rich medium, we prepared a protein extract and analyzed Atg8 modification by Western blot. As shown in Figure 6C, two different bands were present in the wild-type strain and they represent the unmodified and lipidated forms of Atg8, respectively (Ichimura et al., 2000; Suzuki et al., 2001). The atg4Δ deletant is unable to link Atg8 to phosphatidylethanolamine and consequently only the unconjugated species was observed in cell extracts from this strain (Ichimura et al., 2000; Kirisako et al., 2000; Kim et al., 2001a; Suzuki et al., 2001). The ubiquitin-like Atg8 protein was normally lipidated in the act1-159 mutant and that led us to the conclusion that Atg8-phosphatidylethanolamine was not correctly transported to the PAS in this strain (Figure 6C).
We next investigated if pexophagy proceeded normally in the act1-159 mutant. Wild-type and act1-159 cells were transformed with the plasmid expressing GFP-SKL and analyzed as described in Figure 3A. When grown in the oleate-containing medium, both strains revealed the presence of an average of 11-12 fluorescent peroxisomes in each cell (Figure 7). In agreement with our previous results (Figure 3A), most of these organelles were degraded when the wild-type strain was nitrogen starved for 6 h, leaving approximately one peroxisome per cell (Figure 7). Accordingly, some of the fluorescent signal was clearly observed in the vacuole lumen. In contrast, peroxisomes were not eliminated in the presence of the act1-159 allele, and the GFP signal was excluded from the vacuole lumen (Figure 7). We concluded that actin is directly involved in pexophagy.
Figure 7.
Pexophagy is blocked in the act1-159 mutant. (A) The act1-159 allele impairs peroxisome degradation. The wild-type (LWY1412) and act1-159 (DDY1493) strains transformed with a plasmid expressing GFP-SKL were grown in conditions that induce peroxisomes at 24°C for 10 h, washed, and resuspended in SD-N medium for 6 h at the same temperature. Cells were imaged before and after starvation. The merged panels are an overlay of the GFP and DIC, differential interference contrast, images. (B) Quantification of peroxisome accumulation in the act1-159 mutant. Peroxisomes were counted in ∼30 cells prepared for the imaging shown in A. The experiment was repeated twice, and error bars represent the peroxisome number deviation between cells.
Actin Mediates Precursor Ape1 Recruitment to the PAS
The current mechanistic model of the Cvt pathway is the following: After oligomerization of prApe1 in the cytosol, the Ape1 complex binds to the Atg19 receptor to form the Cvt complex, which is subsequently recruited nearby the vacuole surface in an Atg11-dependent step (Shintani et al., 2002). This event triggers the recruitment of the rest of the Atg proteins, leading to PAS formation and subsequent sequestration of the Cvt complex into a double-membrane vesicle (Shintani and Klionsky, 2004b). Once formation of the vesicle is finished, its outer membrane fuses with the vacuole limiting membrane, liberating its contents into the lumen of this organelle. To gain insight into the role of actin in the Cvt pathway, we decided to identify the step at which this component acts.
We first analyzed the ability of act1-159 cells to complete the sequestration process through a protease-protection assay (Figure 8A). If a mutant is blocked during sequestration, before vesicle completion, prApe1 will accumulate in a form sensitive to exogenous protease after osmotic lysis of the spheroplast plasma membrane. However, if a mutant is competent for sequestration, this hydrolase will be protected from exogenous protease, becoming sensitive only upon membrane disruption by the addition of detergent (Harding et al., 1995). Spheroplasts were generated from an act1-159 pep4Δ strain and subjected to osmotic lysis. Efficient separation of Pgk1 into the supernatant fraction indicated efficient lysis of spheroplasts. The pelletable cellular fraction from the act1-159 mutants was subjected to treatment with proteinase K in the absence or presence of detergent. As shown in Figure 8A, the accumulated prApe1 in these cells was accessible to the exogenous proteinase K in the presence or absence of detergent. Proteinase K treatment in the presence of detergent results in a faster migrating band, presumably due to a second uncharacterized cleavage. To verify that this sensitivity is not the result of nonspecific disruption of intracellular membranes, we simultaneously examined the protease sensitivity of endogenous Pho8. Precursor Pho8 is accumulated in the limiting membrane of the vacuole in pep4Δ deletants. The precursor form of Pho8 contains a small cytosolically oriented tail, whereas the major part of the protein, including the C-terminal propeptide, resides within the vacuolar lumen. Treatment with proteinase K in the absence of detergent allowed the cleavage of only the cytosolic tail (Figure 8A), whereas addition of detergent along with the protease resulted in cleavage of the lumenal propeptide as well. This result confirmed that intracellular compartments such as the vacuole or, presumably, Cvt vesicles were not damaged during plasma membrane lysis.
