Abstract
Cellular protein quality control involves a close interplay between molecular chaperones and the ubiquitin/proteasome system. We recently identified a degradation pathway, on which the chaperone Hsc70 delivers chaperone clients, such as misfolded forms of the cystic fibrosis transmembrane conductance regulator (CFTR), to the proteasome. The cochaperone CHIP is of central importance on this pathway, because it acts as a chaperone-associated ubiquitin ligase. CHIP mediates the attachment of a ubiquitin chain to a chaperone-presented client protein and thereby stimulates its proteasomal degradation. To gain further insight into the function of CHIP we isolated CHIP-containing protein complexes from human HeLa cells and analyzed their composition by peptide mass fingerprinting. We identified the Hsc70 cochaperone BAG-2 as a main component of CHIP complexes. BAG-2 inhibits the ubiquitin ligase activity of CHIP by abrogating the CHIP/E2 cooperation and stimulates the chaperone-assisted maturation of CFTR. The activity of BAG-2 resembles that of the previously characterized Hsc70 cochaperone and CHIP inhibitor HspBP1. The presented data therefore establish multiple mechanisms to control the destructive activity of the CHIP ubiquitin ligase in human cells.
INTRODUCTION
The cochaperone and ubiquitin ligase CHIP is a key regulator of protein folding and protein degradation in human cells (reviewed in Höfeld et al., 2001; Esser et al., 2004, 2005). CHIP associates with the molecular chaperones Hsc70 and Hsp90 and attaches a ubiquitin-derived degradation signal onto chaperone-bound client proteins, thereby inducing client degradation by the proteasome. Affected chaperone clients can be broadly divided into two subgroups: 1) proteins that associate with the chaperones during their conformational regulation, such as the glucocorticoid hormone receptor and the oncogenic receptor tyrosine kinase ErbB2 (Connell et al., 2001; Demand et al., 2001; Xu et al., 2002), and 2) aggregation-prone proteins that are recognized by the chaperones during protein quality control, for example, immature forms of CFTR and hyperphosphorylated tau (Meacham et al., 2001; Murata et al., 2001; Shimura et al., 2004). Accordingly, CHIP modulates intracellular signaling and quality control processes.
In light of the central role of CHIP in linking molecular chaperones to the ubiquitin/proteasome system, it seems important to elucidate how the activity of CHIP is regulated in mammalian cells. Recently, a cooperation of CHIP with other cochaperones of Hsc70 emerged as a regulatory principle (Demand et al., 2001; Alberti et al., 2002, 2004; Westhoff et al., 2005). The cochaperone BAG-1, for example, was shown to stimulate the CHIP-mediated degradation of the glucocorticoid hormone receptor (GR; Demand et al., 2001). BAG-1 belongs to a family of proteins, which all share a BAG domain for binding to Hsc70 and which act as a nucleotide exchange factors in the ATP-dependent chaperone cycle of Hsc70 (Höhfeld and Jentsch, 1997; Takayama et al., 1999; Sondermann et al., 2001). Moreover, BAG-1 possesses a ubiquitin-like domain that is utilized for an association of the cochaperone with the proteasome (Lüders et al., 2000a; Alberti et al., 2002). Its domain arrangement enables BAG-1 to recruit Hsc70 complexes to the proteasome, which apparently underlies its stimulating activity in the proteasomal degradation of chaperone clients. The cooperation of BAG-1 and CHIP during proteasomal sorting reflects the ability of the two cochaperones to bind simultaneously to Hsc70 (Demand et al., 2001). BAG-1 interacts with the amino terminal ATPase domain of Hsc70 through its BAG domain and leaves the carboxy terminus of the chaperone available for an association with CHIP (Höhfeld and Jentsch, 1997; Ballinger et al., 1999; Demand et al., 2001).
Importantly, cochaperone cooperation not only provides a means to stimulate chaperone-assisted degradation but also to interfere with it. This was revealed when the Hsc70 cochaperone HspBP1 was identified as an inhibitor of CHIP (Alberti et al., 2004). Like BAG-1, HspBP1 binds to the ATPase domain of Hsc70 and is present in ternary complexes with Hsc70 and CHIP. Intriguingly, HspBP1 inhibits the ubiquitin ligase activity of CHIP in the assembled chaperone/cochaperone complex. As a consequence HspBP1 interferes with the CHIP-induced degradation of CFTR at the cytoplasmic face of the endoplasmic reticulum (ER) and stimulates the maturation of the ion channel (Alberti et al., 2004). HspBP1 apparently exerts an important control function to define the destructive activity of CHIP.
To gain further insight into the function and regulation of CHIP we isolated CHIP-containing protein complexes from human HeLa cells and analyzed their protein composition. We detected the previously uncharacterized BAG-1-related cochaperone BAG-2 as a main component in CHIP complexes and observed that BAG-2 attenuates chaperone-assisted degradation. Our data reveal an unexpected diversity with regard to Hsc70/CHIP complexes that assemble in human cells and identify a novel CHIP inhibitor.
MATERIALS AND METHODS
Antibodies and Proteins
To obtain antibodies against BAG-2, rabbits were immunized with an amino terminal peptide of human BAG-2 (MAQAKINAKANEGRFC). The following antibodies were used for immunoblotting: anti-BAG-2, anti-Hip (Höhfeld et al., 1995), anti-Hop (StressGen, Victoria, British Columbia, Canada), anti-Hsp90 (StressGen), anti-S1 (Affiniti, Golden, CO), anti-C8 (Affiniti), anti-CFTR (Upstate Biotechnology, Lake Placid, NY), anti-CHIP (3), anti-FLAG (Sigma), anti-Hsc/Hsp70 (StressGen), anti-HspBP1 (Delta Biolabs), and anti-Raf-1 (Santa Cruz Biotechnology, Santa Cruz, CA). Rat Hsc70, and wheat E1 were purified as described after recombinant expression in baculovirus-infected SF9 cells (Höhfeld and Jentsch, 1997; Lüders et al., 2000a; Alberti et al., 2002). Hsp40 (Hdj-1), UbcH5b, HspBP1, and CHIP, all of human origin, were expressed in bacteria and purified as described (Höhfeld and Jentsch, 1997; Alberti et al., 2002, 2004). Purified bovine ubiquitin was purchased from Sigma. For recombinant expression of GST-tagged cochaperones Escherichia coli BL21(DE3) cells were transformed with corresponding pGEX4T1 constructs. The bacterial lysate was applied to a glutathione-Sepharose column, and bound proteins were eluted using 10 mM glutathione. For purification of BAG-2 the bag-2 cDNA was cloned into the pTYB12 vector and purified via its self-cutting intein/chitin-binding domain as described by the manufacturer (New England BioLabs, Beverly, MA). The ATPase domain of Hsc70 was purified as described previously (Höhfeld and Jentsch, 1997).
Immunoprecipitation and Binding Experiments
To isolate BAG-2 and CHIP immunocomplexes, HeLa cells were transfected with the plasmid pCMVTag2b-bag-2 and pCMVTag2b-chip, respectively, using the CalPhos transfection kit (Clontech, Palo Alto, CA). When effects of BAG-2 on the association of CHIP with Hsc70 were investigated, cells were cotransfected with pCMVTag2b-chip and pcDNA3.1-bag-2 or an equal amount of empty pcDNA3.1. Two days after transfection HeLa cells were lysed in 25 mM MOPS, pH 7.2, 100 mM KCl, and 0.5% Tween 20 (buffer A) containing 5 mM EDTA and Complete protease inhibitors (Roche, Indianapolis, IN). The lysate was centrifuged at 30,000 × g for 20 min at 4°C and the soluble fraction was used for immunoprecipitation. Samples were incubated with M2-agarose (Sigma-Aldrich) at a concentration of 30 μl of agarose per ml of lysate for 3 h at 4°C. The agarose was collected by centrifugation and washed five times with buffer A and once with buffer A lacking detergent. Next, immunocomplexes were incubated for 20 min at 30°C in detergent-free buffer A containing 2 mM MgCl2, 1 mM ATP, and 1 mM phenylmethylsulfonyl fluoride (PMSF). ATP-eluted proteins were precipitated by the addition of trichloroacetic acid. Agarose-associated proteins were washed once with buffer A, followed by elution with 0.1 M glycine, pH 3.5.
