Abstract
Sp3 is a ubiquitous transcription factor closely related to Sp1. Here we show that Sp3 is a target for SUMO modification in vivo and in vitro. SUMO modification of Sp3 occurs at a single lysine located between the second glutamine-rich activation domain and the DNA-binding domain. Mutational analyses identified the sequence IKXE as essential for SUMO conjugation to Sp3. We identified the protein inhibitor of activated STAT1 (PIAS1) as an interaction partner of Sp3 and Ubc9. Moreover, PIAS1 strongly stimulated SUMO conjugation to Sp3, thus acting as an E3 ligase for SUMO conjugation to Sp3. All mutations that prevented SUMO modification in vitro strongly enhanced the transcriptional activity of Sp3, showing that SUMO modification silences Sp3 activity. SUMO-modified Sp3 bound to DNA with similar specificity and affinity as unmodified Sp3. However, DNA-bound Sp3 did not act as a substrate for SUMO modification.
Keywords: E3 ligase/inhibitory domain/PIAS1/SUMO/transcription factor Sp3
Introduction
The transcription factor Sp3 is a ubiquitously expressed member of the Sp family of transcription factors (Suske, 1999). It contains two glutamine-rich activation domains in the N-terminal moiety and a zinc finger DNA-binding domain at the C-terminus that recognizes the classical GC box present in many promoters. The expression pattern, the structure and the DNA-binding capacity of Sp3 are very similar to Sp1. The physiological role of Sp1 and Sp3, however, appears to be different since Sp3- and Sp1-deficient mice exhibit different phenotypes (Marin et al., 1997; Bouwman et al., 2000). Cell culture experiments also revealed significant differences between Sp1 and Sp3. On many reporter constructs containing multiple Sp-binding sites, Sp3, unlike Sp1, is inactive or acts only as a weak activator (Hagen et al., 1994). The molecular basis for the inactivity of Sp3 under these conditions has been mapped to an inhibitory domain (ID) located between the second glutamine-rich activation domain and the zinc finger region (Dennig et al., 1996). A single lysine residue within this domain is absolutely essential for inhibitor function (Braun et al., 2001).
Lysine residues in proteins can be targets for SUMOylation. SUMO-1, also known as sentrin, GMP1, PIC1, Ubl1 and SMT3, is a small protein that shares 18% identity with ubiquitin (reviewed in Melchior, 2000; Yeh et al., 2000; Hay, 2001; Müller et al., 2001). Together with the two related proteins SUMO-2 and SUMO-3 (also referred to as SMT3a and SMT3b), SUMO-1 forms a small protein family. All three proteins can be covalently linked to the ε-amino group of lysine residues. Enzymes that catalyse the conjugation of SUMO-1 with its substrates are the SUMO-activating E1 enzyme and the SUMO-conjugating E2 enzyme. The E1 enzyme is a heterodimeric protein consisting of the 38 kDa Aos1 and the 72 kDa Uba2 subunits (also known as SAE1 and 2). The SUMO-conjugating enzyme E2 is also referred to as Ubc9. Recent publications show also the existence of SUMOylation-specific E3 ligases that enhance SUMO conjugation to target proteins and might contribute to substrate specificity (Johnson and Gupta, 2001; Kahyo et al., 2001; Sachdev et al., 2001; Takahashi et al., 2001).
The first identified target for SUMO-1 modification was RanGAP1 (Matunis et al., 1996; Mahajan et al., 1997). RanGAP1 is the GTPase activating protein for the Ras-related GTPase Ran that is involved in nucleo-cytoplasmic transport (reviewed in Görlich and Kutay, 1999). More recently, a number of other proteins, mainly transcription factors, including p53 (Gostissa et al., 1999; Rodriguez et al., 1999) and the steroid hormone receptors (Poukka et al., 2000), have also been shown to be modified by SUMOylation. The role of SUMOylation appears to be quite different. In the case of RanGAP1, its association with Ran-binding protein 2 (RanBP2) depends on modification by SUMO-1. As a consequence, RanGAP1 accumulates at the nuclear pore complex (Mahajan et al., 1997). Post-translational PML (promyelocytic leukemia) protein modification by SUMO-1 appears to be required for nuclear body formation and for recruitment of other factors to these structures (Ishov et al., 1999; Zhong et al., 2000). SUMOylation of IκBα occurs at the same lysine residue that can also be modified by ubiquitin. Thus, upon modification by SUMO-1, IκBα becomes resistant to degradation (Desterro et al., 1998). Modification of p53 and IE2-p86 by SUMO-1 leads to enhanced transcriptional potency (Gostissa et al., 1999; Rodriguez et al., 1999; Hofmann et al., 2000). An opposite effect has been described for the steroid hormone receptors, since disruption of the SUMO-1 acceptor site of the androgen receptor enhanced its transcriptional activity (Poukka et al., 2000). In the case of HSF1 and HSF2, SUMO-1 modification was shown to result in increased DNA-binding activity (Goodson et al., 2001; Hong et al., 2001).
Here, we report that the transcription factor Sp3 is a target for the SUMO modification machinery both in vivo and in vitro. Moreover, we identify PIAS1 as an interaction partner of Sp3 that acts as an E3 ligase for SUMO conjugation to Sp3. SUMO modification occurred exclusively at the lysine within the IKEE motif of the ID of Sp3. Mutations of amino acids that were essential for SUMO modification strongly enhanced the transcriptional activity of Sp3. Thus, SUMO modification silences transcriptional capacity of Sp3. The DNA-binding ability of Sp3 is not altered by SUMO modification. However, we found that Sp3 cannot act as substrate for SUMO modification when bound to DNA.
Results
Sp3 is a target for SUMO modification in vivo
Recently, we have identified a single lysine residue within the ID of Sp3 that is essential for silencing the transcriptional activity of Sp3 in cell culture experiments (Braun et al., 2001). In the course of these studies we realized that this lysine lies within a motif (IKEE, amino acids 422–425 according to Kingsley and Winoto, 1992) that could act as a target for SUMO modification (Melchior, 2000).
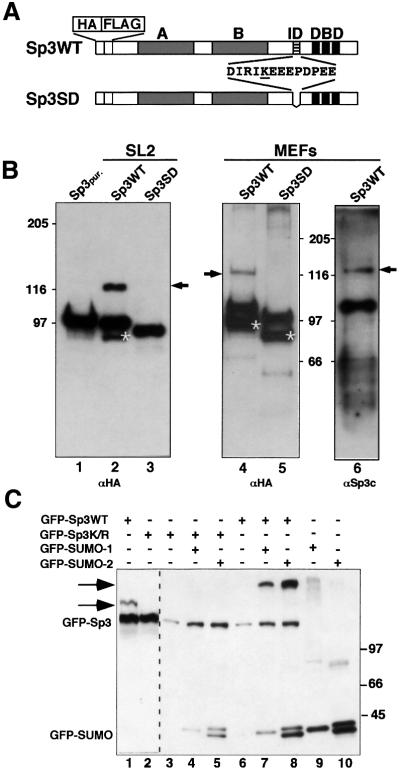
SUMO-modified proteins have an ∼20 kDa higher apparent molecular weight compared with unmodified proteins. However, previously we did not observe such high molecular weight species in immunoblots with anti-Sp3 antibodies. Since hydrolases exist that can readily cleave the isopeptide bond between the C-terminus of SUMO and the lysine residue of target proteins, it might be difficult to detect SUMOylated Sp3 unless special precautions are taken to inactivate these enzymes. In order to avoid the complexity introduced by Sp3 isoforms that all contain the IKEE motif (Suske, 1999), we used stable transfected SL2 cells (Braun and Suske, 1999) and Sp3–/– mouse embryonic fibroblasts (MEFs) expressing the HA/FLAG-epitope-tagged long isoform of Sp3. In addition, the expression level of the long isoform of Sp3 in the transgenic MEFs is comparable to wild-type MEFs (H.Göllner and G.Suske, unpublished data).
