Abstract
This study applies a new quantitative proteomics technology to the analysis of the function of the Myc oncoprotein in mammalian cells. Employing isotope-coded affinity tag (ICAT<! COMMENT -- SGML op. please reinstate sgml trademark code here -- KB>TM) reagent labeling and tandem mass spectrometry, the global pattern of protein expression in rat myc-null cells was compared with that of myc-plus cells (myc-null cells in which myc has been introduced) to generate a differential protein expression catalog. Expression differences among many functionally related proteins were identified, including reduction of proteases, induction of protein synthesis pathways and upregulation of anabolic enzymes in myc-plus cells, which are predicted to lead to increased cell mass (cell growth). In addition, reduction in the levels of adhesion molecules, actin network proteins and Rho pathway proteins were observed in myc-plus cells, leading to reduced focal adhesions and actin stress fibers as well as altered morphology. These effects are dependent on the highly conserved Myc Box II region. Our results reveal a novel cytoskeletal function for Myc and indicate the feasibility of quantitative whole-proteome analysis in mammalian cells.
Keywords: cell growth/cytoskeleton/ICAT™ reagent/Myc oncoprotein/proteomics
Introduction
Myc is one of the most frequently altered genes in human cancer (Nesbit et al., 1999; Oster et al., 2002). Through heterodimerization with Max, Myc binds E-box sequences in DNA and activates transcription. Furthermore, Myc can repress transcription of some genes through inhibition of activators. Myc induces diverse biological activities such as cell proliferation, cell growth, apoptosis, inhibition of differentiation and tumorigenesis, but whether these activities can be fully explained by modulation of target gene expression remains to be clearly established (for reviews, see Amati et al., 2001; Eisenman, 2001; Luscher, 2001; Oster et al., 2002). Indeed, several previous analyses of Myc target gene expression have suggested that Myc influences ribosome biogenesis and translation (Coller et al., 2000; Guo et al., 2000; Boon et al., 2001; Neiman et al., 2001; Schuhmacher et al., 2001), raising the possibility that important effects of Myc may be manifested not only at the transcriptional level, but also through alterations in the types and levels of specific proteins. This notion prompted us to examine changes in global protein expression following expression of Myc.
A very useful tool for the analysis of Myc function has been an immortalized Rat1 fibroblast cell line in which c-myc has been deleted by targeted homologous recombination (Mateyak et al., 1997). There is no detectable expression of N-myc or L-myc in these cells [referred to here as rat myc-null or Myc(–)] (Mateyak et al., 1997). Compared with parental Rat1 cells, the Myc(–) cells display altered morphology, lengthened G1 and G2 phases, and a decreased rate of growth. These differences in Myc(–) cells are suppressed by the reintroduction of Myc to generate Myc(+) cells (Mateyak et al., 1997, 1999).
To gain insight into the molecular basis for the differences between Myc(–) and Myc(+) cells, their global patterns of protein expression were compared using isotope-coded affinity tag (ICAT™) reagent labeling and electrospray-ionization tandem mass spectrometry (Gygi et al., 1999a; Han et al., 2001). Expression differences among many functionally related proteins were identified, which may account for the changes in morphology and proliferation following induction of Myc in many cell types. Based on the identified expression differences, we show that Myc can reduce actin stress fibers and focal adhesions in a manner dependent on the highly conserved Myc Box II region.
Results and discussion
Comparative proteomic analysis of Myc(+) and Myc(–) cells
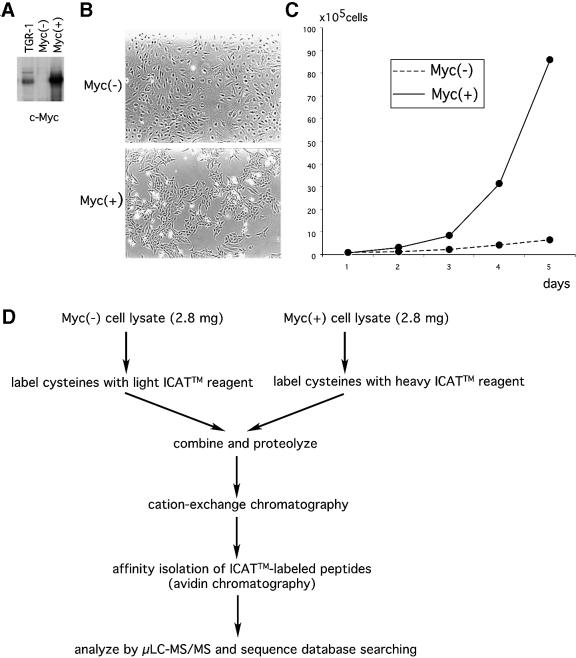
Myc(–) cells display a flattened morphology and proliferate relatively slowly [Figure 1B and C, population doubling (PD) = ∼50 h]. We derived Myc(+) cells from myc-null fibroblasts by infection with c-Myc-expressing retroviruses. Myc(+) cells are refractile, display spindle-shaped morphology and proliferate significantly faster than Myc(–) cells (Figure 1B and C, PD = ∼20 h) (Mateyak et al., 1997). Immunoblot analysis revealed ∼5-fold overexpression of c-Myc in Myc(+) cells when compared with parental TGR-1 cells from which Myc(–) cells were derived (Figure 1A). Most tumor-derived cell lines we have previously surveyed had 4- to 10-fold more c-Myc protein than normal fibroblasts (Hann and Eisenman, 1984), and this 5-fold overexpression in Myc(+) cells is within this range.
