Abstract
Recruitment of TATA-binding protein (TBP) is central to activation of transcription by RNA polymerase II (pol II). This depends upon co-activator proteins including TBP-associated factors (TAFs). Yeast Mot1p was identified as a general transcriptional repressor in genetic screens and is also found associated with TBP. To obtain insight into Mot1p function in vivo, we determined the mRNA expression profile of the mot1-1 temperature-sensitive (Ts) strain. Unexpectedly, this indicated that Mot1p mostly plays a positive role for transcription. For one potential activation target, HXT2, we analyzed promoter recruitment of Mot1p, TBP, Taf1p (Taf130p) and pol II by chromatin immunoprecipitation assays. Whereas TBP becomes stably associated upon activation of the HXT2 and HXT4 promoters, Mot1p showed only a transient association. TBP recruitment was compromised in two different mot1 mutant strains, but was only moderately affected in a taf1 Ts strain. Together, our data indicate that Mot1p can assist in recruitment of TBP on promoters during gene activation in vivo.
Keywords: Mot1p/RNA polymerase II/TBP/TFIID/transcription activation
Introduction
The TATA-binding protein (TBP) is a key factor in eukaryotic transcription (Struhl, 1994). TBP is involved in the early steps of transcriptional initiation by all three nuclear RNA polymerases and its activity is subject to multiple levels of regulation. In yeast cells, TBP is associated with several proteins or protein complexes, such as TFIID, NC2, Mot1p, and in a less stable manner, with the SAGA complex (Lee and Young, 1998; Pugh, 2000). TFIID consists of TBP in complex with 14 different TBP-associated factors (TAFs; Sanders and Weil, 2000) and is considered the major active form of TBP for transcription by RNA polymerase II (pol II). In vivo protein–DNA cross-linking studies showed that yeast promoters have distinct requirements for TAFs (Kuras et al., 2000; Li et al., 2000). Interestingly, TAFs are under-represented in relation to TBP on certain promoters, suggesting that TBP is not present in the form of TFIID on those promoters.
NC2 and Mot1p were initially isolated as general repressors of transcription (Meisterernst and Roeder, 1991; Davis et al., 1992; Wade and Jaehning, 1996). However, recent analyses indicate that NC2 and Mot1p may play a role in activation as well (Collart, 1996; Madison and Winston, 1997; Prelich, 1997; Willy et al., 2000; Geisberg et al., 2001). How the pool of TBP is distributed through Mot1p or NC2 to active promoters remains an open question.
The MOT1 gene was isolated in yeast by screening for mutations that increase the basal transcription of several unrelated promoters, including the core promoters of CYC1 and SUC2 (Davis et al., 1992; Prelich and Winston, 1993). A significant amount of cellular Mot1p is associated with TBP and the Mot1p–TBP complex appears to be of similar abundance as the TFIID complex (Poon et al., 1994). Mot1p contains a C-terminal ATPase domain of the SWI2-superfamily type (Davis et al., 1992) and pairs of putative HEAT repeats within its N-terminal third (Adamkewicz et al., 2001). Analysis of the human ortholog of Mot1p, BTAF1(TAFII170) [we use the new nomenclature for TAFIIs (Tora, 2002)], indicates that these HEAT repeats can mediate interaction with TBP (Pereira et al., 2001). It was shown that Mot1p is able to associate with TBP on TATA elements in vitro and that TBP–TATA interaction is disrupted upon Mot1p-mediated ATP hydrolysis (Auble et al., 1994). This suggests that Mot1p exerts its repressive effect in this way (Darst et al., 2001). Interestingly, experiments performed with crude yeast extracts indicate a moderate stimulatory effect of Mot1p on the transcription of genes like HIS4 or ACT1 (Muldrow et al., 1999). This effect is dependent on Mot1p concentration and was proposed to derive from the redistribution of TBP from non-promoter to promoter sites. Consistent with this result, RNA analysis of mot1 mutant strains suggested a positive role for Mot1p in the transcription of the HIS3, HIS4 and GAL1 genes (Collart, 1996; Prelich, 1997). Genetic links have also been reported between Mot1p and other regulatory factors, such as components of the NOT–CCR4 complex and the Spt3p subunit of SAGA (Collart, 1996; Madison and Winston, 1997). However, the precise in vivo role of Mot1p in transcription activation remains unclear and may result in an indirect manner from a Mot1p-mediated mobilization of TBP within the yeast cell.
An in vivo study into Mot1p function requires identification of bona fide Mot1p target genes. Towards this end, we first determined the mRNA expression profile of a temperature-sensitive (Ts) mot1 mutant strain under non-permissive conditions. From this we selected potential Mot1p target promoters to investigate direct binding of Mot1p to these promoters in vivo using the chromatin immunoprecipitation (ChIP) assay. Under normal vegetative growth conditions, no significant promoter association of Mot1p could be detected. One of the potential Mot1p activation targets is HXT2, which encodes a high-affinity hexose transporter. The gene belongs to the HXT gene family, which in yeast comprises 18 members with different affinities to glucose (Ozcan and Johnston, 1999; Wieczorke et al., 1999). Expression of these genes is dependent upon glucose concentration in the media.
Here we show a transient binding of Mot1p to several HXT promoters, which coincides with their activation and TBP recruitment. Importantly, we have obtained evidence for an impaired association of TBP and reduced transcription activation of HXT2 and HXT4 in two independent mot1 mutant strains. Finally, we investigated the very early steps of activation and found a very rapid co-recruitment of TBP with Taf1p and Mot1p. Unlike Mot1p, Taf1p does not appear to play a critical role in TBP recruitment. Based on these results, we propose that on certain promoters Mot1p can act to escort or deliver TBP during transcription activation.
Results
Expression profiling with DNA microarrays identifies potential Mot1p targets
To investigate Mot1p function in vivo, it was important to identify Mot1p targets. Therefore, we decided to compare the mRNA expression profile of the mot1-1 mutant Ts strain with its isogenic wild-type strain using microarrays containing oligonucleotide probes. The overall analysis of the mot1-1 transcription profile indicates that 7% of yeast genes are downregulated >2-fold in mutant cells shifted to the non-permissive temperature, whereas 4% are upregulated in comparison with an isogenic wild-type strain (Table I; Supplementary figure 1, available at The EMBO Journal Online). Among the MOT1-dependent genes identified in this screen, some had been described previously. For example, the HIS4 gene (3-fold down), several transposons (2- to 3-fold down), the MFA2 gene (2.5-fold up) and the SSA4 gene (3.5-fold up) are known to be affected by Mot1p inactivation (Davis et al., 1992; Collart, 1996; Madison and Winston, 1997). During preparation of this manuscript, Auble and co-workers determined the expression profile of another mot1 Ts strain, mot1-14 (Dasgupta et al., 2002). They employed cDNA microarrays and found that a smaller proportion (3%) is under MOT1 control and only six genes showed a decrease in expression (Dasgupta et al., 2002). As expected, the mot1-1 data display a very significant overlap with the mot1-14 data (Supplementary figure 1). In addition to these, we also identified several new potential targets for Mot1p (Table I; see below). The complete data set is available at http://ruummc.med.uu.nl/research/publications/pub_txpn.html.
