Abstract
Trichome patterning in Arabidopsis is a model for the generation of a spacing pattern from initially equivalent cells. We show that the TRIPTYCHON gene that functions in lateral inhibition encodes a single-repeat MYB-related transcription factor that lacks a recognizable activation domain. It has high sequence simi larity to the root hair patterning gene CAPRICE. Both genes are expressed in trichomes and act together during lateral inhibition. We further show that TRIPTYCHON and CAPRICE act redundantly in the position-dependent cell fate determination in the root epidermis. Thus, the same lateral inhibition mechanism seems to be involved in both de novo patterning and position-dependent cell determination. We propose a model explaining trichome and root hair patterning by a common mechanism.
Keywords: MYB/pattern formation/root hairs/trichomes/TRIPTYCHON
Introduction
In animal systems, gene cassettes controlling pattern formation are often employed in several independent patterning processes, and the particular cell types generated depend on the developmental context. A well-studied example is the receptor–ligand-based Delta/Notch system in Drosophila that governs the epidermal versus neural precursor cell decision in the ectoderm (Campos-Ortega, 1993; Ghysen et al., 1993) as well as the segregation of midgut epithelial cells in the endoderm (Tepass and Hartenstein, 1995).
In Arabidopsis, two patterning systems, trichome (leaf hair) and root hair formation, are known to involve a common set of genes. The underlying mechanisms, however, appear to be different. The root hair pattern seems to be position dependent: root epidermal cells are arranged in adjacent files of root hair cells and non-hair cells. Root hair files are located over the intercellular space between underlying cortical cells; non-hair cells arise over single cortical cells (Masucci et al., 1996). This implies that the underlying cortical cells provide positional cues for the epidermal root hair patterning system. In contrast, trichome patterning is thought to be generated de novo, i.e. all epidermal cells initially are equivalent, and single trichome cells are selected by a self-organizing system (Larkin et al., 1996; Schnittger et al., 1999).
The genetic analysis of trichome and root hair development has enabled the identification of genes that are required for proper pattern formation. Two genes, GLABRA1 (GL1), encoding a MYB-related transcription factor, and TRANSPARENT TESTA GLABRA1 (TTG1), encoding a protein containing a WD40 domain that is thought to mediate protein–protein interactions, function as positive regulators of trichome development (Koornneef, 1981; Marks and Feldmann, 1989; Walker et al., 1999). The corresponding mutants produce no trichomes. In glabra3 (gl3) mutants, only few trichomes are produced, suggesting that GL3 is also a positive regulator of trichome patterning (Hülskamp et al., 1994; Payne et al., 2000). GL3 encodes a basic helix–loop–helix (bHLH)-related transcription factor, and GL3 overexpression causes an increase in trichome density (Payne et al., 2000). The involvement of a bHLH-related transcription factor in this process was postulated previously because the GL3-homologous R gene from maize triggers trichome formation in Arabidopsis when overexpressed (Lloyd et al., 1992, 1994). TRIPTYCHON (TRY) is the only known negative regulator of trichome development. In try mutants, ∼5–10% of the trichomes are arranged in clusters, suggesting that TRY mediates lateral inhibition of cells surrounding an incipient trichome (Schnittger et al., 1998, 1999). The mechanism by which these genes control trichome patterning is unknown. Because GL3 physically interacts with both GL1 and TTG1 in the yeast-two hybrid assay, it is postulated that a multimeric complex of GL1, GL3 and TTG1 promotes trichome development and it is assumed that TRY counteracts their activity (Payne et al., 2000). It is thought that the combinatorial action of the patterning genes regulates trichome-specific expression of GLABRA2 (GL2). GL2 encodes a homeobox transcription factor and is thought to control trichome cell differentiation (Rerie et al., 1994).
Root hair patterning is regarded to be position dependent. During the establishment of cell files, trichoblasts or hair cells (overlying the clefts between cortex cells) and atrichoblasts or non-hair cells (overlying the cortex cells) differ from early on in several morphological features including cell length, cytoplasmic density and vacuolization (Galway et al., 1994; Masucci et al., 1996; Berger et al., 1998). Only two genes, TTG1 and WEREWOLF (WER), a MYB-related transcription factor gene with high similarity to GL1, are known to be required for the initial establishment of these two cell files (Galway et al., 1994; Lee and Schiefelbein, 1999). In wer and ttg1 mutants, all cell files are root hair cell files. Thus both genes can be considered positive regulators of non-root hair file establishment. Similarly to the trichome system, the maize R gene also affects root hair patterning when overexpressed (Lloyd et al., 1994). Overexpression results in a reduced root hair number and counteracts ttg1 mutations but not wer mutations, suggesting that root hair patterning involves an endogenous, as yet unknown bHLH-related transcription factor (Lee and Schiefelbein, 1999). As in trichome development, GL2 governs early differentiation processes. GL2 is expressed exclusively in non-hair cells and suppresses the formation of root hairs (Masucci et al., 1996). Its expression is controlled by WER and TTG1. The CAPRICE (CPC) gene, encoding a MYB-related transcription factor lacking a transactivation domain, is the only known negative regulator of non-root hair cells (Wada et al., 1997). CPC is required for cell file-specific expression of GL2 and genetically counteracts WER and TTG1 (Wada et al., 1997; Lee and Schiefelbein, 1999). Current models postulate that a complex consisting of WER, TTG1 and an R gene-related transcription factor is active in non-hair cells where it promotes expression of GL2. In hair cells, the activity of this complex is inhibited by CPC by lateral inhibition (Lee and Schiefelbein, 1999, 2002). How the underlying cortex cells control this pattern is not known.
A partial overlap in the mechanisms controlling cell fate choice during root hair patterning and trichome patterning is assumed, as several genes function in both processes. In ttg1 and gl2 mutants, root hair differentiation and trichome differentiation are affected in opposite ways: root hair development is promoted whereas trichome development is abolished. In contrast, the mechanism for the actual determination of cell fate seems to be distinct since corresponding genes such as GL1/TRY (trichomes) and WER/CPC (root hairs) are specific for each system. However, the finding that overexpression of the root hair gene CPC abolishes trichome development suggests a genetic overlap with cell fate determination in the leaf epidermis, although this particular finding was interpreted as an unspecific competition with the trichome-promoting factor GL1 (Wada et al., 1997).
In this work, we report the positional cloning and molecular characterization of the TRY gene. TRY encodes a MYB-related protein with high sequence similarity to CPC. Both genes are expressed in young leaf primordia and developing trichome cells. We show that TRY and CPC act not only in trichome or root hair patterning, respectively, but also together in both processes. This suggests that epidermal cell type selection employs the same lateral inhibition mechanism in roots and shoots. Our results lead to a model that explains de novo pattern formation of trichomes and position-biased root hair file establishment by a common mechanism.
