Abstract
Members of the Asm1p, Phd1p, Sok2p, Efg1p, and StuAp (APSES) family of fungal proteins regulate morphogenesis and virulence in ascomycetes. We cloned the Aspergillus fumigatus APSES gene encoding StuAp and demonstrated that stuA transcription is markedly up-regulated after the acquisition of developmental competence. A. fumigatus ΔstuA mutants were impaired in their ability to undergo asexual reproduction. Conidiophore morphology was markedly abnormal, and only small numbers of dysmorphic conidia were produced, which exhibited precocious germination. Whole genome transcriptional analysis during the onset of developmental competence was performed and identified a subset of developmentally regulated genes that were stuA dependent, including a cluster of putative secondary metabolite biosynthesis genes, genes encoding proteins implicated in the regulation of morphogenesis, and genes encoding allergens and other antigenic proteins. Additionally, hyphae of the ΔstuA mutant displayed reduced expression of the catalase gene CAT1 and were hypersusceptible to hydrogen peroxide.
INTRODUCTION
As increasing numbers of patients undergo immunosuppressive medical procedures such as solid organ and bone marrow transplantation, the incidence of invasive fungal infections has increased (Marr et al., 2002). Despite the availability of new therapeutic agents, mortality from infections due to the most common mold, Aspergillus fumigatus, remains at least 50% (Herbrecht et al., 2002). A more comprehensive understanding of the biology of A. fumigatus and its interactions with the host is a critical step in the development of new strategies aimed at the prevention and treatment of infections caused by this opportunistic pathogen.
Aspergillus species have a complex life cycle with distinct developmental stages. Airborne uninucleate conidia are deposited on organic matter where they become metabolically active, swell and germinate, producing filamentous septate hyphae, which grow by apical extension (Adams et al., 1998). After a defined period of growth (∼8-15 h in submerged culture in vitro), these hyphae are able to respond to a variety of stimuli and initiate asexual reproduction or conidiation (Adams et al., 1998). Hyphae after this transition are termed developmentally competent and can be maintained in this phase of development indefinitely if grown in submerged culture. When exposed to a suitable stimulus, such as a static-air interphase, developmentally competent hyphae enter into the final stage of development. This stage is characterized by the production of conidiophores, which are multicellular asexual reproductive structures consisting of highly specialized, morphologically distinct cell types that produce uninucleate conidia via apolar budding (Miller et al., 1992; Adams et al., 1998). These conidia are then dispersed by air currents for the cycle to continue. Humans are infected incidentally by the inhalation of small numbers of spores. In the absence of a robust immune response, these conidia germinate to form hyphae that invade and destroy pulmonary tissue (Latge, 1999). Conidiation does not occur during invasive aspergillosis, and hyphae are the only invasive fungal forms observed by histopathology after the initiation of infection (Fraser, 1993). Sexual reproduction has not been observed in the pathogenic A. fumigatus, but it does occur in some other Aspergillus species, including the well studied model organism A. nidulans.
In fungi, the genes involved in the regulation of fungal development often directly regulate the expression of key virulence factors (Lengeler et al., 2000). In ascomycetes, members of the APSES group of transcription factors have been identified as key regulators of fungal development that often also control virulence traits. This group of proteins was named for the original members (Asm1p, Phd1p, Sok2p, Efg1p, and StuAp) and comprises a family of transcription factors that share a common DNA-binding motif (Dutton et al., 1997; Stoldt et al., 1997; Sonneborn et al., 1999a,b; Doedt et al., 2004). This motif can be structurally modeled as a basic helix-loop-helix (bHLH) protein analogous to eukaryotic Myc and Max proteins (Dutton et al., 1997). Each of these proteins has been shown to play an important role in controlling fungal morphogenesis and development.
The link between the regulation of fungal development and virulence by an APSES protein has been best studied in Candida albicans (Lo et al., 1997; Stoldt et al., 1997). C. albicans Efg1p not only plays a key role in mating and the yeast-to-hypha transition, but it is required for normal adherence to and invasion of host cells as well as virulence in murine models of disseminated and oral candidiasis (Lo et al., 1997; Stoldt et al., 1997; Sonneborn et al., 1999a,b; Doedt et al., 2004; Park et al., 2005).
In the minimally pathogenic Aspergillus nidulans, both sexual and asexual reproduction are controlled by the APSES protein StuAp (Wu and Miller, 1997). Mutants deficient in StuAp produce shortened conidiophores that lack phialides and produce low numbers of conidia by direct budding from the vesicle head (Miller et al., 1992). Although the exact function of StuAp in asexual reproduction is not known, it has been suggested that StuAp functions predominately as a transcriptional repressor, governing the spatial temporal expression of the conidiation genes brlA and abaA (Dutton et al., 1997). However, transcription of stuA is not restricted to conidiation, but rather it is temporally associated with the onset of developmental competence and is further up-regulated at the initiation of conidiation (Miller et al., 1991). In addition, StuAp expression is diffusely distributed in competent hyphae and not restricted to specific cell types (Miller et al., 1992). These findings suggest the hypothesis that in addition to functioning as a transcriptional repressor during conidiation, StuAp may govern other cellular processes in competent hyphae. This possibility is of particular interest given that hyphae are the invasive form of A. fumigatus during human infection, and conidiation is not observed during invasive pulmonary aspergillosis (Fraser, 1993).
In this study, we cloned, disrupted, and characterized the function of the stuA gene of the pathogen A. fumigatus. To understand the role of StuAp in preconidiation competent hyphae, whole genome transcriptional analysis was used to identify the stuA-dependent components of the regulatory program associated with the onset of developmental competence.
MATERIALS AND METHODS
Strains and Growth Conditions
A. fumigatus strain Af293, (a generous gift from P. Magee, University of Minnesota, St. Paul, MN) was used as the basis for molecular manipulations. Except where indicated, strains were propagated on YEPD agar (1% yeast extract, 2% peptone, 2% glucose, solidified with 1.5% agar). For germination time courses, liquid YEPD medium was inoculated with 1 × 106 conidia of the strain of interest per milliliter and grown in shaking culture at 37°C, while exposed to light. To determine the time of acquisition of developmental competence, we used a modification of the method of Axelrod et al. (1973). Briefly, conidia of strain Af293 were germinated in liquid YEPD as described above, and serial aliquots were subcultured to solid YEPD agar and incubated at 37°C. The duration of time required for the production of visible conidiophores for each sample was recorded. Precompetent hyphae require a period of maturation before responding to induction (subculture to solid medium), which manifests as an increase in the delay between induction and conidiophore production. In contrast, competent hyphae can immediately respond to the induction stimulus and therefore produce conidiophores with a fixed delay after induction regardless of the cultures age. Therefore, the acquisition of developmental competence was defined as the time of growth in submerged culture after which the delay between induction and production of conidiophores became constant.
Molecular Genetic Manipulations
Total A. fumigatus RNA and DNA were isolated as described previously (Monroy and Sheppard, 2005). The genomic sequence of stuA was identified by BLAST search of the unfinished A. fumigatus genome sequence assembly (http://tigrblast.tigr.org/er-blast/index.cgi?project=afu1). The predicted open reading frame of stuA was identified via the Institute for Genomic Research annotation pipeline using the Eukaryotic Genome Control software package (Nierman et al., 2005). The stuA gene was cloned by high-fidelity PCR using primers S1 and S2 (Table 1), and the resulting amplicon was cloned into pGEM-T-Easy (Promega, Madison, WI) for sequencing. This product was used as a probe for subsequent Northern and Southern blotting experiments.
Table 1.
