Abstract
Sendai virus (SeV) leader (le) and trailer (tr) RNAs are short transcripts generated during abortive antigenome and genome synthesis, respectively. Recom binant SeV (rSeV) that express tr-like RNAs from the leader region are non-cytopathic and, moreover, prevent wild-type SeV from inducing apoptosis in mixed infections. These rSeV thus appear to have gained a function. Here we report that tr RNA binds to a cellular protein with many links to apoptosis (TIAR) via the AU-rich sequence 5′ UUUUAAAUUUU. Duplication of this AU-rich sequence alone within the le RNA confers TIAR binding on this le* RNA and a non-cytopathic phenotype to these rSeV in cell culture. Transgenic overexpression of TIAR during SeV infection promotes apoptosis and reverses the anti-apoptotic effects of le* RNA expression. More over, TIAR overexpression and SeV infection act synergistically to induce apoptosis. These short viral RNAs may act by sequestering TIAR, a multivalent RNA recognition motif (RRM) family RNA-binding protein involved in SeV-induced apoptosis. In this view, tr RNA is not simply a by-product of abortive genome synthesis, but is also an antigenome transcript that modulates the cellular antiviral response.
Keywords: apoptosis/negative strand RNA virus/RNA-binding protein/small non-coding RNA
Introduction
Successful virus replication requires not only the efficient production and spread of progeny, but also the ability to evade host defenses that limit replication by killing infected cells. In addition to inducing immune and inflammatory responses, virus infection generally triggers apoptosis, the dominant form of programmed cell death (PCD) and an important arm of the innate antiviral immune system. The apoptotic response is thought to be part of the unavoidable counter-measures that cells employ in the face of general signatures of virus infection that result from critical virus replicative functions [e.g. cytoplasmic double-stranded RNA (dsRNA)]. Virus replication thus often depends on the ability of certain viral products to block or delay apoptosis until sufficient progeny have been produced. These viral products generally are proteins that target a variety of strategic points in the apoptotic pathway (Hardwick, 1998; Roulston et al., 1999). Here we report that Sendai virus (SeV), a model paramyxovirus and respiratory pathogen of laboratory mice, encodes small promoter-proximal RNAs that modulate apoptosis, and which until now had been thought to be simply by-products of genome replication.
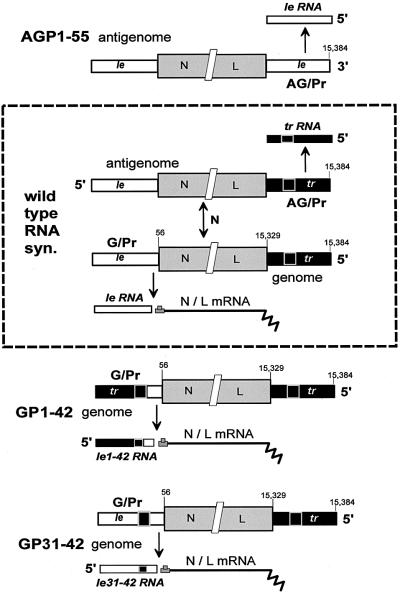
Paramyxoviruses contain non-segmented RNA genomes of negative polarity ([–]), and replicate entirely in the cytoplasm. Their genomes are 15–18 kb in length and contain 6–7 tandemly linked genes (or mRNA units) that are separated by conserved junctional sequences that act as mRNA start and poly(A)/stop sites. The first (N) mRNA of all these viruses starts precisely 56 nucleotides from the 3′ end of the genome, and the 55 nucleotides upstream are called the leader region (le, Figure 1). The last (L) mRNA ends at a similar but variable distance from the 5′ end of the genome, and the ∼50 nucleotides between the end of the L gene and the 5′ end of the genome are called the trailer region (tr, Figure 1). The genomic RNA of [–] RNA viruses functions firstly as a template for synthesis of mRNAs, and then as a template for a full-length complementary copy (the [+] antigenome), which like the genome is found only as nucleocapsids assembled with the viral N protein. Viral RNA synthesis begins at the 3′ end of the [–] genome template (the genomic promoter, G/Pr) with le RNA synthesis. During primary transcription, or when there is insufficient newly made N protein to assemble the nascent le RNA, viral RNA polymerase (vRNAP) terminates near the end of the leader region, and then starts the N mRNA. This vRNAP responds to the junctional signals and synthesizes each mRNA in turn. When sufficient newly made N protein is available to assemble the nascent le RNA before it has terminated, antigenome synthesis and assembly become coupled and this vRNAP now ignores the junctional signals and synthesizes an exact, complementary copy as a fully assembled nucleocapsid (Gubbay et al., 2001). Genome synthesis from antigenome templates is thought to take place in a similar fashion, in that AG/Pr (the antigenomic promoter at the 3′ end of the antigenome) is also always ‘on’ and trailer RNAs (tr RNAs) are made from this region independently of N subunit availability (Figure 1). However, as there are no mRNA start sites on the antigenome template, termination of tr RNAs serves only to recycle vRNAP. The leader and trailer sequences are thought to promote antigenome and genome synthesis, respectively, in two ways. When present at the 3′ ends of the templates, they direct vRNAP initiation, and the complements of these sequences within the nascent [+] le and [–] tr RNAs promote the initiation of nucleocapsid assembly on these promoter-proximal chains, a prerequisite for genome replication (Lamb and Kolakofsky, 2001).
Fig. 1. SeV RNA synthesis from wild-type and promoter-variant viruses. The dotted rectangle encloses a flow diagram of viral RNA synthesis. [–] genomes and [+] antigenomes are shown as a series of three boxes that are not drawn to scale; the leader (le) region in white (nucleotides 1–55), the train of six mRNAs in gray (abbreviated as N/L, nucleotides 56–15 329) and the trailer (tr) region in black (nucleotides 15 330–15 384). Nucleotides 31–42 of the black trailer region (or more properly nucleotides 15 342–15 353 of the genome) are highlighted with a white border. Vertical arrows indicate the flow of viral RNA synthesis. Genome replication requires a continuous supply of newly made N protein subunits, in contrast to the synthesis of le RNA, mRNAs and tr RNA. The sequence exchanges in the leader regions of SeV-GP1–42 and SeV-GP31–42, and in the trailer region of SeV-AGP1–55, and their consequences on the promoter-proximal small RNA transcribed from G/Pr and AG/Pr, are also indicated.
