Abstract
Gene rearrangement in the immune system is always preceded by DNA demethylation and increased chromatin accessibility. Using a model system in which rearrangement of the endogenous immunoglobulin κ locus is prevented, we demonstrate that these epigenetic and chromatin changes actually occur on one allele with a higher probability than the other. It may be this process that, together with feedback inhibition, serves as the basis for allelic exclusion.
Keywords: chromatin/DNA methylation/DNase I sensitivity/double strand breaks/V(D)J rearrangement
Introduction
The development of mature B and T cells in the lymphoid system involves a series of molecular decisions that culminate in the expression of a single antigen receptor on the cell surface. Since every cell has two alleles for each of the seven receptor loci, the ultimate choice of one receptor type must involve a process of allelic exclusion (Pernis et al., 1965; Cebra et al., 1966). It has been proposed that the major pathway for carrying out this choice is by a feedback mechanism in which the generation of a gene product from one allele leads to inhibition of the recombination machinery thereby preventing rearrangement on the remaining germ line allele (for a review, see Rajewsky, 1996).
While this mechanism may certainly play a maintenance role in inhibiting rearrangement on the non-selected allele, recent evidence suggests that the process of allelic exclusion may actually begin early in development, at about the time of implantation when the antigen receptor genes become asynchronously replicating in each cell (Mostoslavsky et al., 2001). This occurs stochastically, such that in some cells it is the maternal allele that replicates early, while in others it is the paternal allele. This epigenetic event, which serves to distinguish between the two alleles, is clonally inherited and appears to provide a signaling mechanism for directing the recombination process, since, at both the µ and κ loci, it is almost always the early allele that is the first to undergo rearrangement in B cells (Mostoslavsky et al., 2001). Although it is not yet known how this chromosomal mark enables the initial selection of only one allele, the process itself must be mediated through monoallelic epigenetic changes.
There is a great deal of evidence suggesting that rearrangement, in general, is controlled by chromatin accessibility (for reviews, see Roth and Roth, 2000; Schlissel, 2000; Krangel, 2001). Even though all lymphoid recombination events are mediated by Rag proteins operating on canonical recombination signal sequences (RSSs), the µ, κ and λ loci undergo rearrangement only in B cells, while TCRα and β become rearranged specifically in T cells (for a review, see Lewis, 1994). Accessibility presumably controls the timing of rearrangement as well, since the µ locus almost always gets activated prior to κ during B cell development. In keeping with this, it has been shown for some of the receptor loci that rearrangement is preceded by an increase in chromatin accessibility and sensitivity to Rag proteins (Stanhope-Baker et al., 1996; Gorman and Alt, 1998; Hesslein and Schatz, 2001; Maës et al., 2001).
In this paper, we demonstrate that allelic exclusion at the κ locus is an instructive process that is mediated through increased chromatin accessibility, initially on a single allele in each cell. This allows the relatively rapid rearrangement reaction to take place on only one allele at a time, thus providing a window of opportunity for executing feedback inhibition of the Rag proteins before the remaining allele can undergo the change in structure required for its rearrangement.
Results
The κ locus undergoes monoallelic demethylation
We have previously shown that the J region of the κ gene is fully methylated on both alleles early in lymphoid development, but undergoes demethylation at later stages (Mostoslavsky and Bergman, 1997). As a result, the rearranged alleles in mature B cells are always found unmodified at this locus (Mostoslavsky et al., 1998). In order to find out whether the two κ alleles undergo monoallelic chromatin changes and to gain more insight into the process of demethylation, we utilized a transgenic mouse (Lκ), which constitutively expresses a pre- rearranged VκJκC gene and therefore brings about feedback inhibition of the Rag proteins, preventing rearrangement of the endogenous κ alleles (Meyer et al., 1990; Betz et al., 1994). About 50–60% of these κ genes were found to be unmethylated in normal B cells from Lκ mice, indicating that demethylation occurs independently of and probably prior to rearrangement (Mostoslavsky et al., 1998). However, these studies did not provide any information about how this demethylation takes place and whether it is restricted to one allele in each cell.
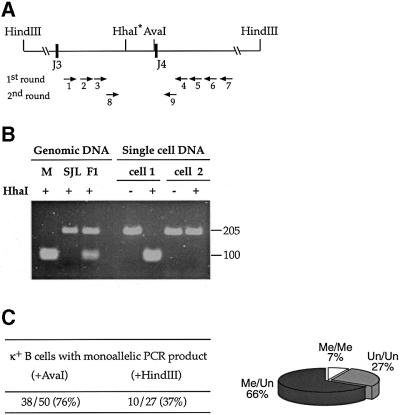
As a first step in understanding whether demethylation of the Jκ locus occurs on a single allele prior to rearrangement, we carried out methyl-sensitive restriction enzyme-dependent PCR (MSRE-PCR) analysis in single cells. To this end, we used FACS to isolate individual κ+ cells from SJL × BALB/c F1 hybrid mice carrying the Lκ transgene. DNA from each cell was digested with the methyl-sensitive restriction enzyme AvaI, and PCR was then carried out using primers specific for the J region (Figure 1A). Under these conditions, only methylated alleles were amplified. In order to distinguish between the SJL and BALB/c alleles, we digested the PCR product with HhaI, which, in this case, is used because it detects a restriction site polymorphism (Figure 1B). Analysis of over 50 individual cells that yielded a PCR product revealed a single methylated allele in 76%, with about half being modified on the maternal copy and the other half on the paternal allele (see legend for Figure 1). To verify that this result is not just a consequence of inefficiencies in the PCR reaction, we carried out the same single-cell assay after digestion with HindIII, an enzyme that does not cut either of the alleles. In contrast to the results obtained using AvaI, these PCR products show a distinctly biallelic (63%) pattern (Figure 1C). Similar results were obtained following AvaI digestion of non-B cell DNA where both alleles are fully methylated (Mostoslavsky et al., 1998).