PCR primers used in this study
| Primer name | Target gene | Sequence 5′–3′ |
|---|---|---|
| F1 | stuA | TCGAGAGCGAACAGTTGGTA |
| F2 | stuA | TCCTGTGTGAAATTGTTATCCGCT |
| CCAACACCTCGGTTGGTAGT | ||
| F3 | stuA | GTCGTGACTGGGAAAACCCTGGCG |
| GGTGTTCTGACCTCGCTGAT | ||
| F4 | stuA | CTCTGTCCTCGCTTTCTTGC |
| M13F | hph | CGCCAGGGTTTTCCCAGTCACGAC |
| M13R | hph | AGCGGATAACAATTTCACACAGGA |
| HY | hph | GGATGCCTCCGCTCGAAGTA |
| YG | hph | CGTTGCAAGACCTGCCTGAA |
| S1 sense | stuA | ACGATTGATCATTGCCTTG |
| S2 antisense | stuA | AACGAATGCAAAGTCGGAAG |
| StuA RT sense | stuA | GAGGACGAAGGGAGTCTCTG |
| StuA RT antisense | stuA | ACCGTTGATCATGTGGTTGT |
| TEF1RT sense | TEF1 | CCATGTGTGTCGAGTCCTTC |
| TEF1RT antisense | TEF1 | GAACGTACAGCAACAGTCTGG |
| CC9 sense | Clock controlled gene 9 | ACGTCTTGGACTCGTATCCC |
| CC9 antisense | Clock controlled gene 9 | GGCGAGATGGGACATTACTT |
| IME2 sense | IME2 | CAGAATTCCGACTTGAACGA |
| IME2 antisense | IME2 | GATTTGGACAGGGACGTTTC |
| CAT1 sense | CAT1 | CGCTGAGACCGAACAAGTTA |
| CAT1 antisense | CAT1 | GCTGAGTGTCCAGGTACGAA |
| IgE sense | IgE binding protein | CGAAGGCACTCAAACTGTTC |
| IgE antisense | IgE binding protein | GAGTGAGCGGTGGTGAACT |
| RosA sense | Repressor of sexual development | ATAGGGACACCAGAGAAGGC |
| RosA antisense | Repressor of sexual development | ATCACACTTCTTCCTTCGCA |
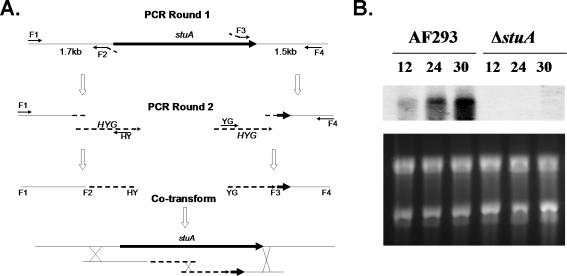
Transformation of A. fumigatus by electroporation is simple and rapid, but it is hampered by a low rate of homologous integration, making targeted integration a challenge (Sheppard et al., 2004). We therefore used a split marker electroporation strategy to generate a stuA-deficient strain of A. fumigatus (Figure 1A) (Catlett et al., 2003). Briefly, A. fumigatus strain Af293 was cotransformed with two DNA constructs, each containing an incomplete fragment of the dominant selection marker HYG (hygromycin phosphotransferase) fused to 1 kb of stuA flanking sequence. These marker fragments shared an approximately 450-bp overlap within the HYG cassette, which served as a potential recombination site during transformation. During transformation, homologous integration of each fragment into the genome flanking stuA allows recombination of the HYG fragments and generation of the intact resistance gene at the site of recombination. Each fragment was generated by two rounds of PCR. First, each flanking region was amplified from Af293 genomic DNA using primer pairs F1-F2 and F3-F4. These produced 1-kb regions of stuA flanking sequence, fused to a 24-bp conserved M13 sequence at their respective internal ends. In parallel, the HYG resistance cassette was amplified from plasmid pAN7-1 by M13F and M13R primers. To generate the final transformation fragments, fusion PCR was performed to combine each flanking fragment with the M13 HYG resistance cassette and amplifying using the F1-HY, and the YG-F4 primer set. This produced two stuA flanking regions, each linked by M13 sequences to incomplete fragments of the HYG resistance cassette. Strain Af293 was then transformed by electroporation using 5 μg of each fragment, and the transformants were retrieved by hygromycin selection. Deletion of stuA was confirmed by PCR (our unpublished data) and Northern blotting to ensure a complete absence of stuA mRNA (Figure 1B). To generate a stuA-complemented strain, an intact stuA insert that overlapped the deletion as well as 1.5 kb of each intact flank was generated using high-fidelity PCR with primers F1 and F4. The resulting amplicon was cloned into pGEM-T and sequence verified. The stuA deletion mutant was then transformed with 10 μg of this insert by spheroplasting and regenerated without selection. The resulting transformants were screened for clones that displayed normal conidiation. Transformants that exhibited normal conidiation were tested by PCR to ensure correct integration of the amplicon at the stuA native locus and subsequent loss of the HYG cassette then by Northern blot to ensure normal restoration of stuA transcription (our unpublished data).
Figure 1.
Disruption of stuA. (A) Split marker strategy for the deletion of stuA. Two deletion fragments were generated by successive rounds of PCR. First flanking regions were amplified containing M13 sequences at the stuA flanking ends. Next, a fusion PCR was performed using each fragment and the HYG resistance cassette, both containing homologous M13 sequences, as template to generate the final deletion fragments. Finally, Af293 was cotransformed with the two deletion fragments. A triple crossover event resulted in replacement of stuA by the HYG resistance cassette. F1, F2, F3, F4, HY, and YG, primers used for amplification. HYG, hygromycin resistance cassette amplified from pAN7-1. (B) Northern blot of Af293 and the stuA null mutant strain. Total RNA from hyphae grown in YPD at 37°C was probed with the entire open reading frame of stuA, confirming an absence of stuA mRNA in the stuA null mutant strain.
Transcriptional Profiling
To identify genes that were differentially regulated during developmental competence, we determined the transcriptional profile of A. fumigatus during germination and hyphal growth in liquid YEPD at 37°C, while exposed to light. RNA was extracted from wild-type Af293, the stuA null mutant, and the stuA complemented strain after 8, 24, and 30 h of growth. After 8 h, the organisms had formed short, precompetent hyphae. At the later time point, the organisms had formed long hyphae that were developmentally competent.
The A. fumigatus Af293 DNA amplicon microarray containing 9516 genes (Nierman et al., 2005) was used in this study. Labeling reactions with RNA, and hybridization were conducted as described in The Institute for Genomic Research (TIGR, Rockville, MD) standard operating procedures found at http://www.tigr.org/tdb/microarray/protocolsTIGR.shtml. In the first set of experiments, the sample from 8 h served as reference in all hybridizations with samples from later time points within each strain to identify genes exhibiting altered transcription in competent hyphae. In addition, in separate experiments, direct hybridizations were performed using RNA pools from wild-type Af293 and the ΔstuA mutant strain after 12, 24, and 30 h of growth. All the hybridizations were repeated in dye-swap sets. Hybridized slides were scanned using the Axon GenePix 4000B microarray scanner and the TIFF images generated were analyzed using TIGR Spotfinder (http://www.tigr.org/software/; TIGR, Rockville, MD) to obtain relative transcript levels. Data from TIGR Spotfinder were stored in MAD, a relational database designed to effectively capture and store microarray data. Data were normalized using a local regression technique LOcally WEighted Scatterplot Smoothing (LOWESS) for hybridizations using a software tool MIDAS (http://www.tigr.org/software; TIGR). The resulting data were averaged from triplicate genes on each array and from duplicate flip-dye arrays for each experiment, taking a total of six intensity data points for each gene. Differentially expressed genes at the 95% confidence level were determined using intensity-dependent Z-scores (with Z = 1.96) as implemented in MIDAS for all experiments. To identify genes that were differentially expressed during the acquisition of developmental competence, we identified all genes exhibiting differential expression in both the wild-type and stuA-complemented strain at 24 or 30 h. Next, the resulting data were organized and visualized using k-means algorithm to find the genes that are differentially expressed between the wild type and the ΔstuA mutant. The results of this analysis were then compared with the list of genes exhibiting differential expression in the direct hybridizations of RNA from the wild type and the ΔstuA mutant at 24 and 30 h of growth. Genes that were found to have significantly different expression in both sets of experiments were then organized based on similar expression vectors using Euclidean distance and hierarchical clustering with average linkage clustering method with TIGR MEV (http://www.tigr.org/software; TIGR).
Real-Time RT-PCR
To test that the genes identified by the microarray studies were both developmentally regulated and stuA dependent, we performed real-time RT-PCR analysis using five genes. The primers used for each gene are shown in Table 1. To obtain RNA, YEPD broth was inoculated with 1 × 106 conidia of either strain Af293 or the ΔstuA mutant and incubated at 37°C. Samples were removed at 8 and 24 h, and total RNA was extracted as in the microarray experiments. Samples were then treated with TurboDNase (Ambion, Austin, TX) before first strand synthesis with RetroScript (Ambion). Real-time PCR was then performed using an ABI 7000 thermocycler (Applied Biosystems, Foster City, CA), and amplification products were detected with SYBR Green (QIAGEN, Valencia, CA). Gene expression was then normalized to A. fumigatus TEF1 expression, and relative expression was estimated using the formula 2ΔCt, where ΔCt = [Cttarget gene - CtTEF1]. To verify the absence of genomic DNA contamination, negative controls were used for each gene set in which reverse transcriptase was omitted from the mix.