Results
TRY encodes a CPC-homologous MYB-related transcription factor
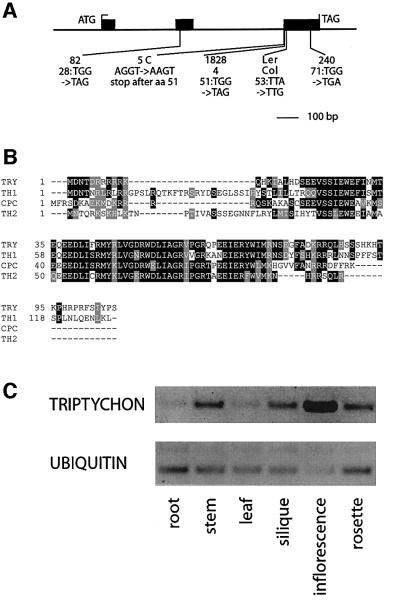
In extensive screens for trichome patterning mutants to date, only one gene, the TRY gene, has been identified that appears to function in lateral inhibition and hence most probably in cell–cell signalling (Hülskamp et al., 1994; Schnittger et al., 1998, 1999). To understand the molecular mechanism underlying lateral inhibition during trichome patterning, we cloned the TRY gene by a positional cloning approach. Initially, the TRY gene was mapped to a 150 kb interval flanked by two newly developed CAPS markers on chromosome V (details are available on request). The annotated genomic sequence in this region revealed one gene that showed high homology to the CPC gene. To test whether this gene encodes TRY, we sequenced its genomic region from several try alleles. Four try alleles exhibited mutations leading to stop codons in the predicted open reading frame (ORF) of the gene. In one allele, the mutation affects the splice acceptor site before the third exon, leading to a stop in the next codon following the splice donor site (Figure 1A). To confirm that this gene encodes TRY, we introduced a 4.2 kb DNA fragment containing the TRY gene into try mutant plants, which rescued the try mutant phenotype.
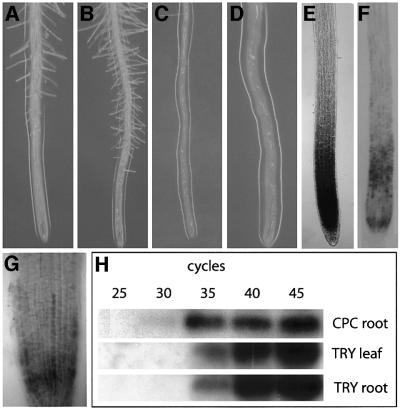
Fig. 1. Molecular characterization of the TRY gene. (A) Exon–intron structure of the TRY gene. The positions of mutations in the different try alleles are indicated. (B) Amino acid sequence comparison with the MYB-related proteins CPC, TRIPTYCHON HOMOLOG1 (TH1) and TRIPTYCHON HOMOLOG2 (TH2). (C) RT–PCR from total RNA extracts of different tissues. The minimal number of PCR cycles that allowed detection of TRY expression was used to enable a comparison of the expression levels between the tissues. RT–PCR amplification of ubiquitin was used as a control.
To determine the genomic structure of the TRY gene, we isolated cDNA fragments by RT–PCR. A comparison of the cDNA with the genomic sequence revealed a structure different from the published annotation. The TRY gene has a 1 kb ORF and contains three exons and two introns (Figure 1A). The 5′ and 3′ ends of the cDNA were determined by RACE–PCR. The 5′ end of the transcript was mapped 139 bp upstream of the start codon, and the 3′ end 81 bp downstream of the stop codon.
The deduced amino acid sequence of TRY revealed a 106 amino acid protein that contains a MYB-related DNA-binding domain but lacks a recognizable activation domain. Overall, TRY shows highest homology to a group comprised of three MYB proteins in Arabidopsis, including CPC; the other two are of unknown function (Figure 1B).
TRY expression was analysed by RT–PCR. TRY mRNA accumulated in all organs analysed, including roots, leaves, siliques and inflorescences (Figure 1C). Thus, TRY is expressed not only in trichome-bearing organs but also in organs lacking trichomes, such as roots and siliques.
A paradoxical situation: ubiquitous expression of TRY suppresses trichome initiation although TRY is expressed in trichomes
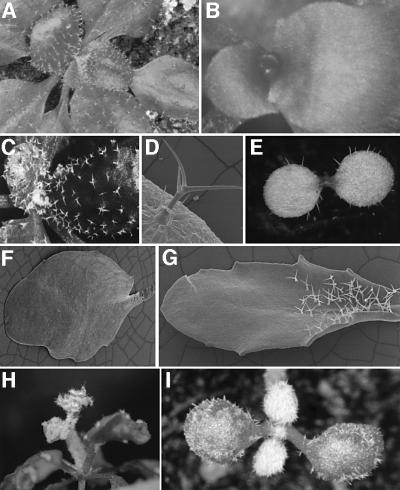
Genetic analysis of try mutants suggests that TRY functions as a negative regulator of trichome development. To test this, we created transgenic lines expressing TRY ubiquitously. A construct containing the TRY coding sequence under the control of the cauliflower mosaic virus (CaMV) 35S promoter was introduced into Ler wild- type. These 35S::TRY plants were completely glabrous (Figure 2B), confirming that TRY is a negative regulator of trichome development.
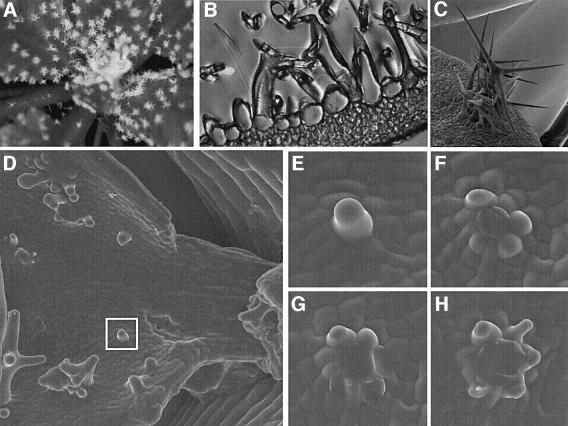
Fig. 2. Overexpression phenotypes of 35S::TRY and 35S::CPC lines. (A) Rosette leaves of a try mutant plant rescued with the genomic 4.2 kb fragment. (B) Glabrous 35S::TRY plant. (C) Rosette leaves of a try mutant plant. (D) Scanning electron micrograph (SEM) of a 35S::GL1 cpc leaf trichome with two accessory cells transformed to trichomes. (E) Cotyledons of a 35S::GL1 cpc plant; many ectopic trichomes are formed. (F) SEM of a glabrous 35S::CPC leaf. (G) SEM of a 35S::CPC 35S::R-GR leaf after R gene induction with dexamethasone. Because in developing leaves only young cells at the base can respond to R gene-induced trichome induction, rescue is found only at the leaf base. (H) Inflorescence of a 35S::GL1 try cpc mutant plant. (I) Primary leaves of a 35S::GL1 try cpc mutant plant.