Fig. 1. Methylation analysis of Lκ single cells. (A) Map of the Jκ 3–4 region showing the location of primers used for MSRE-PCR analysis and relevant restriction enzyme sites. AvaI was used to detect DNA methylation on genomic DNA, whereas HhaI was employed on the PCR product to distinguish between the two alleles. (B) Genomic DNA from Mus musculus BALB/c (M), SJL or F1 mice were PCR amplified with primers P8 and P9, digested with HhaI and analyzed on a 4% agarose gel. The BALB/c gene contains the polymorphic HhaI site, while the SJL gene product is uncut. F1 DNA yields a product that shows 50% digestion. DNA from individual κ+ B cells was digested with AvaI and amplified using two rounds of PCR. Both examples (cells 1 and 2) shown were found to be methylated monoallelically (BALB/c and SJL allele, respectively). (C) Summary of the number of PCR-positive cells that show a monoallelic PCR product in the MSRE-PCR assay, using either AvaI or HindIII (control) digestion. The percentage of biallelic single cells observed following HindIII digestion (63%) is consistent with previous data showing that the efficiency of detection is 1.6 alleles/cell (Muschen et al., 2000). The distribution (%) of single cells containing either 0, 1 or 2 methylated alleles (pie diagram) was derived as follows: 24% of PCR positive cells have a biallelic methylation pattern. Previous experiments had shown that even normal κ+ B cells, which always have at least one allele unmethylated, yield a biallelic pattern in 14% of the cells using this assay, and this is probably due to inefficient AvaI cleavage under the conditions of the reaction (see figure 2 in Mostoslavsky et al., 1998). Subtracting this background (24–14), we conclude that 10% of the PCR positive cells actually have two methylated alleles. In order to calculate the percentage of biallelically methylated cells in the full Lκ B cell population, we utilized Southern blot data showing that only 40% of the κ alleles are methylated (see Figure 2 for an example). Using simple algebra, these data are translated into the numbers shown in the diagram. A simple check shows that out of 100 cells, with seven having two methylated alleles and 66 having one, the fraction of methylated alleles is indeed 40% (80/200). The distribution of methylated alleles can also be ascertained by an alternate method based on first determining the percentage of cells that have two unmethylated alleles. This can be determined by counting the number of single cells that yield no PCR product and correcting for the efficiency of the amplification. After digestion with HindIII alone, 75% of the cells yield a PCR product, but only 54% gave a product when digested with AvaI. As an additional control, we also amplified another region that lacks AvaI sites, and this yielded a product from 73% of the single cells. Thus, in (75–54)/75 = 28% of the Lκ B cells that should have yielded a product, none was obtained, implying that in 28% of the cells, both alleles are unmethylated.
In theory, the monoallelic methylation observed at the κ locus could have come about if both alleles initially underwent demethylation early in B cell development and this was followed by de novo modification of the unrearranged allele as a result of a feedback mechanism. In order to rule out this possibility, we examined DNA methylation in B cells derived from CD19–/– mice. Despite the fact that the feedback process is abrogated in these mice (Shivtiel et al., 2002), κ germline alleles were still found to be fully methylated (data not shown) in a manner similar to normal B cells (Mostoslavsky et al., 1998). Thus, when taken together, our experiments clearly suggest that demethylation of the Jκ region takes place monoallelically and prior to rearrangement.
Allele differential DNase I sensitivity at the κ locus
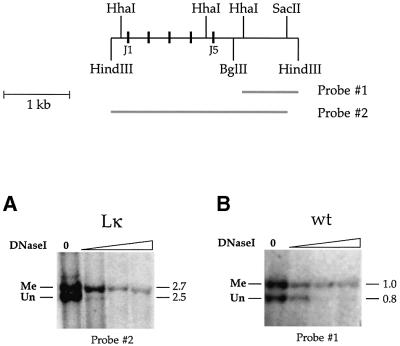
We next asked whether the same alleles that undergo demethylation are also characterized by changes in chromatin accessibility. To this end, we carried out DNase I sensitivity analysis on nuclei from Lκ B220+ B cells, and then restricted the remaining DNA with SacII in order to distinguish between methylated and unmethylated alleles. This experiment clearly demonstrates that unmethylated alleles are far more sensitive to DNase I than the comparable methylated alleles (Figure 2A), and the same is true for both productive and unproductive rearranged copies of the κ locus in normal B cells (Figure 2B). Taken together with the results in Figure 1, we conclude that many individual cells simultaneously harbor one open and one closed allele, and this may represent the molecular basis for allelic exclusion at the κ locus.
Fig. 2. DNase I sensitivity of methylated and unmethylated κ alleles. A map of the Jκ locus showing the J1–5 segments (black rectangles), the relevant restriction enzyme sites and the probes for Southern hybridization. (A) Nuclei were prepared from splenic B cells isolated from Lκ mice treated for 3 days with LPS (10 µg/ml) and reacted with DNase I (0.1–3 µg/ml). Remaining DNA was extracted and digested with HindIII/SacII to distinguish between the 2.7 kb methylated (Me) and 2.5 kb unmethylated (Un) alleles, and subjected to blot hybridization using probe 2. Similar results were obtained using unstimulated ex vivo κ+ or B220+ B cells from Lκ mice. (B) Genomic DNA was isolated from DNase I-treated nuclei purified from splenic B cells taken from wild-type (wt) mice. DNA was digested with BglII/HindIII/HhaI to distinguish between the 1.0 kb methylated and 0.8 kb unmethylated alleles originating from productive and non-productive molecules, respectively, and hybridized with probe 1.