Hydrogen Peroxide Susceptibility Assay
To determine the effects of stuA deletion on susceptibility to oxidative stress, we tested the susceptibility of conidia and hyphae to hydrogen peroxide. Conidia were harvested as described above, and either used directly (for conidia sensitivity) or allowed to germinate in Sabouraud dextrose broth at 37°C for 8 h. Both conidia and germlings were suspended to a concentration of 105 cells/ml, and 10 μl of suspension was added to microtiter wells containing hydrogen peroxide (0-100 mM) and incubated for 30 or 60 min. At each time point, an aliquot was removed and replica-plated to YEPD agar for recovery. Plates were incubated overnight at 37°C and examined for growth.
Virulence Studies
The virulence of the ΔstuA mutant was compared with that of the wild-type Af293 strain and stuA-complemented strain in an intranasal model of invasive murine aspergillosis. Male BALB/c mice (National Cancer Institute, Bethesda. MD), weighing 18-22 g, were immunosuppressed with 250 mg/kg cyclophosphamide (Western Medical Supply, Oklahoma City, OK) and cortisone acetate (Sigma-Aldrich, St. Louis, MO) 2 d before infection. For each strain, 10 mice were then anesthetized with isoflurane and infected by intranasal instillation of 106 conidia of each strain in 25 μl of phosphate-buffered saline (PBS) + 0.1% Tween. A second dose of immunosuppression was given 3 d of infection consisting of 200 mg/kg cyclophosphamide and 250 mg/kg cortisone acetate. Mice were monitored for signs of illness and time to euthanasia was recorded. Differences in survival between experimental groups were compared using the log-rank test. All procedures involving mice were approved by the Institutional Animal Use and Care Committee, according to the National Institutes of Health guidelines for animal housing and care.
RESULTS
Identifying the Onset of Developmental Competence in A. fumigatus
Because stuA expression has been linked to the acquisition of developmental competence (Miller et al., 1991), it was critical to determine the time required for the onset of this developmental stage in A. fumigatus strain Af293. When submerged hyphae were subcultured to solid agar, the delay between induction by exposure to the air-surface interphase, and production of visible conidiophores decreased with increasing hyphal maturation. However, after 9-10 h of hyphal growth, a fixed delay of 4.5 h between exposure to the air-solid interphase and conidiophore production was observed. Thus, for the conditions used in this study, developmental competence occurred after 9-10 h of submerged growth.
Cloning and Transcriptional Characterization of A. fumigatus stuA
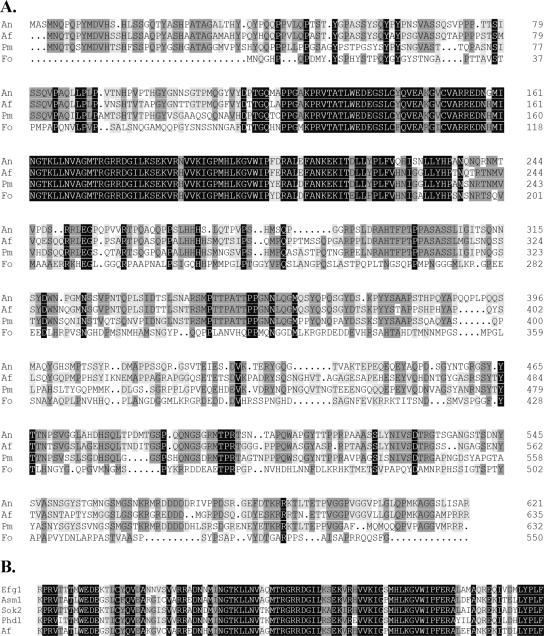
BLAST searches of the ongoing A. fumigatus genome sequencing project identified a single putative open reading frame with high homology to the A. nidulans stuA gene. The predicted protein sequence of this protein contains a bHLH domain that is highly similar to that of the other APSES proteins (Figure 2, A and B). Surprisingly, this domain is slightly more closely related to the Penicillium marneffei stuA (Borneman et al., 2002) rather than that of A. nidulans (Miller et al., 1992), although the overall homology of the predicted proteins is higher between the A. nidulans and A. fumigatus proteins (Figure 2A). This sequence has been subsequently deposited with GenBank by the genome sequence project under accession no. EAL93087.
Figure 2.
Sequence alignments of the members of the APSES protein. (A) Alignment of the predicted protein sequence of the StuA proteins identified from filamentous fungi. An, A. nidulans; Af, A. fumigatus; Pm, Penicilllium marneffei; and Fo, Fusarium oxysporum. (B) Alignment of the basic-helix-loop-helix domains of A. fumigatus StuAp with other APSES family members. Efg1 from C. albicans, Asm1 from Neurospora crassa; and Phd1 and Sok2 from S. cerevisiae.
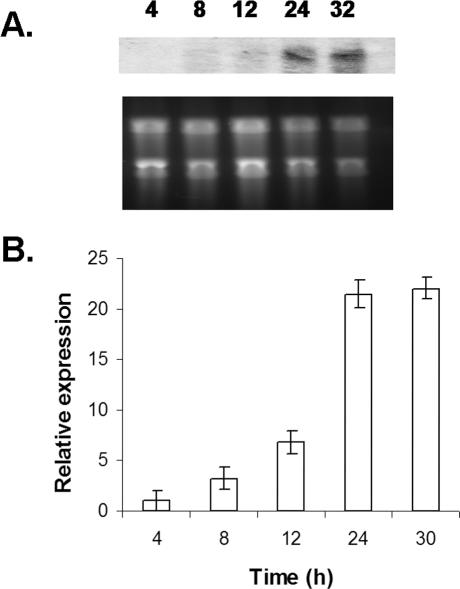
Transcriptional analysis by Northern blotting and realtime RT-PCR during germination and hyphal maturation in submerged culture demonstrated a marked increase in stuA transcription after 12 h (Figure 3), corresponding to the onset of developmental competence. This expression of stuA mRNA was maximal by 24 h of growth and this gene was highly expressed in competent hyphae at all subsequent time points that we have examined. This pattern of transcription is similar to that seen with stuA in A. nidulans (Miller et al., 1992), suggesting that the proteins encoded by these two genes likely share some similarities in function.
Figure 3.
Time course of stuA expression. (A) Northern blot of stuA expression during germination and hyphal development. Total RNA was extracted from organisms growing at 37°C in YEPD broth. Top, blot probed with the entire stuA open reading frame. Bottom, total RNA stained with ethidium bromide. (B) Real-time RT-PCR of stuA expression in an independent experiment. Total RNA was extracted from identical time points, and stuA expression normalized to A. fumigatus TEF1 is displayed on the y-axis. Error bars indicate SD.
Morphological Effects of stuA Deletion
To determine the function of StuAp in A. fumigatus, we constructed a ΔstuA mutant by gene deletion. The resulting ΔstuA mutant was then complemented by transformation with an intact copy of stuA, to ensure that the observed phenotypes were specifically a result of the deletion of stuA.
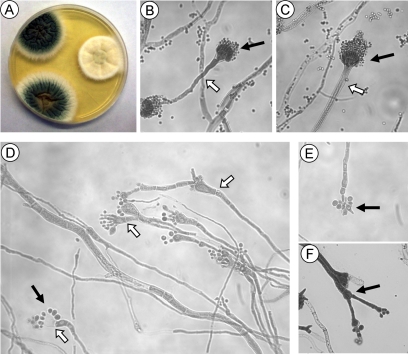
Deletion of stuA produced a dramatic morphological phenotype (Figure 4). Wild-type A. fumigatus colonies produced a vivid green color due to the production of abundant pigmented spores borne on condiophores (Figure 4A). In contrast, strains deficient in stuA produced whitish colonies, which became slightly yellow on prolonged incubation. The explanation for this difference was readily apparent upon microscopic examination. The ΔstuA mutants displayed markedly abnormal conidiophores (Figure 4, D-F). First, the broad, aseptate conidiophore stalk upon which the vesicle is borne was completely absent from the ΔstuA mutant (Figure 4D). The vesicles were produced directly from septate hyphae, and although relatively normal in shape, they were often lacking phialides entirely (Figure 4D). When phialides were present, they were swollen and dysmorphic (Figure 4D). Furthermore, secondary conidiophores were observed emerging from some vesicles, often producing conidia themselves (Figure 4F). The number of conidia was greatly reduced compared with wild-type strain Af293. Conidia were not only produced by phialides as in wild-type A. fumigatus but also were seen budding directly from the vesicle head (Figure 4D), and even from what seemed to be normal hyphae without any detectable vesicle production (Figure 4E). These ΔstuA mutant conidia were approximately twice the size of normal conidia when examined microscopically, and they varied in both shape and size (Figure 4, D-F). Complementation of the ΔstuA mutant with a wild-type allele of stuA restored normal conidiation (Figure 4C), although the conidia produced by this strain were smaller than the original parent Af293.