We performed western blot analyses of HA/FLAG-tagged Sp3 after direct SDS lysis (Figure 1). Under these conditions, we detected an ∼20 kDa slower migrating Sp3 species in both SL2 cells and in MEFs (Figure 1B, lanes 2, 4 and 6). Such an additional Sp3 form does not occur in cell lines expressing an Sp3 mutant protein (Sp3SD) that lacks the IKEE motif (Figure 1B, lanes 3 and 5). This result suggested that wild-type Sp3 can be covalently modified in vivo by SMT3 in Drosophila SL2 cells and SUMO-1 or SUMO-2/3 in mouse fibroblasts. Our attempts to determine which of the three SUMO proteins is attached to Sp3 in MEFs have failed thus far.
Fig. 1. Sp3 is SUMO modified in vivo. (A) Schematic representation of HA- and FLAG-tagged wild-type Sp3 protein Sp3WT and the Sp3 mutant Sp3SD expressed in SL2 cells (Braun and Suske, 1999) and in MEFs deficient of endogenous Sp3 (H.Göllner and G.Suske, unpublished data). Grey boxes indicate the two glutamine-rich activation domains A and B, and three black stripes the zinc fingers of the DNA-binding domain (DBD) of Sp3. The ID of Sp3 is depicted by hatched stripes. Amino acids that are deleted in the Sp3SD mutant protein are shown. The single lysine within this sequence is underlined. (B) Western blot analyses of epitope-tagged Sp3WT and mutant Sp3SD protein. HA- and FLAG-tagged Sp3WT- or Sp3SD-expressing SL2 cells (lanes 2 and 3) and MEFs (lanes 4, 5 and 6) were lysed with SDS-containing buffer. Proteins were separated on 7.5% SDS– polyacrylamide gels and blotted to PVDF membranes. Membranes were subsequently incubated with HA- (αHA) or Sp3-specific (αSp3c) antibodies as indicated. Arrows point to the covalently modified wild-type Sp3WT protein. Asterisks depict C-terminally deleted degradation products of Sp3 and Sp3SD that were detectable with the αHA antiserum but not with the αSp3c antibodies that recognize an epitope at the Sp3 C-terminal end. Lane 1 contains affinity-purified epitope-tagged Sp3 protein (Sp3pur.) (Braun et al., 2001) lacking the covalent modification. (C) GFP fusion vectors (3 µg) for GFP–Sp3WT, GFP–Sp3K/R, GFP–SUMO-1 and GFP–SUMO-2 were transiently transfected in Ishikawa cells as indicated. Cells were lysed and equal amounts of total cellular proteins (20 µg per lane) were separated on a 6.0% SDS–polyacrylamide gel and blotted to PVDF membranes. Detection was by immunoblotting with αGFP antibodies. The arrows point to SUMO-modified GFP–Sp3WT. The occurrence of two GFP–SUMO-2 forms is most likely due to incomplete processing.
To provide further evidence for SUMO modification of Sp3 in vivo, and to test whether the lysine of the IKEE motif is indeed the acceptor site, we transfected gene constructs encoding GFP–Sp3WT or a mutant thereof (GFP–Sp3K/R) in which the lysine residue was replaced by an arginine. Only GFP–Sp3WT but not the GFP–Sp3K/R mutant became modified (Figure 1C, lanes 1 and 2). We next co-transfected constructs coding for GFP–SUMO-1 or GFP–SUMO-2. We have also chosen GFP fusion constructs for SUMO, because they were expressed at a much higher level than corresponding CMV-driven fusions with other tags (our unpublished observation). A slow-migrating form of GFP–Sp3WT was detected in the presence of SUMO-1 or SUMO-2 fusion protein (Figure 1C, lanes 7 and 8). It is similarly abundant in the presence of SUMO-1 or SUMO-2. Thus, both SUMO-1 and SUMO-2 become efficiently conjugated to Sp3 when overexpressed in vivo. In contrast, a slow-migrating protein was absent with the GFP–Sp3K/R mutant. This result clearly demonstrated that wild-type Sp3, but not the Sp3K/R mutant, is a target for SUMO modification in vivo.
In vitro reconstitution of Sp3 SUMO modification
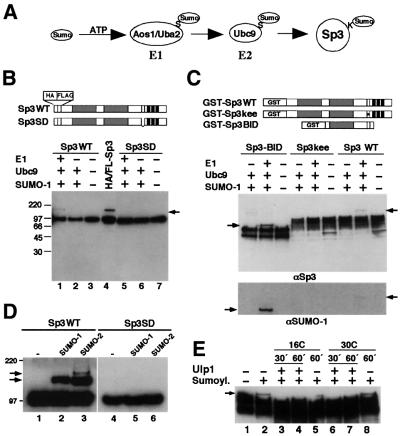
We asked whether Sp3 could also act as a substrate for SUMO modification in vitro. We employed an in vitro SUMOylation assay using purified recombinant E1 (Aos1 and Uba2 subunits) and E2 (Ubc9) enzymes and recombinant SUMO-1 (Figure 2A) (Pichler et al., 2002). As substrates, we used various recombinant epitope-tagged Sp3 fusion proteins that contain either the wild-type Sp3 IKEE sequence (HA/FLAG-tagged Sp3WT, GST–Sp3WT and GST–Sp3BID) or a mutated IKEE sequence (HA/FLAG-tagged Sp3SD and GST–Sp3kee), as outlined schematically in Figure 2B and C. In the GST–Sp3BID protein, an N- and C-terminal truncated Sp3 fragment is fused to GST. The Sp3 part contains the second glutamine-rich activation domain (B domain) and the ID with the IKEE motif. All substrates containing the wild-type IKEE sequence were covalently conjugated with SUMO-1 in vitro in the presence of heterodimeric E1 enzyme, Ubc9 and SUMO-1 (Figure 2). In contrast, all Sp3 mutants that lack the lysine residue of the IKEE motif were not conjugated with SUMO-1 (Figure 2B–D). These results show that the lysine residue within the IKEE motif of Sp3 is the only site for SUMOylation also in vitro. We used SUMO-2 as a substrate for SUMO modification of Sp3 in vitro and found that the efficiency of SUMO-2 conjugation to Sp3 was similar to that of SUMO-1 conjugation (Figure 2D). However, with SUMO-2 as modifying protein, an additional slower-migrating Sp3 conjugate was observed (Figure 2D, lane 3). This protein complex may reflect a dimeric SUMO-2 chain attached to Sp3 since SUMO-2 contains a ΨKXE motif and consequently can form polymeric chains (Tatham et al., 2001). It is unlikely that a second, SUMO-2-specific site becomes modified in vitro, because no Sp3–SUMO-2 complex was detectable with the Sp3SD mutant (Figure 2D, lane 6).
Fig. 2. In vitro SUMOylation and deSUMOylation of Sp3 fragments. (A) Schematic drawing of the conjugation pathway leading to SUMOylation of Sp3. The free carboxyl group of the C-terminal glycine of SUMO forms an isopeptide bond with the ε-amino group of a lysine (K) in Sp3. The reaction is mediated by the ATP-dependent heterodimeric E1 enzyme Aos1/Uba2 and the E2 enzyme Ubc9 that form thioesters (S) with SUMO. (B) Affinity-purified epitope-tagged Sp3WT (lanes 1–3) and Sp3SD (lanes 5–7) were subjected to in vitro SUMOylation reactions in the presence or absence of recombinant E1, Ubc9 and SUMO-1 as indicated. Sp3 and SUMO-modified Sp3 (arrow) were detected by western blot analysis using anti-HA antibodies. Lane 4 (HA/FL-Sp3) contains whole-cell extract from Sp3-expressing SL2 cells. (C) Bacterially expressed GST fusion proteins GST–Sp3WT, GST–Sp3kee and GST–Sp3BID bound to GST–Sepharose were subjected to in vitro SUMOylation reactions in the presence or absence of recombinant E1, Ubc9 and SUMO-1 as indicated. The GST–Sp3BID protein contains the second glutamine-rich activation domain (B domain) and the ID with the IKEE motif lacking the transactivation domain A and the C-terminal DNA-binding domain of Sp3. In the GST–Sp3kee protein, the KEE wild-type sequence of the ID is replaced by three alanine residues. Reaction products were detected by western blot analysis using anti-Sp3 (αSp3) and anti-SUMO-1 (αSUMO-1) antibodies as indicated. Arrows point to the SUMOylated Sp3 fragments. (D) SUMO-1 and SUMO-2 were equally conjugated to Sp3. Epitope-tagged recombinant Sp3 wild-type (Sp3WT) or the Sp3SD mutant was subjected to SUMO modification with equal concentrations of SUMO-1 and SUMO-2 (5 ng/µl each). Detection was by immunoblotting with αHA antibodies. (E) DeSUMOylation of SUMO-1-modified Sp3 by the isopeptidase Ulp1. The GST–Sp3BID fragment (see panel C) bound to glutathione–Sepharose was SUMOylated in vitro and subsequently incubated with recombinant ULP1 isopeptidase at 16 or 30°C for 30 or 60 min, as indicated. Detection was by immunoblotting with αSp3 antibodies.
