Abstract
Group I introns consist of two major structural domains, the P4-P6 and P3-P9 domains, which assemble through interactions with peripheral extensions to fold into an active ribozyme. To assess group I intron folding in vivo, we probed the structure of td wild-type and mutant introns using dimethyl sulfate. The results suggest that the majority of the intron population is in the native state in accordance with the current structural model, which was refined to include two novel tertiary contacts. The importance of the loop E motif in the P7.1-P7.2 extension in assisting ribozyme folding was deduced from modeling and mutational analyses. Destabilization of stem P6 results in a deficiency in tertiary structure formation in both major domains, while weakening of stem P7 only interferes with folding of the P3-P9 domain. The different impact of mutations on the tertiary structure suggests that they interfere with folding at different stages. These results provide a first insight into the structure of folding intermediates and suggest a putative order of events in a hierarchical folding pathway in vivo.
Keywords: DMS probing in vivo/in vivo folding/loop E motif/RNA structure modeling/td group I intron
Introduction
Group I introns have to adopt a defined secondary and tertiary structure to display catalytic properties. Structure models have been derived from phylogeny and biochemical data obtained in vitro (Michel and Westhof, 1990; Michel et al., 1990; Lehnert et al., 1996) as well as from X-ray crystallography (Cate et al., 1996; Golden et al., 1998). A set of structural elements form the core structure common to all group I introns, and peripheral extensions allow their classification. The conserved core is composed of two domains, the P4-P6 (P5-P4-P6) and P3-P9 (P8-P3-P7-P9) domains, formed by coaxially stacked helices. The P3-P9 domain wraps around the P4-P6 domain, forming a cleft into which the substrate helices P1 or P10 dock. Long-range tertiary interactions connecting these domains are required to achieve the globular shape (Michel and Westhof, 1990; Jaeger et al., 1993, 1994; Costa and Michel, 1995; Lehnert et al., 1996; Tanner and Cech, 1997; Tanner et al., 1997).
In vitro RNA folding studies with the Tetrahymena intron indicated that the P4-P6 domain folds first to form a scaffold for the assembly of the P3-P9 domain (Cech, 1993; Murphy and Cech, 1993; Zarrinkar and Williamson, 1994; Laggerbauer et al., 1994; Sclavi et al., 1998). However, recent experiments suggest that the P4-P6 domain stabilizes the folded conformation of the P3-P9 domain, but is not required for its assembly (Doherty et al., 1999; Russell and Herschlag, 1999). The free energy folding landscape of the Tetrahymena group I intron was described as ‘rugged’. This landscape encompasses multiple alternative folding pathways, which consist of distinct intermediates including kinetically trapped conformations. Partitioning between alternative folding pathways is influenced by metal ion and denaturant concentration, temperature, flanking sequence context and specific RNA-binding proteins (Treiber and Williamson, 1999, 2001; Woodson, 2000a,b; Thirumalai et al., 2001; Russell et al., 2002).
Although the td intron is self-splicing in vitro, its folding process is very inefficient and slow. In vivo folding of the intron-containing pre-mRNA is rate limiting and is dependent on the activity of the ribosome (Semrad and Schroeder, 1998; Waldsich et al., 1998). Mutational analyses outlined distinct contributions of defined two-dimensional (2D) elements and three-dimensional (3D) motifs to the overall stability of the td intron (Brion et al., 1999a,b). To address the question whether folding in vivo obeys similar rules as folding in vitro, we analyzed the in vivo folding state of the td wild-type intron and of splicing-defective mutants by probing the structure of intron RNA with dimethyl sulfate (DMS).
The DMS modification pattern suggests that the structure of the td group I intron in vivo is in good agreement with structure models derived from phylogeny and biochemical data in vitro. The new probing data enabled modeling of the P7 extension, in which a loop E motif had been proposed (Leontis and Westhof, 1998b), and its assembly to the core domains. An A-minor type interaction between stem P3 and the P6-P6a A-rich internal loop was deduced and included in the refined 3D model. The structure of two mutants containing point mutations in stem P6 or stem P7 provided first insights into the structure of folding intermediates, and implied that formation of the 3D structure is also cooperative in vivo. Weakening of important structural elements affects the formation of several distant tertiary interactions, leading to an open intermediate conformation, which can be restored to its native state by the group I intron-specific splicing factor Cyt-18.
Results
Structure of the td intron in vivo
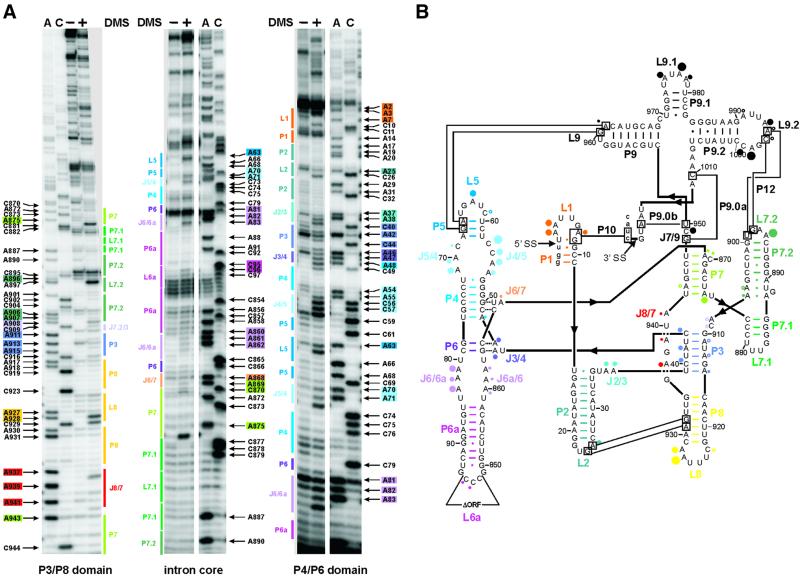
Comparison of the modification pattern of the in vivo DMS-treated intron RNA with the structural model derived from phylogeny and biochemical data obtained in vitro was used to visualize the in vivo folding state of the td intron (Figure 1). Representative gels (Figure 1A) cover the modified sites of almost the entire intron and a summary of all modified residues within the intron is displayed in Figure 1B. The secondary structure elements are all correctly formed as predicted, except for some minor divergence.
