Abstract
Dissection of human centromeres is difficult because of the lack of landmarks within highly repeated DNA. We have systematically manipulated a single human X centromere generating a large series of deletion derivatives, which have been examined at four levels: linear DNA structure; the distribution of constitutive centromere proteins; topoisomerase IIα cleavage activity; and mitotic stability. We have determined that the human X major α-satellite locus, DXZ1, is asymmetrically organized with an active subdomain anchored ∼150 kb in from the Xp-edge. We demonstrate a major site of topoisomerase II cleavage within this domain that can shift if juxtaposed with a telomere, suggesting that this enzyme recognizes an epigenetic determinant within the DXZ1 chromatin. The observation that the only part of the DXZ1 locus shared by all deletion derivatives is a highly restricted region of <50 kb, which coincides with the topo isomerase II cleavage site, together with the high levels of cleavage detected, identify topoisomerase II as a major player in centromere biology.
Keywords: α-satellite/centromere/DT40/DXZ1/topoisomerase IIα
Introduction
In the human genome, centromeres are characterized by vast expanses of satellite and other repeat DNAs, and are often flanked by complex duplications. Despite this complexity, the only repeat DNA found at all normal human centromeres is α-satellite. Therefore α-satellite has presented itself as a strong candidate for a DNA with a functional role in centromere/kinetochore formation. Support for this has come from the transfection of defined arrays of α-satellite DNA into cultured human cells resulting in the reproducible formation of minichromosomes with de novo centromeres (Harrington et al., 1997; Ikeno et al., 1998; Henning et al., 1999; Ebersole et al., 2000; Schueler et al., 2001; Mejia et al., 2002).
The abundance of α-satellite at normal human centromeres raises questions about the functional organization of these domains. Pulsed-field gel electrophoresis (PFGE) analyses and quantitative-fluorescence in situ hybridization (Q-FISH) screens have shown that the amount of α-satellite DNA found on mitotically stable human chromosomes is highly variable (Oakey and Tyler-Smith, 1990; Mahtani and Willard, 1998; Lo et al., 1999). Despite this variation, indirect immunofluorescence studies have suggested that the overall size of the assembled kinetochore is relatively constant (Earnshaw et al., 1989; Fantes et al., 1989; Marzais et al., 1999). Moreover, simultaneous immunofluorescence and FISH (immuno-FISH) has indicated that only part of the α-satellite DNA is actually associated with the inner kinetochore proteins CENP-A and CENP-C (Warburton et al., 1997; Sugimoto et al., 1999; Blower et al., 2002). Given the often immense size of these structurally repetitive DNA arrays, determining whether or not they are also functionally repetitive and whether there are discrete functional domains, has been difficult to address.
To date, the Y is the only human centromere for which a detailed molecular and functional analysis has been undertaken. An extensive collection of abnormal Y chromosomes allowed the minimal DNA sequences required for centromere function to be localized to a 500 kb region encompassing alphoid DNA and adjacent Yp sequences (Tyler-Smith et al., 1993). More recently a region of topoisomerase II (topo II) cleavage activity has been mapped, using PFGE, to the p-arm side of the major Y alphoid array at active, but not inactive, Y centromeres (Floridia et al., 2000). This suggests that α-satellite arrays are not functionally equivalent throughout. However, data for other human chromosomes, for which fewer naturally occurring rearrangements are available, is limited. As an alternative we have undertaken the molecular and functional dissection of a single human X centromere using cloned telomeric DNA.
The organization of centromeric DNA sequences on the X appears relatively simple compared to many human chromosomes. A single major class of α-satellite DNA is present, which forms a vast and largely uninterrupted array designated DXZ1 (polymorphic size range of ∼2.2– 3.7 Mb) (Mahtani and Willard, 1998). Extensive analyses of the pericentromeric regions have revealed a lack of complex genomic duplications and paralogies (Mills et al., 1999; Schueler et al., 2000, 2001; Bailey et al., 2001). Mapping of breakpoints in a series of naturally arising rearranged i(Xq) chromosomes (Lee et al., 2000; Schueler et al., 2001) suggests that all Xp sequences can be eliminated without loss of mitotic stability. Moreover, experimentally generated i(Xp) derivatives and Xq deletion chromosomes have revealed that removal of all Xq sequences is also compatible with mitotic stability (Farr et al., 1992; Higgins et al., 1999). These observations, together with the generation of human artificial chromosomes through transfection, suggest that the DXZ1 α-satellite DNA is sufficient for centromere function (Schueler et al., 2001).
Using immuno-FISH on several independent human X chromosomes we observed a bias in the distribution of CENP-A and -C towards the Xp-side of the DXZ1 signal. We have previously reported the generation of a <3 Mb linear minichromosome based on the human X centromere region (Farr et al., 1992, 1995; Mills et al., 1999). This minichromosome is structurally stable and shows a high level of mitotic stability (Mills et al., 1999). To determine whether the 2.1 Mb DXZ1 array of this chromosome is functionally repetitive, systematic chromosome breakage and de novo telomere formation have been used to generate an extensive series of deletion derivatives following its transfer into the recombination-proficient chicken cell line DT40. The chromosomes recovered have been examined by: (i) detailed molecular analysis of linear DNA organization; (ii) mitotic stability assessments; (iii) immuno-FISH; and (iv) PFGE-mapping of topo II. This has allowed us to co-localize centromere activity, CEN proteins and topo II cleavage activity to a subdomain of the asymmetrically organized DXZ1 locus. Following chromosome breakage and de novo telomere formation in the vicinity of this domain, shifts in the site of topo II cleavage were observed, suggesting that the enzyme recognizes a unique chromatin structure. The high levels of cleavage detected within the DXZ1 array in asynchronously growing cells, together with the retention of the topo II cleavage site on all minichromosomes recovered, suggests a fundamental role in centromere biology.
Results
Asymmetric distribution of kinetochore proteins relative to the DXZ1 DNA array
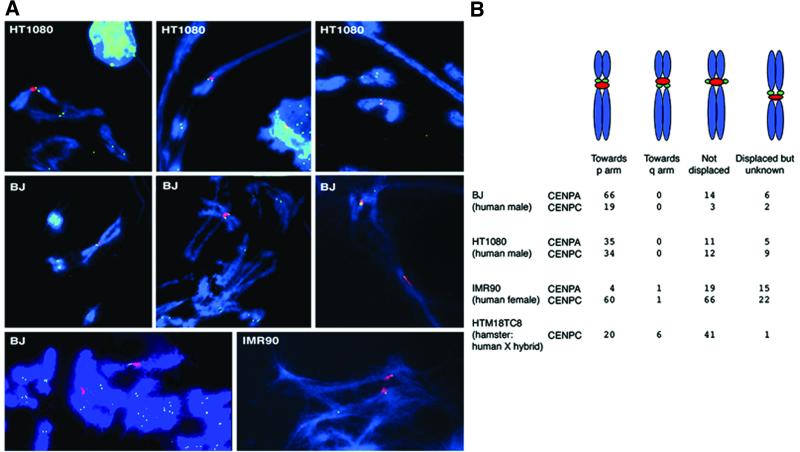
The starting point for this study was an investigation of the distribution of the constitutive centromere proteins CENP-A and -C, relative to the X α-satellite locus DXZ1. Immuno-FISH was undertaken on mechanically stretched metaphase chromosomes from the human cell lines HT1080, BJ and IMR90 and the hamster–human X hybrid HTM18TC8 (examples shown in Figure 1A). Where displacement of the CENP signal was discernible there was a significant bias towards the short arm of the human X chromosome, suggesting an underlying asymmetry in the centromere/kinetochore domain (Figure 1B).