We analysed also whether SUMO-1 could be released from Sp3 in vitro by a SUMO-specific isopeptidase (Li and Hochstrasser, 1999). Yeast Ulp1 was expressed and purified as a GST–Ulp1 fusion protein and incubated with SUMO-1-conjugated GST–Sp3WT. SUMO-1 was completely released from Sp3 after incubation with the Ulp1 enzyme (Figure 2E).
Identification of PIAS1 as an interaction partner of Sp3
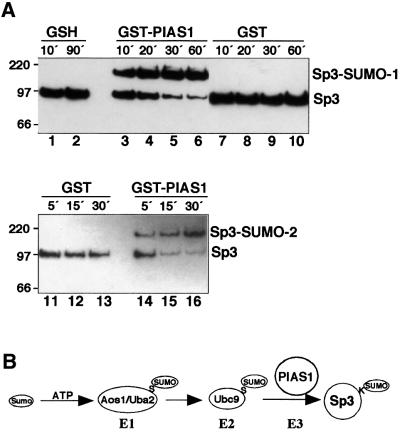
In an initial attempt to study the regulatory mechanism of the ID of Sp3, a yeast two-hybrid screen with the ID of Sp3 fused to the LexA DNA-binding domain (LexA-IDSp3) had been performed. A single clone interacted exclusively with the LexA-IDSp3 wild-type protein but not with LexA on its own or with a mutant in which the KEE sequence of the SUMO motif was replaced by three alanine residues (Figure 3A). Sequencing revealed that the encoded protein was identical to the protein inhibitor of activated STAT1, PIAS1 (Liu et al., 1998). To confirm a direct interaction of PIAS1 and the ID of Sp3, we performed GST pull-down experiments and observed only a very weak direct interaction between PIAS1 and Sp3 (Figure 3B, lane 5).
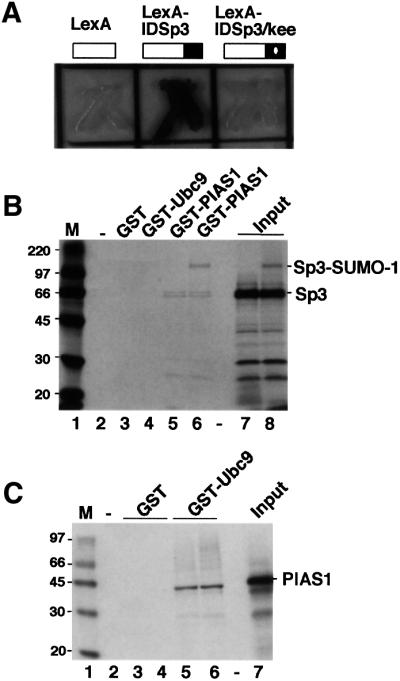
Fig. 3. Identification of PIAS1 as an interaction partner of Sp3 and Ubc9. (A) Interaction of PIAS1 with the ID of Sp3 in Saccharomyces cerevisiae. Yeast cells containing a LexA-driven LacZ reporter construct were transformed with expression constructs for LexA, LexA-Sp3ID or LexA-Sp3ID/kee (baits) along with a construct in which the Gal4 activation domain is fused to the 500 C-terminal amino acids of PIAS1 (Gal4-PIAS1, prey). In the LexA-Sp3ID/kee construct, the KEE sequence of the SUMOylation motif is replaced by three alanine residues. β-galactosidase activity was visualized by addition of 0.5% X-gal to the agar. (B) In vitro association of PIAS1 with Sp3 and SUMO-1-modified Sp3. Sp3 (small isoform) was in vitro translated in the presence of [35S]methionine and subsequently subjected to in vitro SUMO-1 conjugation. The reaction that contained unmodified Sp3 and SUMO-modified Sp3 (lane 8) was incubated with similar amounts of the glutathione matrix (lane 2), immobilized GST (lane 3), GST–Ubc9 (lane 4) or GST–PIAS1 (lane 6). In lane 5, unmodified 35S-labelled Sp3 was incubated with GST–PIAS1. Bound Sp3 proteins were resolved by SDS–PAGE and visualized by fluorography. Lanes 7 and 8 contain 10% of the input 35S-labelled Sp3 protein. Numbers on the left indicate the molecular mass of protein markers in kDa. (C) In vitro association of Ubc9 with PIAS1. PIAS1 was in vitro translated in the presence of [35S]methionine and incubated with glutathione–Sepharose matrix (lane 2) or with ∼2 µg of immobilized GST (lanes 3 and 4) or GST–Ubc9 (lanes 5 and 6). Bound PIAS1 protein was resolved by SDS–PAGE and visualized by fluorography. Lane 7 contains 10% of the input 35S-labelled PIAS1 protein. Numbers on the left indicate the molecular mass of protein markers in kDa.
Since the IKEE motif of the ID of Sp3 is a site for SUMO modification, we reasoned that the selection of the PIAS1 clone in the interaction screen was due to SUMOylation of Sp3 by a yeast SUMO homologue. If so, it seemed quite possible that PIAS1 could interact with SUMO, SUMO-modified Sp3 and/or Ubc9. To address this point, the small isoform of Sp3 was radiolabelled by in vitro translation and subsequently subjected to SUMO modification in vitro. The association of unmodified Sp3 and SUMO-modified Sp3 with PIAS1 was analysed by incubation with immobilized GST–PIAS1. These experiments revealed that PIAS1 bound preferentially SUMO-1-conjugated Sp3 (Figure 3B, lane 6). We next addressed the possibility that PIAS1 interacts with SUMO-1 on its own. No specific interaction of SUMO-1 with PIAS1 was observed (data not shown). However, in the course of our GST interaction studies, we found that PIAS1 could also interact with the E2 enzyme Ubc9 (Figure 3C, lanes 5 and 6).
PIAS1 acts as an E3 ligase for SUMO modification of Sp3
Since PIAS1 interacted specifically with Ubc9, Sp3 and SUMO-conjugated Sp3, we decided to examine whether PIAS1 might be directly involved in the enzymatic process of SUMO conjugation to Sp3. HA/FLAG-tagged Sp3 was subjected to SUMO modification in vitro with recombinant SUMO-1 and limiting amounts of the E1 enzymes Aos1/Uba2 and Ubc9 (one-tenth of the protein used in the experiments described above). Under these conditions, SUMO conjugation to Sp3 was almost undetectable (Figure 4A, lanes 1, 2 and 7–13). However, addition of bacterially expressed GST–PIAS1 allowed for efficient and almost complete conjugation of SUMO-1 or SUMO-2 to Sp3 (Figure 4A, lanes 3–6 and 14–16). Thus, PIAS1 is able to increase dramatically conjugation of both SUMO-1 and SUMO-2 to Sp3. This result identifies PIAS1 as an acting E3 ligase for SUMO conjugation to Sp3 (Figure 4B).
Fig. 4. PIAS1 stimulates SUMO conjugation to Sp3. (A) Purified HA/FLAG-tagged Sp3 from SL2 cells was subjected to in vitro SUMO-1 (lanes 1–10) or SUMO-2 (lanes 11–16) modification in the presence of 10 mM glutathione (GSH; lanes 1 and 2), GST–PIAS1 (lanes 3–6 and 14–16) or GST (lanes 7–10 and 11–13). Reactions contained one-tenth of E1 and Ubc9 enzymes used in the experiments shown in Figure 2. After various time points, reactions were stopped by addition of Laemmli buffer. Proteins were resolved by SDS–PAGE and Sp3 detected by immunoblotting with αHA antibodies. (B) Schematic outline of the conjugation pathway leading to SUMO modification of Sp3. PIAS1 that interacted specifically with Ubc9 and SUMO-modified Sp3 acts as an E3 ligase for SUMO conjugation to Sp3.