Fig. 1. DMS modification of the td intron in vivo. (A) Intron residues accessible to DMS are displayed in these representative gels. Boxed nucleotides correspond to positions within the intron, which are modified by DMS. The P3-P8 domain of the intron core (left panel), the center of the intron core covering the P7 stem, as well as the P6-P6a element (middle panel) and the P4-P6 domain of the intron core and the stem–loops P1-P2 (right panel) are shown. The following color code is used: P1, orange; P2, turquoise; P3, blue-violet; P4 and P5, blue; P6 extension, purple; P7 extension, green; P8, yellow; and J8/7, red. (A) and (C) denote the sequencing lanes. (B) Summary of the td intron residues modified by DMS in vivo. Modified sites are indicated by dots. The size of the dots correlates with the relative modification intensities. The largest dot corresponds to the highest modification intensity [color code for the dots as in (A); modifications in P9 are represented by black dots]. This structure representation is based on that of Cech et al. (1994).
The in vivo DMS modification pattern suggests that stem P3 is in part open. This probably indicates that stem P3 forms late in the hierarchical folding pathway. Stems P4 and P5 are readily formed except A48, which should base pair with U77 and is weakly modified, indicating some breathing at the bottom of stem P4. In the structural model, A875 base pairs with U942 at the bottom of stem P7. However, in vivo residue A875 is strongly modified by DMS, excluding the formation of a Watson–Crick base pair. We therefore suggest that, in vivo, stem P7 is one base pair shorter than predicted by phylogeny. In addition, A869 and A943 in P7 display weak modifications, like A868 in J6/7 and the bulge residue C870 in P7 (Figure 1A, left and middle panels). We conclude from the DMS modification pattern that the secondary structure model accurately describes the structure of the td group I intron in vivo.
Long-range tertiary interactions
For the td intron, several tertiary contacts have been described (Michel and Westhof, 1990; Salvo and Belfort, 1992; Jaeger et al., 1993, 1994). Base triples between P4 and J6/7 as well as between P6 and J3/4 play an important role in orienting the two major domains properly (Michel et al., 1990; Green and Szostak, 1994; Cate et al., 1996). Residues A46 and A47 in J3/4 are moderately modified, implying that the interaction between stem P6 and junction J3/4 takes place, but retains some flexibility. For the second base triplet we can only state that C866 in J6/7 is not modified, indicating the proper formation of its contact with stem P4. In addition, several long-range tertiary interactions contribute to the formation of the native structure. The closing tetraloop of stem P2 contacts stem P8 (Michel and Westhof, 1990; Salvo and Belfort, 1992; Costa and Michel, 1995). This contact is mediated via nucleotide triples involving residue A25 in loop L2, which is moderately modified. Loop L9.2 interacts with loop L7.2 (Michel et al., 1992). Adenine residues involved in this interaction are only very weakly modified. In contrast, most of the neighboring residues within these loops are strongly modified except for A991 in loop L9.2, which is also very weakly modified, and A897 in L7.2, which is not methylated. Another long-range tertiary contact is formed by residues in loop L9, which hydrogen bonds to stem P5 (Jaeger et al., 1994). The adenine of loop L9, which is part of this contact, is only modified very weakly, suggesting that this tertiary contact is formed. The modification pattern of bases involved in tertiary contacts points to the fact that the native structures adopted in vitro and in vivo are indistinguishable. Since DMS modification displays the frequency of the formation of RNA interactions averaged over the ensemble of RNA molecules and over time, the modification intensity we observed here indicates that the majority of the population is in the native state.
Docking of stem P1 in vivo
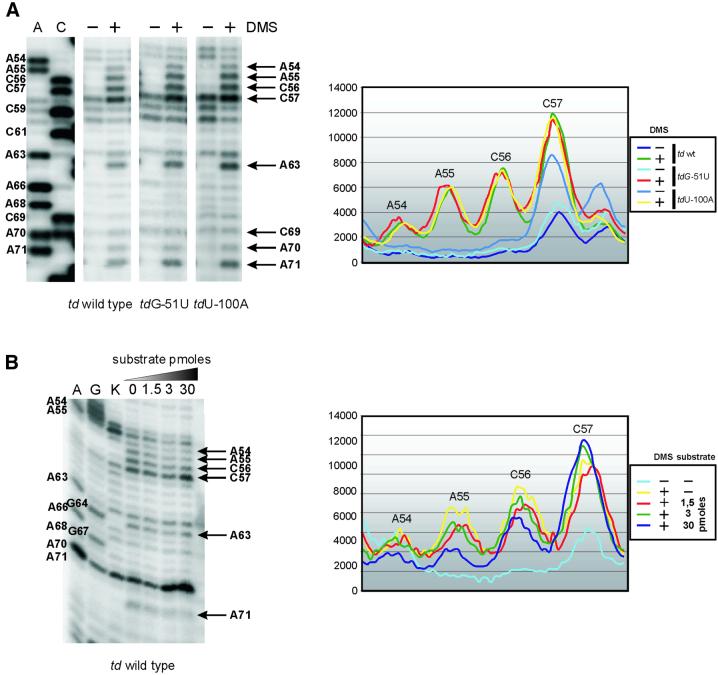
When probing the wild-type td intron construct in vivo, the majority of the td RNA population is composed of spliced intron molecules. To obtain information about folding and docking of stem P1, a population of pre-mRNA molecules is required. We therefore used mutant constructs containing a nonsense codon in the upstream exon, since their intron core structure in vivo is indistinguishable from that of the wild-type intron (Waldsich et al., 2002). Docking of stem P1, which harbors the 5′ splice site, to the catalytic intron core involves several interactions with J8/7 and the P4-P5 internal loop (Pyle et al., 1992; Strobel and Cech, 1995; Ortoleva-Donnelly et al., 1998; Strobel et al., 1998; Szewczak et al., 1998; Strobel and Ortoleva-Donnelly, 1999). The conserved adenines in the P4-P5 internal asymmetric loop form sheared A·A base pairs (Cate et al., 1996). In analogy to the Tetrahymena intron, A54 and A55 in J4/5 base pair with A71 and A70 in J5/4. In vivo, the conserved adenines in the P4-P5 loop are moderately (A54 and A70) or strongly (A55 and A71) modified (Figure 2A). C69 in J5/4 is only weakly modified, whereas the C56 and C57 in J4/5 are strongly modified. Formation of these non-canonical base pairs involves the N3 positions and the 2′ OH groups of the adenines probably leaving the N1 positions accessible to DMS. We nevertheless conclude from the present in vivo DMS modification pattern that the sheared A·A base pairs are formed, because A54 and A70 are less accessible to DMS than A55 and A71.
Fig. 2. Docking of the substrate helix to the P4-P5 A-rich asymmetric internal loop. Comparison of the accessibility of residues in J4/5 and J5/4 in vivo and in vitro. (A) The in vivo DMS modification pattern of the td wild-type intron and of td mutants harboring a nonsense codon in the upstream exon (tdG-51U, tdU-100A) is displayed in the left panel. Positions accessible to DMS are indicated by an arrow and base numbering. A and C are sequencing lanes. The corresponding phosphoimager quantification of these gel segments is shown in the right panel. (B) In vitro DMS modification of the ribozyme td WT-12 was performed in the presence of varying amounts of substrate (picomoles of substrate TDS4). A and G denote the sequencing lanes, and K represents the control lane, which derives from the reverse transcription of an RNA that was not treated with DMS. Quantification of the in vitro modification is shown in the right panel.