To isolate endogenous BAG-2 complexes, HeLa cells were lysed in RIPA-buffer without SDS (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 10% glycerol) containing 5 mM EDTA and Complete protease inhibitors. The lysate was centrifuged at 30,000 × g for 30 min at 4°C and the soluble fraction was used for immunoprecipitation. Samples were incubated with 150 μl BAG-2 antiserum or preimmune serum for 1 h at 4°C. After addition of 40 μl protein G-Sepharose, samples were incubated for another hour. The Sepharose was collected by centrifugation and washed five times with RIPA without SDS and once with washing buffer (25 mM MOPS, 100 mM KCl). Immunocomplexes were incubated for 20 min at 30°C in washing buffer containing 2 mM MgCl2, 1 mM ATP, and 1 mM PMSF. ATP-eluted proteins were precipitated by the addition of trichloroacetic acid. Remaining immunocomplexes were incubated for 30 min at 4°C in washing buffer containing BAG-2 peptide (MAQAKINAKANEGRFC) at a concentration of 150 ng/μl. Eluted proteins were precipitated by the addition of trichloroacetic acid.
To test for interactions of BAG-2 with Hsc70 and CHIP in vitro binding assays were performed. GST-BAG-2, 10 μg, were incubated with glutathione-Sepharose in buffer B (20 mM MOPS, pH 7.2, 100 mM KCl, 0.5 mM EDTA) for 2 h at 4°C with gentle rocking. Samples were washed twice with buffer B, followed by addition of Hsc70, CHIP, and HspBP1 in equimolar ratio to BAG-2. Samples were incubated for additional 2 h at 4°C, followed by four washing steps with buffer B. Proteins were eluted in 20 mM MOPS, pH 7.2, 100 mM KCl, and 10 mM glutathione for 10 min at room temperature and precipitated by the addition of trichloroacetic acid.
To investigate effects of BAG-2 on the interaction of CHIP and Hsc70, GST-CHIP (10 μg) was incubated with glutathione-Sepharose in buffer B for 1 h at 4°C. Samples were washed with buffer B followed by addition of Hsc70 at equimolar concentration to CHIP and addition of BAG-2 at the indicated concentrations. Samples were incubated for additional 2 h at 4°C, followed by two washing steps with 20 mM MOPS, pH 7.2, 100 mM KCl, 0.5% Tween. Sepharose-bound proteins were eluted by boiling in SDS-PAGE sample buffer and were detected by immunoblotting.
Effects of BAG-2 and HspBP1 on the interaction of CHIP with UbcH5b were analyzed by immobilizing 10 μg of GST-CHIP on glutathione-Sepharose in buffer C (20 mM MOPS, pH 7.2, 100 mM KCl, 1% Tween, 2 mM ATP, 3 mM MgCl2) for 1 h at 4°C. Samples were washed with buffer C followed by addition of Hsc70, UbcH5b, and BAG-2 or HspBP1 in equimolar ratio. Hsp40 was added at one tenth of the concentration of GST-CHIP. Samples were incubated for 30 min at room temperature, followed by two washing steps with 20 mM MOPS, pH 7.2, 100 mM KCl, 1% Tween, and elution of Sepharose-bound proteins by boiling in SDS-PAGE sample buffer.
Peptide Mass Fingerprinting
Coomassie-stained protein bands were excised, chopped into cubes, and washed three times with acetonitrile-water (1:1). The gel pieces were shrunk with neat acetonitrile, reswollen in 50 mM NH4HCO3, and dried under vacuum. Dithiothreitol (10 mM; DTT) in 50 mM NH4HCO3 were added to the dried gel pieces and proteins were reduced for 45 min at 56°C. To alkylate reduced cysteine residues, the remaining liquid was removed, an equal volume of 50 mM iodoacetamide in 50 mM NH4HCO3 was added, and the reaction was allowed to proceed for 30 min in the dark. Before in gel digestion the gel pieces were washed and dried as above. The gel pieces were reswollen in an ice-cold solution of 12.5 ng/μl trypsin (sequencing grade, Promega, Madison, WI) in 25 mM NH4HCO3, 10% acetonitrile. After 1 h on ice, excessive trypsin solution was replaced by 5-20 μl of buffer without enzyme and proteins were digested at 37°C over night. The digest was stopped by the addition of 5-20 μl 1% TFA and peptides were extracted for 30 min at 37°C. One microliter of the extracted peptides was mixed with 1.2 μl of 2.5 mg/ml 2,5-dihydroxybenzoic acid in 0.1% TFA-acetonitrile (2:1) and spotted onto a 800-μm anchor target (Bruker Daltonics, Billerica, MA). Positive ion spectra were acquired on a Reflex IV MALDI-TOF mass spectrometer (Bruker Daltonics) in the reflectron mode. A peptide calibration standard was used for external calibration of the mass range from m/z 1046 to m/z 3147. The XMASS 5.1.10 postanalysis software was used for optional internal recalibration on trypsin autolysis peaks and the generation of peak lists. Proteins were identified from MALDI fingerprint data by searching the NCBI nr public database (National Center for Biotechnology Information) using a local installation of MASCOT 1.9.
Gel Filtration Analysis
Subconfluent HeLa cells were sonicated in lysis buffer (20 mM MOPS, pH 7.2, 50 mM KCl) containing 20 mM EDTA and Complete protease inhibitors (Roche). The obtained lysate was centrifuged at 4°C and 100,000 × g for 30 min, and the resulting supernatant was subjected to gel filtration using a Superose 6 column equilibrated in lysis buffer containing 2 mM EDTA. Collected fractions were precipitated with trichloroacetic acid and analyzed by immunoblotting. To analyze protein complexes assembled in vitro, Hsc70, CHIP, and BAG-2 (2 μM of each) were incubated for 1 h at 4°C in 20 mM MOPS, pH 7.2, 50 mM KCl, 2 mM EDTA, 1 mM DTT, 5% glycerol. Samples were centrifuged at 4°C and 100,000 × g for 20 min, and the obtained supernatants were subjected to gel filtration on a Superose 6 column equilibrated in the same buffer.
Yeast Two-hybrid Assay
Dimerization of BAG-2 was examined in a yeast two-hybrid assay. The entire coding region of human bag-2 and a PCR-product that encodes a deletion fragment of BAG-2 without the coiled coil region (ΔCC, aa 69-211) were subcloned into pGAD-C1 and pGBD-C1 vectors. For interaction assays different combinations of corresponding two-hybrid constructs and empty vectors were transformed into yeast strain AH109 and analyzed as described previously (Demand et al., 1998).
In Vitro Ubiquitylation
Bacterially expressed human Raf-1 (Demand et al., 2001) was incubated in the presence of 0.3 μM Hsp40, 3 μM Hsc70, 2 μM UbcH5b, 1 μM CHIP, 0.1 μM E1, and 2.5 mg/ml ubiquitin in 20 mM MOPS, pH 7.2, 100 mM KCl, 5 mM MgCl2, 5 mM ATP, 10 mM DTT, and 1 mM PMSF for 2 h at 30°C. When indicated purified BAG-2 was added. Samples were analyzed with anti-Raf-1, anti-Hsc/Hsp70, and anti-BAG-2 antibodies. To test if BAG-2 is a substrate of CHIP, ubiquitylation assays were performed as outlined above without addition of Raf-1. Autoubiquitylation of CHIP was analyzed by incubating 1 μM CHIP in the presence of 0.3 μM Hsp40, 3 μM Hsc70, 4 μM UbcH5b, 0.2 μM E1, 2.5 mg/ml ubiquitin, and BAG-2 at the indicated concentrations. Samples were analyzed by immunoblotting with an anti-CHIP antibody.