Figure 4.
Morphology of ΔstuA mutant. (A) Colonial morphology of A. fumigatus strains Af293 (top left), ΔstuA mutant (middle right), and stuA-complemented strain (bottom left) grown for 4 d at 37°C on YEPD agar. The ΔstuA mutant strain displays markedly impaired conidiation in vitro. (B-F) 400× magnification view of conidiophores. (B) Wild-type strain Af293 produces long aseptate conidiophore (open arrow) and chaining conidia produced by phialides (solid arrow). (C) The stuA-complemented strain produces normal conidiophores indistinguishable from strain Af293. (D-F) Conidophores of the ΔstuA mutant strain. (D) The ΔstuA mutant produces abnormally short to absent conidiophores (open arrows) and conidia that often emerge directly from the vesicle (solid arrow). (E) Production of condia by the ΔstuA mutant in the absence of a visible vesicle. (F) Compound conidiophores with conidiophores emerging from the vesicle head (solid arrow).
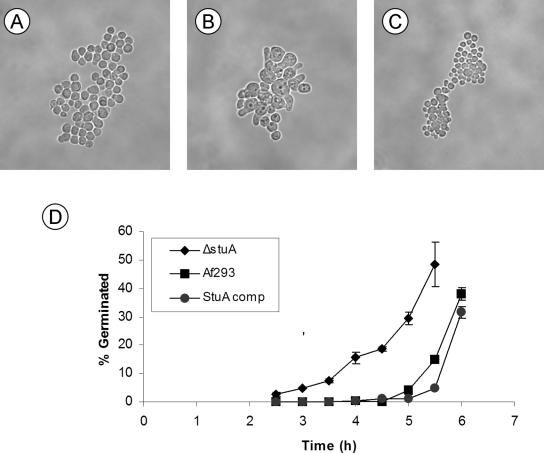
The ΔstuA mutant also had accelerated germination. Wild-type Af293 conidia and the stuA-complemented strain produced visible germ tubes after 6-8 h of incubation at 37°C. In contrast, germ tubes of the ΔstuA were observed as early as 3-4 h of incubation (Figure 5, A and B). The germ tubes and hyphae of the ΔstuA mutant were indistinguishable from that of wild-type Af293 or the complemented strain. Radial growth on solid media was also similar among the three strains.
Figure 5.
Precocious germination of ΔstuA mutants. Photomicrograph (400× magnification) of conidia grown for 4 h in YEPD broth at 37°C. (A) Wild-type Af293. (B) ΔstuA mutant strain showing germination and early hyphal formation. (C) stuA-complemented strain. (D) Time course of germination of conidia in YEPD broth at 37°C. Samples removed at each time interval were examined microscopically, and the number of conidia with visible germ tubes were counted. A minimum of 100 organisms were counted in triplicate for each time point. Error bars indicate SD of three separate experiments.
Given the impaired conidiation of the ΔstuA mutant, and the correlation of stuA transcription with developmental competence, we tested the effects of stuA deletion on the time course of developmental competence. Conidiophore production by the ΔstuA mutant was markedly delayed compared with wild-type Af293 and the stuA-complemented strain. This delay was attributable to a much slower production of conidiophores after induction (8-9 h for the ΔstuA strain compared with 4-5 h for the wild-type and stuA-complemented strain). The time to acquisition of developmental competence by the ΔstuA strain was not significantly different from the wild-type or stuA-complemented strain. Collectively, these data suggest a model whereby StuAp does not govern developmental competence but rather functions as an effector of this process.
Transcriptomes during the Acquisition of Developmental Competence
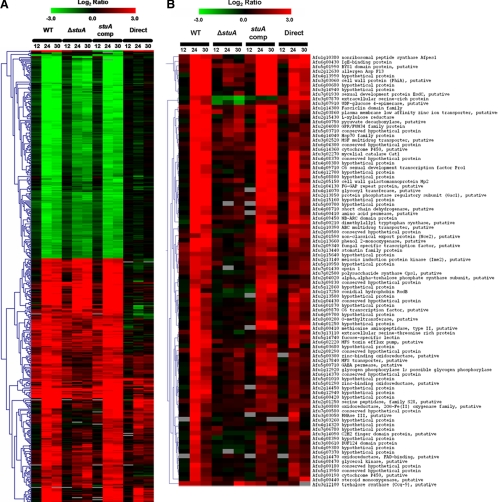
Transcriptional profiling is ideally suited to the study of development in Aspergillus sp. Developmental competence occurs in a highly coordinated manner within growing cultures and is unaffected by nutritional deficiencies (Axelrod et al., 1973; Pastushok and Axelrod, 1976). Furthermore, the majority of known developmental genes in Aspergillus spp. exhibit transcriptional regulation (Adams et al., 1992). Therefore, to identify genes whose expression was linked to developmental competence, we performed whole-genome transcriptional profiling of the wild-type and stuA-complemented strains. Comparing the expression of genes between precompetent (8 h) and postcompetent hyphae (both at 24 and 30 h) identified 720 differentially expressed genes in the wild-type strain Af293, and 597 genes in the stuA-complemented strain. The subset of 445 genes that were found to be differentially expressed in both strains was used in subsequent analysis.
Next, to identify which of these developmentally regulated genes were dependent on StuAp, we performed transcriptional profiling of the ΔstuA mutant using the same time points as described above. Of the 445 competence-associated genes identified in the wild-type hybridizations, 104 genes differed in expression level between the ΔstuA mutant and both the wild-type and complemented strains at 24 and 30 h (when stuA expression is maximal). To confirm these results, we next performed direct hybridizations of RNA isolated from wild-type and ΔstuA hyphae after 24 and 30 h of incubation. Of the 104 candidate stuA-dependent genes identified in the previous experiments, 94 of these again showed a statistically significant difference in expression between the wild-type and the ΔstuA mutant at both 24 and 30 h. Hierarchical cluster analyses of all developmental genes, and of the stuA-dependent developmental genes, are presented in Figure 6, A and 6B.
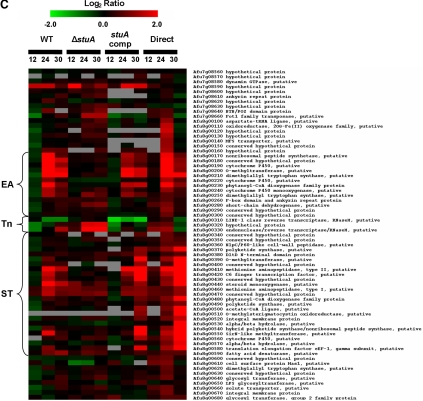
Figure 6.
Transcriptional analysis during the acquisition of developmental competence. (A) Hierarchical clustering of genes displaying significantly different expression (Z > 1.96) after the onset of developmental competence in both wild-type Af293 and the stuA-complemented strain. (B) Hierarchical clustering of genes showing stuA-dependent expression during the onset of developmental competence. (C) Comparative expression of genes comprising the stuA-dependent putative biosynthetic cluster, arranged by genomic location. For all figures, the mean-fold changes (log2) in gene expression are represented by colored squares. Red blocks indicate up-regulation of the gene of interest in the test pool compared with the control pool and green blocks indicate down-regulation. The first nine columns present the relative transcript level at each time point compared with the expression at 8 h for the same strain. The last two columns indicate the relative transcript level comparing the ΔstuA mutant and wild-type Af293 directly at each time point. Cluster groupings on the left (A and B) indicate similar gene expression trends within groups of significant genes. WT, wild-type strain Af293; ΔstuA, ΔstuA mutant strain; stuA comp, stuA-complemented strain; Direct, direct comparison of wild-type and ΔstuA RNA; EA, putative ergot alkaloid synthetic subcluster; Tn, putative retrotransposon; and ST, putative sterigmatocystin/aflatoxin synthetic subcluster.
The stuA-dependent developmental genes identified by the microarray studies could be broadly classified into several groups according to their predicted function (Table 2). As expected, given the role of stuA in conidiation, this included genes encoding putative proteins either known or expected to be involved in the regulation of morphogenesis and development and genes encoding proteins involved in cell metabolism. Interestingly, several genes encoding enzymes involved in secondary metabolite biosynthesis and export were identified as well as the genes encoding proteins that are antigenic during invasive and allergic disease.