Figure 8.
Precursor Ape1 is not correctly recruited in proximity of the vacuole surface in the act1-159 mutant. (A) Precursor Ape1 is protease-sensitive in act1-159 pep4Δ cells. The 13,000 × g pellet (P) fraction was collected from act1-159 pep4Δ (IRA003)-lysed spheroplasts, subjected to treatment with proteinase K (PrK) in the presence or absence of 0.4% Triton X-100 (TX), and analyzed by immunoblot using anti-Ape1 antiserum (Klionsky et al., 1992). Lysis conditions were verified by immunoblot analysis using anti-Pgk1 and anti-Pho8 antibodies. T, total; S, supernatant fraction. (B) Membrane binding of prApe1 is destabilized in the act1-159 mutant. The act1-159 (DDY1493), atg9Δ (JKY007), and atg11Δ (AHY001) strains were grown at 24°C to early log phase and converted to spheroplasts. The spheroplasts were lysed in osmotic lysis PS200 buffer (20 mM PIPES, pH 6.8, 200 mM sorbitol) with or without 5 mM MgCl2. The total lysate (T) was separated into low-speed supernatant (S) and pellet (P) fractions by centrifugation at 2300 × g for 5 min. The fractionated samples were subjected to immunoblot analysis using antiserum to Ape1. (C) Localization of Ape1 to the PAS is defective in the act1-159 strain. Wild-type (LWY1412) and act1-159 (DDY1493) strains cotransformed with plasmids expressing GFP-Ape1 and RFP-Vam3 were grown at semipermissive temperature to early log phase in SMD medium and then imaged. DIC, differential interference contrast.
The protease-protection assay indicated that prApe1 was not sequestered within completed Cvt vesicles. To extend our analysis we used two different approaches to explore whether the Ape1 complex was normally associated with lipid bilayers and correctly transported in proximity of the vacuole in the act1-159 mutant. The Cvt complex binds a pelletable membrane fraction in a salt-dependent manner (Kim et al., 1997). Subcellular fractionation experiments indicate that prApe1 can be recovered entirely in a low-speed pellet fraction in an osmotic lysis buffer containing 5 mM MgCl2, whereas omitting MgCl2 from the lysis buffer results in a largely cytosolic distribution of this hydrolase (Oda et al., 1996). The Ape1 complex association with membranes in the presence of salts is impaired in deletants such as atg11Δ and atg19Δ that are unable to target this oligomer to the PAS (Kim et al., 2001b; Scott et al., 2001; Shintani et al., 2002). We next examined if the act1-159 strain displayed a similar phenotype to this class of mutants.
Cells were grown to early log phase at semipermissive temperature and converted into spheroplasts before osmotic lysis in the presence or absence of 5 mM MgCl2. The lysates were then separated into low-speed supernatant and pellet fractions and analyzed by immunoblots using antiserum to Ape1. The atg9Δ strain was used as a control in which prApe1 displays typical membrane-binding properties, whereas atg11Δ cells provided the negative control. In the absence of salt, prApe1 association with the pellet fraction was severely perturbed (Figure 8B), consistent with previous reports (Oda et al., 1996). As in typical atg mutants such as atg9Δ, addition of MgCl2 stabilized the binding of prApe1 to the membrane fraction (Figure 8B). In contrast, in the atg11Δ strain, prApe1 remained largely in the supernatant fraction even with 5 mM MgCl2. The association of prApe1 with lipid bilayers in the presence or absence of MgCl2 in the act1-159 mutant was almost identical to that observed in the atg11Δ strain; the interaction of prApe1 with the lipid bilayer was compromised even in the presence of 5 mM MgCl2. This result indicates that although actin is not essential for prApe1 membrane binding, the act1-159 allele destabilizes the interactions between the Cvt complex and the membrane structure to which it binds.