Fig. 1. Immuno-FISH showing CENP-A and CENP-C relative to the human X α-satellite array DXZ1. (A) Examples of indirect immunofluorescence to detect CENP-A (green) in combination with FISH to detect DXZ1 (red) on mechanically stretched human X chromosomes present in various cell lines. DNA was counterstained using DAPI (blue). The images show partial chromosome complements and in some instances encompass chromosomes from more than one nucleus and/or show sister chromatids with varying degrees of separation. (B) A tabulated summary of the observed distribution. The arm distribution was based on chromosome morphology. Where displacement was apparent but the morphology too distorted for assignment the signals were counted as ‘displaced, but unknown’. Chi-square analysis of this data indicates a highly significant p-arm bias in the CENP distribution (1 df, probability of a random distribution = <0.0001).
Generation of DXZ1-based minichromosomes by homologous recombination in DT40
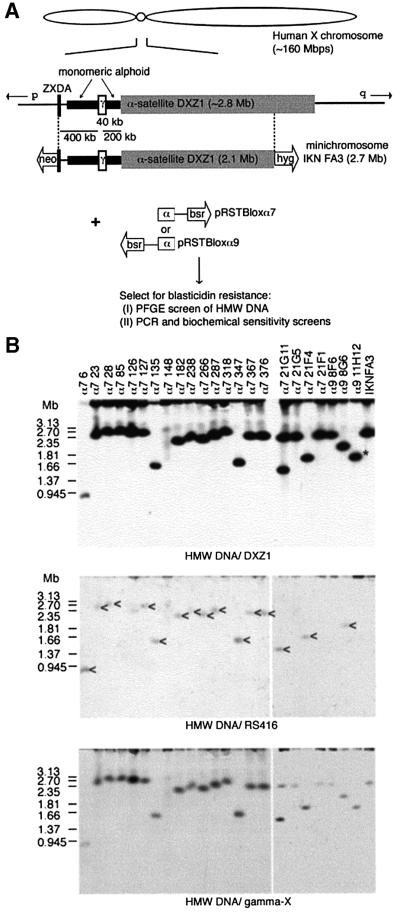
To investigate this asymmetry we have undertaken the systematic dissection of a single human X centromere. The DNA structure of the X centromere-based IKNFA3 minichromosome, relative to the intact X from which it was derived (present in HTM18TC8) is shown in Figure 2A. A telomere-seeding construct based on a blasticidin resistance selectable cassette (bsr), and including a 2 kb block of homology to the DXZ1 locus, was linearized and transfected into a DT40 cell line carrying IKNFA3. Two versions of the construct, differing only in the orientation of the DXZ1 repeat relative to the (TTAGGG)n array, were transfected independently and stable blasticidin-resistant clones isolated.
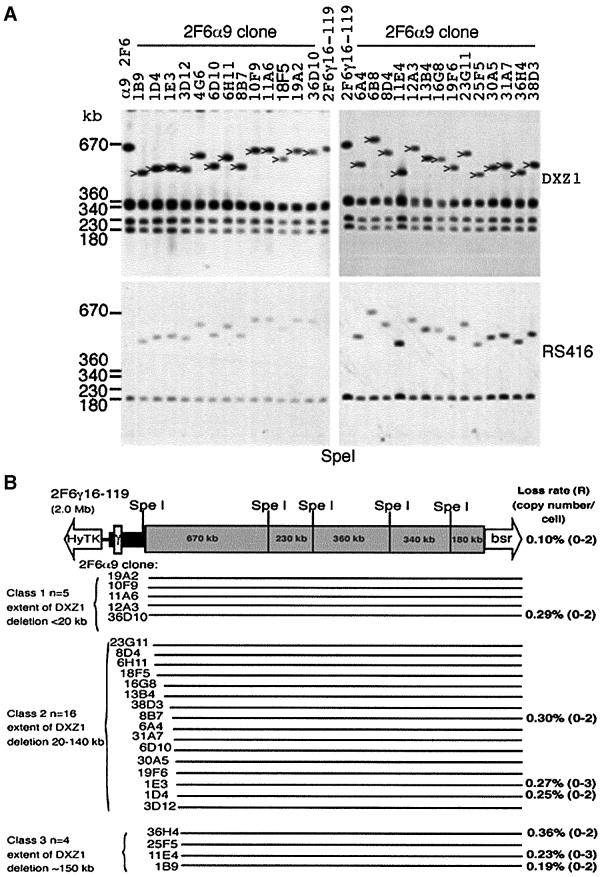
Fig. 2. Strategy for dissecting the human X centromere. (A) The DXZ1 array (grey box) on the IKNFA3 minichromosome appears contiguous with that on the X from which it was derived except for the removal of 700 kb from the Xq-side (Farr et al., 1992; Bayne et al., 1994; Mills et al., 1999). The Xp pericentromeric region is rich in monomeric α-satellite DNA lacking higher order repeat organization (thick black line) (Schueler et al., 2001) and has embedded in it a 40 kb γ-satellite array (white box) (Lee et al., 2000). Experimentally seeded telomeres are represented as open arrows with the selectable marker indicated. To generate smaller derivatives a telomere-seeding construct was designed to target the DXZ1 array (pRSTBloxα). Two versions, differing only in the orientation of the 2 kb block of DXZ1 relative to the ds(TTAGGG)n, were generated (α7 and α9) and independently transfected into a DT40 cell line carrying the IKNFA3 chromosome. Blasticidin-resistant transfectants were screened for new minichromosomes, either by PFGE or for loss of terminal markers. (B) The IKNFA3 minichromosome and examples of derivatives resolved as intact HMW DNA by PFGE, Southern blotted and detected using DXZ1, RS416 (specific for the targeting construct) and γ-satellite DNA (gamma-X). Co-localization of new DXZ1 hybridizing linear DNA molecules with the introduced construct is consistent with targeted breakage events (indicated by an arrowhead for cell lines α7: 6, 23, 28, 127, 135, 182, 238, 266, 287, 347, 367, 376, 21G11, 21F4 and α9 8G6). The minichromosome in cell line α7 148 appears structurally unstable. Cell line α7 21G11 was confirmed as having two minichromosomes per cell. Cell line α9 11H12 contains a ∼1.8 Mb DXZ1 molecule (indicated by an asterisk), but has the construct randomly integrated into the DT40 genome. The other cell lines shown are not targeted.
Two screens were undertaken. In the first, cells from stable transfectants (∼1000) were pooled (six clones/pool), and uncut, high molecular weight (HMW) DNA screened by PFGE and Southern blotting. In total, 10 transfectants with DXZ1-hybridizing linear DNA molecules smaller than the starting minichromosome were identified (five with each construct), of which five were hygromycin-sensitive. A second set of ∼1000 transfectants was PCR-screened for loss of IKNFA3 terminal markers. In total, 28 blasticidin-resistant clones were identified that had lost either the neor (n = 1) or the hygr (n = 27) genes.
Molecular analysis of the minichromosomes as intact HMW DNA
Uncut HMW DNA from all 38 clones was characterized by PFGE. DNA resolved in the 1–3 Mb size range was hybridized with both repeat and single-copy DNA probes (examples are shown in Figure 2B). While some cell lines had α-satellite-hybridizing molecules distinctly smaller than the IKNFA3 minichromosome, several from the terminal marker screen contained linear DXZ1 molecules similar to 2.7 Mb. However, in all of the hygromycin-sensitive clones with 2.5–3 Mb DXZ1 molecules, DNA specific for the targeting construct (RS416) co-localized with the DXZ1 signal. The single G418-sensitive clone recovered (α7 98) also contains a 2.5–3 Mb minichromosome (data not shown) suggesting that targeting of the DXZ1 locus had not occurred.
Of the clones with DXZ1-hybridizing bands significantly smaller than 2.5–3 Mb most co-hybridized with RS416 DNA probe, consistent with targeted truncation (Figure 2B; data not shown). Exceptions were: (i) α9 11H12, a hygromycin-resistant clone isolated in the PFGE screen, which appeared to have undergone a spontaneous deletion (indicated in Figure 2B); and (ii) α7 3D4 and α7 148, two hygromycin-sensitive lines in which a faint smear of DXZ1 hybridization was detected, suggesting structural instability (Figure 2B; data not shown). In addition, three clonal cell lines from the PFGE screen (α9 3B8, α7 21G11 and α7 3A1) were found to each have two DXZ1-containing linear molecules per cell: one the same size as IKNFA3 and one smaller targeted derivative (2.2, 1.5 and 2.1 Mb, respectively) (Figure 2B; data not shown).