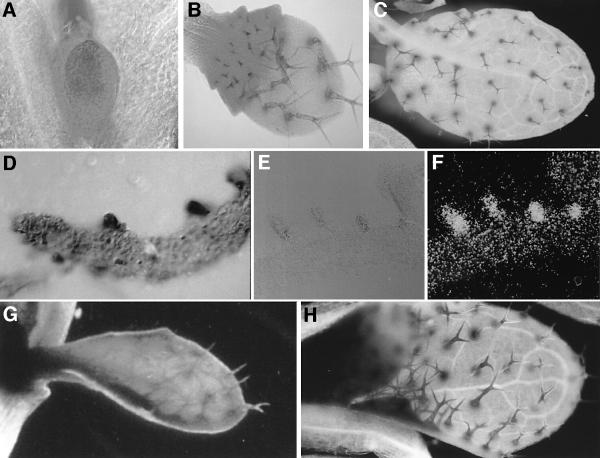
The TRY expression pattern was determined by in situ hybridizations on paraffin-embedded rosette leaf sections with digoxigenin-labelled sense and antisense probes of the TRY cDNA (see Materials and methods). Sections hybridized with the antisense probe revealed staining in developing trichomes (Figure 3D). The same expression pattern was observed in transgenic plants carrying a GUS fusion construct with the 1.4 kb TRY upstream region. Incipient trichome cells at stages when they were still indistinguishable from epidermal cells by their cell size exhibited already stronger GUS activity (Figure 3A). In addition, we observed ubiquitous expression in young leaves. Later, ubiquitous expression is only seen at the leaf base in the trichome initiation zone (Figure 3B). In mature leaves, only trichome cells were stained (Figure 3C).
Fig. 3. Expression analysis of TRY and CPC. (A) Young leaf of a TRY::GUS line showing ubiquitous expression. (B) Slightly older leaf of a TRY::GUS line. Ubiquitous expression is seen at the leaf base. Note that early developmental stages of trichome cells exhibit elevated expression levels. (C) Mature leaf of a TRY::GUS line. TRY expression is limited to mature trichome cells. (D) In situ hybridization of a wild-type leaf section with antisense TRY. Trichome cells display high expression of TRY. (E) Light microscopy micrograph of an in situ hybridization of a wild-type leaf section with antisense CPC. (F) Dark field micrograph of (E). (G) Young leaf of a CPC::GUS line showing ubiquitous expression. (H) Mature leaf of a CPC::GUS line; CPC expression is seen only in trichomes.
The TRY-homologous root hair patterning gene CPC is involved in trichome patterning
Overexpression of the TRY-homologous root hair patterning gene CPC causes a suppression of trichome formation (Wada et al., 1997). Initially, this was interpreted either to indicate that CPC plays also a role in trichome patterning or that it was caused by an unspecific interaction with the GL1 gene. As TRY shares high sequence identity with CPC, CPC might also be able to exert the same inhibitory function as TRY in overexpression studies. In order to test whether CPC is involved in trichome patterning, we analysed its expression pattern in leaves. Sections of paraffin-embedded rosette leaves were hybridized with 3H-labelled sense or antisense probes of the CPC gene. While the sense probe showed weak ubiquitous background staining, the antisense probe revealed a strong distinct signal in trichomes at all developmental stages (Figure 3E and F). The analysis of plants containing an expression construct where the CPC 5′ regulatory sequence was fused to the GUS coding region (CPC:: GUS) revealed an expression pattern indistinguishable from that of TRY. As shown in Figure 3H, CPC expression in mature leaves is confined to trichomes. Younger leaves show high levels of expression in trichomes at all developmental stages and, in addition, ubiquitous expression throughout the young leaf primordium (Figure 3G).
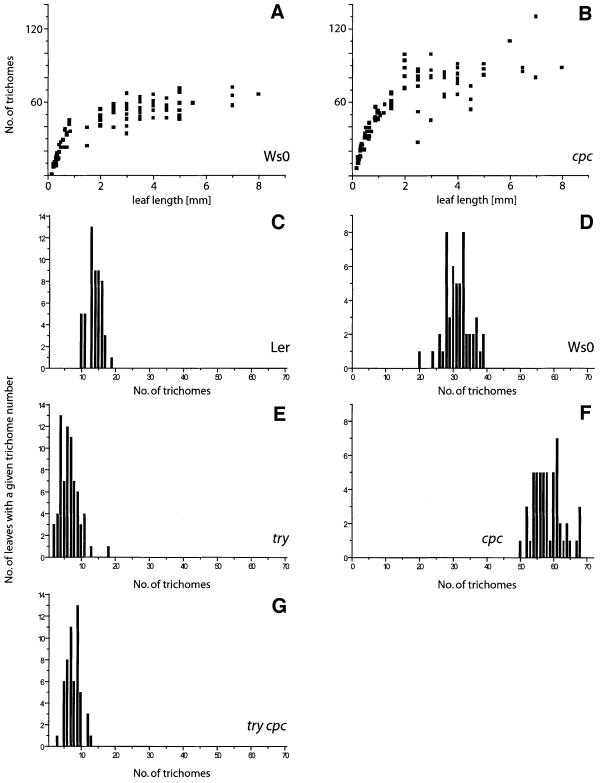
While the trichome-specific expression of CPC suggests a role in trichome development, no obvious trichome phenotype was observed in the initial studies (Wada et al., 1997). In order to assess whether CPC has a role in trichome patterning, we carefully determined two aspects of trichome patterning: the frequency of trichome clusters and the total number of trichomes in cpc mutants. The cluster frequency in cpc mutants showed no deviation from wild-type. However, the total number of trichomes per leaf in cpc mutants was increased up to 2-fold as compared with the corresponding wild-type Ws-0 (Figure 4D and F). A statistical analysis using Wilcoxon rank sum test and Students t-test showed that this difference is highly significant (P < 0.01). This was studied in more detail for leaf number four (Figure 4A and B). Here a difference of >30% in trichome number was found at all developmental stages of leaf development, indicating that the increase in the number of trichomes is due to an increased density rather than to an extended period of time of trichome formation or simply due to the formation of larger leaves (Figure 4A and B).