The decreased ability of the prApe1 oligomer to bind membranes in the atg11Δ and atg19Δ deletants is probably due to the fact that in these mutants, this complex is not correctly transported in proximity of the vacuole surface, where it induces PAS organization and subsequent Cvt vesicle formation (Shintani et al., 2002; Shintani and Klionsky, 2004b). Thus, we took a fluorescence microscopy approach to determine whether the act1-159 cells had a similar defect. Wild-type and act1-159 strains were cotransformed with both a plasmid expressing GFP-Ape1 and a vector carrying the RFP-Vam3 fusion and grown at semipermissive temperature. Vam3 is a SNARE protein that localizes to the vacuole rim and the fusion protein serves as a fluorescent marker for this organelle (Darsow et al., 1997; Reggiori et al., 2000). As shown in Figure 8C and previously reported, GFP-Ape1 is adjacent to the vacuole limiting membrane in the wild-type strain. In contrast, the same fluorescent chimera did not localize in proximity of the vacuole in 10-15% of act1-159 cells (80 cells were scored and the data represent the average of two experiments; Figure 8C). When the same act1-159 culture was then transferred from the semipermissive temperature of 24°C to the restrictive one of 35°C for 30 min, the GFP-Ape1 signal was distanced from the vacuole rim in 25-30% of the cells imaged (80 cells were scored and the data represent the average of two experiments), whereas there was no change in the wild-type cells (our unpublished observations). On the basis of our analyses with the conditional actin mutants, we concluded that functional actin is required for the efficient transport of the Cvt complex to the PAS, similar to the result we found with LatA-treated cells (Figure 4C).
Atg9 Transport to the PAS in Growing Conditions Requires Actin
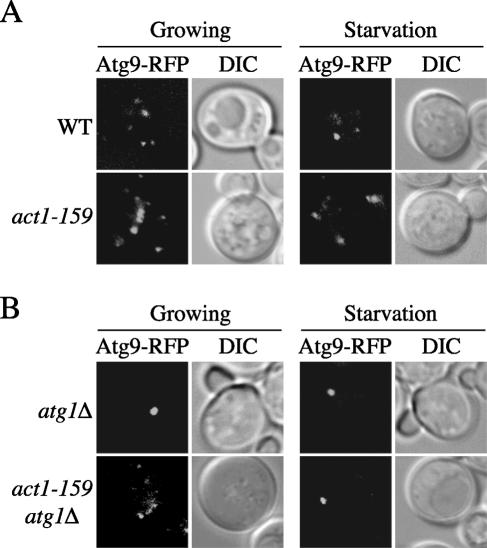
Because LatA disrupts the localization and trafficking of Atg9 (Figure 4, A and B), we decided to determine the subcellular distribution of this protein in the act1-159 mutant. The ATG9 gene was genomically tagged with RFP in the wild-type and act1-159 strains. Cells expressing the fluorescent fusion protein were then grown in rich medium to an early log phase or nitrogen starved for 1 h before imaging. In the wild-type strain, Atg9-RFP was localized to several punctate structures in both conditions as previously shown (Figure 9A). The act1-159 allele did not interfere with this distribution pattern. Based on this observation and on the Atg9-YFP localization result in the presence of LatA (Figure 4A), we concluded that the correct organization of this protein in complexes is only compromised when the mitochondrial morphology is disrupted.
Figure 9.
Atg9 trafficking to the PAS is impaired in the act1-159 mutant. (A) Atg9 distribution in act1-159 cells is normal. Wild-type (FRY245) and act1-159 (IRA004) strains expressing Atg9-RFP were grown at semipermissive temperature to midlog phase in SMD medium or incubated in SD-N for an additional 1 h and then imaged. (B) Atg9 fails to reach the PAS in the atg1Δ deletion background. The atg1Δ (IRA006) and act1-159 atg1Δ (IRA005) strains carrying a genomic Atg9-RFP fusion were grown and imaged as in A. DIC, differential interference contrast.