Fig. 1. Quantitative proteomic analysis of Myc(–) and Myc(+) cells. (A) Expression of the c-Myc protein in parental TGR-1, Myc(–) and Myc(+) cells. Thirty micrograms of each whole-cell lysate was analyzed by anti-c-Myc immunoblotting. (B) Morphology of Myc(–) and Myc(+) cells. (C) Proliferation of Myc(–) and Myc(+) cells. Cells were plated at a density of 1 × 105 cells per 10-cm plate and the mean cell numbers of duplicate plates were determined every 24 h. (D) Outline of ICAT™ reagent labeling procedure.
Whole-cell lysates (2.8 mg each) prepared from ∼2 × 107 cells were labeled [Myc(–), isotopically light ICAT™ reagent; Myc(+), isotopically heavy ICAT™ reagent]. The two labeled lysates were combined, proteolyzed to peptides and fractionated by cation-exchange chromatography. ICAT™ reagent-labeled peptides were purified using the biotin tag present in the reagent and analyzed by microcapillary high performance liquid chromatography-tandem mass spectrometry (µLC-MS/MS) (Figure 1D; Gygi et al., 1999a; Han et al., 2001). Pairs of isotopically light- and heavy-labeled peptides are chemically identical, are easily visualized because they essentially co-elute and display an 8 Da mass difference measured in a scanning mass spectrometer. The relative quantitation of each peptide is determined by the ratio of signal intensities of peptide pairs using the Express software tool (Han et al., 2001). The sequence identity of the proteins in the sample is determined by correlating the collision-induced dissociation spectra with the NCBI protein database using the Sequest algorithm (Eng et al., 1994). Together, Express and Sequest permit determination of the relative expression levels (fold increase or decrease) of proteins in Myc(–) versus Myc(+) cells and the identification of the proteins.
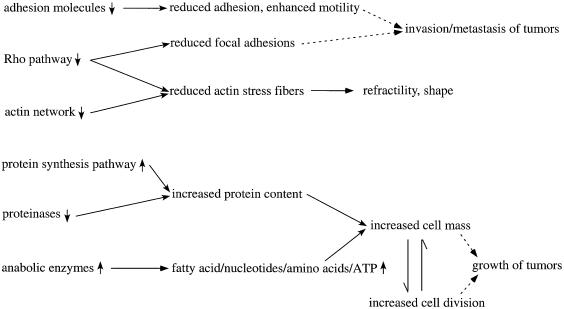
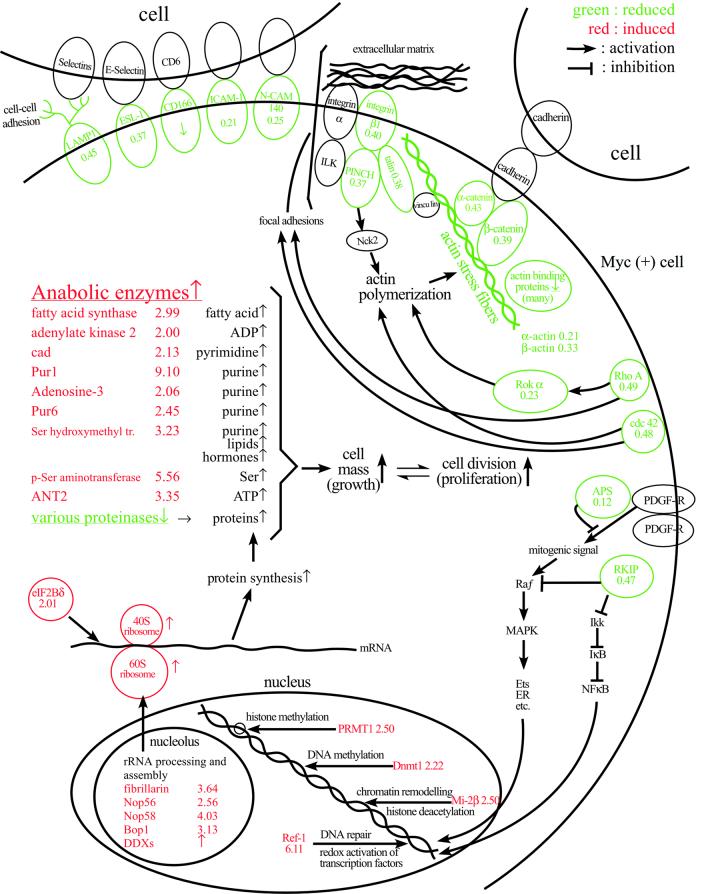
A total of 528 proteins were identified and quantified, and of these 177 displayed >2-fold expression differences. Table I shows a partial list of these proteins (the complete list is available as Supplementary data at The EMBO Journal Online). We found that many proteins displaying coordinated differences in expression between Myc(–) and Myc(+) cells are functionally related (summarized in Figures 2 and 3). These include decreased levels of adhesion molecules, actin binding proteins and Rho pathway proteins in Myc(+) cells relative to Myc(–) cells. Such changes would be expected to lead to reduced cell adhesion and altered morphology. Furthermore, an increase in cell mass would be predicted from the observed reduction in proteases, induction of protein synthesis pathways and induction of anabolic enzymes in Myc(+) cells. We note that nuclear proteins appear to be underrepresented, possibly due to difficulty of extraction or their relatively lower abundance in whole-cell lysates. Preliminary analysis suggests that chromatin-associated regulatory factors can be quantified by analyzing chromatin-enriched fractions.