Table I. Selection of target genes for the Mot1p transcription factor.
| Expression level changed >2-fold | Signals within the linear response range | Change in expression mirrored in rpb1-1 | Change in expression >4-fold | Promoters separated by >1.5 kb | |
|---|---|---|---|---|---|
| Upregulated genes |
258 (4%) |
81 |
38 |
16 |
CMK1, NCA3, PRY1, RPI1 |
| Downregulated genes | 459 (7%) | 207 | 64 | 13 | CBP3, CFT13, HXT2, YOR390W |
Data resulting from comparison of 6365 transcripts of the mot1-1 and its isogenic wild-type strain after a shift of 45 min at non-permissive temperature (37°C). Criteria applied for the selection of potential target genes are indicated in the first row. The number of genes selected at each step is listed. The corresponding percentage of the yeast genome is indicated in parentheses.
Comparison of our mot1-1 data set with other data sets indicated significant overlap with BUR6, encoding one of the NC2 subunits. This overlap was for both down- and upregulated genes, which is in agreement with the proposal that Mot1p and NC2 provide redundant roles in vivo (Prelich, 1997; Lemaire and Collart, 2000). We also noted significant overlap with Taf1p- and Srb10p-regulated genes. Auble and co-workers reported similar overlaps when comparing their mot1-14 data set (Dasgupta et al., 2002).
In conclusion, the mRNA profiling results of the mot1-1 strain are in accordance with the hypothesis that Mot1p is both involved in activation and repression of transcription.
DNA–protein cross-linking indicates transient binding of Mot1p during activation of the HXT2 and HXT4 promoters
A complication in the interpretation of expression profiling data is that changes in mRNA levels may be an indirect consequence of transcription factor inactivation. ChIP (Hecht and Grunstein, 1999) directly determines whether a transcription factor is associated with a particular promoter. We used the mot1-1 expression profile to select potential targets for ChIP analysis. In this, the following stringent criteria were applied: (i) values obtained in the profiling analysis for the target gene should be clearly above the sensitivity threshold; (ii) the target should be affected similarly in the rpb1-1 strain harboring a Ts form of the largest subunit of pol II; (iii) expression should be altered >4-fold; and finally (iv) the potential target promoter should be separated by >1.5 kb from flanking promoters. Applying these criteria, we selected a total of eight potential targets for in vivo Mot1p binding studies (Table I).
To isolate Mot1p–DNA complexes, we employed a yeast strain expressing a HA3-tagged version of Mot1p from the MOT1 chromosomal locus (Poon et al., 1994). This strain was grown under standard rich media conditions. Log-phase cells were cross-linked and processed for ChIP analysis. Using this approach, we failed to detect significant binding of Mot1p to several putative target promoters, such as NCA3, RPI1 and HXT2 (data not shown). This could imply either that these promoters are not bona fide targets for Mot1p action or that Mot1p is not stably associated with the promoters under the conditions employed. The latter possibility is supported by biochemical analyses, which indicate that Mot1p may only interact for a brief period with TBP-bound promoter DNA in the presence of ATP (Darst et al., 2001). Therefore, we decided to follow Mot1p and TBP binding to promoter sequences during changes in transcriptional activity.
The mRNA profiling data indicated that HXT2 gene expression is decreased 5.8-fold in the mot1-1 strain. This gene encodes a high-affinity glucose transporter and belongs to the hexose transporter (HXT) gene family, consisting of 18 members in yeast (Ozcan and Johnston, 1999; Wieczorke et al., 1999). Probably, HXT2 was not detected as a Mot1p-dependent gene in the mot1-14 analysis (Dasgupta et al., 2002), because microarrays containing open reading frames would not discriminate between the homologous members of HXT family. Under low glucose concentrations, HXT2, HXT4 and HXT6 become expressed at increased levels. HXT3 encodes an intermediate affinity transporter, which shows little regulation by glucose. On the other hand, the low-affinity Hxt1p transporter is only expressed at high glucose concentrations (Ozcan and Johnston, 1999). In the mot1-1 data set, HXT3 and HXT4 mRNA levels are reduced 1.8- and 2.0-fold, respectively, and HXT1 is not affected.
Using ChIP, we analyzed TBP and Mot1p recruitment to HXT2 and other HXT promoters after cells had been shifted from 4 to 0.1% glucose in synthetic complete (SC) media (Figure 1A). The PGK1 promoter was included as a positive control. PGK1 is transcribed constitutively at high levels and has been classified as TAF independent (Kuras et al., 2000; Li et al., 2000). All ChIP analyses were performed by multiplex PCR and included a pair of POL1 primers, which amplifies an intragenic fragment of POL1 (Kuras et al., 2000) and serves as a negative control.
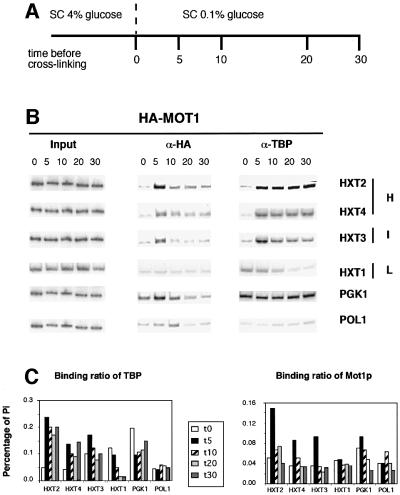
Fig. 1. Association of TBP and Mot1p to promoters in vivo after a glucose concentration shift. (A) Schematic representation of the experimental procedure. HA3-MOT1 cells were grown in 4% glucose SC medium. Log-phase cells were either immediately cross-linked (t = 0) with 1% formaldehyde or rinsed and transferred to a medium containing 0.1% glucose for the indicated times, followed by cross-linking. (B) PCR analysis of input and immunoprecipitated DNA during the glucose concentration shift. In each case, several amounts of input material were used to ensure that PCR signals were in the linear range of the reaction. Immunoprecipitated DNA was analyzed by multiplex PCRs including the POLI pair as the internal control. Panels for individual promoters are shown and represent similar exposures as normalized on the POLI control. The glucose affinity of each HXT transporter (H, high; I, intermediate; L, low) is represented on the right. Time is indicated in minutes. (C) PhosphorImager quantification of the data presented in (B). Pi value represents the immunoprecipitated signal divided by its corresponding input.
When cells are grown in 4% glucose (t = 0 time point), Mot1p association was not above background levels (i.e. POL1) for any of the HXT promoters tested (Figure 1B). Significant binding of TBP is observed only at the HXT3, PGK1 and HXT1 promoters, which are transcriptionally active under these conditions. At 5 min after the glucose shift, TBP was recruited to the HXT2, HXT4 and (to a lesser extent) HXT3 promoters (Figure 1B and C). TBP remained stably associated with these promoters for the duration of the experiment. The increased occupancy of TBP correlated well with the kinetics of transcription induction (see below; data not shown). As expected, TBP levels progressively decrease at the HXT1 promoter. Surprisingly, Mot1p associates with the HXT2, HXT3 and HXT4 promoters at 5 min after the glucose shift. In contrast to TBP, Mot1p binding returns to background levels rapidly after the shift. Similar observations were made in rich media (Supplementary figure 3). Occupancy of Mot1p at the HXT1 promoter does not show any variation. Given the observation that Mot1p removes TBP from promoter DNA in vitro (Auble et al., 1994), one could have expected Mot1p association during HXT1 inactivation. However, HXT1 expression is not affected, as indicated by mRNA profiling of the mot1-1 strain (data not shown), which indicates that it is not a direct Mot1p target. We also detected minor variations of TBP and Mot1p association at the constitutively active PGK1 (Figure 1B) and RPS5 (data not shown) genes. At 5 min after shifting to 0.1% glucose, TBP levels decrease and return to their initial t = 0 level after 30 min. Mot1p displays an inverse pattern on these promoters. Taken together, these observations of Figure 1B indicate that promoter binding by Mot1p is transient and support the hypothesis that Mot1p is directly involved in the activation of specific genes.