Sequence requirement for SUMO modification of Sp3
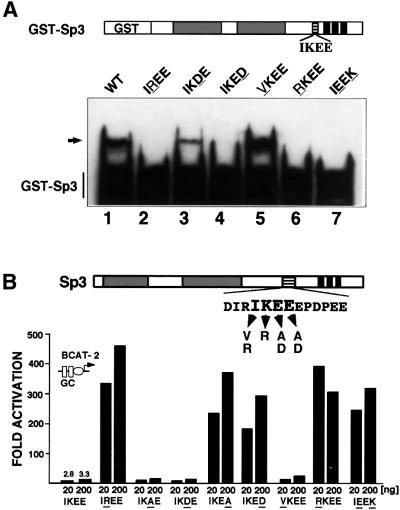
We evaluated the role of each amino acid within the IKEE motif of Sp3 for SUMO-1 conjugation. Single amino acid substitutions (IKEE to VKEE, RKEE, IREE, IKDE and IKED) were introduced into the GST–Sp3 fusion construct and the appropriate mutant proteins were used as substrates for SUMO modification. As shown before, mutation of the lysine residue abrogated conjugation with SUMO-1. However, mutations that leave the lysine residue intact also abrogated SUMOylation in vitro (Figure 5A). The second glutamic acid residue C-terminal to the lysine is also essential for SUMOylation of Sp3 since mutation to an aspartic acid residue (IKED mutant) abolished SUMO modification (Figure 5A, lane 4). In contrast, the glutamic acid immediately adjacent to the lysine seems not to be essential for SUMO conjugation. The mutant protein containing an aspartic acid at this position still became modified (Figure 5A, lane 3). The isoleucine preceding the lysine residue abrogated SUMOylation when mutated to an arginine but not when mutated to a valine (Figure 5A, lanes 5 and 6). In addition, an Sp3 mutant in which the relative position of the lysine and the second glutamic residue was altered (IKEE to IEEK mutation) became not SUMOylated in vitro (Figure 5A, lane 7). Taken together, the amino acid motif in Sp3 that is required for SUMO modification in vitro reads [I/V]KXE.
Fig. 5. Point mutations that prevent SUMO modification in vitro strongly enhance Sp3 activation capacity. (A) Identification of amino acids essential for SUMO modification of Sp3 in vitro. The wild-type IKEE sequence in GST–Sp3 was mutated to IREE, IKDE, IKED, VKEE, RKEE and IEEK (mutated amino acids are underlined) and the recombinant GST–Sp3 mutant proteins were subjected to in vitro SUMO modification. GST–Sp3 and GST–Sp3 mutants were detected by immunoblotting using αGST antibodies. The arrow points to SUMO-modified GST–Sp3. (B) SL2 cells were transfected with 4 µg of BCAT-2 plasmid along with 20 or 200 ng of expression plasmids for Sp3 or Sp3 mutants as indicated. Amino acids in the Sp3 mutants that differ from the wild-type IKEE sequence are underlined. SL2 cells were lysed and CAT activities determined. Fold activation values were calculated from CAT activities in relation to the empty expression vector (pPac), which has been given the arbitrary value of 1.
The IKXE motif is essential for silencing Sp3 transcriptional capacity
We wanted to ascertain whether the amino acids required for SUMO modification are also necessary for silencing Sp3 activity. We introduced essentially the same point mutations that were used for in vitro SUMO modification studies in an Sp3 expression construct designed for transcriptional activation studies in insect SL2 cells (Figure 5B). All mutants were expressed at a similar level after transfection in SL2 cells (data not shown). The transcriptional activity of the Sp3 mutants was analysed by transiently transfecting them along with a reporter (BCAT-2) that contains two Sp1/Sp3-binding sites fused to the E1bTATA box. Wild-type Sp3 was almost inactive, whereas a mutant in which the lysine residue was replaced by an arginine activated the BCAT-2 reporter up to 500-fold (Braun et al., 2001; Figure 5B, IREE mutant). Substitution of the glutamic acid residue adjacent to the lysine by an alanine or an aspartic acid residue (IKAE and IKDE mutants) did not uncover the transactivation capacity of Sp3, indicating that this residue is not critical for silencing function. In contrast, the second glutamic acid residue C-terminal to the lysine residue appeared to be absolutely essential for silencing. Mutations to an alanine or an aspartic acid residue (IKEA and IKED mutants) both rendered Sp3 a strong activator. The isoleucine preceding the lysine also plays a role in inhibition. An Sp3 mutant containing an arginine residue at this position (RKEE mutant) acted as a strong activator. In contrast, an isoleucine to valine mutation (VKEE mutant) showed no significant effect, indicating that the presence of a hydrophobic amino acid preceding the lysine residue is essential for inhibitor function. The Sp3 mutant in which the amino acid sequence composition was unchanged but the relative position of the lysine and the second glutamic residue was altered (IEEK mutant), also rendered Sp3 a strong activator.
Altogether, the results of our mutational analyses show that the central motif responsible for silencing Sp3 activity matches exactly that determined for SUMOylation. All mutations that abrogated conjugation by SUMO-1 in vitro enhanced the transcriptional activity of Sp3 upon transfection. This absolutely tight correlation strongly suggests that conjugation of SUMO to the IKEE motif of the ID of Sp3 silences the Sp3 activation capacity in vivo.
The subcellular localization of Sp3 is not altered upon SUMO modification
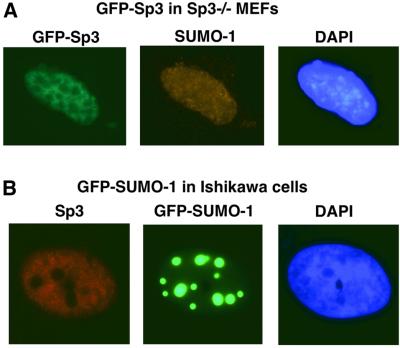
It has been shown that some SUMO-modified proteins like p53, SP100 or IE2-86 can localize to specific subnuclear structures, so-called PML-containing nuclear bodies, although SUMO conjugation was not absolutely essential for their delivery to these structures (Sternsdorf et al.,1999; Fogal et al., 2000; Hofmann et al., 2000). We analysed the subcellular localization of Sp3 in MEFs and in Ishikawa cells. Sp3–/– MEFs were transfected with a GFP–Sp3 construct and localization of GFP–Sp3 and endogenous SUMO-1 was analysed (Figure 6A). Both proteins were localized predominantly in the nucleus. Visualization of GFP–Sp3 showed a sponge-like, diffuse appearance that overlapped with endogenous SUMO-1 (Figure 6A). No dot-like structures were observed. In some cells, SUMO-1 appeared do be accumulated at the rim of the nucleus (not shown). In Ishikawa cells, essentially the same distribution of endogenous Sp3 and SUMO-1 was detected (not shown). However, overexpression of GFP–SUMO-1 revealed a striking redistribution and accumulation of SUMO-1 into punctate structures, whereas the diffuse distribution of Sp3 was not altered (Figure 6B). Sp3 localization also did not alter when Ubc9, in addition to SUMO-1, was overexpressed (data not shown). Taken together, our immunofluorescence experiments revealed that Sp3 is not targeted to nuclear bodies even under conditions where SUMO-1 becomes predominantly accumulated in such nuclear structures.
Fig. 6. Subcellular localization of Sp3 and SUMO-1 in MEFs and Ishikawa cells. (A) Sp3–/– MEFs were transfected with 1 µg of an expression construct for GFP–Sp3. The intracellular distribution of GFP–Sp3 was detected by intrinsic green fluorescence of the GFP tag. Endogenous SUMO-1 localization was detected with a rabbit anti-SUMO-1 antibody and a CY3-conjugated secondary antibody. (B) Ishikawa cells were transfected with 1 µg of an expression construct for GFP–SUMO-1. Visualization was by the intrinsic green fluorescence of the GFP moiety. Endogenous Sp3 localization was detected with a rabbit anti-Sp3 antibody and a CY3-conjugated secondary antibody.