Table I. Partial list of the proteins displaying >2-fold expression differences.
| Category | Protein namea | L [Myc(–)]: H [Myc(+)] ratiob | nb | Notes | Reference |
|---|---|---|---|---|---|
| Adhesion | CD166/HDL-R | L > H | |||
| ICAM-1 | 1:0.21 | 1 | |||
| N-CAM 140 | 1:0.25 | 1 | Guo et al. (2000) | ||
| integrinβ1 | 1:0.40 ± 0.07 | 4 | |||
| PINCH | 1:0.37 ± 0.10 | 3 | Integrin signaling | Guo et al. (2000) | |
| ESL-1 | 1:0.37 ± 0.09 | 16 | E-selectin ligand 1 | ||
| LAMP1 | 1:0.45 | 1 | Presents carbohydrate ligand to selectin | Guo et al. (2000) | |
| Rho pathway | Rho A | 1:0.49 ± 0.04 | 5 | Induces the formation of actin stress fibers and focal adhesions | |
| CDC42 | 1:0.48 ± 0.04 | 2 | Induces the formation of actin stress fibers and focal adhesions | ||
| Rok α | 1:0.23 ± 0.11 | 3 | Induces the formation of actin stress fibers and focal adhesions | ||
| Actin network | α-actin | 1:0.21 ± 0.04 | 3 | ||
| β-actin | 1:0.33 ± 0.06 | 11 | |||
| cofilin | 1:0.48 ± 0.08 | 11 | Actin binding | ||
| profilin | 1:0.46 ± 0.07 | 9 | Actin binding | ||
| calponin | 1:0.24 | 1 | Actin/TPM/CaM binding | ||
| transgelin2 | 1:0.22 ± 0.06 | 7 | Actin crosslinking/calponin family | ||
| t-plastin | 1:0.21 ± 0.10 | 3 | Actin bundling | ||
| sid23p | 1:0.49 ± 0.03 | 9 | Actin depolymerization factor | ||
| f-actin capping protein β | 1:0.26 ± 0.05 | 8 | |||
| Eplin α | 1:0.30 | 1 | Epithelial protein lost in neoplasia, localizes to filamentous actin and suppresses proliferation when overexpressed | ||
| ABP-280 | 1:0.18 ± 0.02 | 2 | Endothelial actin binding protein | ||
| α-actinin 1 | 1:0.34 ± 0.09 | 11 | Actin binding | ||
| α-actinin 4 | 1:0.40 ± 0.08 | 3 | Actin binding | ||
| α-1 catenin | 1:0.43 ± 0.03 | 2 | |||
| β catenin | 1:0.39 ± 0.11 | 4 | |||
| talin | 1:0.38 ± 0.08 | 16 | Involved in connections of major cytoskeletal structures to the plasma membrane | ||
| Protein synthesis | 40S/60S ribosomal proteins (20 subunits) | 1:2–3 | Coller et al. (2000); Guo et al. (2000); Boon et al. (2001); Neiman et al. (2001); Schuhmacher et al. (2001) | ||
| eIF2B δ | 1:2.01 ± 0.26 | 2 | Translation | ||
| DEAD box RNA helicase P47 | 1:3.23 ± 0.10 | 2 | RNA helicase | ||
| DDX5 | 1:2.11 ± 0.03 | 8 | RNA helicase | ||
| DDX17 | 1:2.56 ± 0.00 | 2 | RNA helicase | ||
| DDX21 | 1:5.56 ± 0.00 | 2 | RNA helicase | ||
| fibrillarin | 1:3.64 ± 0.70 | 2 | Ribosome biogenesis | Coller et al. (2000) | |
| Nop56 | 1:2.56 | 1 | rRNA processing and assembly | ||
| Nop58 | 1:4.03 ± 0.58 | 3 | rRNA processing and assembly | ||
| Bop1 (block of proliferation 1) | 1:3.13 | 1 | rRNA processing and assembly | ||
| Protein degradation | Asn endopeptidose (legumain) | 1:0.27 ± 0.00 | 3 | ||
| aminopeptidase PILS | L > H | ||||
| Niban (calpain-like) | 1:0.20 | 1 | |||
| calpain 2 | 1:0.42 ± 0.07 | 2 | |||
| calpain regulatory subunit | 1:0.31 ± 0.00 | 2 | |||
| cathepsin B | 1:0.27 ± 0.06 | 7 | |||
| cathepsin D | 1:0.26 ± 0.00 | 2 | |||
| cathepsin L | 1:0.26 | 1 | |||
| endooligopeptidase | 1:0.29 ± 0.05 | 2 | |||
| prothrombin | L > H | ||||
| Metabolism | Fatty acid synthase | 1:2.99 ± 0.62 | 6 | Schuhmacher et al. (2001) | |
| adenylate kinase 2 | 1:2.00 | 1 | AMP + ATP → 2ADP | ||
| cad* | 1:2.13 | 1 | First, second and third steps of pyrimidine biosynthesis | Schuhmacher et al. (2001) | |
| amidophosphoribosyl transferase (Pur 1) | 1:9.10 | 1 | First step of purine biosynthesis | ||
| adenosine 3 | 1:2.06 ± 0.06 | 2 | Second, third and fifth steps of purine biosynthesis | ||
| Ade 2 (Pur 6) | 1:2.45 ± 1.29 | 13 | Sixth and seventh steps of purine biosynthesis | ||
| Ser hydroxymethyl transferase | 1:3.23 | 1 | Key enzyme in the biosynthesis of purines, lipids, hormones and other components | ||
| phosphoserine aminotransferase | 1:5.56 ± 0.00 | 2 | Biosynthesis of Ser and pyridoxine | ||
| Asn tRNA synthetase | 1:2.04 | 1 | |||
| ANT2 | 1:3.35 ± 0.67 | 4 | Mitochondrial ATP translocase | Guo et al. (2000) | |
| Other Myc targets | Ref-1/Ape | 1:6.11 ± 0.79 | 3 | Redox factor-1/apurinic-apyrimidic endonuclease | Guo et al. (2000) |
| MIF | 1:2.37 ± 0.29 | 5 | Macrophage migration inhibitory factor | Guo et al. (2000) | |
| hsp60* | 1:4.05 ± 1.13 | 9 | Coller et al. (2000); Guo et al. (2000); Boon et al. (2001); Neiman et al. (2001) | ||
| Tropomyosin α | 1:0.26 ± 0.10 | 3 | Coller et al. (2000); Guo et al. (2000) |
aPreviously proposed direct Myc targets are marked with an asterisk. The proteins that were also identified by the previous microarray analyses are in bold and the relevant references are cited.