Table 2.
Subset of StuAp-dependent developmentally regulated genes identified by microarray studies arranged by putative function
| Functional group | Locus | Common name/description |
|---|---|---|
| Development and morphogenesis | Afu3g03060 | PhiA protein |
| Afu7g01930 | ESDC | |
| Afu7g01430 | Opsin 1 | |
| Afu2g13140 | Meiosis induction protein kinase ime2/sme1 | |
| Afu4g09710 | Repressor of sexual development/Pro1 | |
| Antigenic proteins | Afu2g12630 | rAsp f 13 |
| Afu2g05150 | Cell wall galactomannoprotein MP2 | |
| Afu6g00430 | IgE-binding protein | |
| Afu1g17250 | Conidial hydrophobin RodB | |
| Metabolism | Afu2g15430 | l-Xylulose reductase |
| Afu2g04020 | α,α-Trehalose phosphate synthase subunit | |
| Afu2g13850 | Protein phosphatase regulatory subunit (Gac1) | |
| Afu3g12100 | Trehalose synthase (Ccg-9) | |
| Afu4g08710 | Short chain dehydrogenase | |
| Afu2g04080 | GPR/FUN34 family protein | |
| Afu6g08470 | Glycerol kinase | |
| Afu1g12920 | Glycogen phosphorylase 1 | |
| Afu3g07910 | UDP-glucose 4-epimerase | |
| Afu6g00750 | Pyruvate decarboxylase | |
| Secondary metabolite biosynthesis | Afu6g14360 | Cytochrome P450 |
| Afu1g13660 | Phenol 2-monooxygenase | |
| Afu4g14070 | Glycosyl transferase | |
| Afu1g10380 | Nonribosomal peptide synthase Afpes1 | |
| Afu3g00800 | Oxidoreductase, 2OG-Fe(II) oxygenase family, putative | |
| Afu5g00300 | Zinc-binding oxidoreductase, putative | |
| Afu3g02270 | Mycelial catalase Cat1 | |
| Afu5g01290 | Zinc-binding oxidoreductase, putative | |
| Afu2g14470 | Oxidoreductase, FAD-binding, putative | |
| Afu8g00190 | Cytochrome P450 | |
| Afu8g00210 | Dimethylallyl tryptophan synthase | |
| Afu8g00200 | O-Methyltransferase | |
| Afu8g00440 | Steroid monooxygenase | |
| Afu8g00410 | Methionine aminopeptidase, type II | |
| Transport | Afu1g10390 | ATP-binding cassette (ABC) multidrug transporter |
| Afu2g01590 | Nonclassical export protein (Nce2) | |
| Afu2g03860 | Plasma membrane low affinity zinc ion transporter | |
| Afu2g17840 | Major facilitator superfamily (MFS) transporter | |
| Afu3g02520 | MSF multidrug transporter | |
| Afu5g00710 | GABA permease | |
| Afu6g02220 | MFS toxin efflux pump | |
| Afu6g00410 | Amino acid permease |
Six of the stuA-dependent genes identified by the microarray analysis were found to be clustered in proximity on chromosome 8. Examination of this region revealed a putative biosynthetic gene cluster containing at least 36 hypothetical genes. These genes are predicted to encode a variety of proteins required for the biosynthesis of both sterigmatocystin/aflatoxin and ergot alkaloid metabolites (Figure 6C). This cluster is interrupted by the presence of a retrotransposon-like element that is found in at least three other locations within the genome (Afu3g09410-30, Afu4g02620-40, and Afu4g14860-70, identified by BLAST search of the A. fumigatus genome at http://tigrblast.tigr.org/er-blast/index.cgi?project=afu1). With the exception of the putative retrotransposon element, the majority of the genes in this cluster exhibit of stuA-dependent up-regulation during the onset of developmental competence (Figure 6C).
Real-Time Reverse Transcription (RT)-PCR Confirmation of Transcriptomes
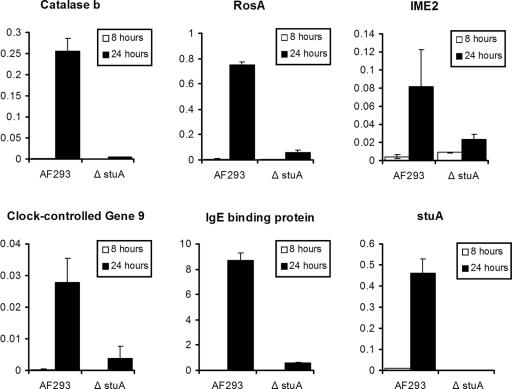
To test the accuracy of the microarray experiments, we used real time RT-PCR to analyze the expression of five putative members of the stuA-regulated developmental program. These results (Figure 7) confirmed that all five of these genes were up-regulated during the acquisition of developmental competence by wild-type A. fumigatus. Furthermore, this induction was significantly reduced or absent for all five of these genes in the ΔstuA mutant strain.
Figure 7.
Real-time RT-PCR of stuA dependent developmental genes. The expression of five candidate stuA-dependent developmental genes was analyzed using RNA extracted from hyphae grown for 8 h (precompetent) and 24 h (postcompetent) in YEPD broth at 37°C. Relative expression (normalized to A. fumigatus TEF1 expression) is shown on the y-axis. Error bars indicate the SD for each result. All five genes exhibited stuA-dependent up-regulation in developmentally competent hyphae. stuA expression in both strains is shown for comparison in the final graph.
Susceptibility of ΔstuA Mutants to Oxidative Stress
The results of the microarray analysis and real-time PCR studies provided strong evidence that competent hyphae of ΔstuA strains are deficient in catalase production. To determine the functional significance of these results, we compared the susceptibility of the ΔstuA strain to the wild-type and complemented strain. Hyphae of the ΔstuA mutant were markedly more susceptible to oxidative stress from hydrogen peroxide than either the wild-type or stuA-complemented strain (Figure 8). Even the lowest concentration of hydrogen peroxide, for the shortest exposure, completely inhibited subsequent growth of the organism. These results mirror the observations from the transcriptional profiling experiments, suggesting that for StuAp governs the response to oxidative stresses in competent hyphae by increasing expression of hyphal catalase.
Figure 8.
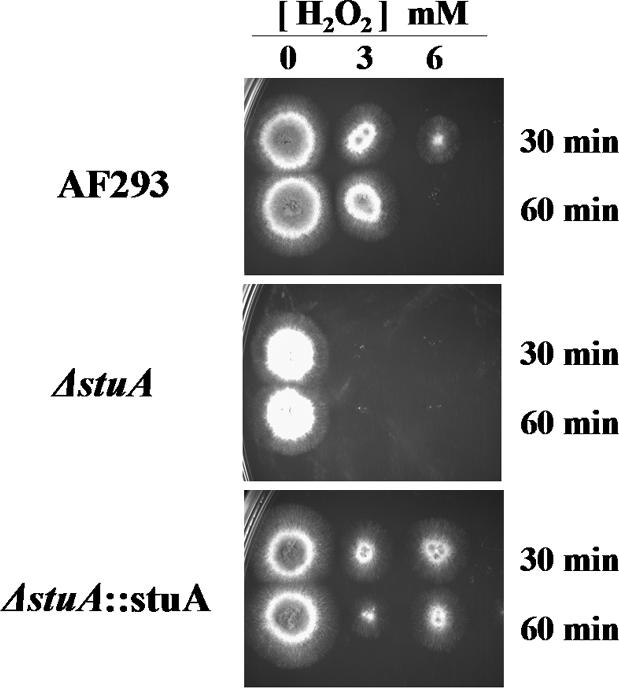
Hyphae from ΔstuA mutants are hypersusceptible to hydrogen peroxide. Hyphae of wild-type Af293, ΔstuA mutant, and stuA-complemented strains were exposed to varying concentrations of hydrogen peroxide for 30 or 60 min. Aliquots were then removed and plated to YEPD agar and incubated overnight at 37°C.
Virulence of the ΔstuA Mutant Strain
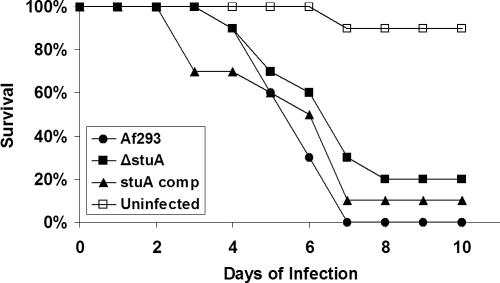
The ΔstuA mutant strain displayed altered regulation of multiple genes that may play a role in virulence. We therefore compared the virulence of the ΔstuA mutant strain to the wild-type Af293 and the stuA complemented strain during experimental murine pulmonary aspergillosis. Mice infected with the stuA mutant exhibited a trend toward increased survival compared with both wild-type and the stuA-complemented strain (Figure 9); however, this difference was not statistically significant (p = 0.1 and 0.4, respectively, by the log-rank test).