SUMOylated Sp3 is capable of binding to DNA
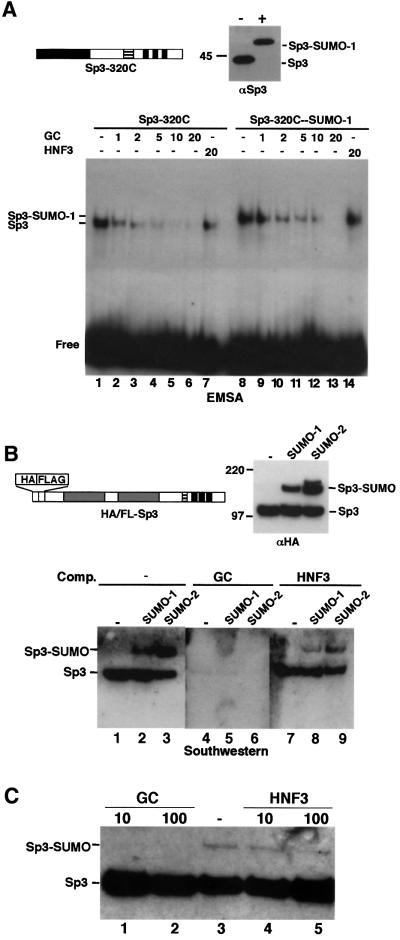
The site of SUMO modification in Sp3 is located close to the zinc finger DNA-binding domain. Thus, it seemed possible that conjugation of SUMO to the ID of Sp3 would abrogate binding of Sp3 to DNA. To analyse the capacity of SUMOylated Sp3 to bind to DNA, we employed electrophoretic mobility shift assays (EMSAs). Recombinant Sp3 from SL2 cells was subjected to in vitro SUMOylation and the reaction was subsequently used for EMSA and immunoblot analysis. Both, the non-conjugated as well as the SUMO-conjugated fractions produced DNA complexes with similar intensities, suggesting that SUMO-modified Sp3 binds DNA with an affinity similar to that of the unmodified protein. However, the protein–DNA complex obtained with the SUMO–Sp3 fraction migrated to a similar position as unmodified Sp3 (data not shown). Therefore, we also used an N-terminal truncated Sp3 fragment containing the 320 C-terminal amino acids for SUMO modification and EMSA (Figure 7A). The SUMO-modified Sp3 fragment bound DNA and the SUMO-modified Sp3–DNA complex migrated slower than the unmodified Sp3–DNA complex. Moreover, competition assays showed that the SUMO-modified Sp3 fragment bound the GC box with similar specificity and affinity as the unmodified protein (Figure 7A).
Fig. 7. SUMO-modified Sp3 binds specifically DNA. (A) A C-terminal Sp3 fragment (Sp3-320C) was subjected to in vitro conjugation with SUMO-1 in the absence (–) or presence (+) of enzymes. Subsequently, reactions were analysed by immunoblotting (top right, αSp3) and EMSA. All DNA-binding reactions contained 0.1 ng of 32P-labelled GC oligonucleotide and various amounts (1- to 20-fold molar excess) of unlabelled GC or HNF3 oligonucleotides, as indicated. (B) Purified HA/FLAG-tagged Sp3 was subjected to in vitro conjugation with SUMO-1 or SUMO-2 and analysed by immunoblotting (top right, αHA) and southwestern analysis. For southwestern analysis, SUMOylation reaction products were separated by 8% SDS–PAGE, transferred to nitrocellulose and subsequently incubated with 32P-labelled GC-box oligonucleotide in the absence (lanes 1– 3) or presence (lanes 4–6) of 100-fold molar excess of a specific competitor (GC box) or an unspecific (HNF3 site) (lanes 7–9) oligonucleotide. (C) Sp3 is not a target for SUMOylation when bound to DNA. Epitope-tagged recombinant Sp3 was subjected to SUMO-1 modification in the absence (lane 3) or presence of 10 and 100 ng of GC-box oligonucleotide (lanes 1 and 2) or HNF3-binding site oligonucleotide (lanes 4 and 5). Reaction products were separated by 8% SDS–PAGE and analysed by immunoblotting with αHA antibodies.
We also performed southwestern analyses with purified, epitope-tagged full-length Sp3 prior and after SUMO conjugation (Figure 7B). The SUMO-1-modified Sp3 protein bound the GC box as efficiently as the unmodified protein (Figure 7B). Moreover, binding to the GC box was specific as it was competed by an excess of unlabelled GC box oligonucleotide (Figure 7B, lanes 4– 6) but not by an oligonucleotide with an HNF3-binding site (Figure 7B, lanes 7–9).
Taken together, the above results show that SUMO-1 conjugation to the ID of Sp3 does not abolish or significantly alter the DNA-binding capacity of Sp3.
Sp3 is not a substrate for SUMO conjugation when bound to DNA
We also asked whether Sp3 could act as a target of SUMOylation when bound to DNA. In vitro SUMOylation of epitope-tagged full-length Sp3 was carried out in the absence or presence of high concentrations of specific GC or unspecific HNF3 oligonucleotides (Figure 7C). Conjugation of SUMO-1 to Sp3 was not impaired in the presence of an HNF3 recognition site (Figure 7C, lanes 4 and 5). In contrast, in the presence of the GC-box oligonucleotide, SUMO-1 modification was abrogated (Figure 7C, lanes 1 and 2). This result shows that Sp3 cannot act as a target for SUMO modification when bound to DNA.
Discussion
Sp3 is a SUMOylated protein
The transcription factor Sp3 is introduced as a novel substrate for SUMO modification. A single lysine residue within the ID of Sp3 becomes covalently SUMO modified in vivo and in vitro. The sequence requirement in Sp3 for SUMOylation (I/VKXE) fits well with the experimentally determined SUMO conjugation motif ΨKXE (Rodriguez et al., 2001), where Ψ represents a large hydrophobic amino acid. It should be noted, however, that not all ΨKXE motifs are targets of SUMO modification. The sequence IKDE present in the N-terminal part of Sp3 also matches the SUMO consensus sequence, but it does not become modified either in vivo or in vitro. Additional parameters must determine whether a potential SUMO target site becomes post-translationally modified. It is likely that the exposure of the sequence on the surface of the protein is essential for the accessibility of the SUMO modification machinery.
PIAS1 acts as an E3 ligase for SUMO conjugation to Sp3
PIAS1 fulfils all requirements to act as an E3 ligase for SUMO conjugation to Sp3. It interacts with the E2 enzyme Ubc9 as well as with Sp3. Most importantly, it strongly stimulates transfer of SUMO-1 and SUMO-2 to Sp3. Very recently, it was shown that the nucleoporin RanBP2 that is not related to PIAS proteins also has E3 ligase activity (Pichler et al., 2002). RanBP2 interacts with the E2 enzyme Ubc9 and enhanced SUMO transfer to Sp100, a component of PML nuclear bodies (Pichler et al., 2002). It is worth mentioning that RanBP2 does not act as an E3 ligase for SUMO conjugation of Sp3 (A.Pichler, G.Suske and F.Melchior, unpublished data).
PIAS1 is a member of a small protein family that also includes PIAS3, PIASx and PIASγ. These proteins were originally identified as interaction partners of various transcription factors, namely STAT1 (Liu et al., 1998), STAT3 (Chung et al., 1997), the androgen receptor (Moilanen et al., 1999; Kotaja et al., 2000; Gross et al., 2001) and Msx2 (Wu et al., 1997). In transfection experiments, PIAS proteins can regulate the activities of these transcription factors either negatively or positively. In particular, PIAS1 and PIAS3 inhibit STAT1- and STAT3-mediated activation, respectively, by blocking their DNA-binding activity (Chung et al., 1997; Liu et al., 1998). On the contrary, PIAS1 enhances the transcriptional activity of the androgen receptor (Gross et al., 2001). Whether the property of PIAS1 to inhibit STAT1 or to stimulate androgen receptor activity is related to its E3 ligase activity remains unclear. The E3 ligase activity of PIAS1 seems not to be essential for activation of p53. A PIAS1 mutant lacking the RING-finger-like domain essential for SUMOylation can still activate p53-mediated gene expression (Megidish et al., 2002). Similarly, PIASγ interacts with the wnt responsive transcription factor LEF1 and stimulates SUMO-1 conjugation to LEF1 (Sachdev et al., 2001). PIASγ also repressed LEF1 activity in cell culture experiments. However, repression of LEF1 activity was independent of the SUMOylation sites within LEF1 (Sachdev et al., 2001). Whether PIAS1 can also regulate Sp3 activity independently of its E3 ligase activity remains unclear at this stage.