A normal Atg9 distribution, however, does not always indicate that this factor is able to cycle through the PAS. For example, when the atg11Δ deletant is grown in rich medium, Atg9 is dispersed in puncta but fails to be transported to the PAS (Shintani and Klionsky, 2004b). For this reason we again used the TAKA assay to monitor the capacity of Atg9 to be delivered to the formation site of double-membrane vesicles. Accordingly, the ATG1 gene was knocked out in wild-type and act1-159 cells expressing Atg9-RFP. The resulting strains were grown at 24°C in rich medium or nitrogen-starved for 1 h at the same temperature. As already shown, Atg9-RFP was concentrated at the PAS in the absence of Atg1 in both conditions (Figure 9B). The accumulation of this chimera at the same location, however, was blocked in growing cells by the presence of the act1-159 allele in the same deletion background. Atg9-RFP condensation at the PAS was reestablished when the act1-159 atg1Δ strain was starved. We conclude that actin is required for Atg9 transport to the PAS in growing conditions.
DISCUSSION
Selective Autophagy Requires the Actin Cytoskeleton
The principal study exploring the role of actin cytoskeleton in autophagy using toxins known to directly affect actin microfilament structure indicates that actin plays a role during autophagosome formation in mammalian cells (Aplin et al., 1992). Because of the lack of clear information regarding the role of actin in yeast autophagy and because our knowledge about autophagy and its molecular machinery has exponentially increased in the last decade, we decided to investigate the involvement of the actin cytoskeleton in this process in the yeast S. cerevisiae, the leading model organism for the study of both selective and nonselective autophagy.
In contrast with the findings in mammalian cells, our experiments using LatA, a potent actin polymerization inhibitor, and act1 point mutants showed that nonspecific autophagy in yeast did not require actin filaments (Figures 1C, 2, and 6B; our unpublished results). The discrepancy between the yeast and mammalian data could be due to differences between organisms or alternatively to the experimental conditions. For example, rat kidney cells were incubated for 5 h in the presence of cytochalasins, resulting in dramatic morphological changes as well as other effects (Aplin et al., 1992). To minimize potential secondary effects, we incubated our cells with LatA for relatively short periods. In addition, the use of the act1-159 mutant has allowed us to avoid dramatic physiological alterations because most actin functions are preserved in this strain (Belmont and Drubin, 1998). Importantly, no published studies have examined the role of the cytoskeleton in specific types of autophagy in higher eukaryotes. In contrast to nonspecific autophagy, the Cvt pathway and pexophagy were severely impaired in LatA-treated cells and certain act1 mutants, revealing that actin is essential for these selective types of autophagy (Figures 1A, 3, 5A, and 7; Table 2). These results are in complete agreement with a recent report showing that specific uptake of the endoplasmic reticulum into autophagosomes was actin-dependent (Hamasaki et al., 2005).
Interestingly, the transport into the vacuole of the major Cvt pathway cargo molecule, prApe1, was reestablished when cells were starved (Figures 1A and 6A). One possibility is that the incorporation of this enzyme into double-membrane vesicles does not rely on actin filaments under these conditions. However, it should be noted that components including Atg11 and Atg19 that are required for the Cvt pathway are presumably still functional after LatA treatment. Both of these proteins are needed for efficient uptake of prApe1 under growing and starvation conditions. In growing conditions, Cvt vesicles are essentially not formed in the absence of the Cvt complex (Shintani and Klionsky, 2004b), which makes sense given that prApe1 appears to be the primary cargo for this pathway. In contrast, autophagosomes can be generated in the absence of prApe1. The presence of a relatively large number of autophagosomes, relative to Cvt vesicles, along with the specificity components Atg11 and Atg19 may allow efficient uptake of prApe1 in starvation conditions even when the actin cytoskeleton is not functional. In other words, actin may normally play a role in the uptake of specific cargo by autophagy, but the increased import capacity after rapamycin treatment or under starvation conditions may partially mask the defect. In contrast to prApe1, peroxisomes were not delivered to the vacuole in the presence of LatA or in the act1-159 mutant even though our pexophagy assay is performed in a nitrogen starvation medium (Figures 3 and 7). This result seems to indicate that actin filaments are indeed required for the sequestration of selected cargoes into double-membrane vesicles even in situations where autophagy is active. It is possible that peroxisomes cannot be randomly enwrapped by autophagosomes because they are much larger than the prApe1 complex; however, we cannot exclude the possibility that different mechanisms are used to incorporate the prApe1 complex and peroxisomes into double-membrane vesicles.