Molecular characterization of the minichromosomes by restriction enzyme analysis
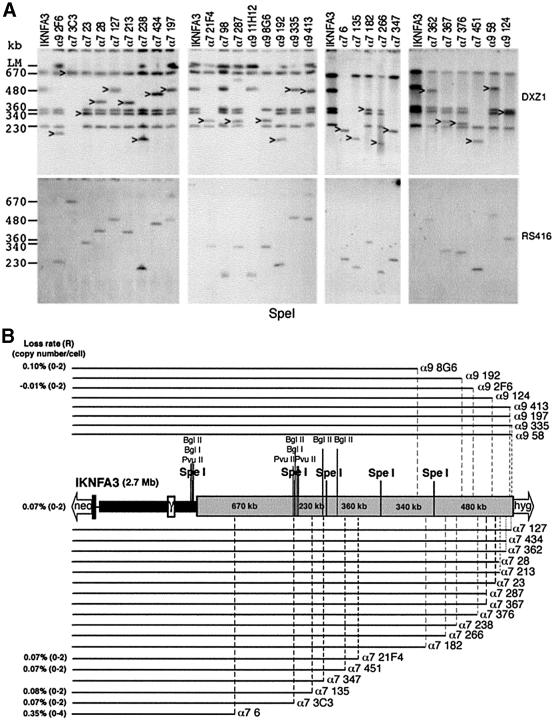
DNA from 32 of the minichromosome-containing cell lines was subjected to further PFGE using restriction enzymes that cut infrequently within the DXZ1 array. Examples of SpeI digests probed using RS416 and DXZ1 are shown (Figure 3A). These analyses enabled us to determine not only the amount of DXZ1 DNA present on each minichromosome, but also its origin relative to the starting array. The structure of the various minichromosomes in which the restriction mapping data are consistent with straightforward targeting and breakage events (n = 26) is summarized in Figure 3B.
Fig. 3. Molecular characterization of minichromosomes by restriction enzyme analysis. (A) Examples of SpeI PFGE analysis. DXZ1-hybridizing fragments that cohybridize with the RS416 DNA probe are indicated. PFGE conditions were 20–60 s pulse at 200 V for 24 h. Sizes of the DXZ1-hybridizing SpeI fragments present in the IKNFA3 DXZ1 array are indicated. (B) The linear DNA structure of IKNFA3 and various DXZ1 truncation derivatives. The structures of all new minichromosomes consistent with a straightforward targeted breakage event is presented. Each derivative is shown as a black line which directly reflects the region of the starting IKNFA3 molecule that is retained. In all cases there has been loss of varying amounts of the DXZ1 array together with replacement of the Xq(hygr)-tagged telomere with a bsr-tagged telomere. Mitotic loss rates per generation (R) and the range in copy number per DT40 cell for various minichromosomes are given.
All minichromosomes recovered share the same basic structure: they retain the proximal Xp DNA and have undergone varying degrees of truncation from the Xq(hygr)-side of the starting chromosome. No chromosomes were recovered which had undergone truncation of the DXZ1 locus from the Xp(neor)-side of the array. Minichromosomes resulting from targeted breakage within the terminal ∼500 kb on the Xq(hygr)-side of IKNFA3 were generated using the homology block in either orientation, suggesting a lack of unidirectionality in this part of the array. We do not know whether this reflects the natural state of the DXZ1 locus, or whether it has arisen during maintenance and manipulation of this particular human X chromosome in cell culture.
The mitotic stability of various minichromosomes in the DT40 background was assessed by FISH following growth in the absence of selection (Figure 3B). Low loss rates and good copy number control (comparable with IKNFA3) were exhibited by minichromosomes down to 1.4 Mb (α7 3C3). The only minichromosome showing a significantly higher loss rate (∼5-fold) and slightly more erratic copy number (0–4) was α7 6 (total size 850 kb with <200 kb of DXZ1 DNA).
Removal of all Xp DNA sequences is compatible with minichromosome recovery
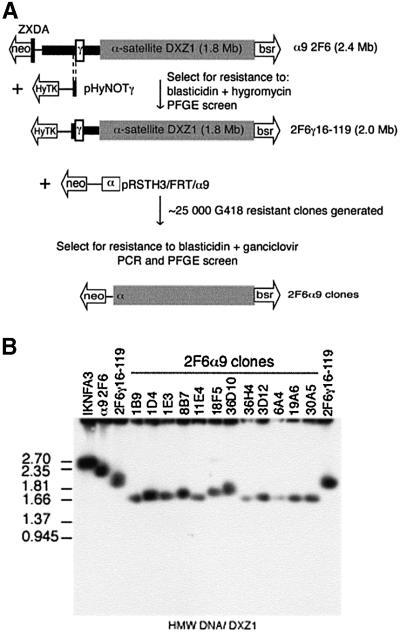
To determine whether the failure to recover minichromosomes truncated from the Xp-side of IKNFA3 reflects a requirement for the ∼650 kb of Xp-derived DNA, an HSV-TK–ganciclovir enrichment strategy was used to increase the number of targeting events analysed (Figure 4A). A bidirectional selectable marker, the hygromycin phosphotransferase–HSV thymidine kinase fusion gene (HyTK) was introduced by targeted-telomere seeding immediately distal to the γ-satellite repeat in α9 2F6 to generate the 2 Mb derivative 2F6γ 16–119 (Figures 3B and 4A). DT40 cells carrying the HSV-TK gene are sensitive to ganciclovir, allowing a large number of transfectants to be screened for events that result in loss of the terminally located HyTK gene. A DXZ1-targeting-telomere seeding construct (pRSTH3/FRT/α9) was generated by replacing the bsr cassette in pRSTBloxα9 by a neor gene. This was linearized, introduced into clone 2F6γ16–119 and ∼25 000 G418-resistant colonies generated and simultaneously selected in ganciclovir (Figure 4A). DNA from the 170 clones recovered was tested by PCR and 48 clones were identified that had lost both the HyTK locus and the γ-satellite array (∼0.2% of all transfectants).
Fig. 4. Xp DNA sequences are dispensible for mitotic chromosome function. (A) Schematic of the two-step targeting strategy used to remove the remaining 650 kb of Xp DNA from α9 2F6 (2.4 Mb). A de novo telomere tagged by a CMV HyTK fusion gene was targeted into the region immediately distal to the γ-satellite array using pHyNOTγ (5/246 transfectants). One correctly targeted cell line 2F6γ16–119, was retargeted using pRSTH3/FRT/Xα9. G418- and ganciclovir-resistant clones (170) were PCR screened for loss of the HyTK locus and γ-satellite DNA. (B) Examples of Xp-truncation derivatives of α9 2F6 resolved as intact HMW DNA by PFGE and Southern blotted for DXZ1.
The status of the DXZ1 array was assessed by PFGE of uncut and SpeI-digested HMW DNA (Figures 4B and 5A; data not shown). In every case the largest DXZ1-hybridizing SpeI fragment had been disrupted and replaced by a new DXZ1-hybridizing band (size range: 520–690 kb). No disruption of the other SpeI fragments (which encompass 1.1 Mb) was observed. For 28 cell lines the data are consistent with a straightforward targeted-truncation event removing part of the 670 kb SpeI fragment (together with the distal DNA sequences) and reducing the overall size of the minichromosome (to 1.6–1.8 Mb) (Figure 4B). The position of the targeted breaks within the DXZ1 array is shown in Figure 5B. The extent of DXZ1 deletion ranges from a few kilobases up to a maximum of 150 kb.