bThe ratio of light ICAT™ reagent-labeled peptide (L) [derived from Myc(–) cells] and heavy ICAT™ reagent-labeled peptide (H) [derived from Myc(+) cells] is shown together with the standard deviation based on the number of independent peptides (n) identified and quantified. For proteins that displayed obvious expression differences, but were difficult to quantify, the ratio is marked L < H or L > H.
The complete list of proteins displaying >2-fold expression differences can be found in the Supplementary data.
Fig. 2. Summary of pathways affected by Myc. Functionally related expression changes in Myc(+) cells and their expected consequences are summarized.
Fig. 3. Summary of functionally related expression changes in Myc(+) cells. The proteins reduced or induced in Myc(+) cells are shown in green or red, respectively. The numbers denote fold expression change. The arrows denote activation and the blocked lines denote inhibition.
Cell growth
Accumulating evidence suggests that Myc influences cell growth (defined as an increase in cell mass). Myc overexpression was shown to increase cell size both in Drosophila (Johnston et al., 1999) and mammalian cells (Iritani and Eisenman, 1999; Schuhmacher et al., 1999; Beier et al., 2000), and a large number of proposed Myc target genes are involved in cell growth (Grandori et al., 2000). Using global protein expression analysis, we have obtained a more comprehensive view of the mode of action of Myc on cell growth. We found that in Myc(+) cells, there is an increase in many proteins implicated in protein biosynthesis, a decrease in different proteases and an increase in several anabolic enzymes. Consistent with the observed augmentation in the levels of many ribosomal protein subunits, proteins implicated in rRNA processing and assembly (fibrillarin, Nop56, Nop58, Bop1, DDX5, DDX17 and DDX21), and a translation initiation factor (eIF2Bδ), we found that the rate of protein synthesis was increased by nearly 3-fold in Myc(+) cells when compared with Myc(–) cells (data not shown), consistent with earlier studies (Mateyak et al., 1997). The increased levels of anabolic enzymes (such as fatty acid synthase, adenylate kinase and cad) should result in increased synthesis of fatty acids, nucleotides, amino acids and ATP. Collectively, the increase in these biomolecules may account for the growth stimulatory effects of Myc (Figures 2 and 3). Selected expression differences were analyzed by immunoblotting (Figure 4A). While the immunoblotting and ICAT™ analyses correlate reasonably well, quantitative differences can most likely be attributed to the fact that immunoblotting analysis is dependent on the quality of the antibodies, effects of denaturation on the protein and linearity of the detection system.
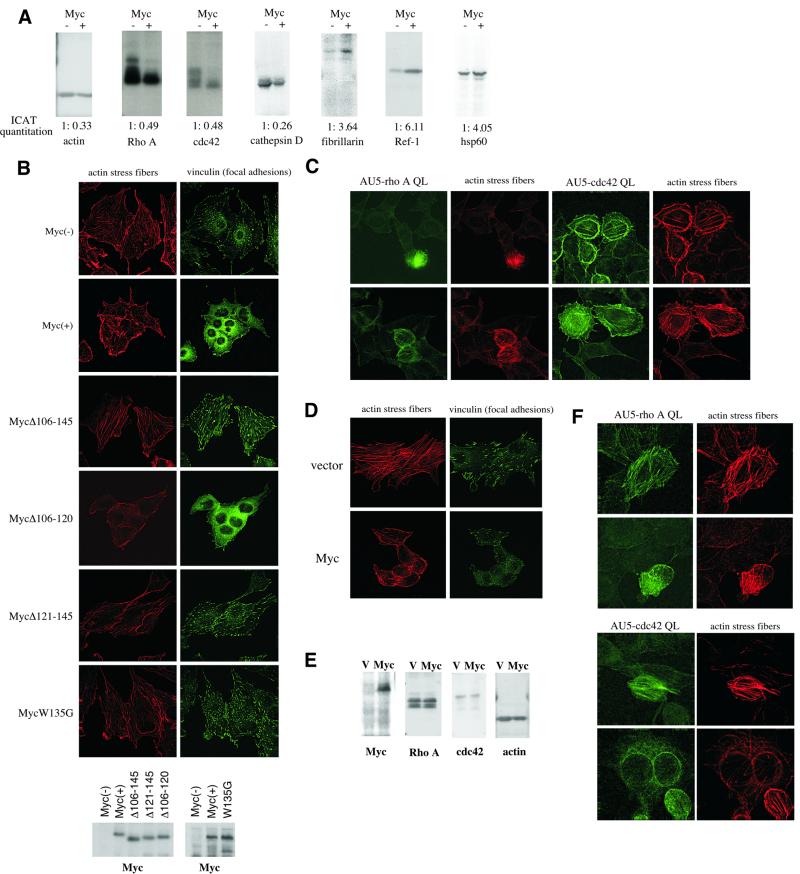
Fig. 4. Myc influences cytoskeletal architecture. (A) Immunoblot analysis. Thirty micrograms of each whole-cell lysate from Myc(–) and Myc(+) cells was analyzed. Mass spectrometric quantitation is shown at the bottom. (B) Reduced actin stress fibers and focal adhesions in Myc(+) cells. Actin stress fibers were stained with rhodamine-conjugated phalloidin (red). Focal adhesions were stained with anti-vinculin (focal adhesion marker) antibody (green). Myc(+) cells have only a few focal adhesions (vinculin is mostly located diffusely in the cytoplasm). Myc(–) cells introduced with Myc deletion mutants (Δ106–145, Δ106–120 and Δ121–145) or a point mutant (W135G) were similarly analyzed for actin stress fibers and focal adhesions. Comparable expression of the Myc proteins was confirmed by anti-c-Myc immunoblotting (bottom). (C) Restoration of actin stress fibers in Myc(+) cells by