Actin Filaments and Atg9 Cycling
It is well established that the actin cytoskeleton is required to maintain proper mitochondrial reticulum organization, and consequently LatA disrupts the morphology of this organelle (Drubin et al., 1993; Belmont and Drubin, 1998). Atg9 localizes in part to mitochondria and shuttles between this location and the PAS (Reggiori et al., 2005). Atg9 is normally localized in clusters that may reflect its ability to self-interact. Analysis of the subcellular distribution and trafficking of Atg9 in LatA-treated cells grown in rich medium shows that actin filament dissolution causes the disassembly of part of the Atg9 clusters and results in a more homogeneous redistribution of this protein along the mitochondrial surface (Figure 4A; our unpublished results). In addition, Atg9 is no longer able to reach the PAS (Figure 4A); however, it is unclear if the two phenomena are connected. It is interesting to note that all the defects observed in cells incubated with LatA were also present in the act1-133 mutant (Figures 5 and 6). The act1-133 allele contains two point mutations that generate a protein where the two alanine residues at positions 24 and 25 are substituted with aspartates (Smith et al., 1995), and these mutations result in an abnormal mitochondrial morphology. The act1-159 mutant, however, has intact mitochondrial morphology and normal Atg9 distribution in puncta, but this protein was unable to leave this organelle and reach the PAS (Drubin et al., 1993; Figures 5C and 9). These results imply that actin plays at least two roles in Atg9 trafficking, that is, clustering on the mitochondrial surface and transport to the PAS.
Atg9 delivery to the PAS was reestablished in LatA-treated cells after starvation (Figure 4B). Under these conditions, Atg9 seemed to be exclusively concentrated in discrete punctate structures and was no longer homogeneously dispersed on the mitochondrial limiting membrane. The ability of mitochondria to undergo fusion and fission reactions plays a key role in establishing and maintaining the architecture of this organelle (Hermann and Shaw, 1998; Jensen et al., 2000). This dynamic property is particularly important during cell division because it guarantees the correct distribution of mitochondria between the mother and the daughter cell (Boldogh et al., 2005). Actin filaments are part of the machinery that catalyzes these fusion and fission events and consequently their absence has a major impact upon the mitochondrial reticulum structure (Boldogh et al., 1998). Starvation provokes the arrest of the cell cycle in the G0 phase and normal Atg9 localization may be due to the resulting reduced dynamic activity of the mitochondrial population. The correct Atg9 organization in puncta, however, cannot alone account for the proper transport of this component to the PAS. In the act1-159 mutant, for example, Atg9 has a normal distribution in growing conditions (Figure 9A) but fails to reach the PAS (Figure 9B), indicating that the assembly of Atg9 per se is not the only requirement for proper transport.
Actin Is Involved in Cvt Complex Recruitment to the PAS
To separate the actin function of maintaining the mitochondrial reticulum organization from other functions more specific for the Cvt pathway, we have screened a collection of 10 actin point mutants (Drubin et al., 1993; Table 2). In support of our results with LatA, we found that all the mutants with abnormal mitochondrial morphology also displayed a prApe1 maturation defect (Table 2; Figure 5, A and C). In addition, we also identified alleles that severely affected the Cvt pathway without altering the structure of this organelle (Table 2; Figure 5, A and C). We have focused our studies on the act1-159 strain because it showed a clear defect in the Cvt pathway even at the semipermissive temperature of 24°C, while maintaining normal mitochondrial morphology even at the nonpermissive temperature. When starved, these cells had normal autophagy, and prApe1 was correctly delivered into the vacuole interior in complete agreement with our experiments using LatA (Figure 6, A and B). Pexophagy, however, was completely blocked under the same conditions (Figure 7). The act1-159 allele blocked prApe1 sequestration into Cvt vesicles (Figure 8A); this block was possibly caused by the fact that the Cvt complex was not correctly targeted to the site, the PAS, where these structures are generated (Figure 8C).