Fig. 5. Molecular characterization of minichromosomes in the Xp-truncation series. (A) Examples of SpeI PFGE analysis, Southern blotting and hybridization with DXZ1 and RS416. Novel DXZ1-hybridizing fragments that co-hybridize with the RS416 DNA probe (present on the DXZ1-targeting constructs) are indicated. The 180 kb DXZ1 fragment that co-hybridizes with RS416 and is present in all clones originates from the construct associated with the Xq(bsr)-tagged telomere of α9 2F6 (see also Figure 3A). Sizes of the DXZ1-hybridizing SpeI fragments present in α9 2F6/2F6γ16–119 are indicated. In clone 6B8 a rearrangement appears to have accompanied the targeted breakage event resulting in a novel SpeI fragment slightly greater than the starting 670 kb. PFGE conditions were 30–70 s pulse at 200 V for 24 h. (B) The molecular structure of the ganciclovir- resistant 2F6γ16–119 derivatives consistent with a straightforward targeted breakage event is presented (n = 25). Mitotic loss rates per generation (R) and minichromosome copy number per DT40 cell for selected minichromosomes are shown.
The mitotic stability of a selection of the chromosomes in the Xp-truncation series was assessed. Removal of the remaining Xp sequences and the Xp-edge of the DXZ1 array was accompanied by a slight loss of mitotic stability (3- to 5-fold) compared with similarly sized chromosomes in the Xq-truncation series (e.g. α7 451 and α7 21F4) (Figures 3B and 5B).
Immuno-FISH reveals localization of centromere proteins to the Xp-edge of the IKNFA3 DXZ1 array
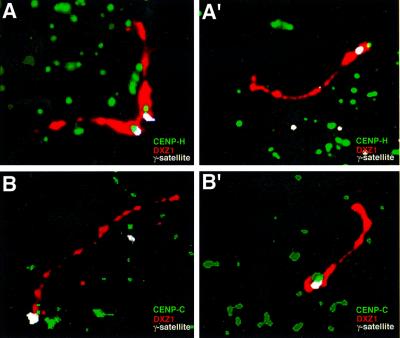
The distribution of the constitutive centromere proteins CENP-C and -H relative to the DXZ1 array was investigated by immuno-FISH on extended chromatin fibres. A DT40 cell line containing the IKNFA3 minichromosome was modified by targeting of the single copy chicken CENP-H gene to produce a CENP-H–green fluorescent protein (GFP) fusion protein (Fukagawa et al., 2001). This cell line (1aH2) proliferates well, with the centromerically located fusion protein readily detectable. Indirect immunofluorescence using anti-GFP was combined with dual FISH to detect both DXZ1 and DNA spanning the γ-satellite array in proximal Xp (BAC 342O19; Schueler et al., 2001). Where the DXZ1 array was sufficiently extended, specific localization of discrete anti-GFP signals was reproducibly at the Xp-end of the DXZ1 array (Figure 6A and A′). The same asymmetric distribution was obtained in the untargeted IKNFA3 cell line, using antibodies to detect endogenous chicken CENP-C (Fukagawa et al., 1999) (Figure 6B and B′).
Fig. 6. Localization of CENP-H and -C to the Xp-side of the DXZ1 array using immuno-FISH. (A and A′) Chromatin fibres from 1aH2, a DT40 line carrying the IKNFA3 minichromosome and producing a CENPH–GFP fusion protein, were generated. The centromerically located fusion protein was detected (on all chromosomes in the cell) using anti-GFP (green). This was followed by dual FISH for the human X centromere-based minichromosome using DXZ1 (red) and BAC 342O19 (which spans the γ-satellite array DNA) (white). (B and B′) Chromatin fibres from the DT40 cell line IKNFA3. CENP-C has been detected using antibodies to chicken CENP-C (green) followed by dual FISH for DXZ1 (red) and BAC342O19 (white).
Mapping a major site of topo II cleavage within the DXZ1 array
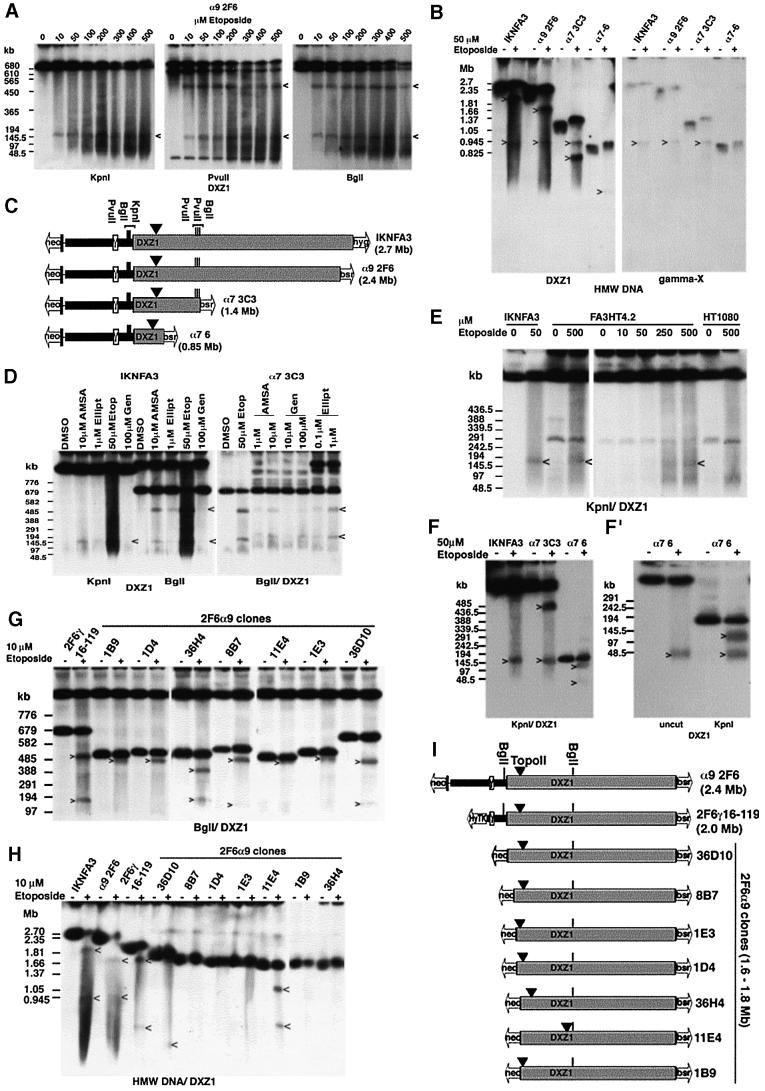
We then set out to characterize the IKNFA3 DXZ1 locus for topo II activity. DT40 cells containing the α9 2F6 minichromosome were incubated with etoposide, which acts to stabilize the enzyme–DNA strand passing intermediate, resulting in double-stranded DNA breaks that can be mapped by PFGE. KpnI digestion of HMW DNA from untreated α9 2F6 releases a single DXZ1-hybridizing fragment of 1.8 Mb. Following etoposide exposure a DXZ1-hybridizing band of ∼150 kb is clearly detectable (Figure 7A). Restriction mapping with BglI and PvuII, which both cut at additional sites within the DXZ1 locus, allowed the topo II cleavage site to be localized to within 200 kb of the Xp-side of the DXZ1 array (Figure 7A and C). All minichromosomes from the original Xq-truncation series were similarly treated with etoposide and the DNA digested with KpnI. In each case, an etoposide-specific ∼150 kb DXZ1-hybridizing fragment was identified (data not shown). DNA from these various etoposide-treated minichromosome lines was also resolved without prior restriction enzyme digestion (examples shown in Figure 7B). This revealed that following etoposide, in addition to the DXZ1 signal characteristic of the intact linear DNA molecule, two other DXZ1-hybridizing fragments are resolved in each cell line. The etoposide-specific 850 kb signal was constant and co-hybridizes with γ-satellite DNA. The other etoposide-specific DXZ1 signal varies in size (correlating with the extent of truncation of the DXZ1 array in the various minichromosomes) (Figure 7C).