CFTR Turnover
To determine the influence of BAG-2 on the steady state level of CFTR HEK 293 cells seeded in six-well plates were transfected with 0.375 μg of pcDNA3.1-cftr DNA using Effectene transfection reagent (Qiagen, Chatsworth, CA). When indicated, 0.375 μg of pcDNA3.1-chip and 0.375 μg or 0.75 μg of pcDNA3.1-bag-2 were cotransfected. Plasmid pcDNA3.1 was added to keep the total amount constantly at 1.5 μg. Twenty-four hours after transfection the cells were washed once with phosphate-buffered saline (pH 7.4, 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.4 mM KH2PO4) and lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Nonidet P40, 0.5% deoxycholate, 0.2% SDS) containing Complete protease inhibitor. The lysate was centrifuged at 30,000 × g for 20 min at 4°C and the supernatant was used as a soluble extract. For immunoblotting 60 μg of total protein was loaded onto an SDS-PAGE gel.
BAG-2 levels in HEK293 cells were depleted using a pSUPER plasmid (OligoEngine, Seattle, WA), directed against the sequence GGAGCTGGTTCCAAAACTC of human bag-2. HEK293 cells were seeded in six-well plates and transfected with 0.5 μg of pcDNA3.1-cftr by using the Effectene transfection reagent (Qiagen). When indicated 0.5 μg of pSUPER-bag-2, the same amount of pSUPER, 0.5 μg of pCDNA3.1, and pCDNA3.1-bag-2 were transfected. Cells were harvested 48 h after transfection and processed as described above.
NBD1 aggregation assays were performed as previously described using the NBD1 of human ΔF508 CFTR (Strickland et al., 1997). Aggregation was initiated by rapid dilution of purified NBD1 (solubilized in 6 M GdnHCl) into prewarmed folding buffer (100 mM Tris-HCl, pH 7.4, 0.385 M l-arginine-HCl, 2 mM EDTA, 10 mM DTT, and 25 mM GdnHCl) to a final concentration of 2 μM. When indicated a final concentration of 1 μM or 2 μM BAG-2 was present during the reaction. Aggregation was followed over 1 h at 37°C by turbidity measured at 400 nm.
Peptide Scans
Cellulose-bound peptide scans were generated as described previously (Saidowsky et al., 2001). Membranes contained 13-mer peptides covering NBD1 of CFTR (aa 389-673), which overlapped by 10 amino acids. Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline (TBS) (25 mM Tris/HCl, pH 7.6, 137 mM NaCl) for 1 h at room temperature, followed by three washes with TBS for 15 min. Purified Hsc70, BAG-2, and CHIP were fluorescently labeled with DY-681 according to the manufacturer's directions (Dyomics, Jena, Germany). Labeled protein, 100 nM, was added to the membranes in Odyssey blocking buffer (Li-Cor Biosciences, Lincoln, NE) diluted 1:1 with TBS, followed by incubation overnight at 4°C with gentle agitation. After washing with TBS/0.05% Tween, the binding of labeled proteins was visualized with a fluorescence scanner (Li-Cor Biosciences). When indicated a fivefold or 10-fold molar excess of the unlabeled Hsc70 ATPase domain over BAG-2 was added during incubation.
RESULTS
BAG-2 Is a Main Interaction Partner of CHIP in HeLa Cells
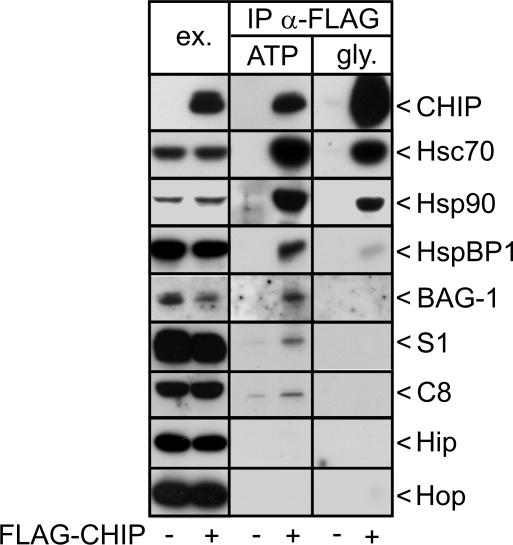
We sought to analyze the composition of CHIP-containing protein complexes. For this purpose a FLAG-tagged form of CHIP was transiently expressed in HeLa cells and complexes were isolated by immunoprecipitation with an anti-FLAG antibody. Initially, antibodies against diverse chaperones, cochaperones, and proteasomal subunits were used to verify their presence in CHIP complexes. Consistent with previous findings (Connell et al., 2001; Demand et al., 2001; Meacham et al., 2001; Alberti et al., 2004), we detected the chaperones Hsc70 and Hsp90, the CHIP-regulating cochaperones BAG-1 and HspBP1, and the proteasomal subunits S1 and C8 in CHIP complexes (Figure 1). In contrast, the cochaperones Hip and Hop, shown to assist Hsc70 during protein folding (Höhfeld et al., 1995; Demand et al., 1998), were absent from CHIP complexes.
Figure 1.
Protein composition of CHIP-containing protein complexes isolated from HeLa cells. HeLa cells were transfected with pCMVTag2b-chip, which encodes a FLAG-tagged form of CHIP (+ FLAG-CHIP), or with an equal amount of a control plasmid (-). CHIP-containing protein complexes were precipitated with an immobilized anti-FLAG antibody (IP α-FLAG). Purified immunocomplexes were treated with ATP (ATP), followed by addition of glycine, pH 3.5, to release CHIP and remaining associated proteins from the antibody (gly.). CHIP immunocomplexes were separated by SDS-PAGE and analyzed for the presence of binding partners by immunoblotting with specific antibodies. To monitor expression levels, 40 μg of protein extracts were loaded (ex.).
All binding partners largely dissociated from CHIP upon treatment of isolated complexes with ATP (Figure 1). In case of Hsc70 and Hsp90 this reflected ATP-regulated chaperone/cochaperone interactions. HspBP1 and BAG-1 were released because their binding to CHIP is mainly mediated through Hsc70 (Demand et al., 2001; Alberti et al., 2004). The interaction between CHIP and the proteasome remains to be elucidated in detail. However, the observed ATP-sensitivity suggests a regulation through AAA ATPases of the regulatory 19S particle of the proteasome. ATP-regulated interactions apparently provide the basis for dynamic associations of CHIP with chaperone complexes and with the proteasome.
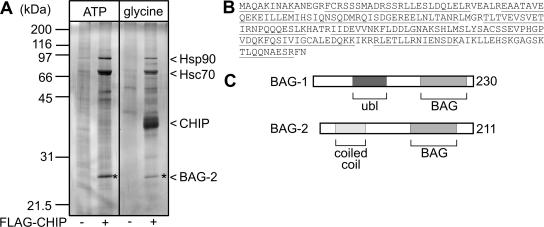
When isolated immunocomplexes were analyzed by SDS-PAGE and silver-staining a prominent polypeptide of 27-kDa apparent molecular mass was specifically detected in association with CHIP (Figure 2A). Peptide mass fingerprinting identified the polypeptide as the BCL2-associated athanogene-2 (BAG-2), a so far largely uncharacterized member of the BAG domain cochaperone family (Takayama et al., 1999; Ueda et al., 2004; Figure 2B). Like BAG-1, BAG-2 associates with the ATPase domain of Hsc70 via its BAG domain (Takayama et al., 1999), but it lacks a ubiquitin-like domain that is essential for the association of BAG-1 with the proteasome (Lüders et al., 2000a) (Figure 2C).
Figure 2.
The cochaperone BAG-2 is a major component of CHIP-containing protein complexes. (A) Analysis of isolated CHIP immmunocomplexes by SDS-PAGE and silver-staining. Hsp90, Hsc70, and CHIP were identified according to their molecular masses. The star denotes a prominent polypeptide of ∼27 kDa. The corresponding polypeptide was identified as the cochaperone BAG-2 by peptide mass fingerprinting. (B) Complete sequence of human BAG-2. Polypeptides of BAG-2 identified by mass spectrometry are underlined. (C) Schematic presentation of the domain structure of BAG-1 and BAG-2. Both cochaperones possess a BAG domain at their carboxyl termini, which mediates Hsc70 binding and regulation. In addition, BAG-1 carries a ubiquitin-like domain (ubl), whereas BAG-2 possesses a coiled-coil domain near the amino terminus.