Figure 9.
Virulence of ΔstuA mutants during murine pulmonary infection. Immunosuppressed mice were infected intranasal with 1 × 106 conidia of each strain and monitored daily. ΔstuA, ΔstuA null mutant; Af293, wild-type parent strain; stuA comp, stuA-complemented strain; and uninfected, sham infection using PBS + 0.1% Tween alone.
DISCUSSION
The initiation of A. fumigatus stuA transcription coincides with the onset of developmental competence, and after 24 h remains relatively constant in competent hyphae during all time points tested. Disruption of stuA produced a dramatic oligo-conidial phenotype with reduced or absent conidiophores, few to no phialides, and dysmorphic conidia. These results parallel the abnormalities seen in stuA-deficient A. nidulans (Miller et al., 1991), although the production of conidia in the absence of vesicle heads, and the production of compound vesicles (conidiophores arising from vesicle heads) has not been described in these mutants. In addition, the rapid germination of A. fumigatus ΔstuA conidia has not been reported in A. nidulans ΔstuA strain, although it is unclear whether this phenomenon has been specifically examined.
A. nidulans has long served as a model system for studying the regulation of conidial development. The majority of this work, however, has focused on the events surrounding conidiation, and not the molecular events underlying the acquisition of developmental competence. In contrast, because A. fumigatus does not undergo conidiation during invasive disease, the preconidiation stage of developmental competence is of great interest from the perspective of pathogenicity. Indeed, recent studies from our group have found that hyphae from mice infected with A. fumigatus express stuA in vivo (Doedt et al., 2005), suggesting that competent hyphae are present during infection
We therefore performed whole-genome transcriptional analysis to identify the genes displaying differential regulation during the acquisition of developmental competence and to identify the subset that exhibit stuA dependence. Although microarrays are a powerful tool for the identification of transcriptional programs, validation of these results is critical. We used multiple complementary strategies to ensure that the results of these experiments were robust, including dye-swaps, in-slide triplication of hybridizations target spots, the use of the complemented and wild-type strain for biological replicates, and an experimental design that used both time-course studies as well as direct hybridizations between strains. Furthermore, several lines of evidence support the validity of these data. In addition, direct verification of expression by real-time PCR using RNA from an independent experiment was performed. These results confirmed that the expression of all five candidate genes was both developmentally regulated and stuA dependent. Finally, the hypersusceptibility of the ΔstuA mutant strain to hydrogen peroxide provides a phenotypic confirmation of stuA-dependent hyphal catalase expression. Collectively, these results suggest that the gene list generated by the microarray experiments is an accurate one. Conversely, it is important to note that this list of stuA-dependent genes is not exhaustive. In particular, by focusing our analyses on the later time points of 24 and 30 h (when stuA expression is maximal), genes with transient alterations in expression coincident with the onset of stuA expression, or other stuA-dependent genes that are not developmentally regulated, would not have been identified.
Because much of the A. fumigatus genome remains either unannotated, or annotated only by automated algorithms, a detailed analysis of genes by functional category from the results of microarray studies is not yet possible. Indeed, almost half (48/94) of the stuA-dependent genes identified in these experiments are of unknown function. However even given these limitations, interesting patterns of gene expression can be discerned. As expected, several stuA-dependent genes were homologous to genes known to be involved in the developmental regulation of other fungi, including a homologue of the Saccharomyces cerevisiae IME2, which positively regulates meiosis during sporulation (Smith and Mitchell, 1989). Opsin 1 is homologous to the Nop-1 protein of Neurospora crassa, which encodes a photo-reactive protein involved in light sensing, and whose expression is highest during conidiation (Bieszke et al., 1999). A homologue of the Blastomyces dermatitidis bys1 gene was also identified. This gene encodes a protein of unknown function whose transcription is tightly linked to morphogenesis and is expressed only during the pathogenic yeast phase of this organism (Burg and Smith, 1994; Bono et al., 2001). Expression of rosA was stuA dependent and associated with developmental competence in A. fumigatus. Interestingly, in A. nidulans, high level expression of rosA was found only after the initiation of conidiation and not in submerged competent hyphae (Vienken et al., 2005). In addition, deletion of rosA was associated with increased stuA expression, suggesting either a direct or feedback regulatory link between these two proteins (Vienken et al., 2005). A novel finding was the decreased expression of a phiA homologue in the stuA mutant during development. The cell wall protein encoded by the phiA gene of A. nidulans plays a critical role in the development of normal phialides, with phiA-deficient mutants displaying abnormal phialides and marked reductions in conidiation (Melin et al., 2003). Whether the abnormal phiA expression in stuA null mutants is responsible for, or results from, the marked abnormalities in conidiphore growth is not known. Finally, clock-controlled gene 9 encodes a trehalose synthase required for normal conidiation in response to light cycling in N. crassa (Shinohara et al., 2002)
Several other stuA-dependent developmental genes are predicted to encode proteins that are involved in the immune response to A. fumigatus during allergic or invasive disease. The elaboration of IgE-binding proteins by A. fumigatus (including rAsp f13, an alkaline serine protease; and IgE-binding protein Afu6g00430) is a key stimulus for the development of allergic bronchopulmonary aspergillosis (Kurup et al., 2002; Stevens et al., 2003), suggesting that StuAp may play an important role in the pathogenesis of this condition. MP2 encodes a surface protein in A. fumigatus that elicits a specific antibody response during invasive disease, although its role in protective or allergic immunity has not yet been defined (Chong et al., 2004). Finally, rodB encodes a hydrophobin that does not mediate rodlet formation or oxidative resistance in conidia of A. fumigatus (Paris et al., 2003a). The function of this protein in developmentally competent hyphae however is unknown. In other filamentous fungi, expression of hyphal hydrophobins is required for the production of aerial hyphae or normal conidiophores (Fuchs et al., 2004). These hydrophobins can also contribute to virulence by mediating adherence to host tissues (Talbot et al., 1996). Collectively, these data suggest that stuA may regulate virulence-associated phenotypes during both invasive and allergic disease.
Perhaps the most interesting finding of the transcriptional analysis studies is the identification of a stuA-dependent putative secondary metabolite biosynthesis cluster. Unlike most other genes in Aspergillus spp., genes encoding proteins that mediate the synthesis of these mycotoxins are often found physically linked in clusters such as the aflatoxin/sterigmatocystin biosynthesis clusters of Aspergillus parasiticus, Aspergillus flavus, and Aspergillus. nidulans (Keller and Adams, 1995; Brown et al., 1996; Yu et al., 2004) and the gliotoxin biosynthesis cluster of A. fumigatus (Gardiner and Howlett, 2005). In A. fumigatus, two biosynthetic clusters involved in the synthesis of ergot alkaloids such as fumigaclavine have been identified (Coyle and Panaccione, 2005; Unsold and Li, 2005) as has a polyketide synthesis cluster responsible for the production of conidial pigment (Tsai et al., 1999). A recent genomic analysis of A. fumigatus found at least 26 of these putative clusters throughout the genome of strain Af293 (Nierman et al., 2005).
The composition of the cluster identified in this study is unique in that it contains both genes encoding synthetic enzymes required for both sterigmatocystin/aflatoxin synthesis as well as ergot alkaloid synthesis with each set of genes clustered at one end of the cluster. The ergot alkaloid synthetic arm of the cluster is composed of two dimethylallyl tryptophan synthetases, three cytochrome p450 enzymes, an o-methyltransferase, a nonribosomal peptide synthetase, and several open reading frames encoding proteins of unknown function (Figure 6C). A similar group of genes is found in both other ergot alkaloid synthesis clusters identified from A. fumigatus as well as the archetypal ergot alkaloid synthesis cluster first described in Claviceps purpurea (Tudzynski et al., 1999; Haarmann et al., 2005). Interestingly, although a homologue of the nonribosomal peptide synthetase in the stuA-dependent A. fumigatus cluster is found in C. pupurea, this gene is not found in either of the other A. fumigatus ergot biosynthesis clusters previously described, suggesting that different mycotoxins are produced by individual clusters.