Fig. 7. Mapping topo II activity within the DXZ1 array. (A) DT40 cells carrying the α9 2F6 minichromosome were incubated with etoposide in vivo for 30 min and embedded in agarose. HMW DNA was cut with KpnI, PvuII or BglI, resolved by PFGE and Southern blotted with DXZ1. Etoposide-specific DXZ1-hybridizing fragments are indicated. PFGE conditions: 20–60 s pulse, 200 V, 24 h. (B) Uncut HMW DNA from various 50 µM etoposide-treated (+) or DMSO only (–) minichromosome-carrying DT40 lines from the Xq-truncation series was resolved by PFGE. The Southern blot filter was hybridized sequentially for X γ-satellite (gamma-X) and DXZ1. Etoposide-specific hybridizing fragments are indicated. (The reason for the slight mobility shift of the intact minichromosomes following etoposide exposure is unknown, but may be due to an effect of the apoptotic genome-wide chromosome fragmentation on HMW DNA migration conditions.) (C) Schematic showing localization of the topo II cleavage site on IKNFA3 and various minichromosomes from the Xq-truncation series. Also indicated are the positions of BglI, PvuII and KpnI sites within and flanking the DXZ1 array (grey box). (D) The IKNFA3 and α7 3C3 DT40 cell lines treated with various topo II inhibitors: AMSA (amsacrine), Ellipt (ellipticine), Etop (etoposide) and Gen (genistein), or with DMSO only. HMW DNA was digested with BglI or KpnI, resolved by PFGE and Southern blotted with DXZ1. PFGE conditions: 40–80 s pulse, 200 V, 24 h. The 520 and 150 kb inhibitor-specific cleavage products are indicated. (E) Preservation of the position of topo II cleavage on the IKNFA3 minichromosome in human and chicken. FA3HT4.2 (minichromosome in HT1080), HT1080 and IKNFA3 (minichromosome in DT40) were treated with etoposide for 60 min before preparation for PFGE. KpnI-cut HMW DNA was Southern blotted with DXZ1. PFGE conditions: 20–60 s pulse, 200 V, 24 h. Etoposide-specific DXZ1-hybridizing KpnI fragments are indicated. (F and F′) Analysis of etoposide-sensitive topo II cleavage in the α7 6 minichromosome using KpnI-cut and uncut HMW DNA. Samples are 50 µM etoposide-treated (+) or DMSO only (–). PFGE conditions for (F): 20–60 s pulse, 200 V, 24 h and for (F′): 10–30 s pulse, 200 V, 18 h in 1.5% agarose. Etoposide-specific DXZ1-hybridizing fragments are indicated. (G) Analysis of topo II cleavage in various 2F6α9 clones from the Xp- truncation series. HMW DNA from 10 µM etoposide-treated (+) and untreated (–) cells was digested with BglI, resolved by PFGE and Southern blotted. Etoposide-specific DXZ1-hybridizing fragments are indicated. PFGE conditions were 40–80 s pulse at 200 V for 24 h. (H) Analysis of topo II cleavage in various 2F6α9 clones from the Xp-truncation series. Uncut HMW DNA from etoposide-treated (+) and untreated (–) cells was resolved by PFGE and the Southern blotted for DXZ1. Etoposide-specific hybridizing fragments are indicated. (I) Schematic showing localization of topo II cleavage. The position of BglI sites relative to the DXZ1 array (grey box) is indicated.
These observations are consistent with a single major site of etoposide-sensitive topo II cleavage on the IKNFA3 minichromosome located ∼150 kb in from the Xp-edge of the DXZ1 array. Moreover, cleavage is readily detectable with the amount of minichromosome molecules present in the cleaved relative to the intact form, after 30 min treatment with 50 µM etoposide, being ∼30% (based on α7 3C3 in which all three DXZ1-hybridizing bands are easily resolvable) (Figure 7C; data not shown). Further more, evidence for the same major site of cleavage in IKNFA3 and α7 3C3 was obtained using three other topo II inhibitors: amsacrine, ellipticine and genistein (Figure 7D).
We also examined whether etoposide-sensitive topo II cleavage is detectable when the IKNFA3 minichromosome is maintained in a human cell background. An HT1080 hybrid (designated FA3HT4.2) (Mills et al., 1999) in which each cell has several copies of the IKNFA3 minichromosome (in addition to the endogenous X) was treated with etoposide (Figure 7E). While no etoposide-specific DXZ1 hybridizing bands were detectable in the parental HT1080 cell line under these PFGE conditions, we reproducibly identified a weakly hybridizing ∼150 kb DXZ1 signal in FA3HT4.2, suggesting that the position of topo II cleavage within the DXZ1 array is preserved between chicken and human.
Finally, we investigated more closely the position of etoposide-sensitive topo II activity in α7 6 (which exhibits the maximum extent of DXZ1 deletion observed from the Xq-side of the array) and in seven clones from the 2F6α9 Xp-truncation series (where the de novo telomere has been targeted to the region of the DXZ1 locus immediately adjacent to the site of topo II cleavage in the starting array). For α7 6, PFGE of uncut and KpnI-digested DNA revealed that the limited region of DXZ1 DNA retained by this minichromosome should just encompass the topo II cleavage site and suggests that the position of cleavage relative to this restriction enzyme site has shifted by a few kilobases compared with its position in IKNFA3 (Figure 7F and F′). PFGE of uncut, BglI and SpeI digests of 2F6α9 clones also revealed topo II cleavage within the DXZ1 array. In most cases the site of cleavage had shifted relative to its starting position (Figure 7G–I; data not shown). In 1E3, 36D10, 8B7, 1D4 and 1B9 the position of cleavage had moved only slightly (by <20 kb), while in 36H4 and 11E4 much larger shifts were detected (of ∼100 and ∼500 kb, respectively). This strongly suggests that the site at which topo II cleaves is determined by chromatin structure rather than primary DNA sequence.
Discussion
Localization of centromere activity to the Xp-side of the DXZ1 locus
The accumulation of vast expanses of tandemly repeated DNA in centromeric regions, together with a condensed chromatin structure, has hampered their functional dissection. To follow up observations suggesting a biased distribution of CENP-A and -C to the short-arm side of the DXZ1 array on various human X chromosomes we have systematically dissected a single human X centromere in DT40. We hypothesized that if the assembled kinetochore demarcates a distinct domain within the repeat array the structure of the minichromosomes recovered would correlate with retention of a specific part of the DXZ1 locus.
Detailed restriction enzyme mapping of the initial truncation series revealed an asymmetric structure in which DNA from the Xp-side of the DXZ1 array was always retained. No truncation derivatives based on the Xq-side of the DXZ1 locus were recovered and in none of the cell lines isolated using PFGE as the primary screen was the reciprocal product of the targeted breakage event retained. These observations suggested that the Xq-side of the DXZ1 array lacks innate centromere/kinetochore function. An HSV TK/ganciclovir screen was then used to isolate events resulting in removal of the Xp-side of the DXZ1 locus. A substantial number of new minichromosomes arising from DXZ1-targeted breakage and loss of Xp proximal DNA was recovered. Strikingly, however, all the truncation events clustered to within 150 kb of the Xp-edge of the DXZ1 locus.
Further support for the idea that the Xp-side of the DXZ1 locus is dominant in conferring centromere function was provided by immuno-FISH on chromatin fibres from DT40 cells containing the IKNFA3 minichromosome, which revealed that both CENP-H and -C have a restricted localization to the Xp-side of the DXZ1 locus.
A major site of topo II cleavage within the DXZ1 locus
Topo IIα has been implicated in a host of cellular processes, but a specific role at the centromere has been suggested by its transient association with this structure from late S–G2 up until anaphase (Taagepera et al., 1993; Rattner et al., 1996; Sumner, 1996; Niimi et al., 2001; Christensen et al., 2002) and by the observation that inhibitors of topo II perturb normal centromere/kinetochore structure and function (Downes et al., 1991; Kallio and Lahdetie, 1996; Rattner et al., 1996; Sumner, 1998).