constitutively activated Rho A and cdc42. Myc(+) cells were transfected with AU5-tagged constitutively active Rho A (Rho QL) or cdc42 (cdc42 QL) and were stained with FITC-conjugated anti-AU5 antibody (green) and rhodamine-conjugated phalloidin (red). Double staining of two different fields is shown. (D) Effect of Myc on actin stress fibers and focal adhesions in NIH 3T3 fibroblasts. NIH 3T3 fibroblasts were infected with pBabepuro vector or pBabepuro-FLAG-Myc retroviruses and the puromycin-selected cells were stained for actin stress fibers and focal adhesions as in (B). (E) Effect of Myc on the expression of Rho A, cdc42 and actin in NIH 3T3 cells. Thirty micrograms of each whole-cell lysate from NIH 3T3 cells infected with pBabepuro or pBabepuro-FLAG-Myc viruses were analyzed by immunoblotting. (F) Restoration of actin stress fibers and focal adhesions in NIH 3T3/Myc cells by active Rho A and cdc42. NIH 3T3/Myc cells were transfected with AU5-Rho A QL or AU5-cdc42 QL and were stained with FITC-conjugated anti-AU5 antibody and rhodamine-conjugated phalloidin. Double staining of two different fields is shown.
Cytoskeletal proteins
Consistent with the mass spectrometric quantitations, the levels of actin, Rho A and cdc42 were significantly reduced in Myc(+) cells (Table I; Figure 4A). The Rho pathway and actin network proteins play pivotal roles in the formation of actin stress fibers and focal adhesions (Schwartz and Shattil, 2000; Ridley, 2001). Figure 4B demonstrates that whereas Myc(–) cells contain numerous widely spread actin stress fibers and many focal adhesions, Myc(+) cells in contrast possess few actin stress fibers and a small number of focal adhesions. Reduction of actin stress fibers and focal adhesions may account for the refractility and spindle-shaped morphology of Myc(+) cells when compared with the flat morphology of Myc(–) cells. To determine whether the Rho pathway is critical for Myc’s effects, we asked if hyperactivation of the Rho pathway can abrogate the cytoskeletal effects of Myc. Figure 4C shows that transfection of a constitutively active form of Rho A (Rho QL) or cdc42 (cdc42 QL) (Chiariello et al., 2001) into Myc(+) cells resulted in a significant restoration of actin stress fibers. Recently, the Rho pathway was also shown to negatively regulate cell and organism size (Sordella et al., 2002), raising the possibility that downregulation of the Rho pathway by Myc plays some role in cell growth in addition to the well established role in cytoskeletal organization.
Myc possesses a transcriptional activation domain at its N-terminus within which lies an evolutionarily conserved subdomain called Myc Box II, which is known to bind the transcriptional co-activator TRRAP (McMahon et al., 1998) and to be essential for all known biological activities of Myc (Henriksson and Luscher, 1996). To determine if Myc Box II is required for the cytoskeletal effects of Myc, Myc deletion mutants lacking Myc Box II (Δ106–145 and Δ121–145) or the sequence adjacent to Myc Box II (Δ106–120) were introduced into Myc(–) cells. As shown in Figure 4B, deletion of Myc Box II, but not the adjacent sequence, abolished Myc’s effects on actin fibers and focal adhesions. Furthermore, a point mutation in Myc Box II (W135G), which was shown to severely compromise the ability of Myc to transform rat embryo fibroblasts in conjunction with activated Ras (Brough et al., 1995), also abolished the cytoskeletal effect of Myc (Figure 4B). Thus, a region within Myc required for its known biological effects is also required for the changes in cytoskeletal proteins observed here. This suggests that these changes share a mechanism in common with other Myc functions.