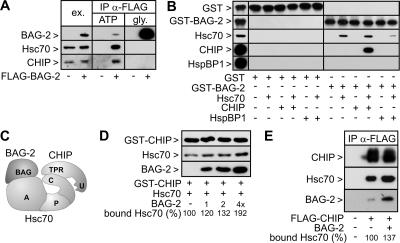
To verify the observed interaction between BAG-2 and CHIP, FLAG-tagged BAG-2 was expressed in HeLa cells and immunoprecipitated. Hsc70 and CHIP were detected in BAG-2 complexes and were released from the precipitated cochaperone upon ATP treatment (Figure 3A). Such elution behavior would be consistent with a role of Hsc70 in mediating the BAG-2/CHIP interaction. BAG-2 associates with the ATPase domain of the chaperone (Takayama et al., 1999), while CHIP binds simultaneously to the carboxy terminus of Hsc70. Addition of ATP dissociates BAG-2 from Hsc70 and in this way also stimulates release of CHIP. Indeed, BAG-2 and CHIP were unable to interact directly with each other in in vitro binding experiments (Figure 3B). Yet, a retention of CHIP on immobilized BAG-2 was observed in the presence of Hsc70, consistent with the formation of a ternary chaperone/cochaperone complex comprising BAG-2 and CHIP bound to Hsc70 (Figure 3C). We also noted that the interaction between BAG-2 and Hsc70 was attenuated by HspBP1, a previously identified inhibitor of CHIP that binds to the Hsc70/CHIP complex through an association with the ATPase domain of the chaperone (Alberti et al., 2004). Clearly, HspBP1 and BAG-2 compete in binding to the Hsc70 ATPase domain.
Figure 3.
BAG-2 forms a complex with Hsc70 and CHIP. (A) HeLa cells were transfected with pCMVTag2b-bag-2, which encodes a FLAG-tagged form of BAG-2 (+ FLAG-BAG-2), or with an equal amount of a control plasmid (-). BAG-2-containing protein complexes were immunoprecipitated with an immobilized anti-FLAG antibody (IP α-FLAG) and analyzed for the presence of Hsc70 and CHIP by immunoblotting. The chaperone and CHIP were detected in ATP-eluates of the isolated complexes. To monitor expression levels, 40 μg of protein extracts was loaded (ex.). Twenty percent of each IP fraction was loaded. (B) Binding of BAG-2 to CHIP is mediated by Hsc70. Purified GST-tagged BAG-2 was immobilized and incubated with Hsc70 and CHIP as indicated. Proteins retained on the affinity resin were detected using specific antibodies. As negative control, proteins were incubated with immobilized GST. The left panel shows 5% of the input. (C) Schematic presentation of the BAG-2/Hsc70/CHIP complex. BAG-2 binds to the ATPase domain of Hsc70 (A) utilizing its BAG domain. At the same time CHIP associates with the carboxy terminus of the chaperone (C) via its TPR domain. P denotes the peptide-binding pocket of Hsc70. (D) BAG-2 stimulates the association of Hsc70 with CHIP. GST-CHIP was immobilized and incubated with Hsc70 and BAG-2 as indicated. BAG-2 was added at 1-, 2-, and 4-fold the concentration of CHIP. Bound proteins were detected after immunoblotting using specific antibodies. Bound Hsc70 was quantified, and values are presented as percentage of control in the absence of BAG-2, which was set to 100%. (E) Overexpression of BAG-2 increases the amount of Hsc70 detectable in CHIP-containing protein complexes. HeLa cells were transfected with pCMVTag2b-chip (+ FLAG-CHIP) and pcDNA3.1-bag-2 (+ BAG-2) as indicated. The total amount of added DNA was kept constant by addition of pcDNA3.1. CHIP-containing protein complexes were isolated using an immobilized anti-FLAG antibody (IP α-FLAG). Immunoprecipitated CHIP was detected in glycine eluates using a specific antibody. BAG-2 and Hsc70 were detected in ATP eluates of the isolated CHIP complexes by immunoblotting.
We investigated whether BAG-2 modulated the interaction of CHIP with Hsc70. Indeed, BAG-2 stimulated the association of Hsc70 with immobilized CHIP in an in vitro binding assay (Figure 3D). Similarly, elevation of BAG-2 levels in HeLa cells led to an increased association between Hsc70 and CHIP (Figure 3E). This suggests cooperative binding of the two cochaperones to Hsc70. Because a direct interaction between the two cochaperones could not be detected (see above), BAG-2 may induce a conformational change of Hsc70 favorable for CHIP binding.
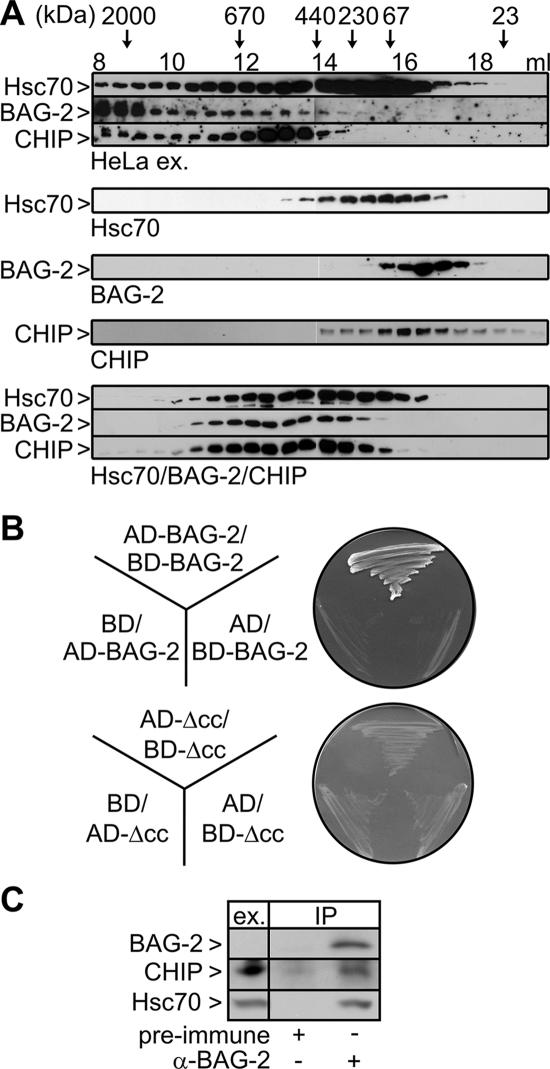
To verify the physiological significance of the observed interactions, a HeLa cell extract was fractionated by gel filtration and the elution profiles of endogenously expressed Hsc70, BAG-2, and CHIP were analyzed (Figure 4A, top panel). Consistent with the observed interactions, the three proteins cofractionated in complexes ranging from 500 to >2000 kDa. This size range partially overlapped with the range detected for Hsc70/BAG-2/CHIP complexes formed from purified components in vitro (Figure 4A, bottom panel, ∼300-700 kDa). Larger complexes detectable in the cell extract might arise from the association of BAG-2 with as yet unidentified additional binding partners. Remarkably, Hsc70 and CHIP coeluted with BAG-2 also in high-molecular-mass fractions (>700 kDa), implying that BAG-2 may recruit the Hsc70/CHIP machinery to distinct protein complexes or subcellular structures.
Figure 4.