Although G/Pr and AG/Pr carry out similar functions, they also have distinct properties, and the exchange of these sequences has interesting consequences on virus infection. A series of recombinant SeV in which the leader region of G/Pr was replaced progressively with the analogous trailer region of AG/Pr were constructed (rSeV-GP1–24 to 1–48; Garcin et al., 1998; Figure 1). All these rSeV were viable and transcribed chimeric tr/le (instead of le) RNAs from G/Pr. Remarkably, rSeV-GP1–42 and 1–48 infections did not kill cultured cells but led to high level persistent infections, whereas rSeV-GP1–24 to 1–36 were wild-type in this respect, i.e. their infections induced PCD (Garcin et al., 1998; Itoh et al., 1998; Bitzer et al., 1999). Moreover, the non-cytopathic phenotype of rSeV-GP1–42/48 was dominant in co-infections with rSeV-wt, i.e. rSeV-GP1–42/48 appeared actively to suppress PCD normally induced by SeV-wt infection. As le or tr/le RNAs are the only known products of this region and are transcribed abundantly, the expression of tr-like RNAs from both G/Pr and AG/Pr appeared somehow to prevent PCD in response to the infection. Since rSeV-GP1–42/48 are ‘gain-of-function’ mutants, this led us to examine whether tr RNA bound cellular proteins that did not interact with le RNA.
Results
tr RNA binds a 40 kDa host protein via the AU-rich sequence 31UUUUAAAUUUU41
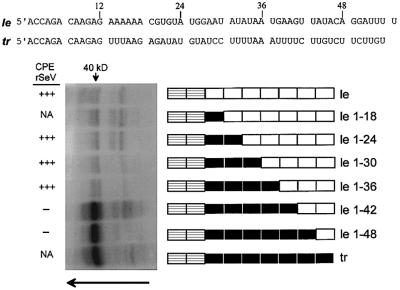
To determine whether host cell proteins interacted with le or tr RNAs, these 55 nucleotide RNAs were transcribed in vitro in the presence of 5-Br-UTP and [α-32P]UTP (Materials and methods). The le or tr RNAs were isolated and added to cytoplasmic extracts of human or murine cells, and irradiated with 312 nm light. Under these conditions, the input 5-Br-UMP containing RNA is excited specifically. The reactions were then treated with RNase A, and the proteins separated by SDS–PAGE. In all cases (human HeLa, A549 and 293 cells, and murine BF cells), tr RNA strongly identified a 40 kDa host protein in a UV-dependent manner, whereas only weakly labeled bands were identified by le RNA (that of HeLa cells is shown in Figure 2). Although only a single 40 kDa band is visible in Figure 2, this band resolves as a doublet on higher resolution (see Figure 5). As le RNA is so non-reactive in this assay as compared with tr RNA, we examined chimeric tr/le RNAs for protein cross-linking in an effort to map the 40 kDa protein-binding sequence. A series of plasmids were constructed, which like rSeV-GP1–24 to 1–48 transcribe chimeric tr/le RNAs (le GP1–18 to 1–48, Figure 2), and these chimeric RNAs were used for UV cross-linking. Only chimeric RNAs that contain the first 42 or 48 nucleotides of the trailer sequence cross-linked to the 40 kDa host protein (Figure 2). As rSeV-GP1–42 and 1–48 infections are non-cytopathic and lead to persistent infections, this phenotype thus correlates with the ability of their le* RNAs to bind the 40 kDa host protein (left side, Figure 2).
Fig. 2. UV cross-linking of wild-type and mutant promoter-proximal RNAs to a 40 kDa host protein. The le and tr RNA sequences are shown above in blocks of six nucleotides, which are shown schematically as boxes below. The first two boxes are always striped, as these two hexamers are conserved between le and tr RNA. The subsequent sequences are poorly conserved, and are shown as white boxes for le RNA, and black boxes for tr RNA. The le and tr RNAs and the various chimeric tr/le RNAs (le 1–18 to 1–48) are shown schematically next to the cross-linking gel. The various promoter-proximal RNAs were mixed with HeLa cell cytoplasmic extracts and irradiated with 312 nm light for 10 min [or kept in the dark as a control (not shown), Materials and methods]. The reactions were then digested with RNase A, and separated on 12.5% protein gels to determine the relative abilities of these small RNAs to cross-link to host proteins. The cytopathic nature (CPE) of rSeV expressing these chimeric tr/le sequences in the leader region of G/Pr (Garcin et al., 1998) is indicated on the left; NA, not applicable as these rSeV were not prepared.
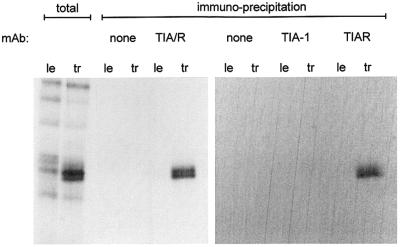
Fig. 5. TIAR-specific mAb immunoprecipitates the 40 kDa host protein. le or tr RNA cross-linked HeLa cell extracts, intentionally over-reacted to also contain non-specific bands (total), were immunoprecipitated with mAb specific for both TIA-1 and TIAR (TIA/R) or mAb specific for each protein, as indicated above. The precipitates and the starting materials (total) were separated by 12.5% SDS–PAGE, and the gel exposed to film.
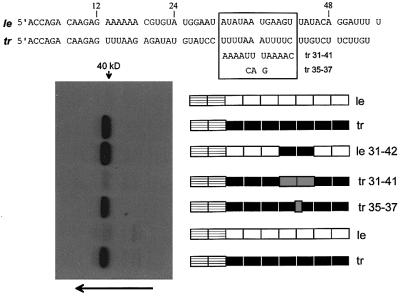
Between the first 36 and 42 nucleotides of the trailer sequence lies the AU-rich sequence 31UUUUAAAU UUU41 that stands out due to its symmetry (Figure 3). As AU-rich sequences are common sites for RNA-binding proteins (Brennan and Steitz, 2001), we examined the effect of exchanging this sequence of tr RNA with its complement (5′ 31AAAAUUUAAAA41, tr 31–41 RNA). This sequence exchange in tr RNA strongly reduced 40 kDa protein binding (Figure 3, lane tr 31–41). To examine whether the central 35AAA37 of the AU-rich sequence was essential, it was mutated to 35CAG37 (tr 35–37, Figure 3). This latter exchange, in contrast, had only a minor effect on 40 kDa protein cross-linking (lane tr 35–37). We next examined whether replacing just the AU-rich sequence 31AUAUAAUGAAG41U of le RNA with the tr sequence 31UUUUAAAUUUU41C (le 31–42) could confer on le 31–42 RNA the ability to cross-link to the 40 kDa protein (we also changed nucleotide 42 from U to C (underlined) because nucleotide 43 is also U). As shown in Figure 3, this exchange of AU-rich sequences in the leader region was indeed sufficient to confer on le 31–42 RNA the ability to cross-link to the 40 kDa protein at wild-type levels (lane le 31–42 versus lane tr). The AU-rich sequence 5′ 31UUUUAAAUUUU41, within the context of the otherwise AU-rich SeV le and tr RNAs, is thus necessary and sufficient for 40 kDa host protein binding.