To determine whether Myc can reduce actin stress fibers and focal adhesions in other cell types, we examined the effect of Myc expression in mouse NIH 3T3 fibroblasts. Here, expression of Myc increased refractility and caused marked reduction of actin stress fibers and focal adhesions (Figure 4D). Immunoblot analysis indicated that the expression of Rho A was not reduced, but the levels of cdc42 and actin were diminished in NIH 3T3 cells expressing Myc (Figure 4E). As in Myc(+) cells, constitutively active Rho A or cdc42 restored actin stress fibers in NIH 3T3/Myc cells (Figure 4F). Therefore, while the specific proteins affected by Myc expression differ between rat myc-null cells and mouse NIH 3T3 cells, the overall phenotypic consequences and the pathways affected are similar.
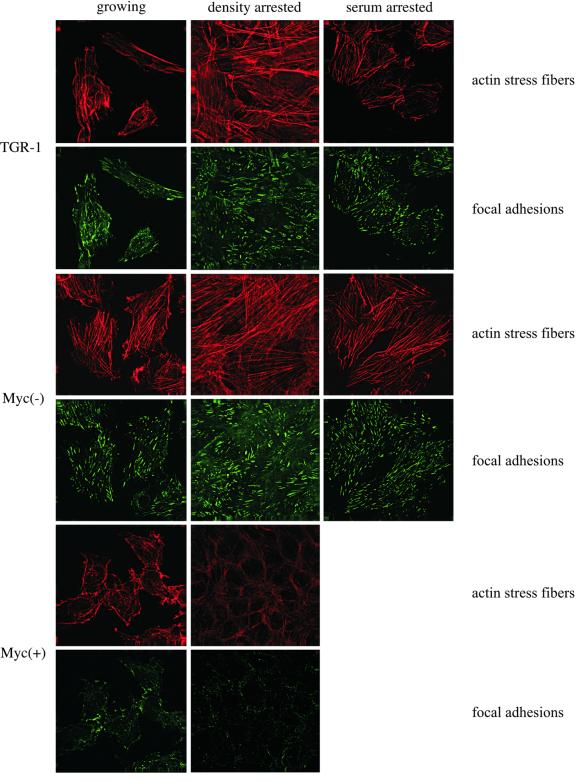
To see if the reduction of actin stress fibers and focal adhesions in Myc(+) cells is due to altered cell cycle progression of Myc(+) cells when compared with Myc(–) cells, we examined the status of actin stress fibers and focal adhesions in parental TGR-1 cells, Myc(–) cells or Myc(+) cells either growing or arrested by contact inhibition or by serum deprivation. As shown in Figure 5, TGR-1 cells and Myc(–) cells contain many actin stress fibers and focal adhesions regardless of the growth conditions. In contrast, Myc(+) cells contain a small number of actin stress fibers and focal adhesions either in a growing or in a density-arrested state. These results demonstrate that the reduction of actin stress fibers and focal adhesions in Myc(+) cells is not a secondary consequence of altered cell cycle profile. The results also indicate that reduction of actin stress fibers and focal adhesions is a function of overexpressed Myc, and may reflect a role for Myc in invasion and metastasis of tumors through reduced adhesion and enhanced motility.
Fig. 5. Effect of growth conditions on actin stress fibers and focal adhesions. Actin stress fibers and focal adhesions of the indicated cell lines either growing, density arrested for 48 h or serum starved for 48 h were stained as in Figure 4B. For Myc(+) cells the serum starvation experiment was omitted because of generalized apoptosis induced by Myc.
Correlation with RNA expression analyses
Since the advent of microarray and SAGE technologies, several laboratories have analyzed global mRNA expression changes induced by Myc (Coller et al., 2000; Guo et al., 2000; O’Hagan et al., 2000; Boon et al., 2001; Neiman et al., 2001; Schuhmacher et al., 2001). As shown in Table I, we find considerable overlap between the genes whose expression changes were identified by the previous RNA expression analyses and our quantitative proteomic analysis. Furthermore, the direction of change was the same for all such overlapping genes. However, we have also detected changes in the levels of specific proteins that were not predicted from the RNA expression studies. Discrepancies between mRNA and protein expression levels in eukaryotes have been noted before (Gygi et al., 1999b; Ideker et al., 2001). In Myc-expressing cells, translation may be generally enhanced as a consequence of stimulation of ribosome and translation-factor biosynthesis. Furthermore, specific transcripts may respond differently to the alteration of translation efficiency caused by Myc, as previously proposed for cyclin D1 protein expression (Rosenwald et al., 1993). Some Myc-induced expression changes that are likely to fall into this category, such as the reduction in actin network proteins and Rho pathway proteins, are noteworthy because Myc function has not previously been strongly linked to cytoskeletal changes. This is in contrast to the well established role of cytoplasmic oncoproteins such as Src and Ras in affecting cytoskeletal structures (Beug et al., 1978; Land et al., 1983). Although cytoskeletal changes are characteristic of transformed cells, introduction of Myc does not transform Rat1 cells (Stone et al., 1987; Blackwood et al., 1994), indicating that these cytoskeletal alterations are unlikely to be a secondary consequence of transformation.
Other protein changes
Some of the protein changes not characterized in detail in the present study may also play important roles in mediating Myc function (Figure 3): induction of Ref-1/Ape (Xanthoudakis and Curran, 1992) by Myc (Figure 4A) may alter the redox regulation of other transcription factors and protect cells from DNA damage. Reduction of APS (Yokouchi et al., 1997) and RKIP (Yeung et al., 1999) by Myc may enhance mitogenic signaling in Myc(+) cells. Induction of PRMT1 (Lin et al., 1996), Dnmt1 (Yoder et al., 1997) and Mi-2β (Zhang et al., 1998) by Myc may affect chromatin function by altering methylation of histones and DNA, and by chromatin remodeling and histone deacetylation. Interestingly, Dnmt1 has also been found to be regulated by the Fos oncoprotein (Bakin and Curran, 1999). Although their meaning is unclear at present, we also noted a number of coordinated protein changes such as reduction of annexins, and induction of molecular chaperones and hnRNPs in Myc(+) cells (see Supplementary data).