TBP and Mot1p co-localize on the HXT2 promoter during transcription activation
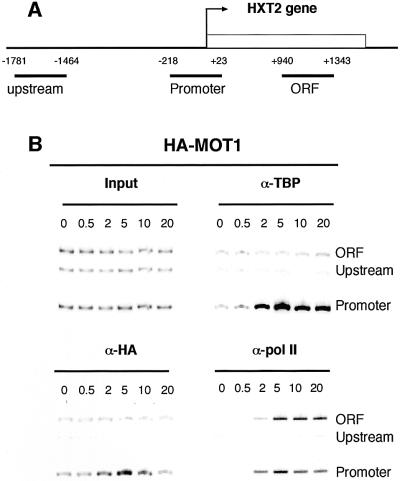
A limitation of the ChIP technique is that it does not determine the DNA binding site of a transcription factor with high resolution. Although we verified that DNA fragments are sheared to a median size of <500 bp, this does not guarantee that TBP and Mot1p associate to the same promoter region. It is also possible that during the shift from 4 to 0.1% glucose, Mot1p serves to clear a repressed promoter before TBP could associate and drive activation. These possibilities were investigated in Figure 2B. Association of Mot1p and TBP was determined very early after the glucose shift, and different parts of the HXT2 locus were scanned for Mot1p, TBP and pol II association. We designed primer pairs for the upstream region and the body of the HXT2 gene (Figure 2A) and employed them in the ChIP assay. Figure 2B shows that within 2 min after glucose shift, binding of Mot1p and TBP can be detected on the HXT2 gene, but only on the promoter fragment spanning the putative TATA box (–122/–116; Ozcan and Johnston, 1996). This indicates that within the resolution of this assay, Mot1p and TBP seem to bind to the same part of the promoter. In this experiment, we modified the glucose-shift procedure to be able to observe changes in transcription factor binding immediately after glucose shift (see Materials and methods). Because of this, 5–7 min should be added to allow a direct comparison of the time course of Figure 1B. Figure 2B shows that promoter binding by Mot1p and TBP already occurs at 2 min after the glucose shift. In addition, we find that TBP arrives to the HXT2 promoter at the same moment as Mot1p. We also find that association of pol II with the HXT2 promoter is very rapid and, as expected, precedes pol II presence on the gene-internal fragment of HXT2 (Figure 2B, compare t = 2 with t = 5). In conclusion, this experiment shows that upon activation of the HXT2 promoter Mot1p arrives at the same place and time as TBP. This suggests that a Mot1p–TBP binary complex might deliver TBP to this gene during transcription activation, which is consistent with their association in cell extracts (Poon et al., 1994; Wade and Jaehning, 1996).
Fig. 2. TBP and Mot1p specifically co-localize on the promoter of HXT2. (A) Location of the PCR products on the HXT2 genomic region used for multiplex PCR analysis. The start site of translation is indicated by +1. (B) Using a slighty modified procedure as compared with Figure 1 (see Materials and methods), cells were transferred from 4 to 0.1% glucose for the indicated times (in minutes) before cross-linking, extract preparation and immunoprecipitation. Analysis of the corresponding input samples indicated that the signals were in the linear range of the PCR.
Stable recruitment of TBP to the HXT2 promoter is impaired by mot1 mutations
To elaborate Mot1p involvement in activation of the HXT2 promoter, we made use of mot1 Ts strains. The growth rate of these strains is significantly reduced at 30°C, indicating that Mot1p function is already disturbed under these conditions. Extracts were prepared for ChIP analysis from mot1 mutant strains and their isogenic controls at different times after glucose shift according to the scheme of Figure 1A. RNA was isolated from parallel cultures for analysis on northern blots.
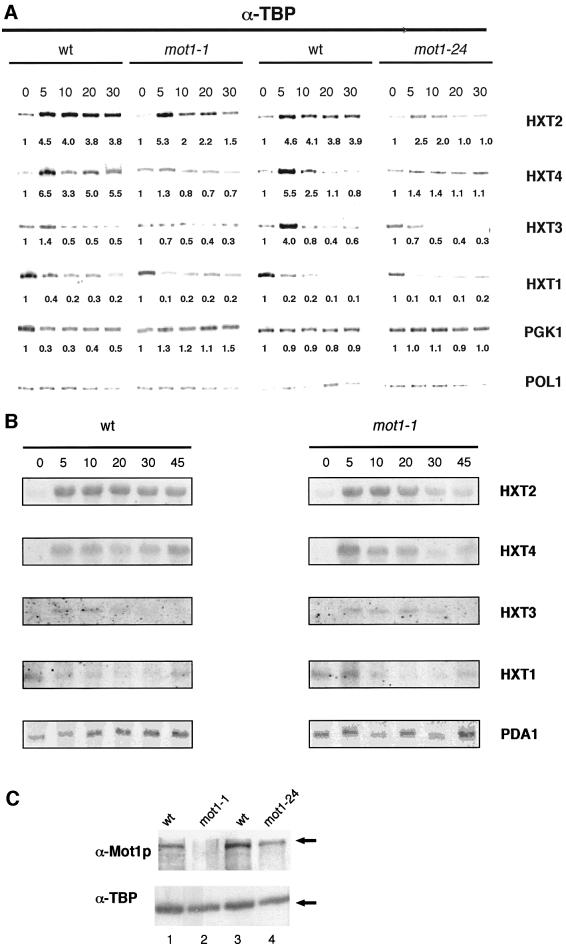
At 5 min after glucose shift, TBP is efficiently recruited to the HXT2 promoter in both the wild-type and the mot1-1 strain (Figure 3A). Although recruited efficiently, TBP did not remain stably associated to HXT2 in the mot1-1 strain, as indicated by the later time points. Paradoxically, this suggests that functional Mot1p is required for a stable association of TBP to HXT2 promoter, even when Mot1p itself has already left the promoter (Figures 1B and 2B). The failure of TBP to stably associate could be indirect as the mot1-1 mutation leads to a strong decrease in the level of wild-type Mot1p protein (J.L.Davis, C.G.F.Mueller, K.E.Hansen, J.I.Adamkewicz and J.Thorner, personal communication; Figure 3C). To test this, we analyzed the mot1-24 allele, which contains a missense mutation (K1509R; Madison and Winston, 1997). In this strain, the transient increase in TBP binding on the HXT2 promoter is impaired (compared with the wild-type control strain, ∼50% of TBP is recruited at 5 min in mot1-24). Also, in this mutant TBP does not become stably associated with the HXT2 promoter (Figure 3A). We have attempted to directly monitor Mot1p occupancy in these mutant strains, but have been unable to obtain Mot1p-specific antibodies functional in ChIP assays. The experiment in Figure 3A also showed that the reduced level of TBP at the PGK1 promoter at 5 min after glucose shift, observed in the wild-type and HA3-MOT1 (Figure 1) strains, does not occur in the mot1-1 strain. Our observations are in agreement with previous results, which indicated that TBP repartition is impaired by the mot1-1 mutation (Li et al., 1999). Furthermore, induction of TBP association to HXT4 is also impaired in mot1-1 and mot1-24. Differences between the wild-type controls could be due to HXT4 promoter polymorphisms between strains, leading to differences in its regulation (Diderich et al., 1999). We have not explored these differences further. TBP association to the HXT1 promoter is largely unaffected by mutations in MOT1. In the case of HXT3, a transient increase in TBP association can be detected at 5 min after glucose shift, which is most pronounced in the wild-type control to mot1-24. Interestingly, the increase in TBP association is absent in both mot1 strains.