Fig. 3. The AU-rich sequence 5′ UUUUAAAUUUU mediates promoter-proximal RNA cross-linking to the 40 kDa protein. The tr and le RNA sequences are shown above, and the fourth and fifth hexamers (nucleotides 31–42) are boxed. The various promoter- proximal RNAs examined are shown schematically as for Figure 2, next to the protein gel used to determine their relative abilities to UV cross-link to the host 40 kDa protein. The alternative sequence exchanges (tr 31–41 and tr 35–37) are shown as gray boxes, and their sequences are shown as well in the box above.
rSeV-GP31–42
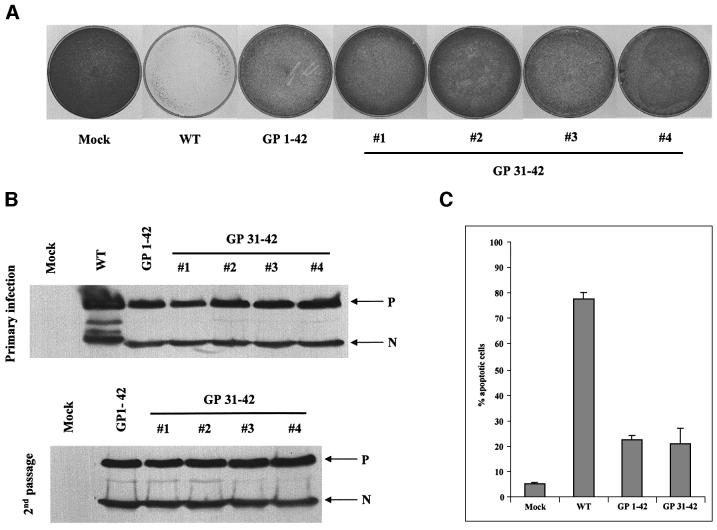
Since rSeV-GP1–42 is non-cytopathic, we were interested in whether rSeV that expressed le 31–42 RNAs from G/Pr (rather than le 1–42 RNAs) would reproduce this phenotype. rSeV-GP31–42 was therefore prepared (Materials and methods; Figure 1) and it was found to grow as well as SeV-wt in eggs and cell culture (see also Figure 7). Four independent isolates of rSeV-GP31–42 showed only transient cytopathic effects in infections of HeLa cells, like rSeV-GP1–42, in marked contrast to rSeV-wt. Moreover, these infected cells could be passaged and grew to confluency almost as rapidly as uninfected HeLa cells (Figure 4A), yet maintained the same intracellular virus load as the primary infection, again, like rSeV-GP1–42 (Figure 4B, Garcin et al., 1998). SeV-GP1/31–42 infections at a 10-fold higher m.o.i. (200 infectious units/cell) still led to persistent infections that could be passaged (data not shown), so the lack of cell death in these infections is not due to less viral replication. We also compared the relative abilities of rSeV-GP31–42 and rSeV-GP1–42 to induce apoptosis using annexin V staining. As shown in Figure 4C, rSeV-GP31–42 was as poorly apoptotic as rSeV-GP1–42 (only 20% of the cells had elevated annexin staining), under conditions where 80% of the SeV-wt-infected cells had elevated annexin staining). Thus, rSeV-GP31–42, like rSeV-GP1–42, is a relatively non-cytopathic virus, and the transfer of the trailer AU-rich sequence by itself to the leader region is sufficient to confer this phenotype to rSeV.
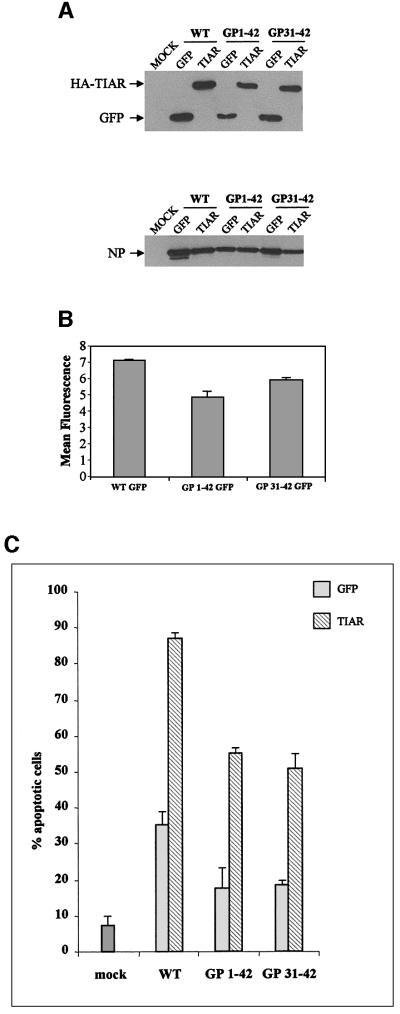
Fig. 7. Transgenic expression of TIAR during SeV infection. Parallel HeLa cell cultures were infected with 10 infectious units of each of the six rSeV indicated (see text), or mock infected, and harvested at 48 h.p.i. Half of the cells were used to determine the intracellular levels of the transgenes and the SeV N protein by immunoblotting with anti-HA, anti-GFP and anti-N (A). The other half was used to determine the level of annexin V staining by FACS (C). For the three rSeV that express GFP, the level of intracellular replication was determined more accurately by following GFP fluorescence directly by FACS (B). The error bars in (C) indicate the results of infecting duplicate cultures.
Fig. 4. rSeV-GP31–42 is non-cytopathic in cell culture, similarly to rSeV-GP1–42. HeLa cells were infected with 20 p.f.u. (or equivalent)/cell of rSeV-wt, rSeV-GP1–42 or rSeV-GP31–42, or mock infected. The cultures were trypsinized at 48 h.p.i. and equal samples of the harvested cells were used to estimate the levels of N and P proteins present intracellularly by western blotting [primary infection, (B)]. Other samples were used to determine the level of annexin V staining by FACS (C). The remaining cells (one-fifth of the total) were replated in Petri dishes and grown for 3 days, and the cultures were stained with methylene blue (A). The second passage cultures were also used to estimate the levels of N and P proteins present intracellularly by western blotting (second passage, B).
The host 40 kDa protein is TIAR
Using UV cross-linking as an assay, the 40 kDa protein could be purified extensively by chromatography on poly(U) Sepharose, but there were no clear candidates at this stage. At least 14 proteins in cell extracts that selectively bind AU-rich sequences found in the 3′-untranslated region (UTR) of certain mRNAs have been identified by UV cross-linking and band-shift assays (reviewed in Brennan and Steitz, 2001). These candidates were also being examined during our 40 kDa protein purification as their antibodies became available. This latter approach proved to be more successful.
TIA-1 (T cell-activated intracellular antigen) and TIAR (TIA-1-related, 80% amino acid identity) are ∼40 kDa proteins containing three RNA recognition motifs (RRMs) and a glutamine-rich (and prion-like) C-terminal domain. Close homologs of these factors exist in Drosophila and Caenorhabditis elegans (Anderson, 1995; Anderson and Kedersha, 2002). TIA-1 and TIAR bind preferentially to short stretches of uridylates as determined by SELEX (Dember et al., 1996). Most intriguingly, TIA-1 induces DNA fragmentation characteristic of apoptosis in permeabilized thymocytes (Tian et al., 1991). A monoclonal antibody that specifically recognizes both TIA-1 and TIAR proteins (mAb 3E6; Taupin et al., 1995), or an mAb specific for each protein [mAb 6E3 (TIAR) and ML-29 (TIA-1)], were used to immunoprecipitate le or tr RNA cross-linked extracts (over-reacted intentionally) (Figure 5). The tr RNA cross-linked 40 kDa doublet band was recognized by mAbs specific for TIA/R or TIAR, but not by an mAb specific for TIA-1. The 40 kDa host protein that specifically interacts with tr RNA is thus TIAR. Alternative splicing of TIAR mRNA in human cells yields 40 and 42 kDa proteins, thus accounting for the doublet band in HeLa cells (Figure 5; and Taupin et al., 1995).