Recent data using the topo II poison etoposide has revealed the presence of a single region of topo II cleavage within the active, but not the inactive, Y centromere (Floridia et al., 2000). In this study, we have mapped a major site of etoposide-sensitive topo II cleavage in the DXZ1 array ∼150 kb from the Xp-edge. This cleavage site is detectable in both chicken and human cells. To rule out the possibility that we may have failed to reveal sites of enzyme activity where this particular drug is inefficient at stimulating cleavage, the effects of three other poisons on in vivo DNA cleavage were investigated: amsacrine, ellipticine and genistein (Burden and Osheroff, 1998). All identified the same single site of cleavage in living cells as etoposide. It therefore appears that this particular region of the DXZ1 domain is highly accessible to topo II.
Co-localization of topo II activity, centromere activity and CEN proteins within the α-satellite array
Why should topo II cleavage activity localize to one specific part of the vast DXZ1 locus when the topo II consensus binding sequence occurs frequently in A+T-rich α-satellite DNA? The observation that the site of topo II cleavage is constant relative to the flanking restriction enzyme sites, except when chromosome breakage and de novo telomere formation occur immediately adjacent to the original site of topo II activity, suggests that the site of action of this enzyme can shift. This observation would be consistent with the idea that topo II recognizes a specific chromatin structure rather than primary DNA sequence. We have demonstrated co-localization of topo II, the CENPs -A, -C and -H and centromere activity to the Xp-side of the DXZ1 array. An attractive explanation, therefore, would be that the enzyme recognizes a unique chromatin structure defined by the centromere/kinetochore (Udvardy and Schedl, 1991).
For the Y chromosome, localization of topo II cleavage to within ∼150 kb of the Yp-edge of the DYZ3 locus is also consistent with this enzyme being a marker for active centromeres: the centromere proteins CENP-A and -C have been shown to localize to one side of the DYZ3 alphoid array (Floridia et al., 2000); work characterizing naturally occurring rearrangements of the Y mapped centromere function to the Y alphoid and adjacent Yp DNA sequences (Tyler-Smith et al., 1993); in several inactive Y centromeres deletions removing portions of the alphoid array near the Yp-edge have been identified (Fisher et al., 1997). Moreover, dissection of the Y centromere using cloned telomeric DNA generated two independent derivatives whose structure was consistent with localization of the kinetochore to the Yp-side of the DYZ3 array (Brown et al., 1994).
Localization of the centromere/kinetochore to the short-arm side of the major α-satellite loci on both human sex chromosomes
The localization of centromere function, CENPs and topo II cleavage to the edge of the major α-satellite arrays on the human sex chromosomes, and to the short-arm side of the arrays in both cases, is intriguing but its significance is as yet unknown. For the Y, assessment of the position of topo II cleavage for multiple chromosomes, showing polymorphic variation in the DYZ3 array size, suggested that it is fixed relative to the edge of the array (Floridia et al., 2000). For the X chromosome restriction enzyme mapping of topo II activity is available only for a single DXZ1 array. However, the Xp bias in the localization of CENP-A and -C on several independent human X chromosomes suggests that the position of the kinetochore on the X is also fixed on the short-arm side of the major α-satellite domain. Analysis of other chromosomes is required to determine whether this asymmetric organization relative to the major block of α-satellite DNA is a general feature of the human centromere.
The function of topo II at the centromere
Although the function of topo II at the centromere is unknown, the observation that the DXZ1 topo II cleavage site is retained on all minichromosomes recovered, together with the substantial proportion of minichromosome DNA molecules that are in the cleaved form following a brief period of etoposide exposure, suggests a fundamental role at the vertebrate centromere (Warburton and Earnshaw, 1997). Even more striking is the observation that the only part of the 2.1 Mb DXZ1 locus that is retained by all deletion derivatives is a region of a few kilobases (<50 kb) that coincides with the position of topo II cleavage in the starting array.
Topo II is essential for disentangling sister chromatids and one early suggestion was that topo II activity also triggers the final decatenation of sister chromatids within centromeric DNA (Murray and Szostak, 1985). Although evidence has since accumulated for an independent cohesion apparatus (Nasmyth et al., 2000), links between topo II and cohesion are emerging (Skibbens et al., 1999; Wang et al., 2000; Losada and Hirano, 2001; Niimi et al., 2001). Thus in vertebrate cells the loading of topo II onto centromeric chromatin may reflect its involvement in a variety of functions, including roles in centromeric DNA replication, in remodelling the specialized centromeric chromatin structure, in facilitating the cohesion that persists at centromeric heterochromatin during mitosis prior to sister separation, as well as in chromatid separation at the metaphase–anaphase transition.
The domain structure of the human centromere
Detailed molecular characterization of this series of X centromere-based linear minichromosomes has revealed that the kinetochore consistently propagates itself on the same part of the DXZ1 α-satellite array, allowing us to assign this domain within an amorphous sea of repeat DNA. Moreover, a substantial amount of the α-satellite DNA, even within this single relatively homogeneous array, lacks innate centromere activity, consistent with the rarity of centric fission in human and other higher eukaryotes. Recent studies in yeast and Drosophila have revealed that eukaryotic centromeres are made up of structurally and functionally distinct molecular domains that show a conserved organizational pattern (Megee et al., 1999; Partridge et al., 2000; Pinto and Winston, 2000; Bernard et al., 2001; Blower et al., 2002). Future characterization of the behaviour of this well defined series of X centromere-based minichromosomes will provide a powerful system for understanding the complex organization of normal higher eukaryotic centromeres.
Materials and methods
Recombinant DNA constructs
The DXZ1 targeting-telomere-seeding plasmids pRSTBloxα7 and pRSTBloxα9 were constructed as follows: an 800 bp array of ds(TTAGGG)n together with a CAG lox71 bsr pA cassette (Araki et al., 1997) were cloned into pRS416 (Stratagene). A 2 kb block of DXZ1 was inserted as a NotI–SstII fragment to give two versions, which differ only in the orientation of the DXZ1 DNA relative to the ds(TTAGGG)n. The final construct was linearized with NotI prior to transfection. A second construct was generated in which CAG bsr was replaced by CMV neor (designated pRSTH3/FRT/Xα7 and pRSTH3/FRT/Xα9).
A telomere-seeding construct designed to target DNA immediately distal to the γ-satellite array was generated as follows: an 11.5 kb HindIII fragment from cosmid CX16-D12 (Lee et al., 1995) was inserted into a plasmid containing SV2HyTK (Lupton et al., 1991) and an 800 bp array of ds(TTAGGG)n. The homology block was orientated so as to allow recovery of a stably truncated DXZ1-carrying linear chromosome following a successful targeting event. The final construct, designated pHyNOTγ (22 kb), has a unique NotI site for linearization.
Transfection and cell culture
DT40 cells were cultured and transfectants generated by electroporation (Dieken and Fournier, 1996; Mills et al., 1999). Selective agents were added as appropriate: 30 µg/ml blasticidin S (ICN); 2 mg/ml G418SO4 (Geneticin; Gibco-BRL); 2 mg (∼2000 U)/ml hygromycin (Calbiochem); 1 µM ganciclovir (Sigma).
For targeted replacement of chicken CENPH with CENPH–GFP the pCENPH–GFP replacement construct (Fukagawa et al., 2001) was linearized with NotI prior to electroporation. G418-resistant clones were expanded and screened for centromeric GFP signals.
Mitotic stability assays
Cells grown in the presence of selection were harvested for FISH and then passaged (1:10–1:20 split every other day) without selective agents for at least 50 generations (doubling time ∼12 h). Cells were then re-harvested, minichromosome content assessed by FISH and loss rates calculated (Ikeno et al., 1998; Mills et al., 1999; Lee et al., 2000).
DNA probes
Southern blot filters were probed with DXZ1, a 2 kb BamHI fragment from pSV2X5; RS416, a 2.1 kb SspI fragment encompassing the ARSH4, CEN6 and URA3 region of pRS416; and gamma-X, a 1.2 kb EcoRI fragment from the X γ-satellite containing cosmid CX16–2D12. In situ hybridization was carried out using DXZ1 and BAC 342O19 (Schueler et al., 2001).