Fig. 4. Comparison of trichome number in different mutants. (A and B) Trichome number counts on the fourth leaf at all developmental stages of leaf development. (C–G) Number of trichome initiation sites on the first leaf. (A) In wild-type (Ws-0), trichome number increases until the leaf reaches a length of 1 mm, after which trichome initiation ceases but leaf growth continues. (B) A cpc mutant. Compared with wild-type, cpc mutants produce more trichomes at all developmental stages. (C) Wild-type ecotype Ler. (D) Wild-type ecotype Ws-0. (E) A try mutant. (F) A cpc mutant. (G) A try cpc double mutant.
Another phenotypic difference between cpc mutants and try mutants was observed for the regulation of endoreduplication. Trichomes in try mutants contain on average twice the DNA (64C) content of wild-type (32C), overexpression of GL1 results in an increased DNA content (64C) and trichomes in the try 35S::GL1 double mutant contain four times the DNA content of wild-type (128C). To determine whether the TRY-homologous CPC gene is also involved in the control of endoreduplication, we studied the cpc single and several double mutants. Neither the single cpc mutant nor the cpc 35S::GL1 double mutant showed an increased DNA content as compared with wild-type or 35S::GL1, respectively (Table I). The DNA content in try cpc double mutant trichomes was not increased as compared with that in try mutants. These data show that CPC, in contrast to TRY, is not involved in the regulation of trichome endoreduplication.
Table I. DNA content in wild-type and mutant trichomes.
| Ws-0 | try | cpc | try cpc | 35S::GL1 | 35S::GL1 cpc | |
|---|---|---|---|---|---|---|
| DNA content [C] | 44.4 | 68 | 45.3 | 49.1 | 68.3 | 61.4 |
| SD | 15.6 | 22.3 | 18 | 21.1 | 38.1 | 33.1 |
| n | 100 | 50 | 100 | 100 | 50 | 100 |
TRY and CPC play different roles in lateral inhibition during trichome patterning
An increase in both cluster frequency in try mutants and trichome density in cpc mutants suggests that both genes act as negative regulators of trichome patterning. How ever, the different phenotypes also suggest separate functions. To assess this further, we generated the cpc try double mutant and analysed its phenotype by two criteria, cluster frequency and number of trichome initiation sites (TIS, defined as a location containing a single trichome or a trichome cluster).
The try cpc double mutant consisted of the strongest try allele available, try-EM1, and a cpc mutant in which the insertion of a T-DNA results in truncated protein that lacks part of the MYB domain. While clustering in cpc mutants was similar to wild-type (0.1%) and not very frequent in try mutants (4.4%, Table II), in the try cpc double mutant 76.3% of all TIS were clusters containing between 20 and 30 trichomes (Figure 5A–C). Cluster formation in try cpc mutants was initiated from single (66%, n = 100) or twin (34%, n = 100) trichomes. We used dental wax impressions to create replicas of subsequent stages of leaf development to follow the ontogeny of trichome clusters. During wild-type development, all trichome-surrounding cells start to elongate and to develop into trichome accessory cells. In try cpc double mutants, all cells in immediate contact with an already existing trichome started to bulge out and developed into trichomes (Figure 5D–H). Several rounds of trichome induction are necessary to account for the large clusters seen on mature leaves.
Table II. Cluster frequency in wild-type and mutants.
| Wild-type (Ler) | try | cpc | try cpc | |
|---|---|---|---|---|
| No. of trichome initiation sites | 1009 | 707 | 1170 | 722 |
| No. of clusters | 1 | 31 | 3 | 551 |
| Cluster frequency | 0.1% | 4.4% | 0.25% | 76.3% |
Fig. 5. try cpc double mutant phenotype. (A) Rosette leaf of a try cpc double mutant plant showing trichome clusters. (B) Section through a try cpc trichome cluster. (C) SEMs of a large try cpc cluster. (D–H) SEMs of wax replicas of developing try cpc trichome clusters. (D) Overview of a young leaf. The square marks the initial stage of the developing trichome cluster shown below. Replicas of the developmental stages were taken at 8 h intervals. Note that the central trichome has been torn away. (E) One initial trichome has emerged from the epidermis. (F) At least three immediately neighbouring epidermal cells appear as bulges. (G) Additional neighbouring cells show trichome development. (H) Surrounding trichomes are elongated.
The analysis of the number of TIS in the try cpc double mutant revealed a significantly (P < 0.1) reduced number of TIS as compared with the respective wild-types Ler and Ws. We also found a reduced TIS number in try mutants. Since cpc mutants show the opposite phenotype, these findings indicate that try is epistatic to cpc with respect to the number of TIS (Figure 4).
TRY and CPC act together in establishing atrichoblast and trichoblast cell files in the root epidermis
The finding that CPC acts together with TRY in lateral inhibition during trichome formation raised the possibility that TRY might be involved in root hair patterning. Consistent with this, TRY expression was detected in roots by RT–PCR (Figure 1). In try mutants, root hair formation was indistinguishable from wild-type. Initial evidence for a role for TRY in root hair patterning came from the finding that 35S::TRY lines produce extra root hairs (Figure 6B). In order to test whether a role for TRY during root hair formation might be masked by a redundant function of TRY and CPC similar to that observed for trichome formation, we compared the root hair phenotype of the cpc mutant and the try cpc double mutant. In cpc single mutants, the number of root hairs is significantly reduced (Wada et al., 1997) (Figure 6C). In the try cpc double mutant, we never found any root hairs (Figure 6D).
Fig. 6. Analysis of root hair phenotypes. (A–G) Light microscopy micrographs of wild-type and mutant roots. (A) Wild-type. (B) A 35S::TRY root displaying extra root hairs. (C) A cpc mutant showing fewer root hairs. (D) A try cpc double mutant devoid of root hairs. (E) TRY:GUS expression in a cpc try background. (F) TRY:GUS expression in 35S:RGR induced roots. (G) TRY:GUS expression in 35S:RGR uninduced roots; expression is found in the tip region of the root. (H) Semi-quantitative RT–PCR of TRY and CPC. Note that CPC expression can be detected ∼5 amplification cycles earlier than TRY.
In wild-type development, root epidermal differentiation is carried out in two steps: First, a distinction between trichoblasts and atrichoblasts is established (patterning). Secondly, root hair differentiation is suppressed in atrichoblasts during further development (morphogenesis). In the wild-type, cell files overlying the longitudinal anticlinal cell walls of the cortical cells are trichoblasts (T) and contain more cells than atrichoblast (A) cell files that are overlying the cortex cells. We used the relative cell number, trichoblast to atrichoblasts (T/A) ratio, to determine whether the initial distinction between these two cell files is made. In wild-type, the T/A ratio is 1.3 in the root hair initiation and elongation regions. To enable a comparison between wild-type and mutants, cell files were defined according to their position relative to the cortex cells, and the T/A ratio was determined (Table III). The initial establishment of atrichoblast files and trichoblast files is normal in both the try and cpc single mutants as indicated by the T/A ratios of 1.27 and 1.25, which correspond to the T/A ratio of 1.27 found in wild-type roots. The try cpc double mutant revealed a T/A ratio of 1.07, indicating that all cell files have approximately the same number of cells. This indicates that TRY and CPC act together early during the initial establishment of atrichoblast and trichoblast cell file identity. Since try cpc double mutants do not form root hairs and 35S::CPC and 35S::TRY plants produce extra root hairs, it is conceivable that all cell files in try cpc mutants develop atrichoblasts whereas those in 35S::CPC and 35S::TRY plants develop trichoblasts.