Residue G9, the G22 equivalent in the Tetrahymena intron, should interact with the N3 positions and the 2′ OH groups of A55 and A71 (A114 and A207 in the Tetrahymena intron) in the shallow groove of the P4-P5 loop (Strobel and Cech, 1995; Cate et al., 1996; Strobel et al., 1998). The strong modification of A55 and A71 implies that the P1 helix is not docked to the P4-P5 loop. This conclusion is emphasized further by in vitro chemical probing data obtained from the P4-P5 loop of the ribozyme td WT-12 in the presence of various substrate concentrations (Figure 2B). Upon binding of the substrate to the internal guide sequence and subsequent docking of the P1 equivalent, residues A54-C56 and A71 become less accessible to DMS. In the absence of the substrate, the in vitro DMS modification pattern of the P4-P5 internal loop is comparable to the in vivo DMS modification pattern (compare the quantification graphs in Figure 2A and B, right panels).
The residues in J8/7 are proposed to form a minor groove triple helix with stem P1 (Szewczak et al., 1998). Each interaction involves at least one 2′ hydroxyl group, and the only contact involving a N1 of A is A302, which accepts a hydrogen from the 2′ OH group of U-3 in the Tetrahymena intron (Pyle and Cech, 1991; Pyle et al., 1992). The corresponding residue in the td intron A937 is moderately accessible to DMS, but it is the most intensively modified residue of J8/7 (Figure 1). Since all other conserved adenines in J8/7 are only very weakly modified, we assume that the internal guide sequence might contact junction J8/7.
From the modification intensities of the bases in P4-P5 and J8/7, we suggest that P1 is docked to J8/7, but is definitely not permanently docked to the P4-P5 internal loop. Thus, P1 is only in part docked to the catalytic core of the td intron. Since the interaction between stem P1 and the P4-P5 internal loop contributes to the chemical transition state (Strobel and Ortoleva-Donnelly, 1999), one might speculate that this interaction only occurs immediately before the splicing reaction in vivo.
In vivo probing of td mutants with reduced structural stability
To access the structural background of splicing deficiency we probed the folding state of three splicing-defective td intron mutants. One mutant, with a change in the semi-conserved bulged nucleotide in P7 (tdC870U), has no severe structural defect, whereas the two other mutants present destabilized essential structural elements in either stem P6 (tdC865U) or stem P7 (tdC873U). The in vitro structural stability of these mutant intron RNAs was determined via ultra-violet (UV) melting. While the tdC870U mutant had a UV melting profile almost indistinguishable from wild type, the other two mutants displayed profiles that indicated a reduced thermal stability (Brion et al., 1999b; Chen et al., 2000).
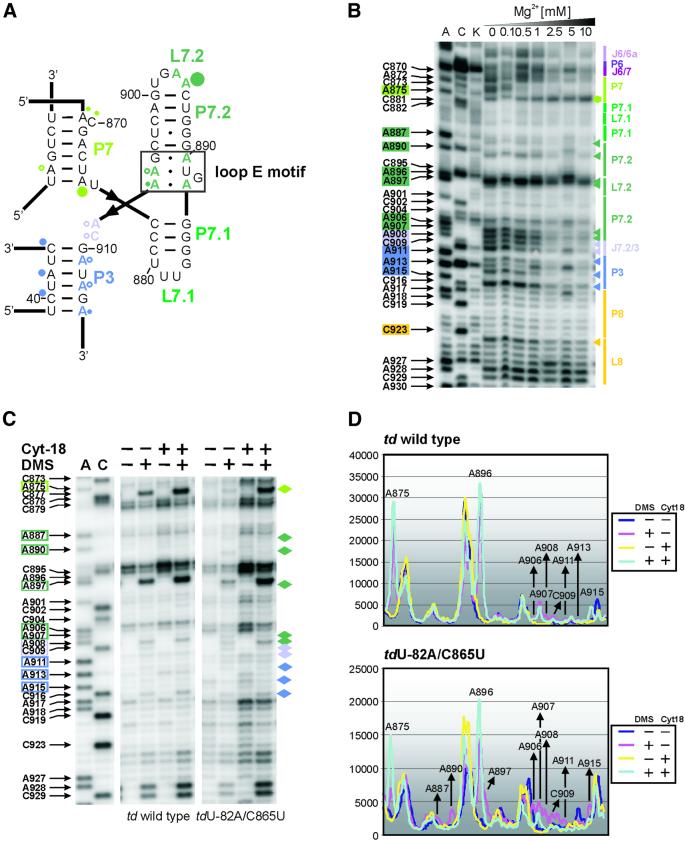
Mutations in stem P6 decrease the splicing activity in vivo and in vitro, and affect the stability of the tertiary structure (Chen et al., 2000). The mutation C865U in stem P6 reduces the splicing activity by 50% (Waldsich et al., 2002). In order to reduce further the splicing activity and to obtain a population composed predominately of pre-mRNA molecules, a mutant with a nonsense codon 82 nucleotides upstream of the 5′ splice site (tdU-82A/C865U) was probed (Figure 3A and C, upper panel). We observed that the modification pattern around J3/4, in which A48 is always less intensively modified than A46 and A47, is the same as in the wild-type td intron, but the modification intensity of these residues is increased in this mutant. The adenines in J3/4 interact with stem P6 to form base triples which are crucial for proper folding of the td intron. The observed changes are thus consistent with a perturbation at the base triples. Another indication that the formation of the base triples is affected in this mutant is the moderate modification at C79, which is part of the second base pair in stem P6 that is involved in the base triplet interaction. In addition to disturbances close to the mutation site, residues in stem P7 show altered accessibility to DMS: A943 becomes more intensively methylated, whereas A875 is less accessible to DMS and becomes partially protected. The adenines in J8/7 as well as A911, A913 and A915 in P3 display enhanced modification compared with the wild-type td intron. In addition, docking of the loop E motif of the P7.1-P7.2 extension (see below) to the intron core is perturbed, as indicated by an enhanced modification of residues A887, A890 and A906-C909 (Figure 3A). As far as the 3D long-range contacts are concerned, A961 in loop L9 is more accessible to DMS, indicating that the tertiary interaction with stem P5 is loosened. The residues A995 and C996 in loop L9.2, which contacts loop L7.2, are also more responsive to DMS (Figure 3A and C). The assumption that this interaction is weakened is strongly supported by the modification of A897 in the tdU-82A/C865U mutant. This residue is not accessible to DMS in the wild-type td intron, whereas A896 is always strongly methylated. The modification pattern of this mutant indicates that the entire intron is in a non-native open state and demonstrates the importance of the base triplet interactions for the formation of the overall 3D structure of the intron in vivo.