Summary
Using quantitative proteomic analysis, we have identified functionally related protein expression differences between Myc(–) and Myc(+) cells that may account for their differences in morphology and proliferation. Our data support previous global RNA expression analyses in showing that Myc influences an extraordinarily wide range of cellular functions. However, the proteomic approach has permitted us to document changes in the abundance of specific gene products that would likely not be detected at the RNA level due to post-transcriptional and -translational regulation. The hypothesis-free, holistic proteomic approach generates interesting hypotheses that may then be addressed by more specific experiments. We believe our study indicates the feasibility of quantitative whole-proteome analysis in mammalian cells and proves the power of systems biology in understanding the molecular functions of pleiotrophic regulators such as Myc.
Materials and methods
Cell culture
Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% calf serum. Rat myc-null fibroblasts [Myc(–)] were infected with pBabepuro-FLAG-Myc (human c-Myc) retroviruses and selected with puromycin to generate myc-plus [Myc(+)] cells. Myc(–) cells were similarly infected with pBabepuro-FLAG retroviruses expressing MycΔ106–145, MycΔ106–120, MycΔ121–145 or MycW135G for the analysis of the requirement of Myc Box II for the cytoskeletal function of Myc. NIH 3T3 cells were infected with pBabepuro vector or pBabepuro-FLAG-Myc retroviruses. Calcium-phosphate co-precipitation was used for transfection of AU5-rho A QL or AU5-cdc42 QL expression vectors.
Cell staining
Cells grown on coverslips were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Actin stress fibers were stained with rhodamine-conjugated phalloidin (Molecular Probes). Focal adhesions were stained with anti-vinculin antibody (Sigma). Expression of AU5-rho A QL or AU5-cdc42 QL was detected with FITC-conjugated anti-AU5 antibody (Covance).
Immunoblotting
Thirty micrograms of each whole-cell lysate made by boiling in SDS–PAGE sample buffer was analyzed. Antibodies were a gift from Dr Tom Curran (anti-Ref-1) or from Santa Cruz Biotechnology (anti-c-Myc, sc-764; anti-actin, sc-1616; anti-rho A, sc-418; anti-cdc42, sc-8401; anti-fibrillarin, sc-11335; anti-cathepsin D, sc-6486; anti-hsp60, sc-1052).
ICAT<! COMMENT -- SGML op. please reinstate sgml trademark code here -- KB>TM reagent labeling and electrospray-ionization tandem mass spectrometry
Whole-cell lysates (2.8 mg each) prepared from ∼2 × 107 cells were labeled [Myc(–), isotopically light ICAT™ reagent; Myc(+), isotopically heavy ICAT™ reagent]. The two labeled lysates were combined, proteolyzed to peptides and fractionated by cation-exchange chromatography. ICAT™ reagent-labeled peptides were purified using the biotin tag present in the reagent and analyzed by µLC-MS/MS as described previously (Gygi et al., 1999a; Han et al., 2001).
A total of 528 proteins were identified and quantified. Of these, 521 were detected in both Myc(–)- and Myc(+)-derived peptides. The remaining seven were those for which only one signal (heavy or light) was detected in the MS scan.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank T.Curran and Santa Cruz Biotechnology for antibodies and J.S.Gutkind for the AU5-rho A QL and AU5-cdc42 QL constructs. We are grateful to P.E.Neiman and C.Grandori for critical readings of the manuscript. This work was supported by grants from NIH (D.R.G., R.A. and R.N.E.), the NCI–Japanese Foundation for Cancer Research Training Program in the US–Japan Cooperative Cancer Committee (Y.S.) and by a gift from Merck and Co. to the ISB. R.N.E. is a Research Professor of the American Cancer Society.