SUMO modification is involved in regulating the transcriptional activity of Sp3
All amino acid substitutions that impaired SUMO modification dramatically enhanced the transcriptional activity of Sp3. The absolute correlation between transcriptional silencing of Sp3 and its ability to become a substrate for SUMO modification strongly suggests that conjugation of a single SUMO molecule is the cause for the very low transcriptional activity of wild-type Sp3. Silencing of the transcriptional activity by post-translational SUMO conjugation has also been reported for two other transcriptional regulators, the androgen receptor (Poukka et al., 2000) and c-jun (Müller et al., 2000). However, silencing of the transcription activity seems not to be a general feature of SUMO modification of transcriptional activators. In the case of p53 (Gostissa et al., 1999; Rodriguez et al., 1999) and the human CMV IE2-p86 protein (Hofmann et al., 2000), transcriptional capacity was enhanced by SUMO modification.
The site of SUMO modification lies immediately N-terminal to the DNA-binding domain of Sp3. Thus, an intriguing idea was that conjugation of SUMO would prevent binding of Sp3 to its recognition sites. We consider this mechanism to be very unlikely since SUMOylated Sp3 still bound specifically to DNA, as shown by EMSA and southwestern blot analyses. An alteration of the DNA-binding activity by SUMO modification has been reported for the heat shock factors HSF1 and HSF2 (Goodson et al., 2001; Hong et al., 2001). In these cases, however, SUMO-1 conjugation increased DNA-binding activity.
Given that SUMOylation at the IKEE motif of the ID is essential for silencing the transcriptional activation capacity of Sp3, one could speculate that SUMOylated Sp3 alters the interaction with other nuclear proteins, which in turn alters the subnuclear distribution of Sp3. Thus far, we have no indication for such a mechanism. A diffuse nuclear distribution of Sp3 was observed even under conditions where a dramatic accumulation of SUMO-1 in punctate nuclear structures occurred.
Yet another mechanism would be that SUMO modification of Sp3 might entail an interaction with co-repressors. Alternatively, SUMOylation may abrogate the interaction with the basal transcription machinery or co-activators. Previously, we have shown that the glutamine-rich activation domains of Sp3 can interact with dTAFII110 (Dennig et al., 1996). This interaction was not influenced by the presence of the ID of Sp3. However, these experiments were performed under conditions where Sp3 was not conjugated with SUMO. In future, these experiments will have to be repeated using SUMO-modified Sp3.
Acetylation versus SUMOylation?
Recently, we showed that Sp3 is also an acetylated protein in vivo. Strikingly, a mutant of Sp3 that lacks the lysine residue of the IKEE sequence exhibits a far weaker acetylation compared with wild type (Braun et al., 2001). This result suggested originally that acetylation of this lysine is responsible for silencing. However, the experiments described here, which identify the lysine of the IKEE sequence as a substrate for SUMO modification, made us reconsider the role of acetylation. Our experimental data strongly suggest that the same lysine residue can be modified by either acetylation or SUMOylation. Since both covalent modifications occur at the ε-amino group of the lysine, acetylation would prevent SUMO modification and vice versa. SUMO modification silences Sp3 activity since all mutations that prevent SUMOylation relieve inhibition. Whether acetylated Sp3 is also inactive or whether its activation potential is similar to that of the unmodified protein is not clear at this stage. If acetylation would keep the Sp3 protein active, one could imagine an intriguing scenario by which the transcriptional activity of Sp3 could be regulated by different modifications at the same lysine residue.
Materials and methods
Plasmid constructions
The construction of the parental expression plasmids for transient (pPacUSp3) and stable expression of Sp3 (pRm-HA/FLAG-Sp3) in Drosophila Schneider SL2 cells as well as for GST fusion proteins (pGEX-2TK-Sp3 and pGEX-2TK-Sp3BID) has been described previously (Dennig et al., 1996; Braun and Suske, 1999; Braun et al., 2001). Point mutations were introduced into the pGEX-2TK-Sp3 plasmid by PCR amplification using appropriate oligonucleotides. Detailed primer information will be provided upon request. The pPacUSp3 expression plasmids that contain point mutations were obtained by replacing the wild-type Sp3 cDNA of pPacUSp3 by the mutated Sp3 cDNA obtained as BamHI–XhoI fragments.
The plasmid pEGFP-C1-Sp3fl encoding wild-type full-length Sp3 fused to GFP was constructed as follows. The Sp3 ORF was amplified by PCR using the following primers: sense Sp3-AUG-T (5′-TAAACGA ATTCTATGGCTGCCTTGGACGTG-3′) and antisense Sp3-STOP (5′-TATTAAGGATCCCTCCATTGTCTCATTTCC-3′). The PCR product was digested with EcoRI and BamHI, and cloned into the corresponding EcoRI- and BamHI-restricted pEGFP-C1 plasmid (Clontech). The pEGFP-C1-Sp3fl-K/R mutant plasmid was obtained by replacing a 750 bp Bst11071–BamHI wild-type fragment with a corresponding fragment obtained by PCR amplification from pPacUSp3K/R plasmid, which contained the lysine to arginine mutation at the IKEE motif.
The vector for in vitro translation of PIAS1 (pBK-CMV-PIAS1) was generated by cloning the 1.7 kb PIAS1 cDNA insert of the yeast plasmid pACT2-PIAS1 (encoding for the 500 C-terminal amino acids of PIAS1) into XhoI- and EcoRI-restricted pBK-CMV (Stratagene). Full-length PIAS1 was obtained by RT–PCR amplification using RNA from H441 cells and the following primers: upstream, 5′-TCCAAGAATT CGGCGAAGTTCACTGCGCTTGCGCTGACA-3′; downstream, 5′-GGTTTCTCGAAAGCGCTGACTGTTGTCTG-3′. The PCR product was cut with EcoRI and Eco47III, and cloned into EcoRI–Eco47III-restricted pBK-CMV-PIAS1-500C. The expression plasmid for GST–PIAS1 was obtained by cloning the full-length PIAS1 cDNA from pBK-CMV-PIAS1 into the SalI–XhoI-restricted pGEX-2TK-P vector (Pharmacia).
Expression and purification of enzymes and substrates for SUMO modification reactions in vitro
Cloning, expression and purification of recombinant E1 (His-Aos1 and Uba2) and E2 enzyme (Ubc9) and SUMO-1 are described in Pichler et al. (2002). The SUMO-2 ORF was amplified from the I.M.A.G.E. consortium cDNA clone IMAGp998D071314, ligated into the NdeI–BamHI sites of pET11a, and purified and expressed as described for SUMO-1. Expression and purification of recombinant HA/FLAG-tagged Sp3 and Sp3SD proteins were performed by affinity chromatography using the αFLAG M2 matrix (Serva) as described previously (Braun et al., 2001).
GST fusion proteins were expressed from pGEX-2TK vectors (Pharmacia) in Escherichia coli strain BL21 after induction with 1 mM IPTG. GST–Sp3 fusion proteins used as substrates for in vitro SUMO modification were bound to glutathione–Sepharose 4B beads (Pharmacia) according to the manufacturer’s instructions and either used directly in enzyme reactions or eluted with 10 mM reduced glutathione.
In vitro SUMOylation and deSUMOylation assay
SUMO modification reactions were typically performed in a total volume of 10 µl with 12.5 ng of Aos1/Uba2, 140 ng of Ubc9, 50 ng of SUMO-1 and recombinant GST–Sp3 proteins bound to beads (1 µg) or purified HA/FLAG-tagged Sp3 proteins (<100 ng). The reaction buffer contained 1 mM ATP, 0.05% Tween-20 and 0.2 mg/ml ovalbumin. Reactions were incubated at 30°C for 30 min and stopped by addition of SDS–PAGE sample buffer.