Table III. Trichoblast/atrichoblast ratio in wild-type and mutant roots.
| Ws-0 | Ler | try | cpc | try cpc | ttg | gl2 | |
|---|---|---|---|---|---|---|---|
| T/A | 1.24 | 1.27 | 1.27 | 1.25 | 1.07 | 1.01 | 1.25 |
| SD | 0.06 | 0.05 | 0.07 | 0.05 | 0.1 | 0.14 | 0.06 |
| n | 6 | 10 | 9 | 9 | 9 | 10 | 8 |
In order to assess whether TRY is expressed in atrichoblasts or trichoblasts, we analysed GUS expression in roots of the TRY::GUS lines. No GUS activity was detected in the wild-type background. A comparison of the TRY and CPC RNA levels by semi-quantitative RT–PCR showed that TRY expression in the root is reduced compared with that of CPC, suggesting that the expression levels of TRY are below the detection limit of the TRY:GUS construct (Figure 6H). However, strong expression of TRY was observed in cpc try double mutants and in a line overexpressing the R gene (Figure 6E–G). These data allow two conclusions. First, TRY levels are controlled by TRY and CPC. Secondly, since in both lines all cell files are atrichoblasts, TRY is likely also to be expressed in atrichoblasts in wild-type.
TRY and CPC exhibit different interactions
Genetic results indicate that TRY acts as a negative regulator of GL1, as overexpression of GL1 is counteracted by TRY, with respect to both trichome initiation and the control of endoreduplication. The overexpression phenotype of GL1 was also enhanced in a cpc mutant background, as ectopic trichomes were found on cotyledons, and accessory cells were transformed into trichomes (Figure 2D and E). However, in contrast to the 35S::GL1 try combination, no ectopic trichomes were found in the subepidermis or in flower tissues, nor were the endoreduplication levels affected (Table I). The 35S::GL1 cpc try triple mutant showed essentially an additive phenotype, consistent with the view that both genes act as inhibitors of GL1 (Figure 2H and I).
Although try and cpc act together in lateral inhibition, their phenotypes show differences in some important features, suggesting that they might interact with different partners, most probably the positive regulators GL1 or GL3. In this case, one might expect that the overexpression phenotype of TRY and CPC can be rescued by overexpression of the specific partner. In an attempt to rescue the overexpression phenotypes of 35S::TRY and 35::CPC by overexpression of the GL1 or the R gene, we found a striking difference. Overexpression of GL1 did not affect the 35S::TRY and 35S::CPC glabrous phenotype. In contrast, overexpression of the GL3-homologous R gene resulted in a complete rescue of the 35S::CPC phenotype but not the 35S::TRY phenotype (Figure 2G).
Discussion
The roles of TRY and CPC in lateral inhibition during trichome patterning
Our genetic model to explain the establishment of the trichome spacing pattern is based on a theoretical model (Meinhardt and Gierer, 1974). In this model, initially equivalent cells compete with each other to become a trichome cell. Only two components are required, an activator and a repressor. The activator stimulates its own activity and that of the negative regulator. The negative regulator represses the activator. Both factors may diffuse, though the negative regulator is predicted to be faster. As a result, cells start to compete and, because the activator is capable of autoactivation (a positive feedback loop), small fluctuations in the amount of one of the components are sufficient to break equivalence. Subsequently, the neighbouring cells are suppressed by the diffusible inhibitor and, eventually, a single cell is selected to become a trichome. Genetic data suggest that three genes, GL1, GL3 and TTG1, together represent what is formally called the activator (Payne et al., 2000). The TRY gene represents the negative regulator. Because mutations in TRY can strongly enhance the overexpression phenotype of GL1, it is thought that GL1 is the target of the inhibitory effect of TRY (Schnittger et al., 1998; Szymanski and Marks, 1998). The TTG1 gene is likely to play an accessory role, e.g. in the postulated self-enhancing loop, because the ttg1 mutant trichome phenotype can be rescued by overexpression of GL1 in the absence of TRY. The GL3 gene acts genetically downstream of TTG1 since overexpression of GL3 can partially rescue ttg1 mutants. It is therefore conceivable that the activity of GL3 is controlled by TTG1.
The try cpc double mutant phenotype described here revealed that the inhibitory function during trichome patterning consists of at least two components. Although these genes encode homologous proteins with indistinguishable expression patterns, the different phenotypes of the single mutants, such as cluster formation and reduction in trichome number in try mutants and an increased trichome number in cpc mutants, point to mechanistically different roles. One possibility to explain the single as well as the double mutant is that the CPC protein diffuses faster than the TRY protein. As a result, TRY would be important in short-range inhibition whereas the lack of CPC would have predominantly a distant effect. This interpretation is consistent with the ontogenesis of the large clusters in the try cpc double mutant. Initial pattern formation is similar to that in try mutants. Only after trichomes become more and more separated due to cell divisions of epidermal cells does CPC become important and result in the formation of additional trichomes around an already existing trichome.
At first glance, the expression pattern of TRY and CPC is surprising. As shown by the mutant phenotypes and the suppression of trichome initiation in overexpressing lines, both genes act as negative regulators of trichome development, and hence the finding that the highest expression is found in cells that are suppressed seems paradoxical. However, the expression patterns fit nicely with the dynamics of the inhibitor as predicted by the theoretical model of Meinhardt. According to the model, the activity of the negative regulator is tightly controlled by the activator. Hence, the expression of the inhibitor is always highest where the activator is highest. Because the diffusion rate of the inhibitor is higher than that of the activator, the inhibitor peak is broader than the activator peak. Therefore, at the point of highest activator activity, which eventually becomes the selected cell, the activity of the activator always exceeds the activity of the inhibitor. Consistent with this model, the spatial distribution of GL1 is very similar to that of TRY/CPC such that initially GL1 is expressed in all epidermal cells to become restricted only to trichome cells later (Larkin et al., 1993). Whether expression levels or activity levels are different is unknown.