Fig. 3. Both TBP occupancy and transcription of HXT genes are impaired in strains carrying mot1 mutant alleles. (A) ChIP analysis of TBP binding in the mot1-1 and mot1-24 strains. The experimental design is similar to Figure 1. The respective cultures grown at the permissive temperature were sampled at t = 0, 5, 10, 20 and 30 min after a shift from 4 to 0.1% glucose media. The numbers below the autoradiograms represent the fold change in TBP occupancy relative to the t = 0 control. (B) RNA expression of HXT genes upon glucose shift. Cells were grown as in (A), harvested and frozen before RNA extraction. An additional time point (45 min after glucose shift) was collected. Specific mRNAs were detected using specific oligonucleotide probes, described previously (Diderich et al., 1999). PDA1 transcripts were used as an internal control. (C) Immunoblot analysis of Mot1p and TBP in chromatin preparations. Cells were cultured in SC 4% glucose and chromatin was prepared from KY804 (lane 1), MY603 (lane 2), FY142 (lane 3) and FY1211 (lane 4). After cross-link reversal, immunoblots were prepared and analyzed with anti-TBP and anti-Mot1p antisera. The arrows indicate positions of TBP and Mot1p.
Analysis of mRNA levels of the HXT genes in the different strains (Figure 3B; data not shown for mot1-24) demonstrated that HXT mRNA levels closely follow TBP recruitment. Induction of HXT2 and HXT4 mRNAs occurs within 5 min after glucose shift. Maximal induction of HXT2 mRNA is 30-fold in wild-type cells and 17-fold in the mot1-1 strain. Importantly, and as expected, accumulation of HXT2 and HXT4 mRNA is affected in the mot1-1 strain (after 45 min, HXT2 mRNA levels are 6.5-fold lower in mot1-1). These observations are in close agreement with previous ChIP studies, which showed a close correlation of promoter binding by TBP with the expression level of the gene (Kuras and Struhl, 1999; Li et al., 1999). We determined levels of Mot1p and TBP in crude extracts (data not shown) and chromatin preparations of the mot1 strains (Figure 3C). As expected, we found that Mot1p can be readily detected in mot1-24 cells, but not in mot1-1 cells. TBP levels are similar in all chromatin preparations.
Taken together, our data support a model invoking direct involvement of Mot1p in the process of activation and stable binding of TBP to the HXT2 and HXT4 promoters.
TFIID participates in the activation of HXT2 and HXT4 genes
TFIID is considered a major active form of TBP for pol II transcription. Previously, it was shown that certain yeast promoters are relatively insensitive to conditional inactivation of TFIID-specific TAFs (for review, see Chao and Young, 1996). Subsequently, at these TAF-independent promoters, TFIID-TAFs are under-represented relative to TBP (Kuras et al., 2000; Li et al., 2000). Given these observations, we tested whether TFIID-TAFs are involved in the activation of the HXT genes. Promoter association of several TFIID-specific TAFs, like Taf1p (Taf130/145p), Taf2p (Taf150p) and Taf11p (Taf40p), is comparable (Kuras et al., 2000; Li et al., 2000) and therefore we focused on Taf1p and used this subunit as a specific marker for TFIID.
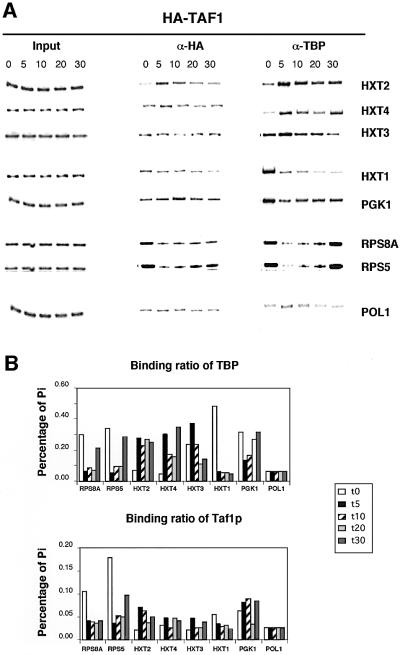
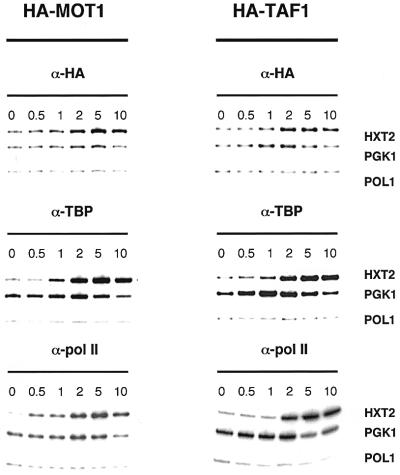
Following the experimental scheme of Figure 1A, we analyzed variation in promoter association of both TBP and Taf1p during a glucose concentration shift using a yeast strain expressing a HA3-tagged version of TAF1. The tagged protein is expressed from its own promoter on a low-copy plasmid in taf1-null cells. The data of Figure 4A show that association of Taf1p to the HXT2 and (to a lesser extent) HXT4 promoter is increased after glucose shift. Binding of Taf1p to the HXT3 promoter does not alter, whereas binding to HXT1 decreases. TBP binding to HXT promoters parallels binding of Taf1p and is followed closely by pol II association (Supplementary figure 2). It is important to note that Taf1p and TBP binding differ in magnitude, which becomes clear after similar analysis of the TAF-dependent RPS5 and RPS8A promoters (Kuras et al., 2000; Li et al., 2000). We detect substantial binding of Taf1p and TBP to both promoters during exponential growth in YPD medium (data not shown) and in SC media (Figure 4, t = 0 time point). When compared with the ratio of Taf1p/TBP on RPS5 and RPS8A promoters (Figure 4B), Taf1p is under-represented on HXT promoters. Accordingly, expression profiling of a taf1 Ts strain indicated that expression of the HXT1, 2, 3 and 4 genes is relatively insensitive to Taf1p inactivation (1.3-, 1.6-, 1.3- and 1.5-fold reduced, respectively), whereas RPS5 and RPS8A decrease 1.9- and 3.8-fold (Holstege et al., 1998). Unexpectedly, TBP binding to RPS5 and RPS8A fell at early time points after glucose shift, before returning to a level roughly equivalent to that prior to the shift. This is reproducibly observed in several experiments. The PGK1 gene was also included in the experiment of Figure 4. Again, we find a transient fall in TBP binding to PGK1 after glucose shift. In rich media, the PGK1 promoter has been reported to be TAF independent (Kuras et al., 2000), but we observe only 3- and 6-fold lower levels of Taf1p than TBP on PGK1 compared with the TAF-dependent RPS5 and RPS8A promoters, respectively. Based on comparison with PGK1, the HXT genes analyzed here could be classified as TAF-independent genes. Remarkably, the Taf1p/TBP ratio increases transiently, which agrees with the observation that TAF dependence can vary during growth conditions changes (Kuras et al., 2000).