Double strand breaks are formed preferentially on unmethylated alleles
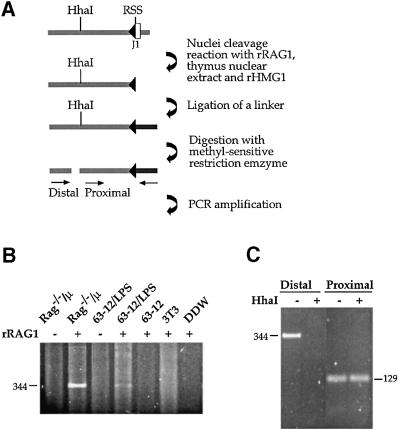
In order to show directly that unmethylated alleles are actually the preferred substrate for the Rag proteins themselves, we analyzed the frequency at which double strand breaks (DSBs) are generated at Jκ RSS sites in normal CD19+ bone marrow (BM) cells that have not yet undergone Igκ locus gene rearrangement. These DSBs are easily identified in both the J1 and J5 region of the κ locus using ligation-mediated PCR (LM-PCR; Figure 3A). However, when this same DNA was pre-digested with the methyl-sensitive restriction enzyme HhaI, at least 10-fold fewer DSBs were observed (distal primer, Figure 3B), even though a control amplification demonstrated that the ligation product is still present in the reaction mix (proximal primer). This suggests that DSBs take place preferentially on unmethylated molecules in vivo. It should be noted that since demethylation is developmentally regulated, unlike κ+ mature spleen cells, only a small percentage of κ alleles have actually undergone this process (Mostoslavsky et al., 1998) in the early lymphoid BM cells examined in this experiment. Taking this into account, it is clear that there must be a very large difference in the Rag accessibility of unmethylated as opposed to methylated DNA in vivo.
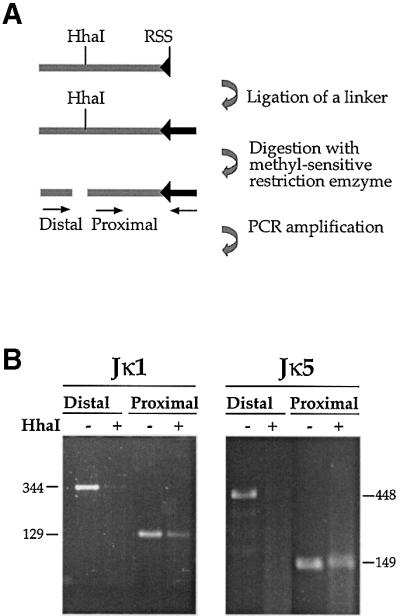
Fig. 3. Pre-existing DSBs in Jκ. (A) Strategy of methylation-sensitive LM-PCR analysis. CD19+ cells were isolated from BM of wild-type mice. These cells actively rearrange the κ locus and therefore contain signal end intermediates which can be detected by ligation of total genomic DNA to a partially double-strand oligonucleotide linker (the darker bar attached to RSS). Following ligation, half of the sample was digested with the methylation-sensitive restriction enzyme, HhaI, and both the digested and undigested samples were PCR analyzed with two sets of primers. In each set, one primer is specific for the linker and the other for the Jκ locus. The proximal set uses a primer located downstream of the HhaI site and thus yields a PCR product irrespective of the methylation state. The distal set contains a primer flanking the HhaI site, making this PCR reaction sensitive to methylation. (B) Genomic DNA purified from CD19+ BM cells of wild-type mice was linker- ligated and analyzed by LM-PCR for DSBs at RSSs flanking the Jκ1 and Jκ5 gene segments (see map in Figure 2).
Allele differential Rag accessibility at the κ locus
As a final test of the idea that allelic exclusion is attained through differential accessibility of the two alleles in each cell, we measured the ability of purified Rag proteins to introduce DSBs on either methylated or unmethylated κ molecules in isolated nuclei in vitro (Figure 4). It should be noted that this reaction is highly specific and only takes place on chromatin templates taken from the correct cell type and stage of lymphoid development (reviewed in Schlissel and Stanhope-Baker, 1997). DSBs are not detected, for example, when the substrate is derived from either fibroblasts or pro-B cells (Stanhope-Baker et al., 1996). In contrast, DSBs are readily observed when Rag proteins are added to nuclei from pre-B cells (line 63–12) pre-treated with the κ transcriptional inducer.
Fig. 4. In vitro Rag cleavage at the Jκ locus. (A) Strategy of methylation-sensitive in vitro Rag cleavage. Nuclei from Rag1–/–/µ BM cells were incubated with fetal calf thymus nuclear extract, recombinant (r) Rag1 and rHMG1 proteins at 30°C for 60 min, and DNA recovered from the reactions was linker ligated (the darker bar attached to RSS) to a partially double strand oligonucleotide. Following ligation, half the DNA was digested with HhaI and both samples then amplified as described in Figure 3A. (B) Nuclei from BM of Rag–/–/µ mice or from Rag2-deficient pro-B cell lines 63–12 and NIH 3T3 were assayed for in vitro Rag cleavage in the presence (+) or absence (–) of rRag1 protein. The specifically amplified 344 bp product was detected by ethidium bromide staining on an agarose gel. (C) Genomic DNA recovered from in vitro Rag-treated Rag–/–/µ BM nuclei was linker ligated, incubated in the presence (+) or absence (–) of HhaI, and amplified with the distal and proximal sets of primers. Amplification using the proximal set of primers serves as a control to confirm the presence of DNA.