Fig. 3. The folding defect of splicing-deficient td intron mutants in vivo. (A) Changes within the modification pattern of the loop E motif and of the joining segment J8/7, which are due to a mutation in stem P6 (mutant tdU-82A/C865U), are displayed. The changes relative to the wild type are indicated as red triangles and the corresponding residue number is boxed in red. (B) Local changes within the td intron structure due to the mutation of the semi-conserved bulge in stem P7 (mutant tdC870U) are shown in the representative gels. Residues with an altered accessibility to DMS when compared with the wild-type intron are marked with blue triangles, and the corresponding nucleotide number is boxed in blue. (C) Quantification of the gel segments is shown for the td wild type and the mutant tdU-82A/C865U in the upper panel, as well as for the td wild type and the mutant tdC870U in the lower panel. (D) Summary of the changes in the DMS modification pattern occurring in the td intron mutants tdU-82A/C865U (red), tdC873U (yellow) and tdC870U (blue). The mutated residue is indicated by boxes labeled in the respective color. These changes are indicated relative to the wild-type td intron. Residues that are modified by DMS within these mutant introns, but whose modification intensity is not altered with respect to the wild-type construct, are not indicated (compare with Figure 1). Different dot sizes correlate to the relative increase in modification intensities compared with the wild type, whereas triangles indicate a reduced accessibility to DMS.
The tdC873U mutant is splicing deficient, showing only 10% splicing and a melting profile of lower thermal stability compared with the wild type (Belfort et al., 1987; Mohr et al., 1992; Brion et al., 1999a,b). Probing the structure of this mutant in vivo revealed that changes within the td intron structure are predominately located in the P3-P8 domain and in the P9 extension. Comparable to the tdU-82A/C865U mutant, the main tertiary structure interactions are perturbed, including the loop E motif (see below) and long-range interactions (Figure 3D). In addition, A868 and A869 are more intensely modified than in the wild-type intron. These results suggest that the mutation C873U globally affects folding of the 3D structure of the td intron, but does not affect formation of the base triplet.
The splicing-deficient mutant tdC870U (Schroeder et al., 1991), which has altered the semi-conserved bulge nucleotide in P7, was analyzed as a source of pre-mRNA with a correct folding state. The analysis of the structure of the mutant tdC870U allows the mapping of a homogenous td pre-mRNA population with a wild-type-like structural stability. As expected from the wild-type-like melting profile, there are only minor differences in the modification pattern compared with the wild type (Figure 3B and C, lower panel). These differences are located upstream of the bulged residue. A868 and A869 are slightly more responsive to DMS in this mutant. In the wild-type td intron, A868, A869 and C870 as well as adenines in J6a/6 are all modified to the same level, whereas in the tdC870U mutant the adenines A868 and A869 are more intensely modified than the adenines in J6a/6. In addition, A48 in stem P4 is more accessible to DMS with a modification intensity similar to A46 and A47 in J3/4 compared with the wild-type modification pattern. However, the long-range tertiary interactions, L9 with P5 and L7.2 with L9.2, as well as the base triples are all correctly formed, explaining the structural stability of the mutant. Mutation of C870 to U was proposed to interfere with binding of a magnesium ion close to the mutation site (Streicher et al., 1996). The modification pattern shown here strongly supports the idea that this mutant is mainly impaired in a chemical rather than a structural step.
Both mutants, tdU-82A/C865U and tdC873U, show enhanced accessibility of adenines in J8/7, indicating that the P1 helix is not docked to the intron core, whereas structural changes within the P4-P5 loop were not detected. In contrast, in the mutant tdC870U, adenines in J8/7 were modified in a comparable manner to the wild-type intron, indicating that P1 is docked to this junction (Figure 3D).
Folding of the loop E motif in the P7.1-P7.2 extension influences the geometry of the base pair at the bottom of stem P7
The loop E motif is a common RNA building block, which organizes the structure of internal loops or multiple junctions and mediates RNA–RNA and RNA–protein interactions (Leontis and Westhof, 1998a,b; Correll et al., 1999). It has been proposed that the peripheral P7 extension of the td group I intron encompasses such a eukaryotic-like loop E motif (Leontis and Westhof, 1998b). As determined by X-ray crystallography, this motif usually consists of a sheared A·G pair, a trans Watson–Crick–Hoogsteen U·A pair, a bulged G located in the deep groove and a trans Hoogsteen–Hoogsteen A·A pair (Figures 4A and 5B). Thus, all N1 positions of the adenines within a free loop E motif are reactive to DMS (Correll et al., 1997; Leontis and Westhof, 1998a).
Fig. 4. Perturbation of the loop E motif. (A) The loop E motif of the P7.1-P7.2 extension of the td group I intron (boxed in black). The modified residues are labeled with dots whose size correlates to the relative modification intensities within the wild-type intron (color code as in Figure 1). (B) In vitro DMS modification of the ribozyme td WT-12 with increasing magnesium concentrations. Junction J6/6a to stem P8 is displayed. The increasing magnesium concentrations are indicated also. Boxed nucleotides on the left side of the gel correspond to positions within the intron whose DMS modification intensity changes with increasing magnesium concentration. The colored triangles represent a decrease in accessibility to DMS, whereas the green hexagon corresponds to an increased accessibility to DMS with increasing magnesium concentration. The color code is as in Figure 1. (C) The representative gels display the modification status of residues which are part of or close to the loop E motif in the context of the wild-type intron and of the mutant tdU-82A/C865U, in the absence and presence of the group I intron-specific splicing factor Cyt-18. The nucleotide numbering highlighted with colored boxes as well as the colored diamonds outline those residues whose accessibility was altered due to the mutation in stem P6. (D) Quantification of the modification intensities in the absence and presence of Cyt-18. The upper panel depicts the relative change in modification intensities of residues that are part of, or that are close to, the loop E motif within the wild-type intron, and in the lower panel within the mutant tdU-82A/C865U. Due to increased levels of td RNA in cells co-expressing Cyt-18, these differences were normalized (see Materials and methods).