Which one of the positive regulators is the target of TRY and CPC? Our finding that overexpression of the R gene but not of GL1 can rescue the glabrous phenotype of CPC-overexpressing lines indicates that CPC and the R gene can titrate each other’s function. An attractive explanation for this finding is that CPC competes with the R gene homolog GL3 in Arabidopsis and, depending on their balance, trichome cell fate is promoted when more GL3 is available, while an excess of CPC promotes the epidermal cell fate. In contrast, TRY did not interact with either of the two overexpressor lines. It seems to interact with TTG1, as suggested by the genetic observation that double heterozygous try/+ ttg1/+ plants showed an increased cluster frequency (Schnittger et al., 1999).
Molecular interactions during trichome patterning
Except for the TTG1 gene, all factors involved in trichome patterning code for putative transcription factors. Thus, it is conceivable that the relative balance or competitive interactions are important. The physical interactions observed in two-hybrid assays suggest that the factors involved can form multimeric complexes. As shown by Payne et al. (2000), GL3 physically interacts with both GL1 and TTG1 in the yeast two-hybrid system, whereas no interaction was found between GL1 and TTG1. Because TRY and CPC encode single MYB repeat proteins lacking a transactivation domain, an attractive scenario is that they compete with the positive trichome initiation factors for binding sites. The CPC gene is likely to counteract the GL3 gene specifically. Competition of TRY or CPC with the positive regulators could occur either by protein–protein interactions resulting in inactive complexes or by a competition at the level of DNA binding. Since the ttg1 mutant trichome phenotype can be rescued in a 35S::GL1 try background, it is likely that TTG1 is not absolutely required for these interactions (Schnittger et al., 1999). One possibility is that TTG1 modifies the activity of the other factors probably by interacting with GL3, since molecular interactions between GL3 and TTG1 were observed in the yeast two-hybrid system (Payne et al., 1999).
One major question is how cell–cell communication is mediated molecularly. Our finding that both genes involved in lateral suppression of trichome fate are eventually expressed exclusively in developing trichomes supports two alternative explanations. Either TRY and CPC switch on an unknown gene that can move into the neighbouring cells, or TRY and CPC proteins can move and act directly in the neighbouring cells. Generally, the transport of transcription factors between cells is well established in plants (Lucas, 1995). Because TRY and CPC proteins are very small and well below the exclusion size of 40 kDa found for plasmodesmata between epidermal cells in tobacco leaves (Oparka et al., 1999), transport could even occur by diffusion. Genetic mosaics and a careful comparison between RNA and protein expression patterns are needed to clarify this.
Root hair cell file establishment by lateral inhibition
The current genetic and molecular data provide a detailed explanation for the control of cell differentiation in atrichoblasts and trichoblasts as well as for the atrichoblast-specific expression of the GL2 gene. The genetic machinery involved is remarkably similar to that known in trichome development except that the logic is reversed such that atrichoblast cells correspond to trichome cells.
The expression of GL2 in atrichoblasts depends on the GL1 homolog WER that is expressed in atrichoblasts (Lee and Schiefelbein, 1999). Also, the TTG1 gene controls the expression of GL2 and, because the maize R gene can suppress the ttg1 root hair phenotype, probably involves an as yet unknown bHLH-related transcription factor. In hair cells, GL2 expression is down-regulated by the CPC gene. However, the finding that GL2 expression during embryo development is not changed in cpc mutants suggested that at least one additional unknown factor is required (Lin and Schiefelbein, 2001). In both systems, a TTG1-controlled bHLH-related transcription factor (GL3 in the shoot and an as yet unknown R gene homolog in roots) together with a MYB-related transcription factor (GL1 in the shoot and its homolog WER in the root) determine epidermal cell differentiation via the GL2 gene (Figure 7). This similarity of the involved factors raises the question of whether a lateral inhibition system is also involved during root hair patterning. Recently, an elegant series of genetic experiments has revealed that CPC is involved in the lateral suppression of WER and GL2 (Lee and Schiefelbein, 2002). Our finding that in try cpc double mutants root hair patterning is completely abolished, leaving only atrichoblast cell files, strongly indicates that TRY and CPC act similarly to the behaviour observed during trichome patterning in a lateral suppression process. By analogy, CPC and TRY would be most strongly accumulated in non-root hair files and be involved in the lateral suppression of GL2 expression in root hair files. Consistent with this idea, we found TRY expression only in genetic situations where root epidermal cells are atrichoblasts. Similarly, CPC was found to be expressed predominantly in atrichoblasts (Lee and Schiefelbein, 2002; T.Wada and K.Okada, unpublished). These data support an attractive model to explain the positioning of root hairs cells relative to the underlying cortex cells (Lee and Schiefelbein, 2002; Figure 7). If the same mutual inhibition mechanism operated during root hair file establishment, extremely small differences biasing the cell fate choice can easily be amplified to create a stable, predictable pattern. A positive cortex cell-derived signal would be slightly weaker in epidermal cells overlying a cleft between cortex cells but sufficient to bias the patterning system towards root hair cell fate.
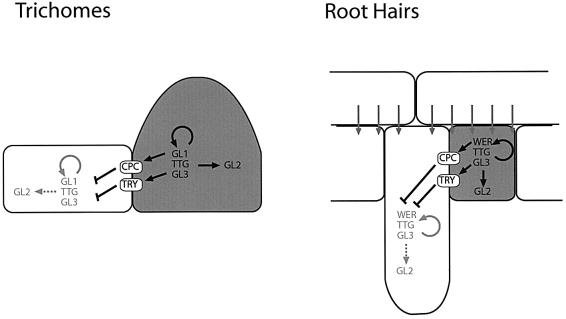
Fig. 7. Genetic model of trichome and root hair patterning. It is postulated that both trichome (left) and root hair patterning (right) are based on a competition mechanism mediated by TRY and CPC. In both systems, this competition results in the activation of a set of positive regulators consisting of TTG1, and a MYB-related (GL1 or WER) and a bHLH-related (GL3 or an unknown factor in root hairs) transcription factor. The biased decision in the root system with respect to the underlying cortex cells is explained by a positive signal from the cortex cells (arrows) which is slightly weaker in epidermal cells overlying a cleft. For clarity, only those regulation events are indicated here that are immediately relevant for the proposed model. These activate the GL2 gene which in the leaf epidermis triggers trichome formation and in the root epidermis promotes non-root hair fate.
Our finding that TRY is expressed at very low levels in wild-type and derepressed in the cpc try double mutant indicates that the regulation of TRY expression involves an auto-repression by the two redundantly acting genes TRY and CPC. According to the model, this would be expected as in the absence of the repressor, the activator concentrations are increased and consequently also the transcription of the repressors.