BM cells obtained from Rag1–/– mice are arrested at an early, pro-B stage of development (Mombaerts et al., 1992). In these cells, the κ locus has not yet become accessible, and its chromatin is therefore a poor substrate for the action of Rag proteins (Figure 4). However, this block in lymphoid development can be partially overcome, by generating Rag1–/– mice carrying a µ transgene (Spanopoulou et al., 1994). When nuclei from these cells are treated with Rag proteins in vitro, DSBs are readily produced (Figure 4B), indicating that the Jκ region has become more accessible, even though these cells do not have the machinery necessary to carry through with rearrangement.
In order to test for allele specificity, we digested all of the unmethylated molecules in this Rag1–/–/µ BM DNA sample with HhaI and repeated the assay for DSBs by LM-PCR. Almost no DSBs are generated on the remaining methylated alleles (distal primer, Figure 4C). As a control for the presence of DNA, we also amplified the linker-ligated DNA using a primer whose binding site is not affected by HhaI digestion (proximal primer). Thus, these studies confirm at the level of chromatin that methylated alleles are poor substrates for the rearrangement machinery, clearly suggesting by extrapolation that this epigenetic effect may serve as the basis for directing differential rearrangement in B cells where the κ alleles are differentially methylated.
Discussion
Allelic exclusion is an integral aspect of the immune system, which serves to guarantee that only a single antigen receptor is expressed on the surface of each B or T cell (Pernis et al., 1965; Cebra et al., 1966). Although the exact control mechanisms are unknown, it has been proposed that the key step in this process involves feedback inhibition (Rajewsky, 1996). It is usually assumed that each locus is initially in an inaccessible closed conformation and only becomes available for rearrangement through developmentally programmed changes in chromatin structure that open up the region to the recombination machinery already present in the cell (for a review, see Sleckman et al., 1996). The rearrangement reaction itself actually begins when the Rag1 and Rag2 proteins recognize and cleave the RSSs which flank rearranging gene segments (Schatz et al., 1989; Oettinger et al., 1990; Gellert, 1996).
In order to explain allelic exclusion, it has been assumed that the rearrangement machinery operates at suboptimal efficiency, cleaving one allele at a time. As soon as rearrangement is complete, newly synthesized receptor molecules make their way to the cell surface and bring about feedback inhibition of the Rag proteins, thereby precluding rearrangement of the remaining allele. In the event that the first rearrangement event does not succeed in generating a genuine receptor molecule, the rearrangement machinery would still be available for enough time to carry out this reaction again on the other allele. Thus, according to this hypothesis, both alleles constitute equal substrates for recombination, with choice of the first allele being stochastic and determined by the kinetics of interaction between Rag proteins and their target sequence.
Our experiments provide a new perspective on the mechanism of allelic exclusion by showing that, in contradistinction to the previous model, the κ locus does not become generally accessible to recombination on both alleles. In this study, we show that in many cells only one allele initially undergoes demethylation and chromatin opening, while the other remains closed and unavailable for rearrangement. Since this monoallelic epigenetic event occurs both in normal B cells, which are constantly subject to rearrangement, as well as in B cells from Lκ mice in which rearrangement is inhibited, we conclude that allelic exclusion takes place primarily at the level of chromatin structure and in a manner that is independent of the rearrangement reaction itself. Several previous studies have already demonstrated that the general programming of receptor locus activation during development is mediated through the control of chromatin accessibility (Stanhope-Baker et al., 1996; Mathieu et al., 2000; McBlane and Boyes, 2000; McMurry and Krangel, 2000; Agata et al., 2001; Chowdhury and Sen, 2001). It now appears that allelic exclusion is also regulated by this mechanism.
Altering the accessibility of the κ locus in pre-B cells involves both a change in chromatin structure, as well as demethylation of CpG sites in the Jκ region (Cedar and Bergman, 1999). By measuring the state of methylation, it is possible to follow the kinetics of this epigenetic event. We thus carried out single cell methylation analysis on B lymphocytes derived from Lκ mice. These cells express an exogenous pre-rearranged Igκ sequence that constitutively inhibits Rag gene expression, thereby preventing rearrangement of the endogenous κ alleles. This system is unique in that it allows one to evaluate the kinetics of demethylation independently of rearrangement, and thus provides a snapshot of the epigenetic events that occur during the lifetime of a B cell.
Lκ B cells are found in three different states of κ methylation: in some cells, both alleles are fully methylated; in others, one allele has undergone demethylation; and in the last group, demethylation is observed on both alleles. If we assume that the probability of the first allele to undergo demethylation during the average lifetime of a B cell is p and that of the second allele is q, the chance of neither allele being unmethylated is then (1 – p)(1 – q), while the probability of both undergoing demethylation is pq. Thus, from the data in Figure 1C, one can derive two independent equations:
(1 – p)(1 – q) = 0.07
and
pq = 0.27
which, when solved, yield the unique values p = 0.9 and q = 0.3.
This calculation indicates that one allele in each cell has a relatively high probability of becoming demethylated (0.9) and accessible to the rearrangement machinery, whereas the second allele undergoes this process with a 3-fold lower probability. Taking into consideration that this kinetic measurement may not be in the linear range, this difference could, in fact, be much larger. It should be noted that this analysis does not, a priori, assume that the probability for each allele to undergo demethylation must be different (p ≠ q). However, the fact that this is the case clearly suggests that the methylation profile shown in Figure 1C is mathematically inconsistent with models suggesting that both alleles become demethylated with the same probability.