DeSUMOylation reactions were performed as follows. GST–Sp3 fusion proteins (1 µg) were immobilized on beads and incubated with 20 µl of reaction mixture containing E1 and E2 enzyme and SUMO-1, as described previously. After incubation at 30°C for 60 min, beads were washed three times with PBS and 0.5% Triton X-100, followed by incubation with GST–Ulp1 (500 ng) as described previously (Li and Hochstrasser, 1999), with the exception that cleavage reactions were performed at either 16 or 30°C. As a control, SUMOylated protein was incubated with reaction buffer in the absence of GST–Ulp1.
Cell culture, transfections and preparation of cell lysates
SL2 cells were transfected as described previously (Dennig et al., 1996; Suske, 2000). Every plate received 4 µg of reporter plasmid BCAT-2 and 2 µg of the β-galactosidase expression plasmid p97b as internal reference. Variable amounts of pPacUSp3 expression plasmids were compensated with the plasmid pPac. Each transfection was repeated at least three times. Chloramphenicol acetyltransferase (CAT) was assayed by ELISA according to the manufacturer’s instructions (Roche). β-galactosidase assays were carried out as described previously (Hall et al., 1983). CAT activity was normalized to β-galactosidase activity.
Ishikawa cells were grown as a monolayer in minimum essential medium supplemented with 10% FCS, l-glutamate and antibiotics. For the immunoblot experiments shown in Figure 1C, cells were transfected with 6 µg of plasmid DNA per 6 cm dish using the FuGENE6 reagent according to the manufacturer’s instructions (Roche). Variable amounts of GFP–SUMO expression plasmids were compensated with the plasmid pRC/CMV.
Lysates for immunoblot analyses were prepared from stable transfected SL2 cells and MEFs, and transiently transfected Ishikawa cells. Cells were washed with PBS and lysed in 200 µl of a 1:3 dilution of buffer I (5% SDS, 150 mM Tris–HCl pH 6.7, 30% glycerol) and buffer II (25 mM Tris–HCl pH 8.2, 50 mM NaCl, 0.5% NP-40, 0.1% SDS, 0.1% azide), supplemented with 5 mM DTT, 0.2 mM PMSF and 0.5× complete protease inhibitor cocktail (Roche). Lysates were boiled for 10 min and spun in a microfuge (Heraeus, Biofuge Pico) for 10 min at 13 000 r.p.m. at 4°C. The protein concentration of the supernatants was determined and equal amounts of protein were subjected to SDS–PAGE.
Immunoblot analysis
Proteins resolved by SDS–PAGE were electrotransferred to PVDF membranes and probed with the appropriate antibodies. The following antibodies and dilutions (in 20 mM Tris–HCl pH 7.6, 137 mM NaCl, 0.1% Tween-20, 1% skim milk) were used: mouse anti-GFP (Clontech) 1:2000; rabbit anti-Sp3 polyclonal IgG (Santa Cruz) 1:2000; rabbit anti-Sp3 serum (Hagen et al., 1994) 1:1000; rabbit anti-HA (Santa Cruz) 1:2000; rabbit anti-SUMO-1 (Santa Cruz) 1:1000; horseradish peroxidase-conjugated sheep anti-mouse IgG (Amersham Life Science) 1:10 000; and horseradish peroxidase-conjugated donkey anti-rabbit IgG (Amersham Life Science) 1:10 000. The enhanced chemiluminescence detection system (Amersham–Pharmacia, Freiburg, Germany) was used to visualize the peroxidase reaction.
Immunofluorescence and microscopy
MEFs or Ishikawa cells were plated on coverslips in 24-well tissue culture plates and, 24 h post-transfection, fixed for 25 min at room temperature with 4% paraformaldehyde in PBS. Cells were washed twice for 5 min with PBS and subsequently permeabilized with 0.2% Triton X-100 in PBS for 15 min. After blocking with 3% BSA in PBS for 1 h, cells were incubated with appropriate antibodies for 1 h at room temperature. Primary antibodies and dilutions used were polyclonal rabbit anti-Sp3 (Santa Cruz) 1:300 and polyclonal rabbit anti-SUMO-1 (Santa Cruz) 1:500. Coverslips were washed three times for 5 min with PBS and incubated with a Cy3-conjugated secondary goat anti-rabbit IgG antibody (Jackson ImmunoResearch) for 1 h at room temperature in a dark cabinet. Subsequently, coverslips were mounted onto glass slides using Vectrashield mounting media with DAPI and sealed with nail polish. Fluorescence images were obtained on Leica DMLB or Leitz Orthoplan microscopes.
EMSAs and southwestern blot analysis
EMSAs were essentially performed as described previously (Garner and Revzin, 1986) using 32P-labelled double-stranded GC box oligonucleotides (Hagen et al., 1992) purified via G50 microcolumns (Amersham). For competition assays, unlabelled GC box or HNF3 oligonucleotide was added in excess.
For southwestern analysis, HA/FLAG-Sp3 was conjugated with SUMO-1 or SUMO-2 as described above. Reactions were terminated with SDS–PAGE sample buffer without β-mercaptoethanol, boiled, fractionated by 8% SDS–PAGE and electroblotted onto nitrocellulose. Filters were incubated with buffer A (10 mM HEPES pH 7.9, 50 mM NaCl, 0.1 mM EDTA, 1 mM MgCl2, 10% glycerol, 1 mM DTT) supplemented with 6 M guanidine hydrochloride for 10 min at 4°C. Incubation with buffer A was repeated five times (5 min each) with stepwise dilutions of the guanidine hydrochloride (3, 1.5, 0.75, 0.375 and 0.18 M). Filters were blocked with buffer B (10 mM HEPES pH 7.9, 50 mM NaCl, 0.1 mM EDTA, 1 mM MgCl2, 10% glycerol, 1 mM DTT, 100 µM ZnSO4) containing 5% skim milk at 4°C over night. Prior to the incubation with radiolabelled oligonucleotides, filters were washed once with binding buffer (buffer B containing 0.25% skim milk, 500 ng/ml salmon sperm DNA) for 10 min. The binding reaction was performed via a 4 h incubation at room temperature in binding buffer containing 1 × 106 c.p.m./ml of 32P-labelled GC-box oligonucleotide. Filters were washed four times with buffer B, dried at room temperature for 1 h and exposed to X-ray film.
Yeast two-hybrid screen
A fusion protein of the ID of Sp3 (230 bp Bst1107I–KpnI fragment of the Sp3 cDNA) and the DNA-binding domain of LexA (LexA-IDSp3) was used as a bait to screen a commercial yeast two-hybrid human WI-38 cDNA library (Clontech). Positive clones were analysed for their interaction with the LexA DNA-binding domain on its own or the LexA-IDSp3kee mutant. A 1.7 kb clone that interacted exclusively with the wild-type LexA-IDSp3 protein when expressed in yeast was sequenced. It encoded the 500 C-terminal amino acids of human PIAS1.
In vitro association assays
Following expression in E.coli, ∼2 µg of GST fusion proteins were bound to glutathione–Sepharose (Amersham) and incubated with 15 µl of 35S-labelled proteins for 4 h at 4°C in binding buffer (25 mM HEPES pH 7.5, 150 mM KCl, 12.5 mM MgCl2, 20% glycerol, 0.1% NP-40, 1 mM DTT, 0.5× protease inhibitor cocktail, 1 mg/ml BSA and 0.1% skim milk). After washing the beads four times with a buffer containing 25 mM HEPES pH 7.5, 200 mM KCl, 12.5 mM MgCl2, 20% glycerol, 0.5% NP-40, 1 mg/ml BSA and 0.1% skim milk, SDS sample buffer was added and proteins were fractionated by 10% SDS–PAGE. Radioactive labelled proteins were visualized by autoradiography.
Acknowledgments
Acknowledgements
We thank Iris Rohner for excellent technical assistance. Martha Kalff-Suske is gratefully acknowledged for critically reading the manuscript. H.Will generously provided us with the plasmids for GFP–SUMO-1 and GFP–SUMO-2, and M.Hochstrasser with GST–Ulp1. This work was supported by a grant of the Deutsche Forschungsgemeinschaft to G.S. and the BMBF (BioFUTURE 0311869) to F.M.