SeV infection and stress granule formation
TIA-1 and TIAR normally are concentrated in the nucleus, but they are also present in the cytoplasm and they shuttle continuously between these two compartments (Taupin et al., 1995; Kedersha et al., 2000). In response to environmental stress such as heat shock or arsenite poisoning, TIA/R co-localize with untranslated mRNA at discrete cytoplasmic foci or stress granules (SGs) (Kedersha et al., 1999). Despite their relatively stable appearance by immunofluorescence, SGs are highly dynamic structures whose constituent proteins and mRNAs are in constant and extremely rapid flux. Due to their dynamic nature, drugs such as cycloheximide that trap mRNAs in polysomes quickly dissolve pre-formed SGs (Kedersha et al., 2000).
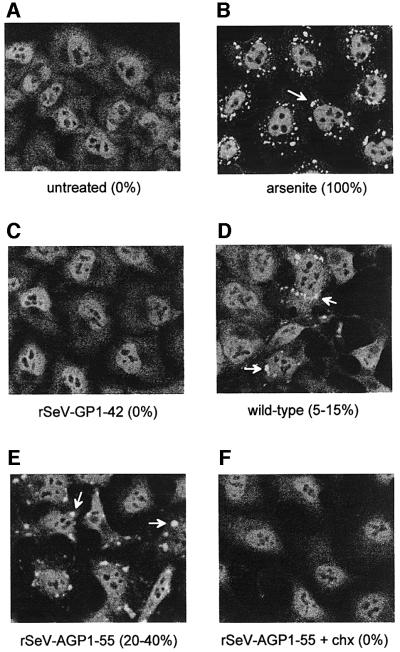
Virus infection is a gradual form of stress, and we examined whether various SeV infections would induce SG formation. For reference, we used harsh but sublethal arsenite poisoning of HeLa cells (500 µM As2O3 for 30 min), which induces (transient) SG formation in almost all cells within 30 min (Figure 6B; Kedersha et al., 1999). During this response, the diffuse and granular cytoplasmic TIA/R staining of the control cells (Figure 6A) largely disappears and is replaced by bright SGs (Figure 6B). SeV-wt infection did indeed induce SG formation in a small but reproducible fraction of these cells by 18 h post-infection (h.p.i.) when viral gene expression is maximal (5–15% in three experiments, Figure 6D). No SGs were visible until 12 h.p.i., and these structures were examined at 18 h.p.i. as the cytopathic effects of SeV-wt infection are too severe beyond this time. Although the diffuse, granular cytoplasmic TIA/R staining does not disappear on SeV infection, this may be due to the degree of stress applied. A concentration of 50 µM As2O3 for 30 min produces immunofluorescent patterns identical to those of SeV-wt infection, i.e. ∼20% of the cells contain SGs in which the granular cytoplasmic staining remains basically unchanged (not shown, compare with panels D and E). Infection with SeV-GP1–42, in contrast, did not induce SG formation in any cells at any time (Figure 6C), even though this virus replicated well in HeLa cells (Figure 4B). The total absence of SGs in infections that express only tr or tr-like RNAs led us to also examine SeV-AGP1–55, whose trailer region of AG/Pr was replaced with the leader sequence (the converse of SeV-GP1–42, see Figure 1). SeV-AGP1–55, which express le RNAs from both promoters, induced SGs in 20–40% of the cells (Figure 6E), even though SeV-AGP1–55 replicates intracellularly similarly to SeV-GP1–42. We also treated the SeV-wt- and SeV-AGP1–55-infected cells at 18 h.p.i. with cycloheximide for 1 h. In both cases, the SGs disassembled completely (rSeV-AGP1–55 is shown in Figure 6F), similarly to SGs in 500 µM arsenite-treated cells (Kedersha et al., 2000). SeV-induced SGs thus also appear to be sites of mRNA accumulation, similarly to SGs in arsenite-treated cells.
Fig. 6. TIA/R immunofluoresence of arsenite-treated and SeV-infected HeLa cells. Replicate cultures of HeLa cells were infected with 20 p.f.u. (or equivalent)/cell of the various SeV indicated and examined for TIA/R immunofluorescence by confocal microscopy at 18 h.p.i. (Materials and methods). Some rSeV-AGP1–55-infected cultures were treated with 10 µg/ml of cycloheximide at 18 h.p.i., and fixed for examination at 19 h.p.i. (bottom right). At least 200 cells from each culture in three independent experiments were examined, and the percentages containing SGs (white arrows) are shown in parentheses. Uninfected cells were treated with 500 µM As2O3 for 30 min and then examined, for reference.
In summary, there is an inverse correlation between the ability of various SeV infections to induce SG formation and the extent to which they express tr or tr-like promoter-proximal RNAs that can bind TIAR. These results are consistent with the notion that some basic aspect of the SeV infection induces SG formation (e.g. cytoplasmic RNA synthesis or dsRNA), and that the 5′ UUUUAAA UUUU-containing/TIAR-binding promoter-proximal RNAs act by sequestering TIAR, thus preventing SG formation.
A role for TIAR in SeV-induced apoptosis
SeV that express hemagglutinin (HA)-tagged TIAR as a supplemental transgene in the wild-type, GP1–42 and GP31–42 promoter backgrounds were prepared, to examine whether overexpression of TIAR would affect SeV cytopathogenicity. Using SeV itself as a transducing vector allows us to examine TIAR overexpression in a homogeneous population of cells, as well as in the natural background of SeV infection. Our working model is that TIAR is somehow required for SeV-induced PCD and that le* and tr RNAs buffer TIAR action by binding to it. Overexpressing TIAR during SeV infection should thus increase the cytopathogenicity of SeV-wt. It should also reverse the non-cytopathic nature of SeV-GP1/31–42 infections, by out-titrating the le/tr RNA that binds TIAR. SeV that carry a green fluorescent protein (GFP) transgene were prepared as a neutral control.