Fig. 4. Kinetics of Taf1p binding to promoters in vivo after a glucose concentration shift. (A) ChIP analyses of TBP and Taf1p promoter occupancy using a yeast strain carrying a HA3-tagged TAF1 gene were performed as described in Figure 1B. RPS5 and RPS8A are used as standards for TAF-dependent genes, whereas PGK1 is TAF independent. (B) PhosphorImager quantification of the immunoprecipitation efficiency. This is expressed as Pi, which was determined as in Figure 1C.
In conclusion, our analysis of the HA3-tagged TAF1 strain shows weak but significant association of Taf1p to the HXT promoters, which suggests involvement of TFIID. However, the ratio of Taf1p to TBP is lower for HXT promoters than for cognate TAF-dependent promoters such as RPS5 and RPS8A.
Analysis of early events of Mot1p, TBP and Taf1p binding to HXT2
Our analyses indicate that transcriptional activation of the HXT2 gene is an extremely rapid process (Figures 1 and 2B). After having shown that TFIID is also recruited to the activated HXT2 promoter, it becomes important to compare the kinetics of recruitment of Taf1p and Mot1p very early after the glucose shift. Possibly, recruitment of these factors is a sequential event. To this end, we employed the ‘rapid glucose shift’ protocol of Figure 2 and compared the HA3-MOT1 and HA3-TAF1 strains in ChIP experiments. Recruitment of TBP, Mot1p and Taf1p to the HXT2 and PGK1 promoters was followed in parallel (Figure 5). Maximal recruitment of all three factors to the HXT2 promoter occurs at 5 min after glucose shift. In two independent experiments and compared with 4% glucose growth, levels for Mot1p, Taf1p and TBP are induced 3- to 4-fold, 3-fold and 8- to 10-fold, respectively. Careful comparison of the timing of transcription factor binding indicates that Mot1p and Taf1p binding coincides with TBP recruitment, which is closely followed by pol II binding. Promoter binding can be detected as early as 1 min after glucose shift. Following the peak induction at 5 min, binding of TBP and pol II to the promoter remains high (Figure 2), whereas Mot1p binding reduces to background levels again (data not shown; Figures 1 and 2). Parallel analysis of the PGK1 promoter indicates a weak induction of Mot1p binding, which precedes a small decrease in TBP and Taf1p association, as observed previously (Figures 1B, 3A and 4).
Fig. 5. Early events of HXT2 promoter activation. ChIP analysis of HA3-Mot1p, HA3-Taf1p, TBP and pol II very rapidly after a glucose concentration shift. This experiment was performed using the HA3-MOT1 (left) and HA3-TAF1 (right) strains. TBP binding was determined to check the correspondence of results between the two strains. Analysis of input extract showed linearity of the PCR (data not shown). The glucose concentration shift procedure was performed as in Figure 2.
In conclusion, this experiment shows that upon activation of the HXT2 promoter, TBP binding increases in concert with binding of both Mot1p and Taf1p to this promoter. These results suggest that TBP is delivered concomitantly with Mot1p and TFIID-TAFs.
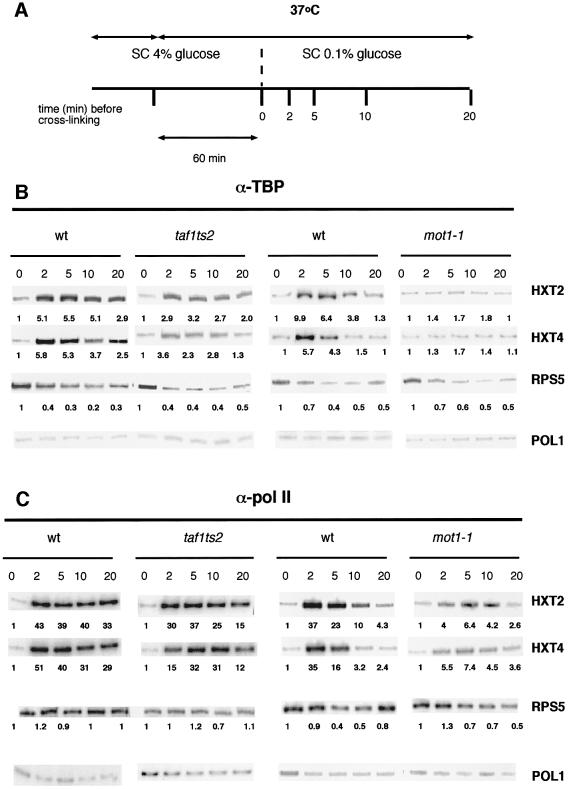
Analysis of mot1 and taf1 Ts strains at the restrictive temperature indicates that Mot1p is primarily responsible for TBP recruitment
To further investigate the relative contributions of Mot1p and Taf1p in TBP recruitment, we made use of mot1 and taf1 Ts strains. If TFIID would be the main source of TBP for HXT2 and HXT4 activation, under non-permissive conditions TBP recruitment would be expected to be blocked in the taf1ts2 strain. This strain was used previously for mRNA profiling (Holstege et al., 1998) and displays a very tight Ts phenotype. Figure 3 indicates that TBP recruitment is already impaired in mot1 mutant strains at 30°C. The mot1-1 strain was included in this experiment to investigate whether TBP recruitment was further inhibited at the non-permissive temperature. Figure 6 shows that TBP recruitment to the HXT2 and HXT4 promoters is reduced ∼2-fold in the taf1 Ts strain. In contrast, TBP recruitment to these promoters in the mot1-1 strain is severely impaired. The RPS5 and RPS8A promoters serve as controls. Binding of TBP to these promoters is reduced by the glucose shift (see also Figure 4) and by the temperature shift, but it is not affected by Mot1p inactivation. In contrast, shifting taf1ts2 to 37°C for 60 min results in a 2-fold decrease in TBP association to RPS5 and RPS8A, which is reflected in a reduced RNA accumulation (data not shown). Analysis of HXT2 and HXT4 mRNA accumulation isolated in parallel showed no reduction in the taf1ts2 strain, and pol II recruitment was only weakly affected (Figure 6C). In the mot1-1 strain, HXT2 and HXT4 mRNA levels were reduced 2- and 4-fold, respectively (data not shown), which is reflected by the impaired recruitment of pol II (Figure 6C). We also observe that TBP association to HXT2 and HXT4 is unstable in the wild-type control for the mot1-1 strain at 37°C. This is not observed in the wild-type control for the taf1ts2 strain. We assume that this relates to particular differences in the strain background as both strains grow well at 37°C.