In the td intron, the loop E motif is part of the extension protruding from J7/3, and consists of residues A887 to A890 and G905 to A907. Formation of the motif enables stacking of helices P7.2 on P7.1 and should favor docking of the P7.1-P7.2 stacked helices to the P3-P7-P8 domain, thereby stabilizing the catalytic core. Indeed, the adenines in the loop E motif are inaccessible to DMS or very weakly modified (Figure 1B). Interestingly, the terminal adenine in P7, residue A875, is strongly methylated. We therefore propose that base pair A875-U942 (as predicted in the secondary structural model) may not form as a Watson–Crick pair in order to enable the association of the P7.1-P7.2 extension with the intron core.
The 3D model of the td intron was revisited in light of the presence of the loop E motif (Figure 5A). The three characteristic base pairs and the bulged nucleotide described above were modeled according to Correll et al. (1999). This implied an enforcement of the right-handed stacking of P7.1 and P7.2. To account for the protection of the adenine residues of the loop E motif, P7.2 was oriented so as to interact with P7. The plausibility of the interaction was checked by examination of the available crystal structures of various interacting loop E motifs in ribosomal RNA. Strikingly, the RNA elements interacting with loop E motifs cluster in the same region of the space and in a similar orientation when the loop E motifs are superimposed (Figure 5B). Docking of the P7.1-P7.2 extension onto P7 was done according to these observations. Of note, the docking is significantly facilitated by a slight opening of the terminal base pair of P7, forming a pair in which the amino group N6 of the A875 interacts with the O2 of the U942, in agreement with probing data presented in this study (Figure 5A).
Fig. 5. The refined 3D model of the td intron. (A) The global view of the intron points to the importance of the loop E motif (balls and sticks in red) both in the organization of the P7 extension and to the contacts between stem P3 and the P6/P6a A-rich internal loop (balls and sticks in orange). The remainder of the P7 extension is colored in green. The P4-P6 domain is marked in blue, while the P9 extension is shown in yellow. Stems P3 and P8 as well as junction J8/7 are labeled in purple. The helices P1 and P2 are outlined in gray. (B) The structure of the eukaryotic loop E motif was taken from Correll et al. (1999). It is described using the nomenclature proposed in Leontis and Westhof (2001): squares indicate that the base interacts via the Hoogsteen edge, triangles via the sugar edge and circles via the Watson–Crick edge. Open symbols are used for trans and full symbols for cis orientations of the glycosidic bonds relative to the hydrogen bonds. The sheared A·G pair involves hydrogen bonding between N6 of A and N3 of G, and between N7 of A and N2 of G. In the universally conserved trans Watson–Crick–Hoogsteen U·A pair, N3 of U interacts with N7 of A, and O2 of U contacts N6 of A. The N1 position of the bulged guanine residue is likely to be hydrogen-bonded to the phosphate oxygen of the cross strand A of the A·A pair. The symmetrical pairing between the trans Hoogsteen–Hoogsteen A·A pair involves hydrogen-bonding of the N7 and N6 positions of one A to the N6 and N7 positions of the other A. A typical example of contacts between a loop E motif and an RNA helix taken from the 23S rRNA of Haloarcula marismortui (Ban et al., 2000) is shown to the left of the one built into the td model. The underlined bases indicate those making the shallow groove contact the loop E motif. (C) The contacts between P3 and J6/6a, and a typical example from the 23S rRNA (Ban et al., 2000), are represented in a secondary-like structure diagram. In J6/6a, the dotted triangles on either side of the A·A pairs are meant to convey that the precise nature of the interactions cannot be fixed at this stage.
The most intriguing change in the modification pattern of the 3D destabilized mutants is the enhanced accessibility of the adenines in the loop E motif, which is coincidental with the protection of adenine A875 from DMS modification and, thus, with the elongation of stem P7 by an additional Watson–Crick base pair. We pursued these observations by probing the fold of this structural element in vitro with DMS in the presence of increasing concentrations of magnesium ions (Figure 4B). The higher the magnesium concentration, the more residues A887, A890 and A906-C909, part of or close to the loop E motif, become protected, whereas nucleotide A875 becomes more and more accessible to DMS. These results indicate that A875 base pairs to U942 when P7 forms, but has to rearrange as the loop E motif containing the P7.1-P7.2 extension associates with the intron core, and they are in line with the in vivo data. The modification intensities of residues A927 and A928 in loop L8 do not change with increasing magnesium concentration and thus serves as control for the interpretation of the in vitro modification pattern of loop E motif residues.
Specific RNA-binding proteins often reduce the magnesium requirement for RNA folding in vitro. Thus, we analyzed further the folding state of the loop E motif in the intronic mutant background, in the presence of the co-expressed group I intron-specific splicing factor Cyt-18 (Figure 4C and D). This protein was shown to rescue splicing-deficient td mutants by stabilizing the tertiary structure of the intron core (Mohr et al., 1992; Caprara et al., 1996a,b). In the mutant tdU-82A/C865U, residues that are part of or close to the loop E motif become protected from DMS modification when Cyt-18 is co-expressed in the cells. According to the interdependence of the accessibility of adenines within or close to the loop E motif and the protection of base A875, the base pair A875-U942 is rearranged in the presence of Cyt-18 as reflected by the enhanced modification intensity of A875 (Figure 4C and D). Thus, the formation of the loop E motif, which is part of the peripheral element P7.2, seems to be a key feature for the stabilization of the overall core of the td intron.
The td intron is stabilized by the interaction between stem P3 and the A-rich junction J6/6a
Since contacts mediated by A-rich motifs are often difficult to detect on the basis of sequence comparisons (Doherty et al., 2001; Nissen et al., 2001), we re-examined the model to identify a structural element capable of interacting with J6/6a. This was done with the aid of the Tetrahymena ribozyme and the 23S ribosomal RNA crystal structures (Golden et al., 1998; Ban et al., 2000). In the Tetrahymena crystal structure, J6/6a is proposed to interact with P3. However, the resolution of the structure (5.0 Å) precludes a thorough analysis of the contacts. The 23S ribosomal RNA (solved to 2.4 Å) was thus scanned for A-rich internal loops capable of interacting with the shallow groove of a neighboring helix. The candidates exhibit a consensus motif usually composed of a sheared A·G pair preceded by a bulging adenine, which stacks onto the adenine of the A·G pair, thus mimicking the interaction of a GNRA tetraloop with a receptor helix (Pley et al., 1994). Various non-canonical pairs sandwich this minimal motif, including other adenine-containing base pairs that increase the number of contacts between the interaction partners. The angle between the interacting helices is constant, at ∼45°. A motif related to the varieties observed in ribosomal RNA was built between the single strands J6/6a and J6a/6 in order to weave interactions with the adjacent P3 stem. The DMS modification data together with sequence analysis suggested a putative base pairing pattern among the great variety of internal loops containing three adenine residues in each strand (Figure 5A and C). In the main, adenine residues in J6/6a were sensitive to DMS to the same extent in vivo, while other residues in J6a/6 were almost fully protected. The minimal motif is composed of a sheared pair between the sugar edge of A862 and the Hoogsteen edge of A82, with A861 stacking on A82. A861 forms another sheared pair with the sugar edge of A83. The sugar edges of A81 and A82 form ribose zippers with U43 and U912. The resulting architecture closely resembles the interaction between P1 and J4/5. It is worth noting that such a motif is likely to take place in most introns, since the connectors between P6 and P6a are usually A-rich.