Demethylation, transcription and the correlated changes in chromatin structure at immune receptor loci are mediated by interactions between trans-acting factors and enhancer elements that operate in cis to increase local chromatin accessibility, thus exposing the RSSs to the rearrangement machinery (for reviews, see Sleckman et al., 1996; Schlissel and Stanhope-Baker, 1997; Hempel et al., 1998). Previous studies have shown that the κ gene replicates asynchronously in all cell types (Mostoslavsky et al., 2001). Since replication timing is undoubtedly a reflection of chromosome structure, we propose that it is this epigenetic pattern that generates a 3-fold difference in the probability that these enhancer-mediated changes in chromatin structure will occur on the early, as opposed to the late, replicating allele. This would be consistent with the observation that the early replicating gene in each cell is usually the first to undergo rearrangement (Mostoslavsky et al., 2001; Skok et al., 2001).
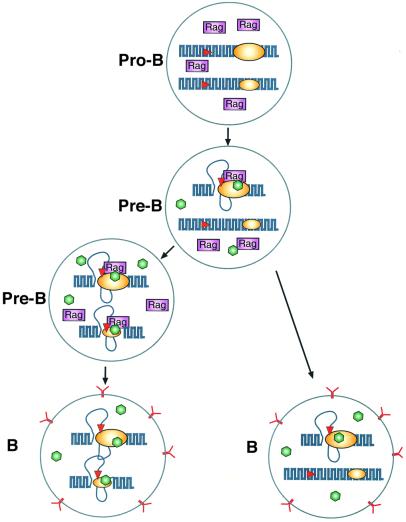
With this understanding of the mechanisms involved in choosing alleles for rearrangement, it is now possible to build an integrative model of how multiple factors contribute to the process of allelic exclusion at the κ locus (Figure 5). Our studies indicate that increased accessibility of a single receptor allele represents the rate-limiting step in V(D)J recombination, and once this is accomplished, rearrangement itself probably occurs very rapidly, consistent with studies showing that DNA methylation and its accompanying closed chromatin structure inhibits the kinetics of recombination by several orders of magnitude (Engler et al., 1991; Hsieh and Lieber, 1992; Cherry and Baltimore, 1999; Figure 4). It is very likely that in most B cells it is the early replicating allele that is first demethylated and made available for rearrangement. Since there is a much lower probability that the second late replicating allele will undergo this process, this provides a window of time for a productive receptor molecule to reach the cell surface and bring about inhibition of Rag gene expression. In the event that rearrangement on the first allele is non-productive, given sufficient time, the second allele can become accessible and competent for rearrangement. It should be noted that although the early allele is the preferred first choice, our model clearly predicts that rearrangement could initially occur on the late allele in a small percentage of cells, and these events can actually be visualized by FISH analysis (Mostoslavsky et al., 2001).
Fig. 5. A model for pre-programmed allelic exclusion. The diagram shows the κ alleles in B cells at different stages of lymphoid development. In pro-B cells, both alleles are in a closed chromatin conformation, and for this reason the Rag proteins present in these cells have no access to the RSS sites (red) of the locus. In pre-B cells, specific factors (green) can activate the Igκ enhancer system (yellow) and open local chromatin. The early replicating allele represents a better target (large yellow ellipse) than the late replicating allele (small yellow ellipse). Once opened, the RSS serves as a substrate for the Rag proteins (purple) that bring about rearrangement. If this results in the generation of a productive cell surface antigen receptor (Y), Rag expression is then inhibited in B cells (right). In the event that the first rearrangement is not productive, with time, the second allele can become accessible and undergo recombination (left), which may again lead to an antibody-producing cell (bottom). It should be noted that in cells containing a productive rearrangement of the first allele (right), the trans-acting enhancer factors (green) are presumably still present, and this could lead to opening of the second allele. Since Rag proteins are not present, however, these will not undergo rearrangement. It is for this reason that one observes unmethylated germ line alleles in a small percentage of mature B cells. The fact that most B cells have only a single accessible κ allele also explains why secondary changes resulting from editing or somatic hypermutation appear to take place preferentially on alleles that have already undergone at least one round of rearrangement.
Our model may also help explain how the original choice of an allele at the µ or κ locus is stably maintained despite the continuing process of fine tuning that takes place during advanced B cell development. κ rearrangement, for example, always takes place in cells that have already selected one productive µ allele at the pro-B cell stage. Yet, despite reactivation of the recombination machinery in these pre-B cells (Nussenzweig, 1998), the DJ-rearranged µ allele remains resistant to rearrangement (Rajewsky, 1996). In a similar manner, antigen-dependent reactivation processes, such as light chain editing (Luning-Prak and Weiger, 1995; Hertz et al., 1998) or somatic hypermutation (Jolly and Neuberger, 2001), appear to occur preferentially on the already rearranged accessible allele. All of these phenomena are better understood if one realizes that the initial rearrangement events involve opening only a single allele at each locus, while the unselected allele remains relatively inaccessible. It was recently shown that these ‘unopened’ alleles are specifically recognized and redirected to heterochromatic regions of the nucleus when B cells are induced to proliferate (Skok et al., 2001), perhaps providing an additional mechanism for augmenting allele stability and preventing unwanted recombination.
Materials and methods
Purification of primary cells
κ+ B cells were purified from the spleen of wild-type and Lκ transgenic (Sharpe et al., 1991; Betz et al., 1994) mice using biotinylated monoclonal anti-κ antibody (PharMingen) and streptavidin paramagnetic beads (MiniMacs system, Miltenyi Biotec), following the manufacturer’s protocol. CD19+ B cells were purified as described previously (Stanhope-Baker et al., 1996) from the BM of Rag1-deficient µ heavy chain transgenic mice (Spanopoulou et al., 1994) using biotinylated anti-CD19 (provided by K.Rajewsky). It should be noted that while these cells are arrested at about the pre-B cell stage, not all of them are necessarily poised for κ recombination.