Fig. 6. Inactivation of Mot1p, but not of Taf1p, abolishes TBP recruitment to the HXT2 and HXT4 promoters. (A) Schematic representation of the experimental procedure. Yeast cultures were rapidly shifted to 37°C prior to the glucose shift, after which cross-linking was initiated at the indicated time points. (B) TBP recruitment to the indicated genes was determined by ChIP analysis very rapidly after glucose shift in mot1-1 and taf1ts2 Ts strains under non-permissive temperatures. Yeast cells were grown in SC 4% glucose at 23°C (taf1ts2) or at 30°C (mot1-1). Log-phase cultures were shifted to 37°C for 1 h, after which the glucose concentration was reduced to 0.1% following the rapid-shift protocol of Figure 2. The numbers below the autoradiograms represent the fold change in TBP occupancy relative to the t = 0 time point. (C) Pol II recruitment to the indicated genes was determined by ChIP analysis very rapidly after glucose shift. The numbers below the autoradiograms represent the fold change in pol II occupancy relative to the t = 0 time point.
The results of Figures 4 and 6 indicate that Taf1p(TFIID) plays only a minor role in the overall level of TBP recruitment to the HXT2 and HXT4 promoters. In contrast, Mot1p plays a role both in recruitment and stabilization of TBP during activation of these promoters.
Discussion
In this study we obtained evidence for an active role of Mot1p in the process of activation of pol II transcription. Support for this comes from mRNA expression profiling using DNA microarrays and in vivo protein–DNA cross-linking assays. We find that a substantial number of genes are reduced in expression upon Mot1p inactivation. From this collection we found that the hexose transporter-encoding HXT2 and HXT4 genes are direct targets for Mot1p action. This conclusion is based on four lines of evidence. First, Mot1p becomes associated with their promoters upon transcription activation by a shift from high to low glucose concentration. Secondly, TBP recruitment to the HXT2 and HXT4 promoters and mRNA accumulation are affected in two independent mot1 mutant strains. Thirdly, we show that recruitment of Mot1p, TBP and Taf1p to the HXT2 promoter coincides with promoter activation and occurs very early after glucose shift, within 1–2 min after transcription induction. Finally, inactivation of Mot1p, but not of Taf1p, severely compromises TBP recruitment to the HXT2 and HXT4 promoters.
Potential involvement of Mot1p in transcriptional repression in vivo
The MOT1 gene has been isolated in screens for elevated expression from certain core pol II promoters (Davis et al., 1992; Prelich and Winston, 1993). Several genes are upregulated in mot1-1 cells (Davis et al., 1992; Auble et al., 1994). Subsequently, Hahn and co-workers discovered that Mot1p dissociated TBP from a TATA box in an ATP-dependent manner (Auble et al., 1994). Recent experiments on Mot1p and its human ortholog, BTAF1, have provided a molecular basis for the TBP–TATA dissociation step. We found that conserved N-terminal parts of BTAF1 can interact with the concave DNA-binding surface of TBP (Pereira et al., 2001). This result and other experiments have led to a model in which these parts of Mot1p/BTAF1 can be inserted as a wedge between TBP and the TATA box, resulting in dissociation of the complex (Darst et al., 2001; Pereira et al., 2001). In agreement with this, we find that upon Mot1p inactivation, a substantial portion of yeast genes is increased in expression. According to the wedge model, dissociation of TBP–TATA is affected in the mot1-1 strain under non-permissive conditions, which would lead to inappropriate or sustained transcription of certain genes. Very recently, Auble and co-workers detected Mot1p association to putative target promoters (Dasgupta et al., 2002). In contrast, we did not obtain in vivo evidence directly linking Mot1p-mediated dissociation to transcriptional repression. First, we failed to detect Mot1p association to the HXT1 promoter when it becomes repressed during shift from high to low glucose (Figure 1B). Secondly, we did not detect stable Mot1p associations to potential repression targets for Mot1p. Our results do not rule out the possibility that Mot1p is actively involved in transcriptional repression. In fact, we reproducibly observe a transient decrease in TBP association with the PGK1 promoter following the glucose concentration shift, which is preceded by a small increase in Mot1p association (Figures 1, 3 and 5). This decrease in TBP occupancy does not occur in the mot1 strains (Figure 3). Detection of Mot1p association during repression may be complicated by the fact that Mot1p-induced TBP–TATA dissociation is a very rapid process (O.Gumbs and P.A.Weil, manuscript in preparation). From these observations, it is clear that further experiments are needed to demonstrate a direct role for Mot1p in transcriptional repression in vivo.
A role for Mot1p in transcriptional activation in vivo
Expression profiling of mot1 Ts strains shows that expression of a significant proportion of yeast genes depends on intact Mot1p function (Table I). These data support recent suggestions that Mot1p is involved in activation of transcription (Collart, 1996; Madison and Winston, 1997; Prelich, 1997; Muldrow et al., 1999). It has been suggested that Mot1p-dependent activation may result from an increase in the pool of available TBP molecules (Muldrow et al., 1999). Our findings suggest a more direct role for Mot1p in transcriptional activation. We find that Mot1p binding to the HXT2 and HXT4 promoters coincides with both TBP and Taf1p recruitment and transcriptional activation. What might be the molecular explanation for this? It is possible that Mot1p’s role in activation concerns clearing of the HXT2 and HXT4 promoters from repressive marks, which could involve Mig1p/Tup1p/Ssn6p-induced repressive chromatin (Ozcan and Johnston, 1996). Mot1p association would allow and precede recruitment of TBP either free or in the form of TFIID. We think that this model is unlikely for several reasons. First, in our kinetic experiments analyzing very early events, we find that Mot1p association exactly coincides with TBP and Taf1p binding. Secondly, according to this model, one could expect a delay in TBP recruitment in mot1 strains, whereas we find a diminished recruitment of TBP, as shown in Figures 3 and 6. Finally, inactivation of Mot1p severely inhibits TBP recruitment, whereas inactivation of Taf1p has a more limited effect on TBP recruitment to HXT2 and HXT4.
Given the observation that a significant portion of cellular TBP exists in a preformed complex with Mot1p (Poon et al., 1994; Wade and Jaehning, 1996), it seems likely that Mot1p is recruited to the HXT2 promoter as a TBP complex. In this case, Mot1p would play a direct role in TBP recruitment. Two observations also indicate Taf1p/TFIID involvement in HXT2 activation: Taf1p also becomes associated to HXT2 during activation and, in contrast to Mot1p, TBP and Taf1p both remain bound to the activated HXT2 promoter. However, Taf1p inactivation leads only to a small decrease in TBP recruitment. Together, this indicates that TBP can be delivered to the HXT2 promoter both by Mot1p and by Taf1p(TFIID). Support for this also comes from the observation that the increase in TBP association seems more pronounced than that of Mot1p or Taf1p alone (Figure 5). ChIP analysis of extracts prepared from Ts strains at the non-permissive temperature, however, indicate that Mot1p and not Taf1p plays a critical role in TBP recruitment.