Discussion
In vivo DMS modification analysis provided first insights into the intracellular structure of the td group I intron. The vast majority of RNA folding experiments reported to date were performed in vitro and it is unknown whether the rules deduced from these experiments apply generally to in vivo conditions. So far there is some evidence that basic features are similar in vitro and in vivo, but in vivo facilitation of the Tetrahymena group I intron folding has also been observed (reviewed in Schroeder et al., 2002). The approach described in this report is a first attempt to elucidate folding intermediates of the td group I intron in vivo. Since kinetic description of RNA folding in vivo is too complex, we chose a genetic approach and analyzed mutant td introns with alterations in essential structural elements.
The thymidylate synthase gene had been submitted to a genetic screen in the search for splicing-deficient mutants. This screen resulted in a collection of mutants carrying mutations mainly at positions involved in the formation of the tertiary fold (Chandry and Belfort, 1987). Mutations in secondary structure elements have a less detrimental effect on the structural stability than mutations in tertiary structure elements (Robineau et al., 1997; Brion et al., 1999a,b). We therefore chose three mutants with very subtle changes for this analysis: in stem P6 or in stem P7, a GC base pair was altered to a GU pair. The tdC870U mutant containing an altered bulged residue in P7 was used as a control for a pre-mRNA with minimal structural but important catalytic defects.
Our results demonstrate that, in vivo, these mutations do not only affect the formation of the mutated structural element but importantly that the folding disturbance spreads to other tertiary structure domains. While mutations in stem P6 affected the global shape including docking of the peripheral P7.1-P7.2 domain, the mutation in stem P7 did not affect the formation of the base triples but still interfered with folding of the long-range interactions and with docking of the loop E motif-containing domain onto stem P7. These two mutants underline the cooperative nature of tertiary structure formation in the td intron in vivo as it had previously been demonstrated in vitro (Jaeger et al., 1993; Brion et al., 1999a). This is also consistent with the effect of Cyt-18 on the overall structure of group I introns. Cyt-18 strengthens the base triplet scaffold, but also contributes to the correct formation of distant structural elements which are not part of the protein binding site (Caprara et al., 1996a,b; Chen et al., 2000).
The efficiency of folding of large RNA molecules in vitro is achieved through the formation of folding intermediates (Treiber and Williamson, 1999, 2001; Woodson, 2000b). Here, we determined the structural defects of two splicing-deficient mutants, showing that their secondary structure is well formed but that they are in different non-native conformations with respect to the tertiary fold. We propose that the conformations of the td mutants represent intermediate folding states. Dependent on the localization of the mutation, the folding process is arrested at different steps in the folding pathway. Thus, folding of the td intron proceeds via intermediates in vivo. Our results further suggest a hierarchical order of folding events. As observed in vitro, secondary structure elements fold first, since they are not affected by mutations in tertiary structure elements. Once the P4-P6 domain is folded, formation of the inter-domain base triples seems to be required for proper folding of P7, including docking of the peripheral P7.1-P7.2 extension together with long-range interactions. Folding and docking of the loop E motif containing a peripheral extension causes an interesting rearrangement in the P7 stem, both in vitro and in vivo. When the P7.1-P7.2 domain is not assembled to the core, the terminal base pair of P7 is closed, while upon docking of this domain the intermediate base pair seems to rearrange, freeing the N1 position. In this study, it is proposed that the loop E motif in the P7 extension docks onto P7 in order to stabilize the latter in its native state. The model of the td intron was revisited (Figure 5) to incorporate this proposed interaction, which involves contacts undetectable by standard comparative sequence analysis.
This work also led to the proposal of an additional interaction between P3 and J6/6a, which is mediated by the sugar-edges of adenine residues towards the shallow groove of the contacting helix, as was originally observed in the crystal structure of the Tetrahymena ribozyme (Golden et al., 1998). Of note, residues from J6/6a suspected to be involved in contacting P3 remain more accessible than expected. This suggestion is consistent with the fact that residues in stem P3 are clearly accessible to DMS, even in the wild-type intron and in the pre-mRNA of the tdC870U mutant, leading to the proposal that the formation of P3 is one of the latest folding events. Thus, the energetic contribution of the newly proposed interaction in the td intron could be responsible for the stabilization of P3 and help the ribozyme reach its native state. This could also be extrapolated to other group I ribozymes, since most of them are likely to present this interaction.
Materials and methods
In vivo DMS modification
The thymine-deficient variant of Escherichia coli strain C600 (F-thr-1 leuB6 thi-1 lacY1 supE44 rfbD1 fhuA21) was transformed with a pTZ18U derivative harboring the td wild-type construct tdP6Δ2 and variants thereof (Waldsich et al., 2002). In vivo DMS modification experiments and the quantification of the gels were performed as described in Waldsich et al. (2002). The plasmid encoding the Neurospora crassa tyrosyl-tRNA-synthetase Cyt18 is a pACYC184 derivative (Mohr et al., 1992).
In vitro DMS modification
Ribozyme RNA of the intron construct td WT-12 was prepared as described in Pichler and Schroeder (2002). RNA (1 pmol) was modified by DMS in the absence and presence of increasing amounts of substrate RNA or increasing MgCl2 concentrations (as indicated in Figures 2 and 4B). The reaction was performed in a total volume of 50 µl. The in vitro-transcribed RNA was mixed with 2× potassium cacodylic acid buffer pH 7.5 (final concentration 50 mM) and MgCl2 was added to a final concentration of 10 mM. Prior to modification, the RNA was renatured by incubation at 56°C for 2 min, and subsequently at 37°C for 20 min. Different amounts of substrate RNA (TDS4; Pichler and Schroeder, 2002) were then added as indicated in Figure 2. DMS (final concentration 21 mM) was added and the samples were incubated at room temperature for 20 min. The modification was quenched by the addition of β-mercaptoethanol (final concentration 29 mM). Finally, the RNA was precipitated, resuspended in 3 µl ddH2O and used for reverse transcription (von Ahsen and Noller, 1993). The oligonucleotides used for reverse transcription hybridized to stem loops P6a or P9: tdP6a-5, 5′-TGTAGA ACCCGGGCAGTC-3′; tdP9-5, 5′-ATCCAGCTGCATGTCACC-3′.