Evolutionary divergence of epidermal cell fate specification
Epidermal cell fate specification in the root, the hypocotyl, the leaf and the seed coat involves a remarkably similar set of genes. The WD40 protein TTG1 and the homeobox transcription factor GL2 are required in all four epidermal tissues (Koornneef, 1981; Galway et al., 1994; Walker et al., 1999). In the root, hypocotyl and the leaf, a bHLH protein similar to the R gene product and functionally equivalent MYB transcription factors (WER and GL1) are important (Oppenheimer et al., 1991; Lloyd et al., 1992; Galway et al., 1994; Hung et al., 1998; Payne et al., 2000). As shown in this work, in addition, two very similar MYB-related transcription factors lacking a transactivation domain are involved in root, hypocotyl (data not shown) and leaf epidermal patterning. In the case of WER and GL1, it was shown that they are functionally equivalent, but do not share a functional overlap (Lee and Schiefelbein, 2001). In contrast, CPC and TRY function in a partly redundant manner. It is therefore tempting to speculate that in this system members of the same gene family such as WER and GL1, and CPC and TRY have evolved from gene duplications of common ancestor genes and that they have adopted new and different functions to variable degrees.
Perspectives
The analysis of trichome and root hair patterning has revealed a common underlying mechanism. The basic functions are carried out by the same genes or by a combination of highly homologous genes. This raises a number of interesting questions. What is the specificity of genes with equivalent functions such as TRY and CPC? Is their function interchangeable as found for GL1 and WER? How can the patterning machinery be modulated by the cortex cells in the root or by environmental factors controlling trichome density in the shoot? The patterning genes identified seem to be responsible only for creating differences between cell types. How is the actual cell fate, root hair versus trichome, determined? Now that all genetically identified genes are cloned, the tools have become available to address these questions.
Materials and methods
Plant material, growth conditions and genetic analysis
Plants were grown on soil at 23 or 18°C under constant illumination. For the analysis of root hairs, plants were grown on 0.6% agar plates containing 1/2 MS medium and 2% sucrose. Wild-type strains used in this work were Landsberg erecta (Ler), Columbia-0 (Col) and Wasilewskaja (Ws-0). The try alleles try-EM1, try-EM2 and try-JC, the single mutants gl2, ttg1 and cpc, and the overexpressing lines 35S::R-GR 1857, 35S::R-1435, 35S::GL1 and 35S::CPC were described previously (Hülskamp et al., 1994; Larkin et al., 1994; Lloyd et al., 1992, 1994; Wada et al., 1997). The try allele try-1828 is a newly identified X-ray allele. The phenotypic analysis of plants overexpressing 35S::TRY or 35S::CPC together with 35S::GL1, 35S::R-GR 1857 or 35S::R-1435 was carried out in the F1 generation. Double mutants were created by crossing the parental lines, screening the F2 generation for new phenotypes and verified by backcrosses to the parental lines. The number of TIS of try, cpc, try cpc, Ws-0 and Ler was counted on the first four leaves of 25 different 21-day-old plants. For a comparison of trichome number in cpc and Ws-0 in developing leaves, leaves smaller than 800 µm were stained with fluorescein diacetate. The trichome number was determined for various leaf stages as defined by their leaf lengths.
Microscopy and cytological methods
4′,6-diamidino-2-phenylindole (DAPI) staining and DNA content measurements of trichome nuclei were done as described (Hülskamp et al., 1994). For light microscopy, we used a Zeiss axiophot and for confocal laser scanning microscopy (CLS) a Leica CLS-microscope and the TCS-NT program. For SEM analysis, critical point dried leaves or Spurr impressions prepared as described below were mounted, sputtered and analysed at 10 or 15 keV.
Images were processed with Adobe Illustrator 9.0 and Adobe Photoshop 5.0 (Adobe Systems Inc., Mountain View, CA) software.
Analysis of root hair patterning
For each line, 7-day-old seedlings were stained with propidium iodide and analysed with a Leica laser confocal microscope. The cell length of at least four neighbouring root cell files was determined by counting the cell number for a defined root section. Each cell file was classified as hair cell files (T) or non-hair files (A) according to their position relative to the underlying cortex.
Wax impressions of leaf surfaces
For monitoring the formation of the large trichome clusters in try cpc double mutants, young developing leaves were covered with dental wax at 6 h intervals. Up to five wax impressions per leaf were filled with SPURR, hardened at 65°C and analysed by SEM as described above.
Molecular biology
New CAPS markers were designed by PCR amplification of random 2 kb fragments in the TRY interval. Fragments with a restriction enzyme polymorphism or a length polymorphism between the ecotypes Ler and Col were used as CAPS markers for further fine mapping. Details are available on request.
For sequencing mutant alleles, a 1.5 kb genomic fragment was amplified and sequenced using the primers 5′-AACGACAAGTCTACACAAAGGG-3′ and 5′-CCTAACCGCATGGATTAAAG-3′.
5′ RACE was carried out with the Gibco-BRL 5′ RACE kit using 5′-TGAATTCTTCAAACTAACAAGAAC-3′ as GSP1 primer and 5′-ACTAGTGACTAGGAAGGATAGATAGAAAAG-3′ as nested primer. For 3′ RACE, an oligo(dT) primer was used together with 5′-TCTAGATAGTAATGGATAACACTGACCGT-3′ and 5′-CGTCGCCGTCGTAAGCAACAC-3′ as nested gene-specific primers. The PCR products were cloned into pGEM (Promega) and the clones sequenced (SP6/T7).
The analysis of TRY and CPC expression by RT–PCR was done as previously described (Kirik et al., 2001). Hybridization experiments using gene-specific probes were included to show that the detected expression is specific for TRY or CPC.
In situ hybridizations with the TRY probe were done as described (Schoof et al., 2000). For the analysis of TRY expression, the TRY coding sequence and the 5′-untranslated region were used to generate the sense and the antisense probe.
The fixation, embedding, sectioning and hybridization to analyse CPC expression were done as described by Drews et al. (1991).
Constructs and plant transformation
To rescue the try mutant phenotype, a 4.2 kb genomic HindIII fragment from the ecotype Ler was cloned into the binary vector pBARB.
For GUS analysis of TRY expression, 1.4 kb upstream untranslated sequence was PCR amplified using 5′-AAGCTTCGAAGAAAACGCAAATCAGG-3′ and 5′-CGTCGACTATTGAAGTAAGAAAAGAAAAATAG-3′ as primers to introduce a 5′ HindIII and a 3′ SalI restriction site, subcloned into pGem and sequenced. This fragment was subcloned in front of the GUS marker gene and a terminator site in the vector pGUS 1. A HindIII and EcoRI fragment containing the TRY promoter::GUS fusion was subcloned further into the HindIII and EcoRI sites of pBARB.