BAG-2 cofractionates with Hsc70 and CHIP in a HeLa cell extract and forms homodimers. (A) Analysis of chaperone/cochaperone complexes by gel filtration chromatography. Top, fractionation of a HeLa cell extract on a Superose 6 column. Proteins were detected by specific antibodies as indicated on the left. The elution behavior of proteins of defined molecular mass is indicated at the top. Bottom, gel filtration analysis of purified components and Hsc70/BAG-2/CHIP complexes formed in vitro. (B) Plasmid constructs encoding BAG-2 or a deletion fragment of BAG-2 (ΔCC) fused to the GAL4-transcription activation domain (AD-BAG-2/-ΔCC) or the GAL4-DNA-binding domain (BD-BAG-2/-ΔCC) and empty vectors (AD and BD) were transformed into yeast and screened for potential interactions in a yeast two-hybrid assay. Dimerization of BAG-2 resulted in growth on minimal medium lacking histidine, leucine, and tryptophan. (C) Coimmunoprecipitation of endogenous BAG-2, CHIP, and Hsc70. HeLa cell extracts were subjected to immunoprecipitation with an anti-BAG-2 antibody. Preimmune serum was used in control reactions. CHIP and Hsc70 were detected in ATP eluates of the isolated immunocomplexes. BAG-2 was eluted with synthetic peptide. Ex. corresponds to 60 μg of HeLa cell extract. Under the chosen conditions endogenous BAG-2 is not detectable in the extract.
We noted that purified BAG-2 alone eluted from the gel filtration column with an apparent molecular mass of ∼60 kDa, approximately twice the size calculated from its primary structure (Figure 4A, middle panel). This suggests that the cochaperone forms homodimers. In fact, BAG-2 was shown to associate with itself in a yeast two-hybrid assay (Figure 4B). Moreover, the amino-terminal coiled-coil region of BAG-2 (see Figure 2) could be identified as the dimerization domain. A BAG-2 fragment that lacked the coiled coil region was unable to dimerize (Figure 4B). The finding that BAG-2 forms homodimers parallels similar observations for CHIP (Nikolay et al., 2004). Both cochaperones could thus possibly contact two Hsc70s at a time, which would lead to the formation of multimeric Hsc70/BAG-2/CHIP complexes significantly larger than expected for a ternary assembly (Figure 4A, bottom panel).
To further confirm the interaction between BAG-2 and CHIP, endogenous BAG-2 was immunoprecipitated from HeLa cell extracts. Endogenous CHIP and Hsc70 were clearly detectable in association with BAG-2 (Figure 4C). The data demonstrate that ternary BAG-2/Hsc70/CHIP complexes exist in human cells.
BAG-2 Inhibits the Ubiquitin Ligase Activity of CHIP
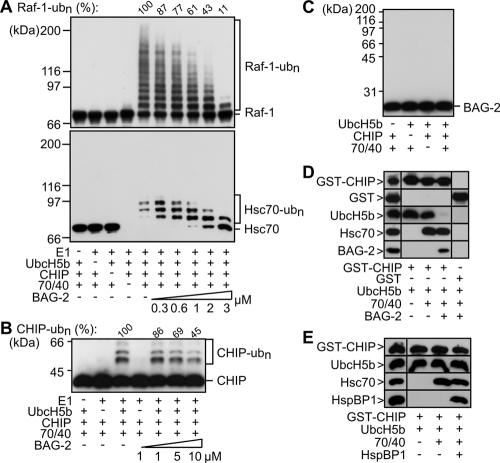
We asked whether association of BAG-2 with the Hsc70/CHIP complex affected chaperone-assisted ubiquitylation using a previously established in vitro assay (Demand et al., 2001; Alberti et al., 2004). In this assay, CHIP mediated the ubiquitylation of Hsc70-bound Raf-1 in the presence of the ubiquitin-activating enzyme E1 and the ubiquitin-conjugating E2 enzyme UbcH5b (Figure 5A). Addition of purified BAG-2 strongly inhibited the CHIP-mediated ubiquitylation of Raf-1.
Figure 5.
BAG-2 inhibits the ubiquitin ligase activity of CHIP when complexed with Hsc70 by abrogating CHIP/E2 cooperation. (A) Ubiquitylation of Raf-1 and Hsc70 was analyzed in vitro in the presence of purified CHIP (1 μM), the ubiquitin-activating enzyme E1, UbcH5b, Hsc70/Hsp40 (70/40), and increasing concentrations of BAG-2, ranging from 0.3 to 3 μM, as indicated. Raf-1, Hsc70, and ubiquitylated forms of both proteins (ub(n)-Raf-1, ub(n)-Hsc70) were detected using specific antibodies. Ubiquitylated Raf-1 was quantified, and obtained values are given as percentage of the amount detected in a control reaction in the absence of BAG-2. Notably, Hsc70 that carried more than three ubiquitin moieties was not recovered under the experimental conditions. (B) Autoubiquitylation of CHIP was investigated in vitro in the presence of purified CHIP (1 μM), the ubiquitin-activating enzyme E1, UbcH5b, Hsc70/Hsp40 (70/40), and increasing concentrations of BAG-2, ranging from 1 to 10 μM. Ubiquitylated forms of CHIP (ub(n)-CHIP) were detected by immunoblotting using specific antibodies and their amount was quantified. (C) BAG-2 is not recognized as a substrate by the CHIP ubiquitin ligase. In vitro ubiquitylation reactions were performed as in A except that the reactions contained purified BAG-2 instead of Raf-1. BAG-2 was detected in immunoblots using a specific antibody. In the presented experiment, CHIP-mediated ubiquitylation of Hsc70 was reduced to 26% in the presence of BAG-2 compared with a control reaction without BAG-2 which was set to 100%. (D) GST-CHIP was immobilized and incubated with UbcH5b, Hsc70/Hsp40 (70/40), and BAG-2 as indicated. Bound proteins were detected after immunoblotting using specific antibodies. As a control proteins were incubated with immobilized GST (right panel). The panel on the left shows 10% of the input. (E) In contrast to BAG-2, HspBP1 does not interfere with the CHIP/E2 interaction. UbcH5b binding to immobilized CHIP was analyzed as described in D in the presence of HspBP1 instead of BAG-2.
In the same assay ubiquitylation of Hsc70 was analyzed. The chaperone was previously identified as a physiological substrate of the CHIP ubiquitin ligase (Jiang et al., 2001). On addition of the CHIP conjugation machinery, Hsc70 was quantitatively modified by ubiquitin attachment (Figure 5A). However, ubiquitylation was accompanied by a loss of signal intensity and forms of Hsc70 carrying more than four ubiquitin moieties could not be detected. We speculate that attachment of a ubiquitin chain of four or more moieties to Hsc70 alters the biophysical properties of the chaperone, resulting in an inability to recover such forms under the assay conditions. In any case, unmodified Hsc70 became detectable upon addition of BAG-2, revealing an inhibitory effect of BAG-2 on the CHIP-mediated ubiquitylation of Hsc70 (Figure 5A). Because BAG-2 forms ternary complexes with Hsc70 and CHIP and promotes the Hsc70/CHIP interaction (see above), the cochaperone seems to inhibit the ubiquitin ligase activity of CHIP within an assembled chaperone/cochaperone complex.
BAG-2 also attenuated the autoubiquitylation of CHIP (Figure 5B), albeit with reduced efficiency compared with the effects of the cochaperone on the CHIP-mediated ubiquitylation of Hsc70 and Raf-1. Because a direct interaction between CHIP and BAG-2 could not be observed (see above), CHIP that is transiently not bound to Hsc70 under the chosen experimental conditions could associate with UbcH5 even in the presence of BAG-2 and could become ubiquitylated, leading to a reduced inhibition.
We investigated whether BAG-2 is a substrate of the CHIP ubiquitin ligase. Intriguingly, BAG-2 was not modified by the CHIP conjugation machinery when added alone or together with Hsc70 (Figure 5C). Substrate competition can thus be excluded as a molecular basis for the inhibitory effect displayed by BAG-2. Another hypothesis may invoke a BAG-2-mediated disruption of the interaction between CHIP and its partner E2 enzyme UbcH5b. In support of this hypothesis we noted that BAG-2 abrogated the binding of UbcH5b to immobilized CHIP (Figure 5D). Apparently, BAG-2 inhibits the ubiquitin ligase activity of CHIP in the assembled chaperone/cochaperone complex by interfering with CHIP/E2 cooperation.