Polymerase chain reaction
The following primer sequences were used. (i) Neomycin-resistance marker gene: CFNEOF3, 5′-TTTTGTCAAGACCGACCTGTCC-3′; CFNEOR3, 5′-CCACAGTCGATGAATCCAGAAAAG-3′ (Ta = 60°C, product size 489 bp). (ii) Hygromycin-resistance marker gene: CFHPHF3, 5′-GAGGTCGCCAACATCTTCTTCTG-3′; CFHPHR3, 5′-TTCTACACAGCCATCGGTCCAG-3′ (Ta = 60°C, product size 288 bp). (iii) Human X γ-satellite DNA array: CFXGAMMAF15, 5′-TTCAACGTACCCCTGAAAGCCTGG-3′; CFXGAMMAR6, 5′-CTATTTTGTCCCAAGCCTGCC-3′ (Ta = 58°C, product size 340 bp).
Immuno-FISH
Centromeric proteins were detected and fixed for subsequent FISH analysis using standard procedures (Sullivan and Warburton, 1999). After fixation in 4% formaldehyde in KCM (120 mM KCl, 20 mM NaCl, 10 mM Tris–HCl pH 7.7, 0.1% Triton X-100) and 3:1 methanol:acetic acid, slides were air dried and aged at room temperature in the dark overnight. Prior to FISH slide denaturation was for 8 min at 80°C.
Preparation of mechanically stretched chromosomes
Actively dividing fibroblast cells were treated with colcemid (0.1 µg/ml) for 90–120 min. IMR90 and BJ cell lines were harvested by mitotic shake-off and HT1080 by trypsinization. After a phosphate-buffered saline (PBS) wash, cells were counted, resuspended in pre-warmed hypotonic solution (75 mM KCl or 0.25× PBS) at 104 cells/ml, and incubated at 37°C for 10–15 min. A 500 µl aliquot of cells was centrifuged onto poly-l-lysine coated slides (BDH) in a Cytospin 3 (Shandon) at 2000 r.p.m. for 10 min. Fixation was in absolute methanol for 10 min at room temperature.
Extended chromatin
DT40 cells were washed with PBS, resuspended in pre-warmed hypotonic (0.2× PBS.A) for 15–20 min and then centrifuged onto slides and fixed as for mechanically stretched chromosomes.
Antibodies
Primary antibodies were: anti-human CENPA raised in rabbit (Valdivia et al., 1998); anti-human CENPC (rabbit 418) (Saitoh et al., 1992); anti-chicken CENPC raised in rabbit (Fukagawa et al., 1999); rabbit polyclonal ab290 to GFP (Abcam).
Digital image capturing
Chromosomes, chromatin fibres and hybridization signals were viewed through appropriate filters using a Zeiss Axioskop 2 microscope equipped with a cooled charge-coupled device (CCD) camera. Genus software was used to view composite images (Applied Imaging Corporation).
Treatment of living cells with topoisomerase inhibitors
Topo II inhibitors (etoposide, amsacrine, genistein and ellipticine; Sigma) were dissolved in 100% DMSO at 10 mM and stored in the dark at –70°C. The inhibitor was added to exponentially growing cells to the specified final concentration and incubated at 37°C for the time indicated. An equivalent volume of 100% DMSO was added as the no drug control. Cells were washed with PBS before embedding in agarose (2 × 107 cells/ml) for PFGE.
Pulsed field gel electrophoresis
Minichromosomes were resolved by PFGE in 0.7% chromosomal grade agarose (Bio-Rad 162–0136), 0.25× TBE, for 72 h at 11°C using the following parameters: a pulse time of 350–50 s (changing logarithmically), a rotor angle of 110–100° (decreasing linearly) and a voltage of 120–50 V (decreasing linearly) (Rotaphor TypeV, Biometra). Elsewhere, 1% agarose (Bio-Rad 162–0125) was used unless otherwise indicated, in 0.5× TBE, and pulsed field conditions were varied according to the DNA fragment sizes separated (see figure legends) (Bio-Rad Chef DRII). Southern transfer and probing were as previously described (Mills et al., 1999; Lee et al., 2000).
Acknowledgments
Acknowledgements
C.J.F. is a MRC Senior Research Fellow. W.C.E. is a Principal Research Fellow of The Wellcome Trust. This work was funded by the MRC and BBSRC of Great Britain.
References
- Araki K., Araki,M. and Yamamura,K.-i. (1997) Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res., 25, 868–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Yavor,A.M., Massa,H.F., Trask,B.J. and Eichler,E.E. (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res., 11, 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne R.A.L., Broccoli,D., Taggart,M.H., Thomson,E.J., Farr,C.J. and Cooke,H.J. (1994) Sandwiching of a gene within 12 kb of a functional telomere and α-satellite does not result in silencing. Hum. Mol. Genet., 3, 539–546. [DOI] [PubMed] [Google Scholar]
- Bernard P., Maure,J.F., Partridge,J.F., Genier,S., Javerzat,J.P. and Allshire,R.C. (2001) Requirement of heterochromatin for cohesion at centromeres. Science, 294, 2539–2542. [DOI] [PubMed] [Google Scholar]
- Blower M.D., Sullivan,B.A. and Karpen,G.H. (2002) Conserved organization of centromeric chromatin in flies and humans. Dev. Cell, 2, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Barnett,M.A., Burgtorf,C., Shaw,P., Buckle,V. and Brown,W.R.A. (1994) Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Hum. Mol. Genet., 3, 1227–1237. [DOI] [PubMed] [Google Scholar]
- Burden D.A. and Osheroff,N. (1998) Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta, 1400, 139–154. [DOI] [PubMed] [Google Scholar]
- Christensen M. et al. (2002) Dynamics of human topoisomerases IIα and IIβ in living cells. J. Cell Biol., 157, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieken E.S. and Fournier,R.E.K. (1996) Homologous modification of human chromosomal genes in chicken B-cell × human microcell hybrids. Methods, 9, 56–63. [DOI] [PubMed] [Google Scholar]
- Downes S., Mullinger,A.M. and Johnson,R.T. (1991) Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc. Natl Acad. Sci. USA, 88, 8895–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Ratrie,H. and Stetten,G. (1989) Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma, 98, 1–12. [DOI] [PubMed] [Google Scholar]
- Ebersole T.A., Ross,A., Clark,E., McGill,N., Schindelhauer,D., Cooke,H. and Grimes,B. (2000) Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet., 9, 1623–1631. [DOI] [PubMed] [Google Scholar]
- Fantes J.A., Green,D.K., Malloy,P. and Sumner,A.T. (1989) Flow cytometry measurements of human chromosome kinetochore labeling. Cytometry, 10, 134–142. [DOI] [PubMed] [Google Scholar]
- Farr C.J., Stevanovic,M., Thomson,E.J., Goodfellow,P.N. and Cooke,H.J. (1992) Telomere-associated chromosome fragmentation: applications in genome manipulation and analysis. Nat. Genet., 2, 275–282. [DOI] [PubMed] [Google Scholar]
- Farr C.J., Bayne,R.A.L., Kipling,D., Mills,W., Critcher,R. and Cooke,H.J. (1995) Generation of a human X-derived minichromosome using telomere-associated chromosome fragmentation. EMBO J., 14, 5444–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A.M. et al. (1997) Centromeric inactivation in a dicentric human Y;21 translocation chromosome. Chromosoma, 106, 199–206. [DOI] [PubMed] [Google Scholar]
- Floridia G., Zatterale,A., Zuffardi,O. and Tyler-Smith,C. (2000) Mapping of a human centromere onto the DNA by topoisomerase II cleavage. EMBO rep., 1, 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Pendon,C., Morris,J. and Brown,W. (1999) CENP-C is necessary but not sufficient to induce formation of a functional centromere. EMBO J., 18, 4196–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T. et al. (2001) CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J., 20, 4603–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington J.J., Van Bokkelen,G., Mays,R.W., Gustashaw,K. and Willard,H.F. (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet., 15, 345–355. [DOI] [PubMed] [Google Scholar]