References
- Bouwman P., Göllner,H., Elsässer,H.P., Eckhoff,G., Karis,A., Grosveld,F., Philipsen,S. and Suske,G. (2000) Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J., 19, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun H. and Suske,G. (1999) Vectors for inducible expression of dual epitope-tagged proteins in insect cells. Biotechniques, 26, 1038–1042. [DOI] [PubMed] [Google Scholar]
- Braun H., Ertmer,A., Nacht,S., Koop,R. and Suske,G. (2001) Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res., 29, 4994–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.D., Liao,J., Liu,B., Rao,X., Jay,P., Berta,P. and Shuai,K. (1997) Specific inhibition of Stat3 signal transduction by PIAS3. Science, 278, 1803–1805. [DOI] [PubMed] [Google Scholar]
- Dennig J., Beato,M. and Suske,G. (1996) An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J., 15, 5659–5667. [PMC free article] [PubMed] [Google Scholar]
- Desterro J.M., Rodriguez,M.S. and Hay,R.T. (1998) SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell, 2, 233–239. [DOI] [PubMed] [Google Scholar]
- Fogal V. et al. (2000) Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J., 19, 6185–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M.M. and Revzin,A. (1986) The use of gel electrophoresis to detect and study nucleic acid–protein interactions. Trends Biochem. Sci., 11, 395–396. [Google Scholar]
- Goodson M.L., Hong,Y., Rogers,R., Matunis,M.J., Park-Sarge,O.K. and Sarge,K.D. (2001) Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem., 276, 18513–18518. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Gostissa M., Hengstermann,A., Fogal,V., Sandy,P., Schwarz,S.E., Scheffner,M. and Del Sal,G. (1999) Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J., 18, 6462–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M., Liu,B., Tan,J., French,F.S., Carey,M. and Shuai,K. (2001) Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene, 20, 3880–3887. [DOI] [PubMed] [Google Scholar]
- Hagen G., Müller,S., Beato,M. and Suske,G. (1992) Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res., 20, 5519–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G., Müller,S., Beato,M. and Suske,G. (1994) Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J., 13, 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C.V., Jacob,P.E., Ringold,G.M. and Lee,F. (1983) Expression and regulation of E.coli lacZ gene fusions in mammalian cells. J. Mol. Appl. Genet., 2, 101–109. [PubMed] [Google Scholar]
- Hay R.T. (2001) Protein modification by SUMO. Trends Biochem. Sci., 26, 332–333. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Floss,S. and Stamminger,T. (2000) Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol., 74, 2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Rogers,R., Matunis,M.J., Mayhew,C.N., Goodson,M.L., Park-Sarge,O.K. and Sarge,K.D. (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem., 43, 40263–40267. [DOI] [PubMed] [Google Scholar]
- Ishov A.M., Sotnikov,A.G., Negorev,D., Vladimirova,O.V., Neff,N., Kamitani,T., Yeh,E.T., Strauss,J.F.,III and Maul,G.G. (1999) PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol., 147, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S. and Gupta,A.A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell, 106, 735–744. [DOI] [PubMed] [Google Scholar]
- Kahyo T., Nishida,T. and Yasuda,H. (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell, 8, 713–718. [DOI] [PubMed] [Google Scholar]
- Kingsley C. and Winoto,A. (1992) Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol. Cell. Biol., 12, 4251–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N., Aittomaki,S., Silvennoinen,O., Palvimo,J.J. and Janne,O.A. (2000) ARIP3 (androgen receptor-interacting protein 3) and other PIAS (protein inhibitor of activated STAT) proteins differ in their ability to modulate steroid receptor-dependent transcriptional activation. Mol. Endocrinol., 14, 1986–2000. [DOI] [PubMed] [Google Scholar]
- Li S.J. and Hochstrasser,M. (1999) A new protease required for cell-cycle progression in yeast. Nature, 398, 246–251. [DOI] [PubMed] [Google Scholar]
- Liu B., Liao,J., Rao,X., Kushner,S.A., Chung,C.D., Chang,D.D. and Shuai,K. (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl Acad. Sci. USA, 95, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Delphin,C., Guan,T., Gerace,L. and Melchior,F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell, 88, 97–107. [DOI] [PubMed] [Google Scholar]
- Marin M., Karis,A., Visser,P., Grosveld,F. and Philipsen,S. (1997) Transcription factor Sp1 is essential for early development but dispensable for cell growth and differentiation. Cell, 89, 619–628. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Coutavas,E. and Blobel,G. (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol., 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megidish T., Xu,J.H. and Xu,C.W. (2002) Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J. Biol. Chem., 277, 8255–8259. [DOI] [PubMed] [Google Scholar]
- Melchior F. (2000) Sumo nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol., 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Moilanen A.M., Karvonen,U., Poukka,H., Yan,W., Toppari,J., Janne,O.A. and Palvimo,J.J. (1999) A testis-specific androgen receptor coregulator that belongs to a novel family of nuclear proteins. J. Biol. Chem., 274, 3700–3704. [DOI] [PubMed] [Google Scholar]
- Müller S., Berger,M., Lehembre,F., Seeler,J.S., Haupt,Y. and Dejean,A. (2000) c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem., 275, 13321–13329. [DOI] [PubMed] [Google Scholar]
- Müller S., Hoege,C., Pyrowolakis,G. and Jentsch,S. (2001) SUMO, ubiquitin’s mysterious cousin. Nat. Rev. Mol. Cell Biol., 2, 202–210. [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast,A., Seeler,J.S., Dejean,A. and Melchior,F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell, 108, 109–120. [DOI] [PubMed] [Google Scholar]
- Poukka H., Karvonen,U., Janne,O.A. and Palvimo,J.J. (2000) Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl Acad. Sci. USA, 97, 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Desterro,J.M., Lain,S., Midgley,C.A., Lane,D.P. and Hay,R.T. (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J., 18, 6455–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont,C. and Hay,R.T. (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem., 276, 12654–12659. [DOI] [PubMed] [Google Scholar]
- Sachdev S., Bruhn,L., Sieber,H., Pichler,A., Melchior,F. and Grosschedl,R. (2001) PIASg, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev., 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen,K., Reich,B. and Will,H. (1999) The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization and modification by small ubiquitin-like modifiers. J. Biol. Chem., 274, 12555–12566. [DOI] [PubMed] [Google Scholar]
- Suske G. (1999) The Sp-family of transcription factors. Gene, 238, 291–300. [DOI] [PubMed] [Google Scholar]
- Suske G. (2000) Transient transfection of Schneider cells in the study of transcription factors. Methods Mol. Biol., 130, 175–187. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Kahyo,T., Toh,E.A., Yasuda,H. and Kikuchi,Y. (2001) Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem., 276, 48973–48977. [DOI] [PubMed] [Google Scholar]
- Tatham M.H., Jaffray,E., Vaughan,O.A., Desterro,J.M., Botting,C.H., Naismith,J.H. and Hay,R.T. (2001) Polymeric chains of Sumo-2 and Sumo-3 are conjugated to protein substrates by Sae1/Sae2 and Ubc9. J. Biol. Chem., 276, 35368–35374. [DOI] [PubMed] [Google Scholar]
- Wu L., Wu,H., Ma,L., Sangiorgi,F., Wu,N., Bell,J.R., Lyons,G.E. and Maxson,R. (1997) Miz1, a novel zinc finger transcription factor that interacts with Msx2 and enhances its affinity for DNA. Mech. Dev., 65, 3–17. [DOI] [PubMed] [Google Scholar]
- Yeh E.T., Gong,L. and Kamitani,T. (2000) Ubiquitin-like proteins: new wines in new bottles. Gene, 248, 1–14. [DOI] [PubMed] [Google Scholar]
- Zhong S., Muller,S., Ronchetti,S., Freemont,P.S., Dejean,A. and Pandolfi,P.P. (2000) Role of SUMO-1-modified PML in nuclear body formation. Blood, 95, 2748–2752. [PubMed] [Google Scholar]