Previously, HspBP1 was identified as an inhibitor of the CHIP ubiquitin ligase (Alberti et al., 2004). In contrast to BAG-2, HspBP1 did not abrogate E2 binding to CHIP (Figure 5E). HspBP1 seems to interfere with ubiquitylation by removing ubiquitin attachment sites from the reach of the conjugation machinery or by masking those sites. The two CHIP inhibitors apparently use distinct mechanisms to control the destructive potential of CHIP.
BAG-2 Stimulates the Maturation of CFTR
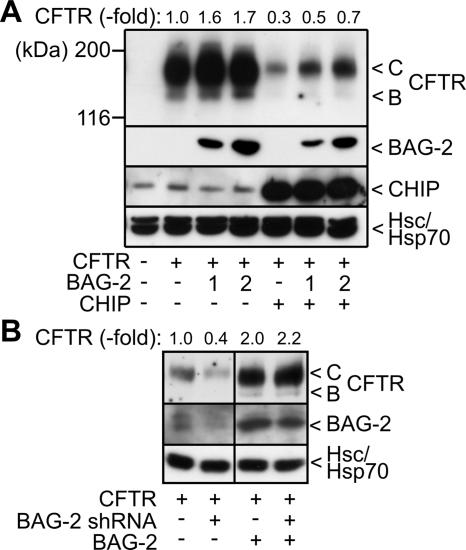
As BAG-2 regulates the ubiquitin ligase activity of CHIP, we analyzed whether the cochaperone affected CHIP-mediated degradation. Previously, CHIP was shown to induce the proteasomal degradation of immature CFTR at the ER membrane (Meacham et al., 2001; Alberti et al., 2004). The ion channel exposes large domains into the cytoplasm that are recognized by cytoplasmic quality control components, including Hsc70 and CHIP. Coexpression of CFTR and BAG-2 in HEK293 cells led to a significant increase of the steady state levels of CFTR (Figure 6A). The increase was observed for the immature ER-localized B form of CFTR as well as for the maturely glycosylated C form that is located in the plasma membrane. BAG-2 apparently stimulates the maturation of CFTR. In contrast, overexpression of CHIP resulted in a strong decrease of CFTR levels, which reflects the degradation-inducing activity of the ubiquitin ligase (Meacham et al., 2001). Notably, BAG-2 counteracted the destructive activity of CHIP during CFTR biogenesis. This further emphasizes a function of BAG-2 as an inhibitor of the CHIP ubiquitin ligase.
Figure 6.
BAG-2 inhibits the chaperone-assisted degradation of CFTR and is essential for CFTR maturation. (A) HEK293 cells were transiently transfected with CFTR-, BAG-2-, and CHIP-encoding plasmids in the indicated combinations. Amounts of pcDNA3.1-bag-2 were 1- and 2-fold of the amount of pcDNA3.1-chip when indicated. The ER-localized, core-glycosylated B-form (B) and the completely glycosylated, plasma membrane-localized C-form of CFTR (C) were detected by immunoblotting. Likewise, BAG-2 and CHIP were detected with specific antibodies. Hsc70 and Hsp70 served as loading control. Each lane represents 60 μg of cellular extract. CFTR levels were quantified from four independent experiments and mean values are presented as fold change compared with the control reaction, which was set to 1. (B) HEK293 cells were transiently transfected with CFTR-, BAG-2 shRNA-, and BAG-2-expressing plasmids as indicated. Cell extracts, 60 μg, were separated by SDS-PAGE. Protein levels were analyzed by immunoblotting using specific antibodies. Hsc70/Hsp70 served as loading control. Samples that did not express BAG-2 shRNA received the pSUPER plasmid without an insert.
To verify whether BAG-2 is essential for CFTR maturation, endogenous BAG-2 was depleted in HEK293 cells by transfection with a corresponding pSUPER plasmid. The plasmid encodes a small hairpin RNA (shRNA), which is subsequently cleaved into small interference RNA (siRNA) in transfected cells. Transient transfection of this plasmid led to a considerable reduction of endogenous BAG-2 levels (Figure 6B). At the same time CFTR levels were significantly reduced, consistent with an essential role of BAG-2 in CFTR maturation. When BAG-2 was overexpressed, shRNA coexpression no longer affected cellular CFTR levels. This demonstrates that the bag-2 shRNA does not nonspecifically decrease CFTR expression. Taken together, the data identify BAG-2 as an inhibitor of the CHIP ubiquitin ligase, which plays an essential role in the maturation of CFTR.
BAG-2 Stabilizes the NBD1 Domain of CFTR
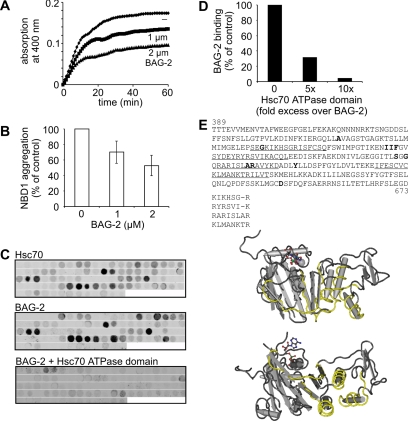
One cause for the misprocessing of CFTR is the inefficient folding of the first of two cytoplasmically exposed nucleotide-binding domains (NBD1) of the membrane protein (Sato et al., 1996; Qu et al., 1997; Kopito, 1999). The cystic fibrosis-causing ΔF508 mutation localizes to NBD1 and further decreases the folding propensity of this domain. During the cotranslational insertion of CFTR into the ER membrane, Hsc70 and its cochaperone Hdj-2 bind to NBD1 and facilitate intramolecular interactions between the domain and other cytoplasmic regions of CFTR (Strickland et al., 1997; Meacham et al., 1999). We investigated whether BAG-2 prevents the aggregation of unfolded ΔF508 NBD1 upon dilution from denaturant. When added at equimolar concentration, BAG-2 reduced the aggregation of NBD1 by ∼50% (Figure 7, A and B). This indicates that the stimulating activity of BAG-2 in the maturation of CFTR is not only caused by an inhibition of chaperone-assisted degradation, but that BAG-2 also stabilizes nonnative conformations of CFTR. To further elucidate the interactions of BAG-2 and Hsc70 with NBD1 a cellulose-bound peptide scan of the domain was generated. 13-mer peptides, which covered the complete NBD1 from amino acid 389-673 of human CFTR and which overlapped by 10 amino acids, were synthesized and spotted onto a nitrocellulose membrane (Figure 7C). Peptide scans were incubated with fluorophor-labeled BAG-2 and Hsc70, respectively, and binding was visualized with a fluorescence scanner. BAG-2 and Hsc70 recognized a very similar pattern of NBD1 peptides (Figure 7C). Main binding regions were located to residues 478-493, 511-526, 553-565, and 586-604. One of these regions (553-565) overlapped with a previously identified Hsc70-binding site within NBD1 (Strickland et al., 1997). Peptides that were recognized by Hsc70 and BAG-2 were enriched in hydrophobic and positively charged amino acids. Moreover, highaffinity binding peptides displayed primary structure motifs similar to the previously defined Hsc70 recognition motif comprising a core of hydrophobic residues flanked by positively charged amino acids (Figure 7E). Apparently, the ability of BAG-2 to recognize hydrophobic patches within NBD1 underlies its intrinsic chaperone activity. Binding of BAG-2 to CFTR peptides was abolished in the presence of the ATPase domain of Hsc70 (Figure 7, C and D), suggesting that the client- and chaperone-binding sites on BAG-2 are at least overlapping.
Figure 7.