Regulation of Mot1p activity
Several important questions remain in order to understand the molecular mechanism of the involvement of Mot1p in recruitment of TBP and how this event results in promoter activation. First, how is the rapid dissociation of the TBP–TATA complex by Mot1p counteracted in vivo in the presence of ATP? In this respect, it is important to note that B-TFIID, the human equivalent of the Mot1p–TBP complex, supports in vitro pol II transcription with an efficiency equal to both TBP and TFIID (Timmers and Sharp, 1991). Possibly, other transcription factors like TFIIA (Auble et al., 1994) can repress or control Mot1p-mediated TBP–TATA dissociation activity or, alternatively, formation of the pol II pre-initiation complex in vivo is more rapid than the rate of Mot1p-mediated TBP–TATA dissociation. A second important question is to determine the transcription factor(s) responsible for gene-specific recruitment of the Mot1p–TBP complex. Unfortunately, factors involved in HXT2 activation have not been characterized in detail (Ozcan and Johnston, 1996). In vitro assays for transcription activation have shown that human B-TFIID does not respond to a number of transcription activators (Timmers and Sharp, 1991). But the mRNA expression profiling also indicates that only a selected set of activators is responsive to Mot1p function. Alternatively, cofactors essential for Mot1p/BTAF1 action may have been missing from the in vitro assays. Importantly, MOT1 and SPT3 have been found to interact genetically (Collart, 1996), and it is possible that Mot1p recruitment to HXT2 involves the SAGA complex. In contrast to Mot1p, association of SAGA (or other co-activator complexes) to the activated HXT2 promoter could be stable and thereby allow sustained TBP binding and transcription. We observed that acetylation of histone H3 tails in HXT2 chromatin increases after glucose shift, but this is independent of functional GCN5 (our unpublished data). Our identification of HXT2 as a direct in vivo target for Mot1p allows a further detailed analysis of Mot1p-dependent transcriptional activation.
Taken together, the data presented in this paper provide support for a role of Mot1p in activated transcription. Our data also provide a framework for future experiments directed at understanding the molecular details and contributions of other complexes and factors to Mot1p-mediated regulation of transcription.
Materials and methods
Strains, reagents and growth conditions
The MOT1 (DPY107) and TAF1 (YBY838) HA3-tagged strains and their parental strain (YPH 252) have been described previously (Poon et al., 1994; Bai et al., 1997). For DNA microarray analysis, the FY1214 (mot1-1) and FY98 (isogenic wild-type) strains (Madison and Winston, 1997) were employed. For ChIP analysis, we used the yeast strains MY603 (mot1-1) and KY804 (isogenic control; Collart, 1996); FY1211 (mot1-24) and FY142 (isogenic control; Madison and Winston, 1997); YSW93 (taf1ts2) and YSW87 (isogenic control; Walker et al., 1996).
Antibodies used in this study were polyclonal affinity-purified anti-yTBP, 12CA5 monoclonal antibody (mAb) (anti-HA) and 8WG16 (anti-CTD).
For the glucose concentration shift experiment, cells were grown in 2.5 l of SC medium containing 4% glucose to OD600 = 0.55. The culture (500 ml) was cross-linked immediately by addition of 1% formaldehyde (corresponding to time t = 0). The remaining cells were harvested by centrifugation for 3 min at 6000 r.p.m. in a Sorvall H6000A rotor, rinsed once in 500 ml of SC (without glucose), resuspended in 2 l of SC with 0.1% glucose and split into four 500 ml cultures, which were incubated for 5, 10, 20 and 30 min at 30°C before cross-linking.
This procedure was slightly modified for Figures 2, 5 and 6 to analyze very early events after glucose shift. Cells were centrifuged only once, resuspended in a small volume of SC 4% glucose and further diluted in SC to adjust the glucose concentration to 0.1%.
DNA microarrays analysis
RNA isolation, sample preparation, hybridization and data analysis were performed as described previously (Holstege et al., 1998). In short, two wild-type (FY98) and two mot1-1 mutant (FY1214) cultures were grown to mid-log phase at 30°C in YPD medium. The temperature was abruptly shifted to 37°C. Incubation at this temperature was continued for 45 min, after which cells were quickly harvested. Total RNA was isolated as described below. Poly(A)-tagged external control RNAs were added to equivalent amounts of total RNA. mRNA enrichment was achieved with the Oligotex buffer kit (Qiagen). Oligo(dT)-enriched RNA (2 µg) was used for generating cRNA target, which was fragmented and hybridized to Affymetrix GeneChip arrays. Initial data analysis for the potential targets was performed as described previously (Holstege et al., 1998).
ChIP
Chromatin extracts (CE) were prepared essentially as described previously (Kuras and Struhl, 1999). Sonication was set to yield an average size of genomic fragments of 300–500 bp. CE (200 or 400 µl) were used for immunoprecipitation (IP) depending on the antibody and 20 µl used for input control preparation. IPs were performed with 25 µl of protein G–agarose beads (Roche) pre-bound to 10 µg of anti-yTBP, 30 µg of anti-HA antibodies or 10 µg of anti-CTD mAb. Typical IPs were performed for 2 h at room temperature. Immune complexes were washed as described previously (Kuras and Struhl, 1999) and chromatin complexes were released from the beads by incubation in 100 µl of 10 mM Tris–HCl pH 8.0, 1 mM EDTA and 1% SDS for 15 min at 65°C. Cross-links were reversed by incubating the eluates with 400 µg of proteinase K for 2 h at 37°C and overnight at 65°C. DNA from input samples was prepared similarly. Finally, DNAs were purified from the eluates and input sample using PCR purification kit columns (Qiagen). Multiplex PCRs (including POL1 primers as internal controls and containing 3 µCi of [α-32P]dATP per reaction) were performed for 26 or 27 cycles. Reactions were carried out using 0.7 U of Taq DNA polymerase and 40 pmol of each oligonucleotide. Primer sequences are available upon request.
RNA extraction
Total RNA was purified using the hot phenol extraction procedure. Briefly, 50 ml of yeast culture (OD600 = 0.55) were pelleted by centrifugation for 2 min at 8000 r.p.m. Cell pellets were frozen in liquid nitrogen. Frozen cells were resuspended in 500 µl of acid hot phenol:chloroform (5:1, pH 4.7, 65°C) and 500 µl of TES buffer (10 mM Tris–HCl, 1 mM EDTA, 0.5% SDS). Cells were incubated for 1 h at 65°C and vortexed every 10 min for 20 s. The aqueous solution was extracted with phenol:chloroform and with chloroform:isoamyl alcohol (25:1). Finally, RNA was collected by ethanol precipitation.