Because we observed a block at an early stage of Cvt vesicle formation, we cannot exclude that the actin cytoskeleton is required for successive steps of specific autophagy such as membrane assembly, vesicle sealing, retrieval transport from the PAS and/or fusion with the vacuole. The act1-159 allele contains one point mutation that generates a protein where the valine at position 159 is substituted with an asparagine residue (Belmont and Drubin, 1998). This modified actin is able to form filaments but they depolymerize slowly resulting in reduced actin dynamics (Figure 5B; Belmont and Drubin, 1998). Thus, cells expressing this mutated protein have more actin cables and they are partially polarized (Figure 5C; Belmont and Drubin, 1998). This hyperstable actin is apparently able to promote the proper organization of the mitochondrial reticulum. The act1-159 mutant, however, has a severe endocytosis defect because this process requires rapid actin filament turnover, which is necessary for the movement of molecules along these structures (Belmont and Drubin, 1998; Winder and Ayscough, 2005). Our data suggest that a similar type of transport, involving rapid actin filament turnover, may take place during selective autophagy.
Atg9 cycling is also blocked in the act1-159 mutant when this strain is kept in rich medium but not when starved (Figure 9B). This result seems to indicate that actin is required for both Atg9 trafficking and cargo recruitment in growing cells. An alternative hypothesis is that Atg9 transport to the PAS is coupled with that of the cargo molecules. In fact, Atg9 is unable to reach the PAS in the absence of prApe1, Atg19, or Atg11 (Shintani and Klionsky, 2004b).
The actin cytoskeleton plays an essential role in the organized movement and transport of large structures and organelles to precise subcellular locations. With our work, we have revealed a similar function of this large protein scaffold in the yeast selective autophagy pathways. In particular, we have shown that actin is required for the recruitment of the Cvt pathway cargo to the PAS and for correct cycling of Atg9. Additional studies in yeast are needed to uncover the mechanistic connections between actin, specific cargoes and also Atg proteins such as Atg9 and Atg11 that will potentially provide conceptual support to understand the diverse selective types of autophagy that are currently being unveiled in higher eukaryotic cells.
Acknowledgments
We thank Drs. David Drubin, Yoshinori Ohsumi, Roger Tsien, and Lois Weisman for strains and plasmids, and Dr. Keith Kozminski for technical advice. We also thank Dr. Phil Crews (supported by National Institutes of Health Grant CA47135) for supplying LatA. This work was supported by the NIH Public Health Service Grant GM53396 (to D.J.K.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0629) on October 12, 2005.