Southern blot analysis
Southern blotting was performed as described previously (Southern, 1975). Generally, 10 µg of cellular DNA was subjected to enzyme digestion for 14 h, electrophoresed in native (Tris–acetate) agarose gels, and transferred to nitrocellulose. The filters were then hybridized with [32P]dCTP random-primed probes and analyzed by autoradiography.
Single-cell methylation analysis
Spleen cells from an F1 progeny of a cross between a BALB/c Lκ transgenic mouse and a SJL mouse were stained with a rat anti-mouse κ light chain monoclonal antibody (R5-240, PharMingen, FITC conjugated). Individual κ+ cells were FACS sorted directly into 25 µl of lysis buffer (50 mM KCl, 2.5 mM MgCl2, 0.5% Tween-20, 0.5% NP-40, 10 mM Tris–HCl pH 8.3) in individual PCR tubes. The genomic DNA was liberated using proteinase K (0.001 mg/ml at 60°C for 2 h) that was then inactivated with PMSF (0.002 mg/ml at room temperature for 1 h). The genomic DNA was subjected to digestion overnight at 37°C with 20 U of AvaI or HindIII (NEB). PCR was performed with Taq DNA polymerase (RedHot, ABgene) under standard conditions (50 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2, 10 mM Tris–HCl pH 8.3, 200 mM each of four dNTPs, 50 ng of each primer). The first-round PCR reaction was carried out in 50 µl using primers P1 (5′-TTGCTCAACT GCTTGTGAAG-3′), P2 (5′-TGTGAAGTTTTGGTCCATTGTGTC C-3′), P3 (5′-TACTGTACAAGCTGAGCAAACAGAG-3′), P4 (5′-AT GCACAGGTTGCCAGGAAT-3′), P5 (5′-TGGGGGATCTTCTATT GATGCAC-3′), P6 (5′-CACAGAAGCCTAAGACGAGTTTTTC-3′) and P7 (5′-CTCAGAAGCCTAAGACGA-3′), which flank the AvaI site. Amplification was performed as follows: 4 min at 94°C; 40 cycles of 10 s at 94°C, 10 s at 56°C and 20 s at 72°C; and, finally, 3 min at 72°C. Second-round PCR conditions were identical to the first round except that 5 µl of the first-round amplification product was added to the PCR mixture containing primers P8 (5′-TGTCAGATTTGTGGGAGA AATG-3′) and P9 (5′-ACCCAAACAGAACCAAAACG-3′). The second-round PCR product was subjected to digestion with HhaI and analyzed by agarose gel electrophoresis. To control for AvaI digestion, a region that does not contain an AvaI site and is located upstream to Jκ2 was amplified. The first-round PCR reaction was carried out using primers P20 (5′-CCCACTGCTCTGTTCCTCTT-3′), P23 (5′-CAC TGGTGTCCCTTCACTCA-3′), P24 (5′-CGAACGTGTACACACAC TGGT-3′), P22 (5′-CCTTCACTCAACCCCCATAC-3′), P21 (5′-TG AGGAGGGTTTTTGTACAGC-3′) and P27 (5′-GCCTACCCACT GCTCTGTTC-3′). Second-round PCR was carried out with primers P26 (5′-CAGGCTACCCTGCTTCTTTG-3′) and P25 (5′-GGGTTTTT GTACAGCCAGACA-3′).
DNase I treatment
Typically, 108 splenic B cells from either wild-type or Lκ transgenic mice were prepared, washed in cold PBS, centrifuged at 1000 r.p.m. for 5 min and lysed in NP-40 containing lysis buffer (20 mM Tris pH 7.0, 3 mM CaCl2, 2 mM MgCl2, 0.3% NP-40). The mixture was kept on ice for 10 min and centrifuged at 800 g for 5 min. The resulting nuclei were resuspended in a solution of 10 mM Tris pH 7.0, 3 mM MgCl2, 10 mM NaCl to a final concentration of 108 nuclei per ml. Aliquots of 10–150 µl nuclei were then digested with increasing concentrations of DNase I (0.1–3 µg/ml) for 15 min at 37°C and digestion was stopped by adding an equal amount of 2× lysis buffer containing 20 mM Tris pH 7, 200 mM NaCl, 2 mM EDTA and 2% SDS. The lysates were treated with proteinase K (200 µg/ml) overnight at 37°C, and DNA isolated by phenol–chloroform extraction (Liu et al., 1988).
Methylation-sensitive ligation-mediated PCR
Double strand linker was prepared by mixing 5 nmol of BW1 (5′-GCGGTGACCCGGGAGATCTGAATTC-3′) and BW2 (5′-GAA TTCAGATC-3′) oligonucleotides in a 100 µl reaction containing 250 mM Tris pH 7.9. The reaction was incubated successively in 95°C for 5 min, 70°C for 15 min, and then allowed to cool to room temperature. Wild-type BM genomic DNA (2 µg) was subjected to linker ligation for 18 h at 14°C in a 50 µl reaction mixture containing ligation buffer (Boehringer), 40 pmol of linker, and 2 U of T4 DNA ligase (Boehringer). The reaction mixture was then heated to 65°C for 15 min. Half of the ligation volume (1 µg of DNA) was then subjected to digestion overnight at 37°C with 200 U of HhaI (NEB). Control samples were incubated without the restriction enzyme. Following digestion, the DNA was extracted successively with phenol–chloroform (1:1), and chloroform. The DNA was precipitated with 20 µg glycogen (Boehringer) and 2.5 vols of ethanol, and then resuspended in 40 µl of H2O. Typically, 2 µl of each sample was analyzed in each PCR reaction, which was performed with Taq DNA polymerase (RedHot, ABgene) under standard conditions (50 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2, 10 mM Tris–HCl pH 8.3, 200 mM each of four dNTPs, 50 ng of each primer). The PCR reaction was carried out in 50 µl using primers J1 (5′-GCAG CGAGTGCCACTAACTG-3′) and BW-H (5′-CCGGGAGATCTG AATTCCAC-3′) for detection of Jκ1 signal ends, and J5 (5′-CAGTTCTCTGAACTTAGCC-3′) for detection of Jκ5 signal ends.