These six rSeV (expressing GFP or TIAR, in the wild-type, GP1–42 and GP31–42 promoter backgrounds) were recovered from DNA, and virus stocks were prepared in embryonated chicken eggs. SeV–GFP grew in eggs to levels similar to SeV-wt, whereas SeV-TIAR grew to ∼3-fold lower levels, independently of promoter background. Overexpression of TIAR during replication in eggs thus has a modest negative effect on virus growth. This relative disadvantage was not pursued further. HeLa cells were infected with 10 infectious units/cell of each of the six SeV, and virus replication was followed by immunoblotting of the accumulated viral N protein and the transgene product (Figure 7A). For the three rSeV expressing GFP, the relative levels of intracellular virus replication were determined more accurately by fluorescence-activated cell sorting (FACS) analysis (Figure 7B). Parallel infections of SeV-GP31–42/GFP and SeV-GP1–42/GFP accumulated almost as much GFP as SeV-wt/GFP infections, consistent with the levels of N protein present in their cell extracts (Figure 7A, lower panel). These N levels were similar to those found in SeV-wt infections (not shown); thus, all these transgenic SeV replicate well in HeLa cells.
The kinetics of the cytopathic effects and the fraction of cells judged apoptotic by annexin V staining during SeV infection depend on the amount of serum (growth factors) present, which protects cells against apoptosis. In the absence of serum (Figure 4C), ∼80% of the cells are apoptotic due to the infection, whereas in 10% serum apoptotic cells were rarely >2-fold above the mock-infected control. In 2% serum, SeV-wt/GFP infection induced apoptosis in 35% of the cells, versus 8% in the mock-infected control (Figure 7C). Transgenic expression of TIAR in otherwise wild-type SeV was clearly pro-apoptotic (relative to GFP). SeV-GP31–42/GFP and SeV-GP1–42/GFP infections were less apoptotic than SeV-wt/GFP infections, as expected. TIAR expression in these promoter backgrounds was again pro-apoptotic, and more than overcame the anti-apoptotic effects of le* RNA expression. Moreover, in contrast to SeV-GP1/31–42/GFP infections that can be subcultured and maintain persistent high level infections, SeV-GP1/31–42 viruses expressing TIAR led to cell death and these cultures could not be continued (data not shown). Thus, transgenic expression of TIAR during SeV infection of HeLa cells promotes apoptosis, and can more than overcome the anti-apoptotic effects of le* RNA expression.
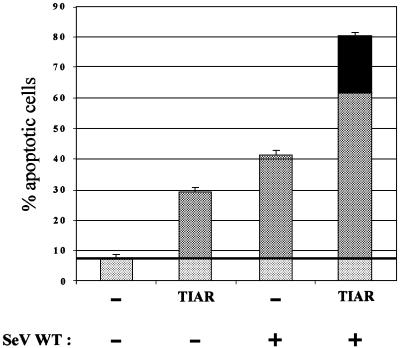
To examine further whether SeV-induced apoptosis operates via TIAR, we determined whether infection and TIAR overexpression acted synergistically in inducing apoptosis. Parallel cultures were transfected with Epstein–Barr (EBV)-based plasmids (pEBS; Bontron et al., 1997) expressing TIAR, or an empty plasmid control. The cells were also co-transfected with pEBS– GFP to control for transfection efficiency, which was 80–90% in all cases. The cells were then infected (or not) 24 h later with SeV-wt for a further 48 h (in 2% serum). TIAR overexpression by itself induced apoptosis in 21% of the cells (over a background of 7.5% for the empty plasmid-transfected control), and SeV infection alone yielded a further 33% apoptotic cells (Figure 8). However, TIAR overexpression coupled with SeV infection caused apoptosis in a greater fraction of the cells than did the simple sum of the two (indicated by the black region of the bar graph). The synergy between TIAR overexpression and infection in inducing apoptosis is consistent with the notion that SeV induces apoptosis via TIAR.
Fig. 8. Synergy of TIAR overexpression and SeV infection in inducing apoptosis. Duplicate cultures of HeLa cells were transfected with 1 µg of pEBS plasmids expressing TIAR or empty pEBS (as indicated), along with 0.2 µg of pEBS–GFP to follow transfection efficiency, with fuGENE (Roche). The cultures were then infected 24 h later with 10 p.f.u./cell of SeV-wt, or mock infected. All cultures were harvested at 48 h.p.i., and the percentage of apoptotic cells was determined by annexin V staining and FACS. The horizontal line indicates the background level of apoptosis, i.e. independent of TIAR expression and SeV infection, which is subtracted from the other bars to determine the level of synergy. The synergy between TIAR overexpression and SeV infection is indicated by the black segment at the top of the bar.
Discussion
UV cross-linking studies have identified the cellular protein TIAR specifically associated with tr RNA. tr RNA binds TIAR via the symmetrical AU-rich sequence 5′ 31UUUUAAAUUUU41. The duplication of this site within the leader region of G/Pr, such that both le* and tr RNAs bind TIAR, yields rSeV that are non-cytopathic and these infections persist whereas SeV-wt infection leads to PCD. We have also found that transgenic overexpression of TIAR during SeV infection promotes apoptosis, and can more than overcome the anti-apoptotic effects of le* RNA expression. Moreover, SeV infection and TIAR overexpression act synergistically in inducing apoptosis, consistent with the notion that SeV induces apoptosis via TIAR.
Paramyxovirus genome promoters G/Pr and AG/Pr are always ‘on’ during intracellular replication, and le and tr RNAs are thus transcribed abundantly, but with different kinetics (Figure 1). Intracellular replication begins with le RNA synthesis, whereas tr RNA synthesis requires the accumulation of viral antigenones that begins 8 h.p.i. Our working model is that SeV-GP1/31–42 infections are non-cytopathic partly because their le* RNAs can now bind TIAR, i.e. that additional TIAR is being sequestered during infection. More importantly, SeV-GP1/31–42 infections are non-cytopathic because TIAR-binding le* RNA is expressed precociously, i.e. during primary transcription (Figure 1). Consistent with this, the dominance of SeV-GP1–42 in co-infections with SeV-wt, i.e. its ability to prevent cell death due to SeV-wt replication, depends on SeV-GP1–42 being present from the start of the co-infection. A delay of only 75 min completely abrogates the anti-cytopathic effect, as if the cellular decision to trigger PCD in response to the infection is taken very early (Garcin et al., 1998).
Despite our best attempts, we have been unable to provide direct support for the notion that tr RNA expressed after the onset of genome replication also acts in an anti-apoptotic fashion. These failed attempts may be due to the complexity of the tr region. SeV in which only the tr RNA TIAR-binding sequence 5′ 31UUUUAAAUUUU has been changed (to 31AAAAUUUAAAA so that its tr RNA no longer binds TIAR, Figure 3) have been prepared. However, these viruses are highly debilitated and replicate too poorly relative to SeV-wt for comparison. Why these minimal changes in the tr region are so deleterious to SeV replication is intriguing, and contrasts sharply with similar changes in the le regions that are well tolerated [SeV-GP31–42/GFP replicates almost as well as SeV-wt/GFP (Figure 7)]. SeV-AGP1–55, in which the entire tr region has been exchanged, on the other hand, grows similarly to GP1–42. This virus is not more apoptotic than SeV-wt, but it also has several unexpected properties that can themselves influence PCD. For example, SeV-AGP1–55 infection leads to abnormal genome to antigenome ratios (le Mercier et al., 2002), as well as to increased levels of interferon β mRNA, in contrast to GP1–42 that is wild-type in these respects (data not shown). The tr region of AG/Pr obviously is complex and presumably contains sites for other RNA-binding proteins that are not detected by UV cross-linking. Our inability to demonstrate a loss-of-function phenotype of viruses expressing tr RNAs that do not bind TIAR may be confounded by this complexity. However, there is little doubt that the acquisition of TIAR binding by le* RNA is anti-apoptotic. Moreover, tr RNA–TIAR interactions late in infection do suppress SG formation due to the infection (Figure 6). It remains possible that these interactions also affect the apoptotic response of the cell, even though SG formation and apoptosis do not appear to be tightly linked. Successful virus replication often depends on the ability of viral products to block or delay apoptosis (Roulston et al., 1999).