References
- Amati B., Frank,S.R., Donjerkovic,D. and Taubert,S. (2001) Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta, 1471, M135–M145. [DOI] [PubMed] [Google Scholar]
- Bakin A.V. and Curran,T. (1999) Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science, 283, 387–390. [DOI] [PubMed] [Google Scholar]
- Beier R. et al. (2000) Induction of cyclin E-cdk2 kinase activity, E2F-dependent transcription and cell growth by Myc are genetically separable events. EMBO J., 19, 5813–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beug H., Claviez,M., Jockusch,B.M. and Graf,T. (1978) Differential expression of Rous Sarcoma virus-specific transformation parameters in enucleated cells. Cell, 14, 843–856. [DOI] [PubMed] [Google Scholar]
- Blackwood E.M., Lugo,T.G., Kretzner,L., King,M.W., Street,A.J., Witte,O.N. and Eisenman,R.N. (1994) Functional analysis of the AUG- and CUG-initiated forms of the c-Myc protein. Mol. Biol. Cell, 5, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K. et al. (2001) N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J., 20, 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough D.E., Hofmann,T.J., Ellwood,K.B., Townley,R.A. and Cole,M.D. (1995) An essential domain of the c-myc protein interacts with a nuclear factor that is also required for E1A-mediated transformation. Mol. Cell. Biol., 15, 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiariello M., Marinissen,M.J. and Gutkind,J.S. (2001) Regulation of c-myc expression by PDGF through Rho GTPases. Nat. Cell Biol., 3, 580–586. [DOI] [PubMed] [Google Scholar]
- Coller H.A., Grandori,C., Tamayo,P., Colbert,T., Lander,E.S., Eisenman,R.N. and Golub,T.R. (2000) Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl Acad. Sci. USA, 97, 3260–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman R.N. (2001) Deconstructing myc. Genes Dev., 15, 2023–2030. [DOI] [PubMed] [Google Scholar]
- Eng J., McCormack,A.L. and Yates,J.R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom., 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley,S.M., James,L.P. and Eisenman,R.N. (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol., 16, 653–699. [DOI] [PubMed] [Google Scholar]
- Guo Q.M. et al. (2000) Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res., 60, 5922–5928. [PubMed] [Google Scholar]
- Gygi S.P., Rist,B., Gerber,S.A., Turecek,F., Gelb,M.H. and Aebersold,R. (1999a) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol., 17, 994–999. [DOI] [PubMed] [Google Scholar]
- Gygi S.P., Rochon,Y., Franza,B.R. and Aebersold,R. (1999b) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol., 19, 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.K., Eng,J., Zhou,H. and Aebersold,R. (2001) Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol., 19, 946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann S.R. and Eisenman,R.N. (1984) Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol. Cell. Biol., 4, 2486–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson M. and Luscher,B. (1996) Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res., 68, 109–182. [DOI] [PubMed] [Google Scholar]
- Ideker T. et al. (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science, 292, 929–934. [DOI] [PubMed] [Google Scholar]
- Iritani B.M. and Eisenman,R.N. (1999) c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl Acad. Sci. USA, 96, 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.A., Prober,D.A., Edgar,B.A., Eisenman,R.N. and Gallant,P. (1999) Drosophila myc regulates cellular growth during development. Cell, 98, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H., Parada,L.F. and Weinberg,R.A. (1983) Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature, 304, 596–602. [DOI] [PubMed] [Google Scholar]
- Lin W.J., Gary,J.D., Yang,M.C., Clarke,S. and Herschman,H.R. (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem., 271, 15034–15044. [DOI] [PubMed] [Google Scholar]
- Luscher B. (2001) Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene, 277, 1–14. [DOI] [PubMed] [Google Scholar]
- Mateyak M.K., Obaya,A.J., Adachi,S. and Sedivy,J.M. (1997) Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ., 8, 1039–1048. [PubMed] [Google Scholar]
- Mateyak M.K., Obaya,A.J. and Sedivy,J.M. (1999) c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol. Cell. Biol., 19, 4672–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon S.B., Van Buskirk,H.A., Dugan,K.A., Copeland,T.D. and Cole,M.D. (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell, 94, 363–374. [DOI] [PubMed] [Google Scholar]
- Neiman P.E., Ruddell,A., Jasoni,C., Loring,G., Thomas,S.J., Brandvold,K.A., Lee,R., Burnside,J. and Delrow,J. (2001) Analysis of gene expression during myc oncogene-induced lymphomagenesis in the bursa of Fabricius. Proc. Natl Acad. Sci. USA, 98, 6378–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbit C.E., Tersak,J.M. and Prochownik,E.V. (1999) MYC oncogenes and human neoplastic disease. Oncogene, 18, 3004–3016. [DOI] [PubMed] [Google Scholar]
- O’Hagan R.C. et al. (2000) Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat. Genet., 24, 113–119. [DOI] [PubMed] [Google Scholar]
- Oster S.K., Ho,C.S., Soucie,E.L. and Penn,L.Z. (2002) The myc oncogene: MarvelouslY Complex. Adv. Cancer Res., 84, 81–154. [DOI] [PubMed] [Google Scholar]
- Ridley A.J. (2001) Rho family proteins: coordinating cell responses. Trends Cell Biol., 11, 471–477. [DOI] [PubMed] [Google Scholar]
- Rosenwald I.B., Lazaris-Karatzas,A., Sonenberg,N. and Schmidt,E.V. (1993) Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol. Cell. Biol., 13, 7358–7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M., Staege,M.S., Pajic,A., Polack,A., Weidle,U.H., Bornkamm,G.W., Eick,D. and Kohlhuber,F. (1999) Control of cell growth by c-Myc in the absence of cell division. Curr. Biol., 9, 1255–1258. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M. et al. (2001) The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res., 29, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.A. and Shattil,S.J. (2000) Signaling networks linking integrins and rho family GTPases. Trends Biochem. Sci., 25, 388–391. [DOI] [PubMed] [Google Scholar]
- Sordella R. et al. (2002) Modulation of CREB activity by the Rho GTPase regulates cell and organism size during mouse embryonic development. Dev. Cell, 2, 553–565. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange,T., Ramsay,G., Jakobovits,E., Bishop,J.M., Varmus,H. and Lee,W. (1987) Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol. Cell. Biol., 7, 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S. and Curran,T. (1992) Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J., 11, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung K. et al. (1999) Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature, 401, 173–177. [DOI] [PubMed] [Google Scholar]
- Yoder J.A., Soman,N.S., Verdine,G.L. and Bestor,T.H. (1997) DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol., 270, 385–395. [DOI] [PubMed] [Google Scholar]
- Yokouchi M., Suzuki,R., Masuhara,M., Komiya,S., Inoue,A. and Yoshimura,A. (1997) Cloning and characterization of APS, an adaptor molecule containing PH and SH2 domains that is tyrosine phosphorylated upon B-cell receptor stimulation. Oncogene, 15, 7–15. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]