References
- Albertini, M., Rehling, P., Erdmann, R., Girzalsky, W., Kiel, J.A.K.W., Veenhuis, M., and Kunau, W. H. (1997). Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89, 83-92. [DOI] [PubMed] [Google Scholar]
- Aplin, A., Jasionowski, T., Tuttle, D. L., Lenk, S. E., and Dunn, W. A., Jr. (1992). Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J. Cell. Physiol. 152, 458-466. [DOI] [PubMed] [Google Scholar]
- Ayscough, K. R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D. G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, M., Osumi, M., Scott, S. V., Klionsky, D. J., and Ohsumi, Y. (1997). Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139, 1687-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont, L. D., and Drubin, D. G. (1998). The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J. Cell Biol. 142, 1289-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankson, H., Holen, I., and Seglen, P. O. (1995). Disruption of the cytokeratin cytoskeleton and inhibition of hepatocytic autophagy by okadaic acid. Exp. Cell Res. 218, 522-530. [DOI] [PubMed] [Google Scholar]
- Boldogh, I., Vojtov, N., Karmon, S., and Pon, L. A. (1998). Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J. Cell Biol. 141, 1371-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I. R., Fehrenbacher, K. L., Yang, H. C., and Pon, L. A. (2005). Mitochondrial movement and inheritance in budding yeast. Gene 354, 28-36. [DOI] [PubMed] [Google Scholar]
- Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, H., Yorimitsu, T., Reggiori, F., Legakis, J. E., Wang, C.-W., and Klionsky, D. J. (2005). Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell 16, 3438-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow, T., Rieder, S. E., and Emr, S. D. (1997). A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 138, 517-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B. L. et al. (2001). A protein interaction map for cell polarity development. J. Cell Biol. 154, 549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D. G., Jones, H. D., and Wertman, K. F. (1993). Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol. Biol. Cell 4, 1277-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen, G., Wang, L., Thorngren, N., and Wickner, W. (2002). Remodeling of organelle-bound actin is required for yeast vacuole fusion. J. Cell Biol. 158, 669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers, M. E., Hohfeld, J., Kunau, W. H., Harder, W., and Veenhuis, M. (1991). Physiological studies on the utilization of oleic acid by Saccharomyces cerevisiae in relation to microbody development. FEMS Microbiol. Lett. 69, 73-78. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher, K. L., Yang, H. C., Gay, A. C., Huckaba, T. M., and Pon, L. A. (2004). Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr. Biol. 14, 1996-2004. [DOI] [PubMed] [Google Scholar]
- Fengsrud, M., Roos, N., Berg, T., Liou, W., Slot, J. W., and Seglen, P. O. (1995). Ultrastructural and immunocytochemical characterization of autophagic vacuoles in isolated hepatocytes: effects of vinblastine and asparagine on vacuole distributions. Exp. Cell Res. 221, 504-519. [DOI] [PubMed] [Google Scholar]
- Funakoshi, T., Matsuura, A., Noda, T., and Ohsumi, Y. (1997). Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192, 207-213. [DOI] [PubMed] [Google Scholar]
- Guan, J., Stromhaug, P. E., George, M. D., Habibzadegah-Tari, P., Bevan, A., Dunn, W. A., Jr., and Klionsky, D. J. (2001). Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell 12, 3821-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki, M., Noda, T., Baba, M., and Ohsumi, Y. (2005). Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6, 56-65. [DOI] [PubMed] [Google Scholar]
- Harding, T. M., Morano, K. A., Scott, S. V., and Klionsky, D. J. (1995). Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G. J., and Shaw, J. M. (1998). Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14, 265-303. [DOI] [PubMed] [Google Scholar]
- Huang, W.-P., Scott, S. V., Kim, J., and Klionsky, D. J. (2000). The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275, 5845-5851. [DOI] [PubMed] [Google Scholar]
- Hutchins, M. U., Veenhuis, M., and Klionsky, D. J. (1999). Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112, 4079-4087. [DOI] [PubMed] [Google Scholar]
- Ichimura, Y. et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488-492. [DOI] [PubMed] [Google Scholar]
- Jensen, R. E., Hobbs, A. E., Cerveny, K. L., and Sesaki, H. (2000). Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc. Res. Tech. 51, 573-583. [DOI] [PubMed] [Google Scholar]
- Kim, J., Huang, W.-P., and Klionsky, D. J. (2001a). Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152, 51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Huang, W.-P., Stromhaug, P. E., and Klionsky, D. J. (2002). Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277, 763-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kamada, Y., Stromhaug, P. E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S. V., Ohsumi, Y., Dunn, W. A., Jr., and Klionsky, D. J. (2001b). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Scott, S. V., Oda, M. N., and Klionsky, D. J. (1997). Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 137, 609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., Noda, T., and Ohsumi, Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., Ohsumi, M., Takao, T., Noda, T., and Ohsumi, Y. (2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard, K., Taylor, M. P., and Jackson, W. T. (2004). Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2, 301-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J. et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539-545. [DOI] [PubMed] [Google Scholar]