Three-dimensional modeling
Modeling was performed as described previously (Westhof, 1993). Docking of the various elements presented in this paper was performed using the program manip (Massire and Westhof, 1998) and started from the model of the intron presented in Streicher et al. (1996). After assembly, the model was subjected to a number of geometrical least-squares refinements based on the algorithm of Konnert and Hendrickson (1980), implemented in the program NUCLIN/NUCLSQ (Westhof et al., 1985). The model was subjected to iterative modeling/refinement cycles until satisfactory rendering of the in vivo DMS probing. Figure 5 was prepared using DRAWNA (Massire et al., 1994).
Acknowledgments
Acknowledgements
We acknowledge Oliver Mayer, Andrea Pichler, Nicolas Piganeau, Herbert Wank and Michael Werner for comments on the manuscript. This work was supported by a grant from the Austrian science foundation (FWF), project number F1703, to R.S., and E.C. grant ERBFMRXCT 970154 to R.S. and E.W.
References
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Belfort M., Chandry,P.S. and Pedersen-Lane,J. (1987) Genetic delineation of functional components of the group I intron in the phage T4 td gene. Cold Spring Harb. Symp. Quant. Biol., 52, 181–192. [DOI] [PubMed] [Google Scholar]
- Brion P., Michel,F., Schroeder,R. and Westhof,E. (1999a) Analysis of the cooperative thermal unfolding of the td intron of bacteriophage T4. Nucleic Acids Res., 27, 2494–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion P., Schroeder,R., Michel,F. and Westhof,E. (1999b) Influence of specific mutations on the thermal stability of the td group I intron in vitro and on its splicing efficiency in vivo: a comparative study. RNA, 5, 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprara M.G., Lehnert,V., Lambowitz,A.M. and Westhof,E. (1996a) A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell, 87, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Caprara M.G., Mohr,G. and Lambowitz,A.M. (1996b) A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol., 257, 512–531. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- Cech T.R. (1993) Structure and mechanism of the large catalytic RNAs: group I and group II introns and ribonuclease P. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 239–269.
- Cech T.R., Damberger,S.H. and Gutell,R.R. (1994) Representation of the secondary and tertiary structure of group I introns. Nat. Struct. Biol., 1, 273–280. [DOI] [PubMed] [Google Scholar]
- Chandry P.S. and Belfort,M. (1987) Activation of a cryptic 5′ splice site in the upstream exon of the phage T4 td transcript: exon context, missplicing and mRNA deletion in a fidelity mutant. Genes Dev., 1, 1028–1037. [DOI] [PubMed] [Google Scholar]
- Chen X., Gutell,R.R. and Lambowitz,A.M. (2000) Function of tyrosyl-tRNA synthetase in splicing of group I introns: an induced-fit model for binding to the P4-P6 domain based on analysis of mutations at the junction of the P4-P6 stacked helices. J. Mol. Biol., 301, 265–283. [DOI] [PubMed] [Google Scholar]
- Correll C.C., Freeborn,B., Moore,P.B. and Steitz,T.A. (1997) Metals, motifs and recognition in the crystal structure of a 5S rRNA domain. Cell, 91, 705–712. [DOI] [PubMed] [Google Scholar]
- Correll C.C., Wool,I.G. and Munishkin,A. (1999) The two faces of the Escherichia coli 23S rRNA sarcin/ricin domain: the structure at 1.11 Å resolution. J. Mol. Biol., 292, 275–287. [DOI] [PubMed] [Google Scholar]
- Costa M. and Michel,F. (1995) Frequent use of the same tertiary motif by self-folding RNAs. EMBO J., 14, 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty E.A., Herschlag,D. and Doudna,J.A. (1999) Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry, 38, 2982–2990. [DOI] [PubMed] [Google Scholar]
- Doherty E.A., Batey,R.T., Masquida,B. and Doudna,J.A. (2001) A universal mode of helix packing in RNA. Nat. Struct. Biol., 8, 339–343. [DOI] [PubMed] [Google Scholar]
- Golden B.L., Gooding,A.R., Podell,E.R. and Cech,T.R. (1998) A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science, 282, 259–264. [DOI] [PubMed] [Google Scholar]
- Green R. and Szostak,J.W. (1994) In vitro genetic analysis of the hinge region between helical elements P5-P4-P6 and P7-P3-P8 in the sunY group I self-splicing intron. J. Mol. Biol., 235, 140–155. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Westhof,E. and Michel,F. (1993) Monitoring of the cooperative unfolding of the sunY group I intron of bacteriophage T4. The active form of the sunY ribozyme is stabilized by multiple interactions with 3′ terminal intron components. J. Mol. Biol., 234, 331–346. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Michel,F. and Westhof,E. (1994) Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol., 236, 1271–1276. [DOI] [PubMed] [Google Scholar]
- Konnert J.H. and Hendrickson,W.A. (1980) Restrained parameters thermal factors refinement procedures. Acta Crystallogr. A, 36, 344–349. [Google Scholar]
- Laggerbauer B., Murphy,F.L. and Cech,T.R. (1994) Two major tertiary folding transitions of the Tetrahymena catalytic RNA. EMBO J., 13, 2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert V., Jaeger,L., Michel,F. and Westhof,E. (1996) New loop–loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol., 3, 993–1009. [DOI] [PubMed] [Google Scholar]
- Leontis N.B. and Westhof,E. (1998a) The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA, 4, 1134–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N.B. and Westhof,E. (1998b) A common motif organizes the structure of multi-helix loops in 16S and 23S ribosomal RNAs. J. Mol. Biol., 283, 571–583. [DOI] [PubMed] [Google Scholar]
- Leontis N.B. and Westhof,E. (2001) Geometric nomenclature and classification of RNA base pairs. RNA, 7, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massire C. and Westhof,E. (1998) MANIP: an interactive tool for modelling RNA. J. Mol. Graph. Model., 16, 197–205. [DOI] [PubMed] [Google Scholar]
- Massire C., Gaspin,C. and Westhof,E. (1994) DRAWNA: a program for drawing schematic views of nucleic acids. J. Mol. Graph., 12, 201–206. [DOI] [PubMed] [Google Scholar]
- Michel F. and Westhof,E. (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- Michel F., Ellington,A.D., Couture,S. and Szostak,J.W. (1990) Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature, 347, 578–580. [DOI] [PubMed] [Google Scholar]