In vitro cleavage reaction
Nuclei were purified as described previously (Stanhope-Baker et al., 1996) and resuspended at a final concentration of 50 000 µl in 20 mM HEPES pH 7.6, 2 mM MgCl2, 70 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 25% glycerol and frozen in aliquots at –80°C. Sources of nuclei included the A-MuLV-transformed cell lines 63–12 (derived from Rag2-deficient mice (Shinkai et al., 1992), the murine NIH 3T3 cell line, and primary cells from bone marrow of either Rag1-deficient (Mombaerts et al., 1992) or Rag1-deficient µ heavy chain transgenic mice (Spanopoulou et al., 1994). For nuclei prepared from lipopolysaccharide (LPS)-treated 63–12 cells, the cells were grown for 12 h in 10 µg/ml LPS (Difco) prior to harvest.
In vitro cleavage reactions were performed as previously described (Stanhope-Baker et al., 1996). Briefly, reactions were performed in a volume of 20 µl containing 100 000 nuclei, ∼0.6 µg rRag1 (van Gent et al., 1995), ∼2 µg fetal cow nuclear extract (Parker and Topol, 1984) and ∼2 µg of rHMG1 proteins (Ge and Roeder, 1994). Reactions containing 40 mM HEPES pH 7.6, 2.5 mM Tris pH 8.0, 110 mM KCl, 1 mM MnCl2, 2.5 mM DTT, 0.2 mM MgCl2, 0.2 mM PMSF, 0.025 mM EDTA and 7% glycerol (v/v) were incubated for 30 min on ice followed by 60 min at 30°C, and stopped by adding 80 µl of lysis buffer (50 mM Tris pH 8.0, 1 mM EDTA, 0.1% SDS, 100 µg/ml proteinase K).
Acknowledgments
Acknowledgements
This research was supported by grants from the NIH (H.C. and Y.B.), the Israel Academy of Sciences (Y.B.), the German Israel Foundation (Y.B.), the European Community 5th Framework Quality of Life Program (Y.B.) and the Israel Cancer Research Fund (H.C.).
References
- Agata Y., Katakai,T., Ye,S.K., Sugai,M., Gonda,H., Honjo,T., Ikuta,K. and Shimizu,A. (2001) Histone acetylation determines the developmentally regulated accessibility for T cell receptor γ gene recombination. J. Exp. Med., 193, 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A.G., Milstein,C., Gonzalez-Gernandez,A., Pannel,R., Larson,T. and Neuberger,M.S. (1994) Elements regulating somatic hypermutation of an immunoglobulin κ gene: critical role for the intron enhancer/matrix attachment region. Cell, 77, 239–248. [DOI] [PubMed] [Google Scholar]
- Cebra J.J., Colberg,J.E. and Dray,S. (1966) Rabbit lymphoid cells differentiated with respect to α-, γ- and µ-heavy polypeptide chains and to allotypic markers Aa1 and Aa2. J. Exp. Med., 123, 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H. and Bergman,Y. (1999) Developmental regulation of immune system gene rearrangement. Curr. Opin. Immunol., 11, 64–80. [DOI] [PubMed] [Google Scholar]
- Cherry S.R. and Baltimore,D. (1999) Chromatin remodeling directly activates V(D)J recombination. Proc. Natl Acad. Sci. USA, 96, 10788–10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D. and Sen,R. (2001) Stepwise activation of the immunoglobulin µ heavy chain gene locus. EMBO J., 20, 6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler P., Haasch,D., Pinkert,C.A., Doglio,L., Glymour,M., Brinster,R. and Storb,U. (1991) A strain-specific modifier on mouse chromosome 4 controls the methylation of independent transgene loci. Cell, 65, 1–20. [DOI] [PubMed] [Google Scholar]
- Ge H. and Roeder,R.G. (1994) The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem., 269, 17136–17140. [PubMed] [Google Scholar]
- Gellert M. (1996) A new view of V(D)J recombination. Genes Cells, 1, 269–275. [DOI] [PubMed] [Google Scholar]
- Gorman J.R. and Alt,F.W. (1998) Regulation of immunoglobulin light chain isotype expression. Adv. Immunol., 69, 113–181. [DOI] [PubMed] [Google Scholar]
- Hempel W.M., Leduc,I., Mathieu,N., Tripathi,R.K. and Ferrier,P. (1998) Accessibility control of V(D)J recombination: lessons from gene targeting. Adv. Immunol., 69, 309–352. [DOI] [PubMed] [Google Scholar]
- Hertz M., Kouskoff,V., Nakamura,T. and Nemazee,D. (1998) V(D)J recombinase induction in splenic B lymphocytes is inhibited by antigen-receptor signaling. Nature, 394, 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslein D.G. and Schatz,D.G. (2001) Factors and forces controlling V(D)J recombination. Adv. Immunol., 78, 169–232. [DOI] [PubMed] [Google Scholar]
- Hsieh C.L. and Lieber,M.R. (1992) CpG methylated minichromosomes become inaccessible for V(D)J recombination after undergoing replication. EMBO J., 11, 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C. and Neuberger,M. (2001) Somatic hypermutation of immunoglobulin κ transgenes: association of mutability with demethylation. Immunol. Cell Biol., 79, 18–22. [DOI] [PubMed] [Google Scholar]
- Krangel M.S. (2001) V(D)J recombination becomes accessible. J. Exp. Med., 193, F27–F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.A. (1994) The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv. Immunol., 56, 27–150. [DOI] [PubMed] [Google Scholar]
- Liu J.K., Bergman,Y. and Zaret,K.S. (1988) The mouse albumin promoter and a distal upstream site are simultaneously DNase I hypersensitive in liver chromatin and bind similar liver-abundant factors in vitro. Genes Dev., 2, 528–541. [DOI] [PubMed] [Google Scholar]