- Klionsky, D. J., Cueva, R., and Yaver, D. S. (1992). Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119, 287-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, B., and Klionsky, D. J. (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463-477. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Nakagawa, I. et al. (2004). Autophagy defends cells against invading group A Streptococcus. Science 306, 1037-1040. [DOI] [PubMed] [Google Scholar]
- Noda, T., Kim, J., Huang, W.-P., Baba, M., Tokunaga, C., Ohsumi, Y., and Klionsky, D. J. (2000). Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Matsuura, A., Wada, Y., and Ohsumi, Y. (1995). Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126-132. [DOI] [PubMed] [Google Scholar]
- Oda, M. N., Scott, S. V., Hefner-Gravink, A., Caffarelli, A. D., and Klionsky, D. J. (1996). Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J. Cell Biol. 132, 999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., Black, M. W., and Pelham, H.R.B. (2000). Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell 11, 3737-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., and Klionsky, D. J. (2002). Autophagy in the eukaryotic cell. Euk. Cell 1, 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., and Klionsky, D. J. (2005). Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 17, 415-422. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., Shintani, T., Nair, U., and Klionsky, D. J. (2005). Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 1, 101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., Tucker, K. A., Stromhaug, P. E., and Klionsky, D. J. (2004a). The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79-90. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., Wang, C.-W., Nair, U., Shintani, T., Abeliovich, H., and Klionsky, D. J. (2004b). Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 2189-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., Wang, C.-W., Stromhaug, P. E., Shintani, T., and Klionsky, D. J. (2003). Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 278, 5009-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. S., Klionsky, D. J., Banta, L. M., and Emr, S. D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein, D. C., DiFiglia, M., Heintz, N., Nixon, R. A., Qin, Z.-H., Ravikumar, B., Stefanis, L., and Tolkovsky, A. (2005). Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 1, 11-22. [DOI] [PubMed] [Google Scholar]
- Schlumpberger, M., Schaeffeler, E., Straub, M., Bredschneider, M., Wolf, D. H., and Thumm, M. (1997). AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J. Bacteriol. 179, 1068-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S. V., Baba, M., Ohsumi, Y., and Klionsky, D. J. (1997). Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 138, 37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S. V., Guan, J., Hutchins, M. U., Kim, J., and Klionsky, D. J. (2001). Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell 7, 1131-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S. V. et al. (2000). Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 275, 25840-25849. [DOI] [PubMed] [Google Scholar]
- Seglen, P. O., Berg, T. O., Blankson, H., Fengsrud, M., Holen, I., and Stromhaug, P. E. (1996). Structural aspects of autophagy. Adv. Exp. Med. Biol. 389, 103-111. [DOI] [PubMed] [Google Scholar]
- Shintani, T., Huang, W.-P., Stromhaug, P. E., and Klionsky, D. J. (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., and Klionsky, D. J. (2004a). Autophagy in health and disease: a double-edged sword. Science 306, 990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., and Klionsky, D. J. (2004b). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889-29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. G., Simon, V. R., O'Sullivan, H., and Pon, L. A. (1995). Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangement in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 6, 1381-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, I., Shochet, N. R., Kashman, Y., and Groweiss, A. (1983). Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219, 493-495. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169-174. [DOI] [PubMed] [Google Scholar]
- Wang, C.-W., Kim, J., Huang, W.-P., Abeliovich, H., Stromhaug, P. E., Dunn, W. A., Jr., and Klionsky, D. J. (2001). Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem. 276, 30442-30451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman, K. F., Drubin, D. G., and Botstein, D. (1992). Systematic mutational analysis of the yeast ACT1 gene. Genetics. 132, 337-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder, S. J., and Ayscough, K. R. (2005). Actin-binding proteins. J. Cell Sci. 118, 651-654. [DOI] [PubMed] [Google Scholar]
- Wolven, A. K., Belmont, L. D., Mahoney, N. M., Almo, S. C., and Drubin, D. G. (2000). In vivo importance of actin nucleotide exchange catalyzed by profiling. J. Cell Biol. 150, 895-904. [DOI] [PMC free article] [PubMed] [Google Scholar]