TIA-1 and TIAR (TIA/R) share a domain structure of three RRMs and a C-terminal glutamine-rich protein interaction domain, and their functions are thought to be diverse and mostly redundant (Forch et al., 2000; Piecyk et al., 2000). Proteins with combinations of similar motifs are found in a variety of factors involved in RNA expression and metabolism, including ELAV, a regulator of alternative splicing in Drosophila, and HelN1, a regulator of mRNA stability in humans (Keene, 2001). Besides inducing DNA fragmentation in digitonin- permeabilized thymocytes (Kawakami et al., 1992), TIA/R are known to act in two ways. In the nucleus where it is normally concentrated, TIA/R acts as an alternative splicing factor (del Gatto et al., 2000; Forch et al., 2000). Alternative splicing is a common mechanism to generate proteins with antagonistic activities in apoptosis, affecting receptors, caspases and bcl-type modulators (Jiang and Wu, 1999), and TIA/R overexpression alters the ratio of mRNAs encoding fas receptor and soluble receptor in favor of apoptosis (Forch et al., 2000). TIA/R also shuttles between the nucleus and the cytoplasm (Taupin et al., 1995). Cytoplasmic TIA/R binds to an AU-rich element (ARE) in the 3′-UTR of tumor necrosis factor-α (TNF-α) mRNA composed of AUUUA repeats (Gueydan et al., 1999). TNF-α is a key pro-inflammatory cytokine (Vassalli, 1992), and a variety of proteins bind to this ARE and their interplay determines the stability and trans latability of TNF-α transcripts. There is evidence that the presence of TIA/R complexes bound to the ARE translationally silence this mRNA (Piecyk et al., 2000). More generally, TIA is thought to target the 5′ ends of mRNAs and promote the assembly of eIF2-deficient and translationally inactive mRNP complexes (Kedersha et al., 2002). It has been suggested that the pro-apoptotic activity of TIA/R is a consequence of translational silencing beyond a threshold time. Finally, TIAR translocates from the nucleus to the cytoplasm during Fas-mediated apoptosis (Taupin et al., 1995), and fas signaling activates FAST (fas-activated ser/thr kinase) which specifically associates with TIA-1 (Tian et al., 1995). Given this plethora of associations with apoptosis, the possibilities of how the proposed decrease in TIAR availability during SeV-GP1/31–42 infection might prevent PCD are rich, and remain to be explored.
Viral products that target strategic points in the apoptotic pathway generally are proteins, but some viruses also use small RNAs for this purpose. Two DNA viruses encode small RNAs that interfere with PCD [adenovirus VA RNAs (Mathews and Shenk, 1991) and EBV (γ-herpesvirus)-encoded small RNAs (EBERs; Sharp et al., 1993)]. VA RNAs and EBERs are highly transcribed by cellular RNA polymerase III, and thus, like SeV le and tr RNAs, contain neither 5′ caps nor 3′ poly(A) tails. They presumably are not associated with cellular cap- and poly(A)-binding proteins, and the various other factors they recruit to regulate mRNA metabolism. VA RNAs and EBERs are partially double-stranded and bind with high affinity to dsRNA-dependent protein kinase (PKR) and prevent its activation (Hovanessian, 1989). Since overexpression of dominant-negative mutants of PKR can protect cells from PCD induced by a wide variety of apoptogenic stimuli (Srivastava et al., 1998), the inhibition of PKR activation is one way in which these small RNAs can interfere with PCD. However, none of our SeV infections shut off host protein synthesis, the hallmark of PKR activation (data not shown).
Another example of small viral RNAs that appear to modulate apoptosis is that of γ-herpesvirus saimiri, which is associated with aggressive T-cell lymphomas in primates. Herpesvirus saimiri encodes seven U-rich snRNAs of the Sm-class called HSURs (herpesvirus saimiri U-rich RNAs) that are expressed abundantly in transformed T cells (Myer et al., 1992). Like cellular snRNAs, HSURs are transcribed by RNA polymerase II but contain tri-methylated 5′ G caps (Lee et al., 1988). Three HSURs have AU-rich 5′ ends containing the mRNA-destabilizing AUUUA sequence motif. HSUR1 interacts with HuR and hnRNP D (two ARE-binding proteins that affect mRNA stability, both RRM family RNA-binding proteins) by in vivo cross-linking, and HSUR1 is degraded in an AUUUA-specific pathway. It has been suggested that HSURs act in a sacrificial manner to improve the stability of cellular AUUUA-containing mRNAs (some involved in PCD) by titrating the AUUUA-specific interacting proteins such as HuR and hnRNP D (Fan et al., 1997). Our results suggest that SeV tr RNA may act in a similar manner to HSURs, by sequestering TIAR, a multivalent RRM family RNA-binding protein that is important in SeV-induced apoptosis.
Non-coding RNAs are known to have roles in a great variety of processes, including transcriptional regulation, chromosome replication, RNA processing and modification, and mRNA stability and translation (Storz, 2002). Small, promoter-proximal paramyxovirus RNAs that are crucial elements in controlling viral genome replication can now be added to this repertoire. These viral RNAs, probably including wild-type le RNA, may well also bind other cellular RNA-binding proteins that remain to be identified by other methods.
Materials and methods
Virus levels and virus infection
SeV was titered by plaque formation on LLC-MK2 cells covered with 0.3% agarose containing 1.2 µg/ml acetyl-trypsin, for 3–4 days at 33°C. For non-cytopathic viruses such as SeV-GP1–42 or GP31–42, an amount of virus particles equivalent to that present in the titered SeV-wt stock was used, as determined by SDS–PAGE and Coomassie Brilliant Blue staining of virus pelleted from allantoic fluid stocks. HeLa cells at 60% confluence in 3.5 cm Petri dishes were infected with various virus stocks for 1 h in 1 ml of Dulbecco’s modified Eagle’s medium (DMEM) without fetal calf serum (FCS), followed by incubation (all at 33°C) in medium containing either 0 or 2% FCS. The mock-infected control otherwise was treated identically.