- Luning-Prak E. and Weiger,M. (1995) Light chain replacement: a new model for antibody gene rearrangement. J. Exp. Med., 182, 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maës J., O’Neill,L.P., Cavelier,P., Turner,B.M., Rougeon,F. and Goodhardt,M. (2001) Chromatin remodeling at the Ig loci prior to V(D)J recombination. J. Immunol., 167, 866–874. [DOI] [PubMed] [Google Scholar]
- Mathieu N., Hempel,W.M., Spicuglia,S., Verthuy,C. and Ferrier,P. (2000) Chromatin remodeling by the T cell receptor (TCR)-β gene enhancer during early T cell development: implications for the control of TCR-β locus recombination. J. Exp. Med., 192, 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane F. and Boyes,J. (2000) Stimulation of V(D)J recombination by histone acetylation. Curr. Biol., 10, 483–486. [DOI] [PubMed] [Google Scholar]
- McMurry M.T. and Krangel,M.S. (2000) A role for histone acetylation in the developmental regulation of VDJ recombination. Science, 287, 495–498. [DOI] [PubMed] [Google Scholar]
- Meyer K.B., Sharpe,M.J., Surani,M.A. and Neuberger,M.S. (1990) The importance of the 3′-enhancer region in immunoglobulin κ gene expression. Nucleic Acids Res., 18, 5609–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P., Lacomini,J., Johnson,R.S., Herrup,K., Tonegawa,S. and Papaioannou,V.E. (1992) RAG-1-deficient mice have no mature B and T lymphocytes. Cell, 68, 869–877. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R. and Bergman,Y. (1997) DNA methylation: regulation of gene expression and role in the immune system. Biochim. Biophys. Acta, 1333, F29–F50. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R., Singh,N., Kirillov,A., Pelanda,R., Cedar,H., Chess,A. and Bergman,Y. (1998) κ chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev., 12, 1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R. et al. (2001) Asynchronous replication and allelic exclusion in the immune system. Nature, 414, 221–225. [DOI] [PubMed] [Google Scholar]
- Muschen M., Re,D., Jungnickel,B., Diehl,V., Rajewsky,K. and Kuppers,R. (2000) Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J. Exp. Med., 192, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig M.C. (1998) Immune receptor editing: revise and select. Cell, 95, 875–878. [DOI] [PubMed] [Google Scholar]
- Oettinger M.A., Schatz,D.G., Gorka,C. and Baltimore,D. (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science, 248, 1517–1523. [DOI] [PubMed] [Google Scholar]
- Parker C.S. and Topol,J. (1984) A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell, 36, 357–369. [DOI] [PubMed] [Google Scholar]
- Pernis B., Chiappino,G., Kelus,A.S. and Gell,P.G. (1965) Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J. Exp. Med., 122, 853–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K. (1996) Clonal selection and learning in the antibody system. Nature, 381, 751–758. [DOI] [PubMed] [Google Scholar]
- Roth D.B. and Roth,S.Y. (2000) Unequal access: regulating V(D)J recombination through chromatin remodeling. Cell, 103, 699–702. [DOI] [PubMed] [Google Scholar]
- Schatz D.G., Oettinger,M.A. and Baltimore,D. (1989) The V(D)J recombination activating gene, RAG-1. Cell, 59, 1035–1048. [DOI] [PubMed] [Google Scholar]
- Schlissel M.S. (2000) Perspectives: transcription. A tail of histone acetylation and DNA recombination. Science, 287, 438–440. [DOI] [PubMed] [Google Scholar]
- Schlissel M.S. and Stanhope-Baker,P. (1997) Accessibility and the developmental regulation of V(D)J recombination. Semin. Immunol., 9, 161–170. [DOI] [PubMed] [Google Scholar]
- Sharpe M.J., Milstein,C., Jarvis,J.M. and Neuberg,M.S. (1991) Somatic hypermutation of immunoglobulin κ may depend on sequences 3′ of C κ and occurs on passenger transgenes. EMBO J., 10, 2139–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y. et al. (1992) RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell, 68, 855–867. [DOI] [PubMed] [Google Scholar]
- Shivtiel S., Leider,N., Sadeh,O., Kraiem,Z. and Melamed,D. (2002) Impaired light chain allelic exclusion and lack of positive selection in immature B cells expressing incompetent receptor deficient of CD19. J. Immunol., 168, 5596–5604. [DOI] [PubMed] [Google Scholar]
- Skok J.A. et al. (2001) Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol., 2, 848–854. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Gorman,J.R. and Alt,F.W. (1996) Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol., 14, 459–481. [DOI] [PubMed] [Google Scholar]
- Southern E.M. (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol., 98, 503–517. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E. et al. (1994) Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev., 8, 1030–1042. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P., Hudson,K.M., Shaffer,A.L., Constantinescu,A. and Schlissel,M.S. (1996) Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell, 85, 887–897. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., McBlane,J.F., Ramsden,D.A., Sadofsky,M.J., Hesse,J.E. and Gellert,M. (1995) Initiation of V(D)J recombination in a cell-free system. Cell, 81, 925–934. [DOI] [PubMed] [Google Scholar]