BAG-2 inhibits the aggregation of the ΔF508 NBD1 domain of CFTR by binding hydrophobic and positively charged segments in NBD1. (A) Aggregation of denatured NBD1 was followed over time at 37°C by turbidity measurement at 400 nm. NBD1, 2 μM, was incubated either alone or in the presence of purified BAG-2 at the indicated concentrations. (B) Data obtained in A are presented as the relative amount of aggregated NBD1 domain after a 60-min incubation period. The amount of aggregated NBD1 in the control reaction was set as 100%. Values represent the mean of 14 independent experiments plus SD. (C) Fluorophor-labeled Hsc70 and BAG-2 were incubated with peptide scans covering NBD1, and retained protein was visualized with a fluorescence scanner. When indicated, binding of labeled BAG-2 to NBD1 peptides was performed in the presence of the Hsc70 ATPase domain at a 10-fold molar excess over the cochaperone. (D) Binding of fluorophor-labeled BAG-2 to aa 589-601 of the NBD1 domain either alone or in the presence of a 5- or 10-fold molar excess of the Hsc70 ATPase domain was quantified. The amount of BAG-2 bound to aa 589-601 in the absence of the ATPase domain was set to 100%. (E) Primary structure of NBD1 of human CFTR. Amino acids shown to be mutated in cystic fibrosis patients are highlighted in bold. Sequences recognized by BAG-2 and Hsc70 are underlined. Comparison of identified sequences reveals the presence of an interaction motif that is characterized by a core of hydrophobic amino acids flanked by positively charged amino acids. Identified binding regions are shown in yellow in the resolved NBD1 structure (1XMJ_A of the NCBI structure database).
DISCUSSION
Here we identify the Hsc70 cochaperone BAG-2 as an inhibitor of the CHIP ubiquitin ligase and regulator of chaperone-mediated protein quality control. BAG-2 associates with the Hsc70/CHIP chaperone machinery through binding to the ATPase domain of Hsc70 and interferes with CHIP-mediated ubiquitylation by abrogating the cooperation between CHIP and its partner E2 enzyme.
Previously, the cochaperone HspBP1 was identified as an inhibitor of the CHIP ubiquitin ligase (Alberti et al., 2004). Like BAG-2, HspBP1 interferes with CHIP-mediated ubiquitylation in an assembled chaperone/cochaperone complex. This may enable CHIP to participate in the regulation of the Hsc70 chaperone cycle without inducing client degradation, consistent with recently established degradation-independent functions of CHIP (Cardozo et al., 2003; Dai et al., 2003; Jiang et al., 2003; Kampinga et al., 2003). Remarkably, BAG-2 and HspBP1 seem to achieve CHIP inhibition by distinct mechanisms. Although BAG-2 abrogates the CHIP/E2 cooperation, HspBP1 does not and may therefore induce conformational changes of the chaperone complex, which interfere with CHIP-mediated ubiquitylation, as previously suggested (Alberti et al., 2004). In any case, the finding that two distinct Hsc70 cochaperones act as inhibitors of the CHIP ubiquitin ligase illustrates the necessity to define the destructive activity of CHIP. The inhibitors may have distinct but nonetheless overlapping functions. Indeed, BAG-2 and HspBP1 both stimulated the maturation of CFTR at the ER membrane (Alberti et al., 2004; Figure 6). The Hsc70 chaperone machinery is intimately involved in the folding and degradation of CFTR (Yang et al., 1993; Strickland et al., 1997; Meacham et al., 1999, 2001; Farinha et al., 2002; Kostova and Wolf, 2003; Alberti et al., 2004; Younger et al., 2004). Hsc70 associates cotranslationally with the first nucleotide-binding domain of the ion channel (NBD1), which is exposed into the cytoplasm, and facilitates the folding or degradation of CFTR dependent on the cochaperones Hsc70 interacts with. Using cellulose-bound peptide scans, we determined the regions in NBD1 that are recognized by Hsc70. Chaperone-binding sites are mainly located in the α-subdomain of NBD1, which also harbors the most common cystic fibrosis-causing mutations (Lewis et al., 2004). Binding sites comprise patches of hydrophobic amino acids flanked by positively charged residues, largely consistent with previous findings for recognition motifs of Hsp70 family members (Rüdiger et al., 1997). Remarkably, a recognition pattern similar to that observed for Hsc70 was also obtained when peptide scans were incubated with purified BAG-2. The ability to recognize hydrophobic patches in NBD1 obviously enables the cochaperone to display a chaperone activity on its own. This might be important for client loading onto Hsc70 and for the stabilization of clients upon ATP-induced release from Hsc70. Notably, binding of BAG-2 to CFTR peptides was abolished in the presence of the ATPase domain of Hsc70 (i.e., the contact region of BAG-2 on Hsc70). Client- and chaperone-binding sites on BAG-2 are apparently overlapping. In this regard it is noteworthy that BAG-2 forms homodimers. Conceivably, one subunit of the dimer may engage in Hsc70 binding and regulation, whereas the other subunit stabilizes the nonnative client protein. In any case, the stimulating activity of BAG-2 in CFTR maturation seems to reflect not only its ability to inhibit the CHIP-mediated ubiquitylation of CFTR, but also to stabilize immature CFTR conformations.
Another BAG domain cochaperone previously shown to regulate CHIP activity is BAG-1 (Lüders et al., 2000a). In contrast to BAG-2, BAG-1 stimulates CHIP-mediated degradation (Demand et al., 2001). In part this might be explained by the ability of BAG-1 to associate with the proteasome through its ubiquitin-like domain and to act as a substrate release factor of Hsc70 in the vicinity of the proteasome. Moreover, BAG-1, but not BAG-2, is efficiently ubiquitylated by the CHIP conjugation machinery, which further promotes binding of BAG-1 to the proteolytic complex. Thus, BAG-1 and BAG-2 might be viewed as antagonistic regulators of the CHIP ubiquitin ligase. However, differences in substrate specificity have to be taken into account here. BAG-2 is an essential component in CFTR maturation, whereas BAG-1 does not affect the CHIP-mediated degradation of the ion channel (Mecham et al., 2001). The two cochaperones may thus cooperate with Hsc70 in the biogenesis of distinct sets of chaperone substrates. Taken together, a functional diversity among BAG domain cochaperones becomes apparent. These cochaperones were often described as inhibitors of Hsc70, because they were found to abrogate the chaperone-mediated stabilization of some model chaperone clients such as heat-denatured firefly luciferase in in vitro and cell culture experiments (Takayama et al., 1999; Nollen et al., 2001). However, depending on the experimental conditions used (i.e., cochaperone concentration, chaperone client, and particular BAG family member investigated), folding-stimulating activities also were detected for BAG cochaperones (Gässler et al., 2001; Lüders et al., 2000b). A recent study even identifies BAG-1 as a neuroprotective protein that enhances Hsc70 chaperone function (Liman et al., 2005). This illustrates that BAG family members cannot be generally described as inhibitors of Hsc70. Rather, the acceleration of nucleotide exchange on Hsc70 in the presence of BAG cochaperones may result in the abrogation or the stimulation of chaperone-assisted folding dependent on the individual properties of each client protein and the physiological situation. The data presented here clearly identify BAG-2 as a cochaperone that stimulates the maturation of CFTR in conjunction with Hsc70. The full repertoire of chaperone clients that are affected by BAG-2, however, remains to be elucidated. Intriguingly, functional impairment of the C. elegans homologue of BAG-2, UNC-23, causes fragile skeletal muscle attachment, consistent with a role of the cochaperone in the assembly or positioning of proteins in the muscle attachment complex (Plenefisch et al., 2000). It is therefore tempting to speculate that the cooperation between BAG-2 and CHIP established here exerts regulatory functions in diverse chaperone-assisted processes in healthy and diseased cells.
While this work was under review, Patterson and coworkers independently identified BAG-2 as an inhibitor of the CHIP ubiquitin ligase (Dai et al., 2005).
Acknowledgments
We thank Karen Himmelberg for expert technical assistance. Franz-Georg Hanisch, Stefan Müller, and Udo Roth from the ZMMK, University Cologne, are acknowledged for performing peptide mass fingerprinting. The described work was supported by the Deutsche Forschungsgemeinschaft (SFB 635 TP 8).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0660) on October 5, 2005.