Similarly, the ∼25 putative proteins encoded by the genes in the sterigmatocystin/aflatoxin subcluster are very similar to members of the conserved ∼25 gene sterigmatocystin/aflatoxin synthetic cluster of A. flavus, A. nidulans, and A. parasiticus (Keller and Adams, 1995; Yu et al., 1995, 2004; Brown et al., 1996). All four clusters include monooxygenases, reductases, a polyketide synthase, and a Zn(II)2, Cys6 domain protein homologous to aflR, the pathway-specific transcription factor required for sterigmatocystin/aflatoxin biosynthesis (Figure 6C) (Yu et al., 1996; Fernandes et al., 1998). Interestingly, the presence of an associated ergot alkaloid synthetic cluster in association with this sterigmatocystin cluster has not been reported in any of these Aspergillus species and seems to be unique to A. fumigatus. The specific synthetic products elaborated by both these subclusters remain unknown, and are under study in our laboratory.
The identification of a stuA-dependent biosynthetic cluster provides a new link between development and secondary metabolite production. The mechanisms underlying this regulation are not yet elucidated; however, as noted above, the A. fumigatus sterigmatocystin/aflatoxin biosynthesis subcluster contains an aflR homologue, which exhibits stuA-dependent expression (Figure 6C). In other Aspergillus spp., aflR is required for sterigmatocystin/aflatoxin biosynthetic gene activation (Ayer and Eisenman, 1993; Yu et al., 1996; Fernandes et al., 1998). Thus, stuA-mediated control of the sterigmatocystin/aflatoxin biosynthetic subcluster is likely mediated at least in part by aflR. Further studies will be necessary to confirm how stuA integrates with aflR and other regulatory pathways governing development and secondary metabolite production including G protein-dependent signaling (Yu and Keller, 2005), the global regulator laeA (Bok and Keller, 2004) and the FluG-brlA pathway (Yu and Keller, 2005).
In addition to the identification of individual genes involved in development, the results of the A. fumigatus microarray studies provide insight into the overall role of stuA in the developmental program governing competence. Genes dependent on stuA made up >20% (94/445) of the total number of genes displaying differential regulation during development. Thus, although other regulatory pathways likely contribute significantly to the acquisition of developmental competence, StuAp plays a central role in the regulation of this biological process.
The transcriptional analysis performed in this study also suggests that stuA may function as a transcriptional activator during the acquisition of developmental competence. Indeed, although almost an identical number of genes were up- and down-regulated during the acquisition of developmental competence (Figure 6A), it is striking that all of the stuA-dependent genes identified by these analyses were up-regulated at these time points (Figure 6B) and none were down-regulated. Indeed, approximately one-half of the genes showing increased expression during developmental competence exhibit stuA dependence. These data contrast with studies of conidiation in A. nidulans in which StuAp has been shown to function as a transcriptional repressor (Dutton et al., 1997). One explanation is that StuAp may function as a transcriptional activator at the level of expression seen in competent hyphae and act as a transcriptional repressor at the higher levels seen during conidiation. Similar threshold effects for StuAp function have been shown in A. nidulans, with sexual reproduction requiring higher StuAp expression than asexual reproduction (Wu and Miller, 1997). Furthermore, a similar model of concentration-dependent transcriptional activation and repression has been proposed for other bHLH proteins, including Efg1p in C. albicans (Stoldt et al., 1997) and Myc in mammalian cells (Ayer and Eisenman, 1993; Ayer et al., 1993; Li et al., 1994). Alternately, StuAp may repress an intermediate regulatory element that is itself normally responsible for repression of the rest of the stuA-dependent program. By this model, StuAp would still function as a transcriptional repressor and yet produce upregulation of many downstream elements.
We found that hyphae of ΔstuA mutants were hypersusceptible to hydrogen peroxide and exhibited markedly reduced levels of catB (CAT1) mRNA. Previously, Paris and colleagues found that disruption of catB (CAT1) was not sufficient to render hyphae susceptible to hydrogen peroxide and that disruption of the second mycelial catalase (CAT2) was required (Paris et al., 2003b). Although these results may reflect methodological differences in determining hydrogen peroxide susceptibility, it is likely that the observed increased susceptibility of the ΔstuA mutant to hydrogen peroxide reflects the contribution of other stuA-dependent proteins. Indeed, examination of the microarray data reveals another putative catalase-peroxidase (locus Afu8g01670) that exhibits significant stuA-dependent expression but that was only significant at 12 h of growth and fell below the z-score cutoff by 24 and 30 h (z-scores of 4.67 at 12 h, 1.82 at 24 h, and 1.12 at 30 h in strain Af293), which resulted in exclusion from our initial analysis of stuA-dependent genes.
Finally, when tested in vivo, the ΔstuA mutant exhibited only a nonsignificant trend toward reduced virulence compared with the wild-type parent or the stuA-complemented strain. This result is somewhat surprising in light of the number of putative virulence factors found to have reduced expression in this strain. One possible explanation for these findings is that the accelerated germination of the ΔstuA mutant strain may have contributed to increasing the virulence of this strain despite the loss of several other virulence factors. Indeed, in other fungi, such as C. albicans, the rate of growth and germination is an important independent predictor of virulence (Rieg et al., 1999). Deletion of genes critical for individual stuA-dependent processes, such as those controlling the expression of secondary metabolites, may be helpful to resolve this issue.
In summary, we have demonstrated that not only does the APSES protein StuAp play an important role in governing conidiation of A. fumigatus but also that it is required for the normal expression of a large subset of the genes that are differentially expressed between competent and precompetent hyphae. StuAp is not required for normal virulence in a mouse model of experimental aspergillosis. In contrast to its role as a transcriptional repressor during conidiation, StuAp functions, either directly or indirectly, as a transcriptional activator in developmentally competent hyphae. In addition, we have found evidence for stuA-mediated expression of a unique mycotoxin biosynthetic cluster predicted to mediate production of ergot alkaloid and sterigmatocystin/aflatoxin.
Supplementary Material
Acknowledgments
We are grateful to Bruce Miller for helpful discussions and advice. This project was supported in part with federal funds from the National Institute of Allergy and Infectious Diseases, under Contract No. N01-AI-30041. Construction of the Af293 microarray was funded by National Institute of Allergy and Infectious Diseases Grant R21 AI052236-01A1 (to W.C.N.). D.C.S. is supported by a Clinician Scientist Award from the Canadian Institutes of Health Research and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0617) on October 5, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adams, T. H., Hide, W. A., Yager, L. N., and Lee, B. N. (1992). Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol. Cell. Biol. 12, 3827-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, T. H., Wieser, J. K., and Yu, J. H. (1998). Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod, D. E., Gealt, M., and Pastushok, M. (1973). Gene control of developmental competence in Aspergillus nidulans. Dev. Biol. 34, 9-15. [DOI] [PubMed] [Google Scholar]
- Ayer, D. E., and Eisenman, R. N. (1993). A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 7, 2110-2119. [DOI] [PubMed] [Google Scholar]
- Ayer, D. E., Kretzner, L., and Eisenman, R. N. (1993). Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72, 211-222. [DOI] [PubMed] [Google Scholar]
- Bieszke, J. A., Braun, E. L., Bean, L. E., Kang, S., Natvig, D. O., and Borkovich, K. A. (1999). The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. USA 96, 8034-8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok, J. W., and Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3, 527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono, J. L., Jaber, B., Fisher, M. A., Abuodeh, R. O., O'Leary-Jepson, E., Scalarone, G. M., and Smith, L. H., Jr. (2001). Genetic diversity and transcriptional analysis of the bys1 gene from Blastomyces dermatitidis. Mycopathologia 152, 113-123. [DOI] [PubMed] [Google Scholar]
- Borneman, A. R., Hynes, M. J., and Andrianopoulos, A. (2002). A basic helix-loop-helix protein with similarity to the fungal morphological regulators, Phd1p, Efg1p and StuA, controls conidiation but not dimorphic growth in Penicillium marneffei. Mol. Microbiol. 44, 621-631. [DOI] [PubMed] [Google Scholar]
- Brown, D. W., Yu, J. H., Kelkar, H. S., Fernandes, M., Nesbitt, T. C., Keller, N. P., Adams, T. H., and Leonard, T. J. (1996). Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93, 1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg, E. F., 3rd, and Smith, L. H., Jr. (1994). Cloning and characterization of bys1, a temperature-dependent cDNA specific to the yeast phase of the pathogenic dimorphic fungus Blastomyces dermatitidis. Infect. Immun. 62, 2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett, N. L., Lee, B. N., Yoder, O. C., and Turgeon, B. G. (2003). Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 50, 9-11. [Google Scholar]
- Chong, K. T., Woo, P. C., Lau, S. K., Huang, Y., and Yuen, K. Y. (2004). AFMP2 encodes a novel immunogenic protein of the antigenic mannoprotein superfamily in Aspergillus fumigatus. J. Clin. Microbiol. 42, 2287-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle, C. M., and Panaccione, D. G. (2005). An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl. Environ. Microbiol. 71, 3112-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedt, T., Chiang, L. Y., Sheppard, D. C., and Filler, S. G. (2005). In vivo analysis of Aspergillus fumigatus gene expression determined by real-time RT-PCR. 105th General Meeting of the American Society for Microbiology, Atlanta, GA, F-066.