To create the 35S::TRY construct, the TRY cDNA was PCR amplified from a Col root cDNA library with 5′-TCTAGATAGTAATGGATAACACTGACCGT-3′ and 5′-ACTAGTGACTAGGAAGGATAGATAGAAAAG-3′ primers containing a 5′ XbaI and a 3′ XmaI restriction site. This fragment was subcloned into the pGem vector, sequenced and introduced between the CaMV 35S promotor and a terminator site into the XbaI site of the vector pRT103, for which the NcoI site was deleted by digestion with SmaI and BalI and religation. A HindIII fragment containing the 35S promotor::TRY cDNA::terminator fusion was subcloned into the HindIII site of the binary vector pBARB.
Plant transformations and the selection of transformed plants were done as previously described (Clough and Bent, 1998).
Acknowledgments
Acknowledgements
We thank the members of our laboratory for critically reading the manuscript, Jürgen Berger (SEM) and Paul Grini (microscopy and computers) for expert advice, Ulrike Schöbinger for excellent technical assistance, Wolfgang Lukowitz, Ulrike Mayer and John Larkin for providing us with new try alleles, and Mandy Walker and John Larkin for sharing unpublished results. This work was supported by the Volkswagen Stiftung and the SFB446.
References
- Berger F., Hung,C., Dolan,L. and Schiefelbein,J. (1998) Control of cell division in the root epidermis of Arabidopsis thaliana. Dev. Biol., 194, 235–245. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J.A. (1993) Early neurogenesis in Drosophila melanogaster. In Bate,M. and Martinez,A. (eds), The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1091–1129.
- Clough S. and Bent,A. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Drews G.N., Weigel,D. and Meyerowitz,E.M. (1991) Floral patterning. Curr. Opin. Genet. Dev., 1, 174–178. [DOI] [PubMed] [Google Scholar]
- Galway M.E., Masucci,J.D., Lloyd,A.M., Walbot,V., Davis,R.W. and Schiefelbein,J.W. (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol., 166, 740–754. [DOI] [PubMed] [Google Scholar]
- Ghysen A., Dambly-Chaudiere,C., Jan,L.Y. and Jan,Y. (1993) Cell interactions and gene interactions in peripheral neurogenesis. Genes Dev., 7, 723–733. [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Misera,S. and Jürgens,G. (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell, 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Hung C., Lin,Y., Zhang,M., Pollock,S., Marks,M.D. and Schiefelbein,J. (1998) A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol., 117, 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Bouyer,D., Schobinger,U., Bechtold,N., Herzog,M., Bonneville,J.M. and Hulskamp,M. (2001) CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr. Biol., 11, 1891–1895. [DOI] [PubMed] [Google Scholar]
- Koornneef M. (1981) The complex syndrome of ttg mutants. Arabidopsis Inf. Serv., 18, 45–51. [Google Scholar]
- Larkin J.C., Oppenheimer,D.G., Pollock,S. and Marks,M.D. (1993) Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell, 5, 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J.C., Oppenheimer,D.G., Lloyd,A.M., Paparozzi,E.T. and Marks,M.D. (1994) The roles of the GLABROUS1 and TRANPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell, 6, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J.C., Young,N., Prigge,M. and Marks,M.D. (1996) The control of trichome spacing and number in Arabidopsis. Development, 122, 997–1005. [DOI] [PubMed] [Google Scholar]
- Lee M.M. and Schiefelbein,J. (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell, 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lee M.M. and Schiefelbein,J. (2001) Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development, 128, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Lee M.M. and Schiefelbein,J. (2002) Cell patterning in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell, 14, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. and Schiefelbein,J. (2001) Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development, 128, 3697–3705. [DOI] [PubMed] [Google Scholar]
- Lloyd A.M., Walbot,V. and Davis,R.W. (1992) Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science, 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Lloyd A.M., Schena,M., Walbot,V. and Davis,R.W. (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by steroid-inducible regulator. Science, 266, 436–439. [DOI] [PubMed] [Google Scholar]
- Lucas W.J. (1995) Plasmodesmata: intercellular channels for macromolecular transport in plants. Curr. Opin. Cell Biol., 7, 673–680. [DOI] [PubMed] [Google Scholar]
- Marks M.D. and Feldmann,K.A. (1989) Trichome development in Arabidopsis thaliana. I. T-DNA tagging of the glabrous1 gene. Plant Cell, 1, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J.D., Rerie,W.G., Foreman,D.R., Zhang,M., Galway,M.E., Marks,M.D. and Schiefelbein,J.W. (1996) The homeobox gene GLABRA 2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development, 122, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. and Gierer,A. (1974) Applications of a theory of biological pattern formation based on lateral inhibition. J. Cell Sci., 15, 321–346. [DOI] [PubMed] [Google Scholar]
- Oparka K.J. et al. (1999) Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell, 97, 743–754. [DOI] [PubMed] [Google Scholar]
- Oppenheimer D.G., Herman,P.L., Sivakumaran,S., Esch,J. and Marks,M.D. (1991) A myb gene required for leaf trichome differentiation in Arabiopsis is expressed in stipules. Cell, 67, 483–493. [DOI] [PubMed] [Google Scholar]
- Payne T., Zhang,F. and Lloyd,A. (1999) GLABROUS3 is a bHLH transcription factor that interacts with other regulators of trichome development. 10th International Conference on Arabidopsis Research, Melbourne, Australia.
- Payne C.T., Zhang,F. and Lloyd,A.M. (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics, 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann,K.A. and Marks,M.D. (1994) The glabra 2 gene encodes a homeo domain protein required for normal trichome development in Arabidobsis. Genes Dev., 8, 1388–1399. [DOI] [PubMed] [Google Scholar]
- Schnittger A., Jürgens,G. and Hülskamp,M. (1998) Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development, 125, 2283–2289. [DOI] [PubMed] [Google Scholar]
- Schnittger A., Folkers,U., Schwab,B., Jürgens,G. and Hülskamp,M. (1999) Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell, 11, 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard,M., Haecker,A., Mayer,K.F.X., Jürgens,G. and Laux,T. (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell, 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Szymanski D.B. and Marks,M.D. (1998) GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell, 10, 2047–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U. and Hartenstein,V. (1995) Neurogenic and proneural genes control cell fate specification in the Drosophila endoderm. Development, 121, 393–405. [DOI] [PubMed] [Google Scholar]
- Wada T., Tachibana,T., Shimura,Y. and Okada,K. (1997) Epidermal cell differentiation in Arabidopsis determined by a myb homolog, CPC. Science, 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Walker A.R., Davison,P.A., Bolognesi-Winfield,A.C., James,C.M., Srinivasan,N., Blundell,T.L., Esch,J.J., Marks,M.D. and Gray,J.C. (1999) The TTG1 (transparent testa, glabra1) locus which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis encodes a WD40-repeat protein. Plant Cell, 11, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]