References
- Alberti, S., Demand, J., Esser, C., Emmerich, N., Schild, H., and Höhfeld, J. (2002). Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 277, 45920-45927. [DOI] [PubMed] [Google Scholar]
- Alberti, S., Böhse, K., Arndt, V., Schmitz, A., and Höhfeld, J. (2004). The co-chaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell 15, 4003-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger, C. A., Connell, P., Wu, Y., Hu, Z., Thompson, L. J., Yin, L.Y., and Patterson, C. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo, C. P., Michaud, C., Ost, M. C., Fliss, A. E., Yang, E., Patterson, C., Hall, S. J., and Caplan, A. J. (2003). C-terminal Hsp-interacting protein slows androgen receptor synthesis and reduces its rate of degradation. Arch. Biochem. Biophys. 410, 134-140. [DOI] [PubMed] [Google Scholar]
- Connell, P., Ballinger, C. A., Jiang, J., Wu, Y., Thompson, L. J., Höhfeld, J., and Patterson, C. (2001). The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93-96. [DOI] [PubMed] [Google Scholar]
- Dai, Q. et al. (2003). CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22, 5446-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Q. et al. (2005). Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J. Biol. Chem. (in press). [DOI] [PubMed]
- Demand, J., Lüders, J., and Höhfeld, J. (1998). The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol. Cell. Biol. 18, 2023-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demand, J., Alberti, S., Patterson, C., and Höhfeld, J. (2001). Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11, 1569-1577. [DOI] [PubMed] [Google Scholar]
- Esser, C., Alberti, S., and Höhfeld, J. (2004). Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta 1695, 171-188. [DOI] [PubMed] [Google Scholar]
- Esser, C., Scheffner, M., and Höhfeld, J. (2005). The chaperone associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J. Biol. Chem. 280, 27443-27448. [DOI] [PubMed] [Google Scholar]
- Farinha, C. M., Nogueira, P., Mendes, F., Penque, D., and Amaral, M. D. (2002). The human DnaJ homolgue (Hdj)-1/heat-shock protein (Hsp) 40 cochaperone is required for the in vivo stabilization of the cystic fibrosis transmembrane conductance regulator by Hsp70. Biochem. J. 366, 797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gässler, C. S., Wiederkehr, T., Brehmer, D., Bukau, B., and Mayer, M. P. (2001). Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 276, 32538-32544. [DOI] [PubMed] [Google Scholar]
- Höhfeld, J., Minami, Y., and Hartl, F. U. (1995). Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell 83, 589-598. [DOI] [PubMed] [Google Scholar]
- Höhfeld, J., and Jentsch, S. (1997). GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 16, 6209-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld, J., Cyr, D. M., and Patterson, C. (2001). From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2, 885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Ballinger, C. A., Wu, Y., Dai, Q., Cyr, D. M., Höhfeld, J., and Patterson, C. (2001). CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276, 42938-42944. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Cyr, D. M., Babbitt, R. W., Sessa, W. C., and Patterson, C. (2003). Chaperone-dependent regulation of endothelial nitric-oxide synthase intracellular trafficking by the co-chaperone/ubiquitin ligase CHIP. J. Biol. Chem. 278, 49332-49341. [DOI] [PubMed] [Google Scholar]
- Kampinga, H. H., Kanon, B., Salomons, F. A., Kabakov, A. E., and Patterson, C. (2003). Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells. Mol. Cell. Biol. 23, 4948-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito, R. R. (1999). Biosynthesis and degradation of CFTR. Physiol. Rev. 79, S167-S173. [DOI] [PubMed] [Google Scholar]
- Kostova, Z., and Wolf, D. H. (2003). For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22, 2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, H. A. et al. (2004). Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 23, 282-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman, J., Ganesan, S., Dohm, C. P., Krajewski, S., Reed, J. C., Bahr, M., Wouters, F. S., and Kermer, P. (2005). Interaction of BAG-1 and Hsp70 mediates neuroprotectivity and increases chaperone activity. Mol. Cell. Biol. 25, 3715-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders, J., Demand, J., and Höhfeld, J. (2000a). The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275, 4613-4617. [DOI] [PubMed] [Google Scholar]
- Lüders, J., Demand, J., Papp, O., and Höhfeld, J. (2000b). Distinct isoforms of the cofactor BAG-1 differentially affect Hsc70 chaperone function. J. Biol. Chem. 275, 14817-14823. [DOI] [PubMed] [Google Scholar]
- Meacham, G. C., Lu, Z., King, S., Sorscher, E., Tousson, A., and Cyr, D. M. (1999). The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 18, 1492-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham, G. C., Patterson, C., Zhang, W., Younger, J. M., and Cyr, D. M. (2001). The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3, 100-105. [DOI] [PubMed] [Google Scholar]
- Murata, S., Minami, Y., Minami, M., Chiba, T., and Tanaka, K. (2001). CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2, 1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolay, R., Wiederkehr, T., Rist, W., Kramer, G., Mayer, M. P., and Bukau, B. (2004). Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem. 279, 2673-2678. [DOI] [PubMed] [Google Scholar]
- Nollen, E. A., Kabakov, A. E., Brunsting, J. F., Kanon, B., Höhfeld, J., and Kampinga, H. H. (2001). Modulation of in vivo HSP70 chaperone activity by Hip and Bag-1. J. Biol. Chem. 276, 4677-4682. [DOI] [PubMed] [Google Scholar]
- Plenefisch, J. D., Zhu, X., and Hedgecock, E. M. (2000). Fragile skeletal muscle attachments in dystrophic mutants of Caenorhabditis elegans: isolation and characterization of the mua genes. Development 127, 1197-1207. [DOI] [PubMed] [Google Scholar]
- Qu, B. H., Strickland, E. H., and Thomas, P. J. (1997). Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J. Biol. Chem. 272, 15739-15744. [DOI] [PubMed] [Google Scholar]
- Rüdiger, S., Germeroth, L., Schneider-Mergener, J., and Bukau, B. (1997). Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidowsky, J., Dodt, G., Kirchberg, K., Wegner, A., Nastainczyk, W., Kunau, W. H., and Schliebs, W. (2001). The di-aromatic pentapeptide repeats of the human peroxisome import receptor PEX5 are separate high affinity binding sites for the peroxisomal membrane protein PEX14. J. Biol. Chem. 276, 34524-34529. [DOI] [PubMed] [Google Scholar]
- Sato, S., Ward, C. L., Krouse, M. E., Wine, J. J., and Kopito, R. R. (1996). Glycerol reverses the misfolding phenotype of the most common cystic fibrosis mutation. J. Biol. Chem. 271, 635-638. [DOI] [PubMed] [Google Scholar]
- Shimura, H., Schwartz, D., Gygi, S. P., and Kosik, K. S. (2004). CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 279, 4869-4876. [DOI] [PubMed] [Google Scholar]
- Sondermann, H., Scheufler, C., Schneider, C., Höhfeld, J., Hartl, F. U., and Moarefi, I. (2001). Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553-1557. [DOI] [PubMed] [Google Scholar]
- Strickland, E., Qu, B. H., Millen, L., and Thomas, P. J. (1997). The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 272, 25421-25424. [DOI] [PubMed] [Google Scholar]
- Takayama, S., Xie, Z., and Reed, J. C. (1999). An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J. Biol. Chem. 274, 781-786. [DOI] [PubMed] [Google Scholar]
- Ueda, K., Kosako, H., Fukui, Y., and Hattori, S. (2004). Proteomic identification of Bcl2-associated athanogene 2 as a novel MAPK-activated protein kinase 2 substrate. J. Biol. Chem. 279, 41815-41821. [DOI] [PubMed] [Google Scholar]
- Westhoff, B., Chapple, J. P., van der Spuy, J., Höhfeld, J., and Cheetham, M. E. (2005). HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr. Biol. 15, 1058-1064. [DOI] [PubMed] [Google Scholar]
- Xu, W., Marcu, M., Yuan, X., Mimnaugh, E., Patterson, C., and Neckers, L. (2002). Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA 99, 12847-12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Janich, S., Cohn, J. A., and Wilson, J. M. (1993). The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl. Acad. Sci. USA 90, 9480-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger, J. M., Ren, H. Y., Chen, L., Fan, C. Y., Fields, A., Patterson, C., and Cyr, D. M. (2004). A foldable CFTR deltaF508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J. Cell Biol. 167, 1075-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]