Preparation of HeLa cytoplasmic extracts
HeLa cytoplasmic extracts were prepared as previously described (Garcin et al., 1998). Briefly, confluent HeLa monolayers in 9 cm Petri dishes (6 × 106 cells) were washed in buffer 1: 150 mM sucrose, 30 mM HEPES pH 7.4, 33 mM NH4Cl, 7 mM KCl, 4.5 mM magnesium acetate. A 1 ml aliquot of lysolecithin (250 µg/ml) in buffer 1 was then added for 1 min, and immediately removed. The cells were then scraped in 300 µl of cold buffer 2: 20 mM HEPES pH 7.4, 7 mM KCl, 50 mM NH4Cl, 4.5 mM magnesium acetate, 1 mM spermidine, 1 mM dithiothreitol (DTT), 10% glycerol, 0.5 mM aminoethylbenzene sulfonyl fluoride (AEBSF). After pipetting up and down 30 times, the extracts were centrifuged for 10 min at 5000 r.p.m. (4°C). The supernatant was recovered and stored at –70°C.
UV cross-linking and immunoprecipitation
Purified [α-32P]UTP and 5-Br-UTP containing promoter-proximal RNAs (100 000 c.p.m. Cerenkov) were added to 30 µl of HeLa cytoplasmic extract and incubated for 15 min at 20°C, and irradiated with 312 nm light (750 µW/cm2) for 10 min. RNA was digested with RNase A (1 mg/ml) for 30 min at 37°C, and the samples were then electrophoresed on a 12.5% SDS–polyacrylamide gel. The gel was exposed to a BIOMAX MS film at –70°C.
For immunoprecipitation, 220 µl of RIPA buffer (150 mM NaCl, 0.5% NP-40, 0.01% DOC, 50 mM Tris pH 7.4) was added to the samples after RNase A digestion. A 2 µl aliquot of either mAb 3E6 (recognizing both TIA-1 and TIAR), mAb 6E3 (TIAR) or mAb ML-29 (TIA-1), or 2 µl of buffer was added, and binding reactions were carried out at 4°C on a rotation wheel for 15 h. A 40 µl aliquot of protein A–Sepharose beads (50:50 slurry) was added to the samples, and incubated for 90 min at 4°C. After four washes in 500 µl of RIPA, the pelleted beads were resuspended in 15 µl of protein sample buffer, boiled for 5 min, and the proteins separated by 12.5% SDS–PAGE.
Preparation of rSeV-GP31–42 and rSeV-AGP1–55
pFL4, a wild-type infectious clone, was digested with SphI and KpnI and this fragment encoding the beginning of the N gene to the end of L gene was removed and replaced with a linker, to generate pFL4-del. All sequence exchanges and mutations described in the text were carried out by site-directed mutagenesis on pFL4-del, and infectious cDNA was regenerated by re-inserting the SphI–KpnI fragment. Virus recovery was carried out as described previously (Schnell et al., 1994; Garcin et al., 1995), except that BSR T7 cells (expressing cytoplasmic T7 RNA polymerase; Bucholtz et al., 1999) were transfected with the full-length cDNA and IRES-containing pTM1 plasmids encoding the N, P and L proteins for 48 h at 37°C. Acetyl-trypsin (1.2 µg/ml) was then added to the cells for 24 h (this activates the fusion protein and appears to act as an amplification step). Virus was then innoculated into 8-day-old embryonated chicken eggs, and allantoic fluid was recovered after 3 days at 33°C. After a second passage in eggs, the allantoic fluid stock was titered and stored at –70°C.
Preparation of transgenic rSeV
pFL5, a wild-type infectious clone containing an MluI cloning site at the M-F junction, was used to prepare rSeV-wt, rSeV-GP31–42 and rSeV-GP1–42 carrying supplemental transgenes. Genes expressing either GFP, or full-length TIAR, were inserted in the three promoter backgrounds, and rSeV were recovered as above. The GFP-expressing SeV were titered both by counting infectious centers and by Coomassie Blue staining after SDS–PAGE. We found that a certain amount of viral proteins contain the same amount of infectious particles for these viruses. All virus stocks were quantitated by Coomassie Blue staining and normalized to SeV-wt or SeV–GFP.
In vitro transcription of promoter-proximal RNAs
The plasmid (pTM1) vectors used for le or tr RNA expression contained a T7 RNAP promoter upstream of the le or tr sequence (with an additional 5′ GGG to ensure efficient use of the T7 promoter), followed by the HDV antigenome (84 nucleotides) ribozyme that forms the 3′ cyclic phosphate end of the le/tr RNA (Perrota and Been, 1991). The various plasmids for chimeric RNAs used in this study were constructed from the two plasmids described above. Linearized plasmids were transcribed in vitro in the presence of 250 µM ATP, CTP and GTP, and 50 µM 5-Br-UTP plus 50 µCi of [α-32P]UTP for 90 min at 37°C, and the products were separated on a 10% sequencing gel. In all cases, roughly half of the T7 transcripts were self-cleaved, generating free le or tr RNAs and ribozyme. The promoter-proximal RNAs were eluted from the gel and recovered by ethanol precipitation.
Measurement of phosphatidylserine exposure by annexin V fluorescence
Medium from 3.5 cm Petri dishes containing adherent HeLa cells was removed at 72 h.p.i. The cells were rinsed with 0.5 ml of 0.05% trypsin, and 0.5 ml of 0.05% trypsin was added for 5 min. Once the cells had lifted off, 3 ml of DMEM + 5% FCS was added, and the cells were recovered by centrifugation at 500 g for 5 min. The cells were resuspended gently in 1 ml of cold phosphate-buffered saline (PBS), and 0.5 ml of these cells were recentrifuged as above. The cell pellet was resuspended in 100 µl of annexin V binding buffer (10 mM HEPES/NaOH pH 7.4, 140 mM NaCl, 5 mM CaCl2) containing 2 µl of annexin V-Alexa™568 reagent (Roche) and 2 µl of 50 µg/ml BOBO™-1 (B-3582) (Boerhinger). After 15 min, the cells were analyzed by FACS.
Immunofluorescence
HeLa cells were plated onto 11 mm glass coverslips in 24-well plates and grown for 24 h. Cells were then infected with various SeV at an m.o.i. of 20 for 18 h. For reference, stress was also induced by treating the cells with 0.5 mM As2O3 for 30 min. The cells were rinsed briefly in PBS and incubated for 10 min in 2% paraformaldehyde in PBS. The cells were then immersed in methanol at –20°C for 10 min, rinsed in PBS and incubated in blocking buffer [5% bovine serum albumin (BSA) in PBS] for 1 h before the addition of the primary antibodies. mAb 3E6 (recognizing both TIA-1 and TIAR) was added to the fixed cells and incubated for 1 h. After three washes in PBS, cells were incubated for 1 h with the isotype-specific secondary antibody [fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG]. Cells were washed three times in PBS, and viewed through a Zeiss LSM 410 confocal microscope. To determine the extent of SG formation, at least five fields were examined in each of three experiments.