- Michel F., Jaeger,L., Westhof,E., Kuras,R., Tihy,F., Xu,M.-Q. and Shub,D.A. (1992) Activation of the catalytic core of a group I intron by a remote 3′ splice junction. Genes Dev., 6, 1373–1385. [DOI] [PubMed] [Google Scholar]
- Mohr G., Zhang,A., Gianelos,J.A., Belfort,M. and Lambowitz,A.M. (1992) The Neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell, 69, 483–494. [DOI] [PubMed] [Google Scholar]
- Murphy F.L. and Cech,T.R. (1993) An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry, 32, 5291–5300. [DOI] [PubMed] [Google Scholar]
- Nissen P., Ippolito,J.A., Ban,N., Moore,P.B. and Steitz,T.A. (2001) RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA, 98, 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortoleva-Donnelly L., Szewczak,A.A., Gutell,R.R. and Strobel,S.A. (1998) The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA, 4, 498–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A. and Schroeder,R. (2002) Folding problems of the 5′ splice-site containing P1 stem of the group I td intron: substrate binding inhibition in vitro and mis-splicing in vivo. J. Biol. Chem., 277, 17987–17993. [DOI] [PubMed] [Google Scholar]
- Pley H.W., Flaherty,K.M. and McKay,D.B. (1994) Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature, 372, 111–113. [DOI] [PubMed] [Google Scholar]
- Pyle A.M. and Cech,T.R. (1991) Ribozyme recognition of RNA by tertiary interactions with specific ribose 2′-OH groups. Nature, 350, 628–631. [DOI] [PubMed] [Google Scholar]
- Pyle A.M., Murphy,F.L. and Cech,T.R. (1992) RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature, 358, 123–128. [DOI] [PubMed] [Google Scholar]
- Robineau S., Bergantino,E., Carignani,G., Michel,F. and Netter,P. (1997) Suppressors of cis-acting splicing-deficient mutations that affect the ribozyme core of a group II intron. J. Mol. Biol., 267, 537–547. [DOI] [PubMed] [Google Scholar]
- Russell R. and Herschlag,D. (1999) New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol., 291, 1155–1167. [DOI] [PubMed] [Google Scholar]
- Russell R., Zhuang,X., Babcock,H.P., Millett,I.S., Doniach,S., Chu,S. and Herschlag,D. (2002) Exploring the folding landscape of a structured RNA. Proc. Natl Acad. Sci. USA, 99, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvo J.L. and Belfort,M. (1992) The P2 element of the td intron is dispensable despite its normal role in splicing. J. Biol. Chem., 267, 2845–2848. [PubMed] [Google Scholar]
- Schroeder R., von Ahsen,U. and Belfort,M. (1991) Effects of mutations of the bulged nucleotide in the conserved P7 pairing element of the phage T4 td intron on ribozyme function. Biochemistry, 30, 3295–3303. [DOI] [PubMed] [Google Scholar]
- Schroeder R., Grossberger,R., Pichler,A. and Waldsich,C. (2002) RNA folding in vivo. Curr. Opin. Struct. Biol., 12, 296–300. [DOI] [PubMed] [Google Scholar]
- Sclavi B., Sullivan,M., Chance,M.R., Brenowitz,M. and Woodson,S.A. (1998) RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science, 279, 1940–1943. [DOI] [PubMed] [Google Scholar]
- Semrad K. and Schroeder,R. (1998) A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev., 12, 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher B., Westhof,E. and Schroeder,R. (1996) The environment of two metals ions surrounding the splice site of a group I intron. EMBO J., 15, 2556–2564. [PMC free article] [PubMed] [Google Scholar]
- Strobel S.A. and Cech,T.R. (1995) Minor groove recognition of the conserved G·U pair at the Tetrahymena ribozyme reaction site. Science, 267, 675–679. [DOI] [PubMed] [Google Scholar]
- Strobel S.A. and Ortoleva-Donnelly,L. (1999) A hydrogen-bonding triad stabilizes the chemical transition state of a group I ribozyme. Chem. Biol., 6, 153–165. [DOI] [PubMed] [Google Scholar]
- Strobel S.A., Ortoleva-Donnelly,L., Ryder,S.P., Cate,J.H. and Moncoeur,E. (1998) Complementary sets of noncanonical base pairs mediate RNA helix packing in the group I intron active site. Nat. Struct. Biol., 5, 60–66. [DOI] [PubMed] [Google Scholar]
- Szewczak A.A., Ortoleva-Donnelly,L., Ryder,S.P., Moncoeur,E. and Strobel,S.A. (1998) A minor groove RNA triple helix within the catalytic core of a group I intron. Nat. Struct. Biol., 5, 1037–1042. [DOI] [PubMed] [Google Scholar]
- Tanner M.A. and Cech,T.R. (1997) Joining the two domains of a group I ribozyme to form the catalytic core. Science, 275, 847–849. [DOI] [PubMed] [Google Scholar]
- Tanner M.A., Anderson,E.M., Gutell,R.R. and Cech,T.R. (1997) Mutagenesis and comparative sequence analysis of a base triple joining the two domains of group I ribozymes. RNA, 3, 1037–1051. [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D., Lee,N., Woodson,S.A. and Klimov,D. (2001) Early events in RNA folding. Annu. Rev. Phys. Chem., 52, 751–762. [DOI] [PubMed] [Google Scholar]
- Treiber D.K. and Williamson,J.R. (1999) Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol., 9, 339–345. [DOI] [PubMed] [Google Scholar]
- Treiber D.K. and Williamson,J.R. (2001) Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol., 11, 309–314. [DOI] [PubMed] [Google Scholar]
- von Ahsen U. and Noller,H.F. (1993) Footprinting the sites of interaction of antibiotics with catalytic group I intron RNA. Science, 260, 1500–1503. [DOI] [PubMed] [Google Scholar]
- Waldsich C., Semrad,K. and Schroeder,R. (1998) Neomycin B inhibits splicing of the td intron indirectly by interfering with translation and enhances missplicing in vivo. RNA, 4, 1653–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich C., Grossberger,R. and Schroeder,R. (2002) The RNA chaperone StpA loosens interactions within the tertiary structure of the td intron in vivo. Genes Dev., 16, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E. (1993). Modelling the three-dimensional structure of ribonucleic acids. J. Mol. Struct. Dyn., 286, 203–210. [Google Scholar]
- Westhof E., Dumas,P. and Moras,D. (1985) Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol., 184, 119–145. [DOI] [PubMed] [Google Scholar]
- Woodson S.A. (2000a) Compact but disordered states of RNA. Nat. Struct. Biol., 7, 349–352. [DOI] [PubMed] [Google Scholar]
- Woodson S.A. (2000b) Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol. Life Sci., 57, 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar P.P. and Williamson,J.R. (1994) Kinetic intermediates in RNA folding. Science, 265, 918–924. [DOI] [PubMed] [Google Scholar]