- Doedt, T., Krishnamurthy, S., Bockmuhl, D. P., Tebarth, B., Stempel, C., Russell, C. L., Brown, A. J., and Ernst, J. F. (2004). APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15, 3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton, J. R., Johns, S., and Miller, B. L. (1997). StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 16, 5710-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, M., Keller, N. P., and Adams, T. H. (1998). Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 28, 1355-1365. [DOI] [PubMed] [Google Scholar]
- Fraser, R. S. (1993). Pulmonary aspergillosis: pathologic and pathogenetic features. Pathol. Annu. 28, 231-277. [PubMed] [Google Scholar]
- Fuchs, U., Czymmek, K. J., and Sweigard, J. A. (2004). Five hydrophobin genes in Fusarium verticillioides include two required for microconidial chain formation. Fungal Genet. Biol. 41, 852-864. [DOI] [PubMed] [Google Scholar]
- Gardiner, D. M., and Howlett, B. J. (2005). Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett. 248, 241-248. [DOI] [PubMed] [Google Scholar]
- Haarmann, T., Machado, C., Lubbe, Y., Correia, T., Schardl, C. L., Panaccione, D. G., and Tudzynski, P. (2005). The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry 66, 1312-1320. [DOI] [PubMed] [Google Scholar]
- Herbrecht, R., et al. (2002). Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N. Engl. J. Med. 347, 408-415. [DOI] [PubMed] [Google Scholar]
- Keller, N. P., and Adams, T. H. (1995). Analysis of a mycotoxin gene cluster in Aspergillus nidulans. SAAS Bull. Biochem. Biotechnol. 8, 14-21. [PubMed] [Google Scholar]
- Kurup, V. P., Xia, J. Q., Shen, H. D., Rickaby, D. A., Henderson, J. D., Jr., Fink, J. N., Chou, H., Kelly, K. J., and Dawson, C. A. (2002). Alkaline serine proteinase from Aspergillus fumigatus has synergistic effects on Asp-f-2-induced immune response in mice. Int. Arch Allergy Immunol. 129, 129-137. [DOI] [PubMed] [Google Scholar]
- Latge, J. P. (1999). Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12, 310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler, K. B., Davidson, R. C., D'Souza, C., Harashima, T., Shen, W. C., Wang, P., Pan, X., Waugh, M., and Heitman, J. (2000). Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. H., Nerlov, C., Prendergast, G., MacGregor, D., and Ziff, E. B. (1994). c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 13, 4070-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, H. J., Kohler, J. R., DiDomenico, B., Loebenberg, D., Cacciapuoti, A., and Fink, G. R. (1997). Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939-949. [DOI] [PubMed] [Google Scholar]
- Marr, K. A., Carter, R. A., Boeckh, M., Martin, P., and Corey, L. (2002). Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood 100, 4358-4366. [DOI] [PubMed] [Google Scholar]
- Melin, P., Schnurer, J., and Wagner, E. G. (2003). Characterization of phiA, a gene essential for phialide development in Aspergillus nidulans. Fungal Genet. Biol. 40, 234-241. [DOI] [PubMed] [Google Scholar]
- Miller, K. Y., Toennis, T. M., Adams, T. H., and Miller, B. L. (1991). Isolation and transcriptional characterization of a morphological modifier: the Aspergillus nidulans stunted (stuA) gene. Mol. Gen. Genet. 227, 285-292. [DOI] [PubMed] [Google Scholar]
- Miller, K. Y., Wu, J., and Miller, B. L. (1992). StuA is required for cell pattern formation in Aspergillus. Genes Dev. 6, 1770-1782. [DOI] [PubMed] [Google Scholar]
- Monroy, F., and Sheppard, D. C. (2005). Taf 1, A class II transposon of Aspergillus fumigatus. Fungal Genet Biol. 42, 638-645. [DOI] [PubMed] [Google Scholar]
- Nierman, W., et al. (2005). Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature (in press). [DOI] [PubMed]
- Paris, S., Debeaupuis, J. P., Crameri, R., Carey, M., Charles, F., Prevost, M. C., Schmitt, C., Philippe, B., and Latge, J. P. (2003a). Conidial hydrophobins of Aspergillus fumigatus. Appl. Environ. Microbiol. 69, 1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris, S., Wysong, D., Debeaupuis, J. P., Shibuya, K., Philippe, B., Diamond, R. D., and Latge, J. P. (2003b). Catalases of Aspergillus fumigatus. Infect Immun. 71, 3551-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H., Myers, C. L., Sheppard, D. C., Phan, Q. T., Sanchez, A. A., Edwards, J. E., Jr., and Filler, S. G. (2005). Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell Microbiol. 7, 499-510. [DOI] [PubMed] [Google Scholar]
- Pastushok, M., and Axelrod, D. E. (1976). Effect of glucose, ammonium and media maintenance on the time of conidiophore initiation by surface colonies of Aspergillus nidulans. J. Gen. Microbiol. 94, 221-224. [DOI] [PubMed] [Google Scholar]
- Rieg, G., Fu, Y., Ibrahim, A. S., Zhou, X., Filler, S. G., and Edwards, J. E., Jr. (1999). Unanticipated heterogeneity in growth rate and virulence among Candida albicans AAF1 null mutants. Infect. Immun. 67, 3193-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, D. C., Ibrahim, A. S., and Edwards, J. E., Jr. (2004). Human mycoses: the role of molecular biology. In: Advances in Fungal Biotechnology for Industry, Agriculture and Medicine, ed. J. S. Tkacz and L. L., New York: Kluwer Academic, 361-384.
- Shinohara, M. L., Correa, A., Bell-Pedersen, D., Dunlap, J. C., and Loros, J. J. (2002). Neurospora clock-controlled gene 9 (ccg-9) encodes trehalose synthase: circadian regulation of stress responses and development. Eukaryot. Cell 1, 33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. E., and Mitchell, A. P. (1989). A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn, A., Bockmuhl, D. P., and Ernst, J. F. (1999a). Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67, 5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn, A., Tebarth, B., and Ernst, J. F. (1999b). Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun. 67, 4655-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, D. A., et al. (2003). Allergic bronchopulmonary aspergillosis in cystic fibrosis-state of the art: Cystic Fibrosis Foundation Consensus Conference. Clin. Infect Dis. 37 (suppl 3), S225-S264. [DOI] [PubMed] [Google Scholar]
- Stoldt, V. R., Sonneborn, A., Leuker, C. E., and Ernst, J. F. (1997). Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16, 1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, N. J., Kershaw, M. J., Wakley, G. E., De Vries, O., Wessels, J., and Hamer, J. E. (1996). MPG1 Encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8, 985-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H. F., Wheeler, M. H., Chang, Y. C., and Kwon-Chung, K. J. (1999). A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181, 6469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski, P., Holter, K., Correia, T., Arntz, C., Grammel, N., and Keller, U. (1999). Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol. Gen. Genet. 261, 133-141. [DOI] [PubMed] [Google Scholar]
- Unsold, I. A., and Li, S. M. (2005). Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 151, 1499-1505. [DOI] [PubMed] [Google Scholar]
- Vienken, K., Scherer, M., and Fischer, R. (2005). The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture. Genetics 169, 619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., and Miller, B. L. (1997). Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 17, 6191-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Bhatnagar, D., and Cleveland, T. E. (2004). Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 564, 126-130. [DOI] [PubMed] [Google Scholar]
- Yu, J., Chang, P. K., Cary, J. W., Wright, M., Bhatnagar, D., Cleveland, T. E., Payne, G. A., and Linz, J. E. (1995). Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol. 61, 2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. H., Butchko, R. A., Fernandes, M., Keller, N. P., Leonard, T. J., and Adams, T. H. (1996). Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet 29, 549-555. [DOI] [PubMed] [Google Scholar]
- Yu, J. H., and Keller, N. P. (2005). Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43, 437-458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.