Acknowledgments
Acknowledgements
We thank Jean-Baptiste Marq for expert technical assistance, and Klaus Conzelmann, Munich, for the BSR T7 cells. F.I. is supported by a long-term EMBO fellowship, and a grant from the Roche Research Foundation. This work was supported by a grant from the Swiss National Science Fund.
References
- Anderson P. (1995) TIA-1: structural and functional studies on a new class of cytolytic effector molecule. Curr. Top. Microbiol. Immunol., 198, 131–143. [DOI] [PubMed] [Google Scholar]
- Anderson P. and Kedersha,N. (2002) Stressful initiations. J. Cell Sci., 115, 3227–3234. [DOI] [PubMed] [Google Scholar]
- Bitzer M., Prinz,F., Bauer,M., Spiegel,M., Neubert,W.J., Gregor,M., Schulze-Osthoff,K. and Lauer,U. (1999) Sendai virus infection induces apoptosis through activation of caspase-8 (FLICE) and caspase-3 (CPP32). J. Virol., 73, 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontron S., Ucla,C., Mach,B. and Steimle,V. (1997) Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol. Cell. Biol., 17, 4249–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.M. and Steitz,J.A. (2001) HuR and mRNA stability. Cell. Mol. Life Sci., 58, 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U.J., Finke,S. and Conzelmann,K.K. (1999) Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol., 73, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Gatto-Konczak F., Bourgeois,C.F., Le Guiner,C., Kister,L., Gesnel,M.C., Stevenin,J. and Breathnach,R. (2000) The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol., 20, 6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember L.M., Kim,N.D., Liu,K.Q. and Anderson,P. (1996) Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem., 271, 2783–2788. [DOI] [PubMed] [Google Scholar]
- Fan X.C., Myer,V.E. and Steitz,J.A. (1997) AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev., 11, 2557–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch P., Puig,O., Kedersha,N., Martinez,C., Granneman,S., Séraphin,B., Anderson,P. and Valcarcel,J. (2000) The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell, 6, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Garcin D., Pelet,T., Calain,P., Roux,L., Curran,J. and Kolakofsky,D. (1995) A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J., 14, 6087–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D., Taylor,G., Tanebayashi,K., Compans,R. and Kolakofsky,D. (1998) The short Sendai virus leader region controls induction of programmed cell death. Virology, 243, 340–353. [DOI] [PubMed] [Google Scholar]
- Gubbay O., Curran,J. and Kolakofsky,D. (2001) Sendai virus genome synthesis and assembly are coupled: a possible mechanism to promote viral RNA polymerase processivity. J. Gen. Virol., 82, 2895–2903. [DOI] [PubMed] [Google Scholar]
- Gueydan C., Droogmans,L., Chalon,P., Huez,G., Caput,D. and Kruys,V. (1999) Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem., 274, 2322–2326. [DOI] [PubMed] [Google Scholar]
- Hardwick J.M. (1998) Viral interference with apoptosis. Semin. Cell. Dev. Biol., 9, 339–349. [DOI] [PubMed] [Google Scholar]
- Hovanessian A.G. (1989) The double stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J. Interferon Res., 9, 641–647. [DOI] [PubMed] [Google Scholar]
- Itoh M., Hotta,H. and Homma,M. (1998) Increased induction of apoptosis by a Sendai virus mutant is associated with attenuation of mouse pathogenicity. J. Virol., 72, 2927–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z. and Wu,J. (1999) Alternative splicing and programmed cell death. Proc. Soc. Exp. Biol. Med., 220, 64–72. [DOI] [PubMed] [Google Scholar]
- Kawakami A., Tian,Q., Duan,X., Streuli,M., Schlossman,S.F. and Anderson,P. (1992) Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl Acad. Sci. USA, 89, 8681–8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta,M., Li,W., Miller,I. and Anderson,P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of elF-2α to the assembly of mammalian stress granules. J. Cell Biol., 147, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Cho,M.R., Li,W., Yacono,P.W., Chen,S., Gilks,N., Golan,D.E. and Anderson,P. (2000) Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol., 151, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Chen,S., Gilks,N., Li,W., Miller,I.J., Stahl,J. and Anderson,P. (2002) Evidence that ternary complex (eIF2–GTP– tRNA(i)(Met))-deficient preinitiation complexes are core con stituents of mammalian stress granules. Mol. Biol. Cell, 13, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D. (2001) Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl Acad. Sci. USA, 98, 7018–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.A. and Kolakofsky,D. (2001) Paramyxoviridae: the viruses and their replication. In Knipe,D.M. et al. (eds), Fields Virology, 4th edn. Lippincott–Raven Publishers, Philadelphia, PA, pp. 1305–1340.
- Lee S.I., Murthy,S.C., Trimble,J.J., Desrosiers,R.C. and Steitz,J.A. (1988) Four novel U RNAs are encoded by a herpesvirus. Cell, 54, 599–607. [DOI] [PubMed] [Google Scholar]
- le Mercier P., Garcin,D., Hausmann,S. and Kolakofsky,D. (2002) Ambisense Sendai viruses are inherently unstable but are useful to study viral RNA synthesis. J. Virol., 76, 5492–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews M.B. and Shenk,T. (1991) Adenovirus virus-associated RNA and translation control. J. Virol., 65, 5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer V.E., Lee,S.I. and Steitz,J.A. (1992) Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc. Natl Acad. Sci. USA, 89, 1296–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta A.T. and Been,M.D. (1991) A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature, 350, 434–436. [DOI] [PubMed] [Google Scholar]
- Piecyk M. et al. (2000) TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J., 19, 4154–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulston A., Marcellus,R.C. and Branton,P.E. (1999) Viruses and apoptosis. Annu. Rev. Microbiol., 53, 577–628. [DOI] [PubMed] [Google Scholar]
- Schnell M.J., Mebatsion,T. and Conzelmann,K.K. (1994) Infectious rabies viruses from cloned cDNA. EMBO J., 13, 4195–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp T.V., Schwemmle,M., Jeffrey,I., Laing,K., Mellor,H., Proud,C.G., Hilse,K. and Clemens,M.J. (1993) Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein–Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res., 21, 4483–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S.P., Kumar,K.U. and Kaufman,R.J. (1998) Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem., 273, 2416–2423. [DOI] [PubMed] [Google Scholar]
- Storz G. (2002) An expanding universe of noncoding RNAs. Science, 296, 1260–1263. [DOI] [PubMed] [Google Scholar]
- Taupin J.L., Tian,Q., Kedersha,N., Robertson,M. and Anderson,P. (1995) The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc. Natl Acad. Sci. USA, 92, 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Streuli,M., Saito,H., Schlossman,S.F. and Anderson,P. (1991) A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell, 67, 629–639. [DOI] [PubMed] [Google Scholar]
- Tian Q., Taupin,J., Elledge,S., Robertson,M. and Anderson,P. (1995) Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J. Exp. Med., 182, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli P. (1992) The pathophysiology of tumor necrosis factors. Annu. Rev. Immunol., 10, 411–452. [DOI] [PubMed] [Google Scholar]