Northern blotting
RNA (10 µg) was separated by electrophoresis on 1% agarose gel containing 10 mM Na phosphate pH 6.7. RNA was then transferred to a nylon membrane (Sambrook et al., 1989) and cross-linked to the membrane by UV-light irradiation. Oligonucleotide sequences and the labeling procedure used for HXT genes have been described previously (Diderich et al., 1999). A PDA1 oligonucleotide probe (5′-TTG AATGAAGCAGCAAGCATTGGC-3′) was used as internal control for mRNA variations (Wenzel et al., 1995). Pre-hybridization was carried out for 2–4 h at 45°C in 10 ml of 6× SSC, 0.1% SDS, 5× Denhardt’s solution and 100 µg/ml sonicated denatured herring sperm DNA. Hybridization was performed for 4 h to overnight using radiolabeled oligonucleotide probes. Blots were washed twice for 3 min in 6× SSC, 0.1% SDS at room temperature and once or twice for 3 min in 6× SSC, 0.1% SDS at 45°C if required. Membranes were exposed to PhosphorImager screens for analysis and quantification with a Storm 820 scanner (Molecular Dynamics) using ImageQaNT 4.2a software.
Acknowledgments
Acknowledgements
We are indebted to L.Kuras and K.Struhl for advice on the ChIP technique. We are grateful to F.Winston, J.Thorner and M.A.Collart for providing yeast strains. We also thank G.Thireos for supplying the GCN5 disruption cassette and J.Thorner for sharing unpublished information. We thank R.Young for support and encouragement of the microarray experiments. We acknowledge J.Kirchner, L.Pereira, B.Winkler and M.Klejman for comments on the manuscript. Grants from the EC to H.Th.M.T. (ERB-FMRX-CT96-0064), the NIH to P.A.W. (GM52461), the HFSPO to P.A.W. and H.Th.M.T. (RG196/98), and the Netherlands Organization for Scientific Research (NOW-MW) to C.J.C.V.O. and H.Th.M.T. (Pionier 900-98-142) supported this study.
References
- Adamkewicz J.I., Hansen,K.E., Prud’homme,W.A., Davis,J.L. and Thorner,J. (2001) High affinity interaction of yeast transcriptional regulator, Mot1, with TATA-box binding protein (TBP). J. Biol. Chem., 276, 11883–11894. [DOI] [PubMed] [Google Scholar]
- Auble D.T., Hansen,K.E., Mueller,C.G.F., Lane,W.S., Thorner,J. and Hahn,S. (1994) Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev., 8, 1920–1934. [DOI] [PubMed] [Google Scholar]
- Bai Y., Perez,G.M., Beechem,J.M. and Weil,P.A. (1997) Structure– function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N-terminus of yeast TAFII130. Mol. Cell. Biol., 17, 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao D.M. and Young,R.A. (1996) Activation without a vital ingredient. Nature, 383, 119–120. [DOI] [PubMed] [Google Scholar]
- Collart M.A. (1996) The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol., 16, 6668–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst R.P., Wang,D. and Auble,D.T. (2001) MOT1-catalyzed TBP– DNA disruption: uncoupling DNA conformational change and role of upstream DNA. EMBO J., 20, 2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Darst,R.P., Martin,K.J., Afshari,C.A. and Auble,D.T. (2002) Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl Acad. Sci. USA, 99, 2666–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.L., Kunisawa,R. and Thorner,J. (1992) A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderich J.A. et al. (1999) Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem., 274, 15350–15359. [DOI] [PubMed] [Google Scholar]
- Geisberg J.V., Holstege,F.C.P., Young,R.A. and Struhl,K. (2001) Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol., 21, 2736–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A. and Grunstein,M. (1999) Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol., 304, 399–414. [DOI] [PubMed] [Google Scholar]
- Holstege F.C.P., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Kuras L. and Struhl,K. (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature, 399, 609–613. [DOI] [PubMed] [Google Scholar]
- Kuras L., Kosa,P., Mencia,M. and Struhl,K. (2000) TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science, 288, 1244–1248. [DOI] [PubMed] [Google Scholar]
- Lee T.I. and Young,R.A. (1998) Regulation of gene expression by TBP-associated proteins. Genes Dev., 12, 1398–1408. [DOI] [PubMed] [Google Scholar]
- Lemaire M. and Collart,M.A. (2000) The TATA-binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4–Not complex and physically interacts with the Not5 subunit. J. Biol. Chem., 275, 26925–26934. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Virbasius,A., Zhu,X. and Green,M.R. (1999) Enhancement of TBP binding by activators and general transcription factors. Nature, 399, 605–609. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Bhaumik,S.R. and Green,M.R. (2000) Distinct classes of yeast promoters revealed by differential TAF recruitment. Science, 288, 1242–1244. [DOI] [PubMed] [Google Scholar]
- Madison J.M. and Winston,F. (1997) Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterernst M. and Roeder,R.G. (1991) Family of proteins that interact with TFIID and regulate promoter activity. Cell, 67, 557–567. [DOI] [PubMed] [Google Scholar]
- Muldrow T.A., Campbell,A.M., Weil,P.A. and Auble,D.T. (1999) MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol. Cell. Biol., 19, 2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S. and Johnston,M. (1996) Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol. Cell. Biol., 16, 5536–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S. and Johnston,M. (1999) Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev., 63, 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L.A., van der Knaap,J.A., van den Boom,V., van den Heuvel,F.A. and Timmers,H.T.M. (2001) TAFII170 interacts with the concave surface of TATA-binding protein to inhibit its DNA binding activity. Mol. Cell. Biol., 21, 7523–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon D., Campbell,A.M., Bai,Y. and Weil,P.A. (1994) Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)–TBP-associated factor complex distinct from transcription factor IID. J. Biol. Chem., 269, 23135–23140. [PubMed] [Google Scholar]
- Prelich G. (1997) Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2a homolog that has both positive and negative roles in transcription. Mol. Cell. Biol., 17, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G. and Winston,F. (1993) Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics, 135, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh B.F. (2000) Control of gene expression through regulation of the TATA-binding protein. Gene, 255, 1–14. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanders S.L. and Weil,P.A. (2000) Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem., 275, 13895–13900. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1994) Duality of TBP, the universal transcription factor. Science, 263, 1103–1104. [DOI] [PubMed] [Google Scholar]
- Timmers H.T.M. and Sharp,P.A. (1991) The mammalian TFIID protein is present in two functionally distinct complexes. Genes Dev., 5, 1946–1956. [DOI] [PubMed] [Google Scholar]
- Tora L. (2002) A unified nomenclature for TATA box binding protein (TBP) associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev., 16, 673–675. [DOI] [PubMed] [Google Scholar]
- Wade P.A. and Jaehning,J.A. (1996) Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol. Cell. Biol., 16, 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.S., Reese,J.C., Apone,L.M. and Green,M.R. (1996) Tran scription activation in cells lacking TAFIIs. Nature, 383, 185–188. [DOI] [PubMed] [Google Scholar]
- Wenzel T.J., Teunissen,A.W. and de Steensma,H.Y. (1995) PDA1 mRNA: a standard for quantitation of mRNA in Saccharomyces cerevisiae superior to ACT1 mRNA. Nucleic Acids Res., 23, 883–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorke R., Krampe,S., Weierstall,T., Freidel,K., Hollenberg,C.P. and Boles,E. (1999) Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett., 464, 123–128. [DOI] [PubMed] [Google Scholar]
- Willy P.J., Kobayashi,R. and Kadonaga,J.T. (2000) A basal transcription factor that activates or represses transcription. Science, 290, 982–985. [DOI] [PubMed] [Google Scholar]