- Henning K.A., Novotny,E.A., Compton,S.T., Guan,X.Y., Liu,P.P. and Ashlock,M.A. (1999) Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human α-satellite and single-copy DNA sequences. Proc. Natl Acad. Sci. USA, 96, 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins A.W., Schueler,M.G. and Willard,H.F. (1999) Chromosome engineering: generation of mono- and dicentric isochromosomes in a somatic cell hybrid system. Chromosoma, 108, 256–265. [DOI] [PubMed] [Google Scholar]
- Ikeno M., Grimes,B., Okazaki,T., Nakano,M., Saitoh,K., Hoshino,H., McGill,N.I., Cooke,H. and Masumoto,H. (1998) Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol., 16, 431–439. [DOI] [PubMed] [Google Scholar]
- Kallio M. and Lahdetie,J. (1996) Fragmentation of centromeric DNA and prevention of homologous chromosome separation in male mouse meiosis in vivo by the topoisomerase II inhibitor etoposide. Mutagenesis, 11, 435–443. [DOI] [PubMed] [Google Scholar]
- Lee C., Li,X., Jabs,E.W., Court,D.R. and Lin,C.C. (1995) Human γ X satellite DNA: an X chromosome specific centromeric DNA sequence. Chromosoma, 104, 103–112. [DOI] [PubMed] [Google Scholar]
- Lee C., Critcher,R., Zhang,J.G., Mills,W. and Farr,C.J. (2000) Distribution of γ-satellite DNA on the human X and Y chromosomes suggests that it is not required for mitotic centromere function. Chromosoma, 109, 381–389. [DOI] [PubMed] [Google Scholar]
- Lo A.W., Liao,G.C., Rocchi,M. and Choo,K.H. (1999) Extreme reduction of chromosome-specific α-satellite array is unusually common in human chromosome 21. Genome Res., 9, 895–908. [DOI] [PubMed] [Google Scholar]
- Losada A. and Hirano,T. (2001) Intermolecular DNA interactions stimulated by the cohesin complex in vitro: implications for sister chromatid cohesion. Curr. Biol., 11, 268–272. [DOI] [PubMed] [Google Scholar]
- Lupton S.D., Brunton,L.L., Kalberg,V.A. and Overall,R.W. (1991) Dominant positive and negative selection using a hygromycin phosphotransferase–thymidine kinase fusion gene. Mol. Cell. Biol., 11, 3374–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani M.M. and Willard,H.F. (1998) Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome Res., 8, 100–110. [DOI] [PubMed] [Google Scholar]
- Marzais B., Vorsanova,S.G., Roizes,G. and Yurov,Y.B. (1999) Analysis of alphoid DNA variation and kinetochore size in human chromosome 21: evidence against pathological significance of alphoid satellite DNA diminutions. Citol. Genet., 33, 25–31. [PubMed] [Google Scholar]
- Megee P.C., Mistrot,C., Guacci,V. and Koshland,D. (1999) The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell, 4, 445–450. [DOI] [PubMed] [Google Scholar]
- Mejia J.E., Alazami,A., Willmott,A., Marschall,P., Levy,E., Earnshaw,W.C. and Larin,Z. (2002) Efficiency of de novo centromere formation in human artificial chromosomes. Genomics, 79, 297–304. [DOI] [PubMed] [Google Scholar]
- Mills W., Critcher,R., Lee,C. and Farr,C.J. (1999) Generation of an ∼2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Hum. Mol. Genet., 8, 751–761. [DOI] [PubMed] [Google Scholar]
- Murray A.W. and Szostak,J.W. (1985) Chromosome segregation in mitosis and meiosis. Annu. Rev. Cell. Biol., 1, 289–317. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Peters,J.-M. and Uhlmann,F. (2000) Splitting the chromosome: cutting the ties that bind sister chromatids. Science, 288, 1379–1384. [DOI] [PubMed] [Google Scholar]
- Niimi A., Suka,N., Harata,M., Kikuchi,A. and Mizuno,S. (2001) Co-localization of chicken DNA topoisomerase IIα, but not β, with sites of DNA replication and possible involvement of a C-terminal region of α through its binding to PCNA. Chromosoma, 110, 102–114. [DOI] [PubMed] [Google Scholar]
- Oakey R. and Tyler-Smith,C. (1990) Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics, 7, 325–330. [DOI] [PubMed] [Google Scholar]
- Partridge J.F., Borgstrom,B. and Allshire,R.C. (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev., 14, 783–791. [PMC free article] [PubMed] [Google Scholar]
- Pinto I. and Winston,F. (2000) Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J., 19, 1598–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J.B., Hendzel,M.J., Furbee,C.S., Muller,M.T. and Bazett-Jones,D.P. (1996) Topoisomerase IIα is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol., 134, 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel,J.E., Cooke,C.A., Ratrie,H.R., Maurer,M., Rothfield,R.F. and Earnshaw,W.C. (1992) CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell, 70, 115–125. [DOI] [PubMed] [Google Scholar]
- Schueler M.G., Higgins,A.W., Nagaraja,R., Tentler,D., Dahl,N., Gustashaw,K. and Willard,H.F. (2000) Large-insert clone/STS contigs in Xq11-q12, spanning deletions in patients with androgen insensitivity and mental retardation. Genomics, 66, 104–109. [DOI] [PubMed] [Google Scholar]
- Schueler M.G., Higgins,A.W., Rudd,M.K., Gustashaw,K. and Willard, H.F. (2001) Genomic and genetic definition of a functional human centromere. Science, 294, 109–115. [DOI] [PubMed] [Google Scholar]
- Skibbens R.V., Corson,L.B., Koshland,D. and Hieter,P. (1999) Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev., 13, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Tsutsui,M., AuCoin,D. and Vig,B.K. (1999) Visualization of prekinetochore locus on the centromeric region of highly extended chromatin fibers: does kinetochore autoantigen CENP-C constitute a kinetochore organizing center? Chromosome Res., 7, 9–19. [DOI] [PubMed] [Google Scholar]
- Sullivan B.A. and Warburton,P.E. (1999) Studying the progression of vertebrate chromosomes through mitosis by immunofluorescence and FISH. In Bickmore,W.A. (ed.), Chromosome Structural Analysis: A Practical Approach. Oxford University Press, Oxford, UK, pp. 81–101.
- Sumner A.T. (1996) The distribution of topoisomerase II on mammalian chromosomes. Chromosome Res., 4, 5–14. [DOI] [PubMed] [Google Scholar]
- Sumner A.T. (1998) Induction of diplochromosomes in mammalian cells by inhibitors of topoisomerase II. Chromosoma, 107, 486–490. [DOI] [PubMed] [Google Scholar]
- Taagepera S., Rao,P.N., Drake,F.H. and Gorbsky,G.J. (1993) DNA topoisomerase IIα is the major chromosome protein recognized by the mitotic phosphoprotein antibody MPM-2. Proc. Natl Acad. Sci. USA, 90, 8407–8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C. et al. (1993) Localization of DNA sequences required for human centromere function through an analysis of rearranged Y chromosomes. Nat. Genet., 5, 368–375. [DOI] [PubMed] [Google Scholar]
- Udvardy A. and Schedl,P. (1991) Chromatin structure, not DNA sequence specificity, is the primary determinant of topoisomerase II sites of action in vivo. Mol. Cell. Biol., 11, 4973–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia M.M., Figueroa,J., Iglesias,C. and Ortiz,M. (1998) A novel centromere monospecific serum to a human autoepitope on the histone H3-like protein CENP-A. FEBS Lett., 422, 5–9. [DOI] [PubMed] [Google Scholar]
- Wang Z., Castano,I.B., De Las Penas,A., Adams,C. and Christman,M.F. (2000) Pol κ: a DNA polymerase required for sister chromatid cohesion. Science, 289, 774–779. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. and Earnshaw,W.C. (1997) Untangling the role of DNA topoisomerase II in mitotic chromosome structure and function. BioEssays, 19, 97–99. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. et al. (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol., 7, 901–904. [DOI] [PubMed] [Google Scholar]