Abstract
We have identified four different mutations causing leukocyte adhesion deficiency (LAD) in the ITGB2 gene of patients from a highly inbred population. Two were novel single-bp deletions (1497delG and 1920delG) causing frame shift and the two others were the missense mutations G284S and R593C. In our study, the G284S was a recurrent mutation while the R593C occurred de novo. We have also characterized a novel Xba1 polymorphic site located at the 5′ end of the ITGB2 locus. Family studies showed that the 1497delG mutation segregated with this marker and the intragenic AvaII polymorphic marker, suggesting the presence of a founder effect. The observation of a heterogeneous spectrum including de novo and recurrent mutations causing LAD in a highly inbred population is rather unexpected. In view of the literature published on the molecular genetics of LAD and considering the ethnic origin of the patients studied, our findings confirm the heterogeneity of the mutations causing LAD and point out potential mutational hot spots in the ITGB2 gene.
INTRODUCTION
Leukocyte adhesion deficiency (LAD) is a primary immunedeficiency inherited as a rare autosomal, recessive genetic disorder. Clinically LAD is characterized by a delayed umbilical cord separation, recurrent life-threatening infections, impaired pus formation, poor wound healing, and persistant leukocytosis. These clinical features are due to defective leukocyte adhesion to endothelial cells, absence of transmigration into inflammed tissues as well as deficient phagocytosis and chemotaxis [1]. Patients with LAD have impaired cell surface expression of the leukocytes specific adhesion molecules or β2 integrin heterodimers [2]: LFA-1 (CD11a/CD18), Mac-1 or CR3 (CD11b/CD18), p150/95 (CD11c/CD18) and probably the newly described CD11d/CD18 molecule [3], due to heterogeneous defect in the common β2(CD18) subunit. LAD clinical expression is however heterogeneous and the disease course can be severe or moderate depending on the level of β2 integrins the patient leukocytes express [4]. The assessment of the ITGB2 (CD18) gene mRNA size and expression level along with the study of the CD18 precursor protein define five different types of LAD [5]. Mutational studies of β2 integrins deficient patients of different ethnic origins have disclosed molecular genetic heterogeneity underlying the disease and helped getting new insights into β2 integrin structure-function. Several mutations scattered throughout the ITGB2 gene have been described, some of which resulted in single amino acid changes while others led to the generation of frame shifts, premature stop codons or splicing defects [6, 7, 8].
In this study, we identified the spectrum of mutations causing LAD in five patients from Tunisia (North Africa) where the population is highly inbred [9] and described two novel mutations in the ITGB2 gene leading to the severe form of the disease. Both mutations are single-bp deletions resulting in frame shifts and premature ermination of the molecule. The other mutations encountered (G284S and R593C) were already reported in patients of different ethnic origin except that the R593C substitution seems to have occurred de novo in one of our patients. These two mutations are probably truly recurrent mutations that with other already reported mutations might define hot spots mutations in the ITGB2 gene.
MATERIALS AND METHODS
Patients
The five patients studied (K, I, S, R, M) were children (one boy and four girls). They belong to four unrelated families and were all born from related healthy parents. Patients S, R, M, and I had the severe form and patient K the moderate form of the disease. They all had delayed umbilical cord separation and suffered recurrent infections.
Epstein-Barr virus transformed lymphoblasts were obtained from each patient, their parents and few available relatives. These cell lines were maintained in RPMI 1640 supplemented with 20% FCS, 2 mM glutamine and 50 μg/ml Gentamicin at 37°C in a humidified atmosphere of 5% CO2.
Flow cytometry analysis
Control and patient peripheral blood mononuclear cells (PBL) obtained by Ficoll-Hypaque centrifugation of heparinized venous blood, were incubated with the anti CD18 antibody (Immunotech, France) and the anti CD11a (Dako M782). Expression was measured using the Becton-Dickinson Vantage cytofluorometer.
Northern blot
Total cellular RNA was prepared from the patients EBV-transformed B cells as described in [10]. Northern blot was carried out following standard procedures [11]. A full length normal CD18 cDNA was used as probe [12].
Oligonucleotides used
Oligonucleotides were synthesized using a Beckman DNA synthesizer. PCR primers used to generate ITGB2 specific DNA fragments are shown in Table 1. The following primers were used for DNA sequencing:
Table 1.
PCR primers used to generate ITGB2 cDNA or genomic specific DNA fragments. Nucleotide numbering is according to position + 1 being the first nucleotide of the initiation codon ATG in the normal CD18 cDNA sequence. 5′UTR = 5′ untranslated region.
| Primer Sequence | Nucleotide position | Fragment Amplified | Size of the PCR Product | |
| 1 | S: 5′CTACTCCATGCTTGATGACCTA 3′ | 402-423 | CD18 extracellular | 1.6 kb |
| R: 5′CATCCACATAGATGAAGGTAGCG 3′ | 1973-1995 | coding region | ||
| 2 | S: 5′GAGGAAATCGGCTGGCGAAC 3′ | 742-761 | Exon 7 | 156 bp |
| R: 5′ATGTTCGCTCGTTCCTTAAG 3′ | 877-896 | |||
| 3 | S: 5′TGTGACACTGGCTACATTGGGA 3′ | 1412-1433 | Exon 12 | 245 bp |
| R: 5′AGACCTGGCCGTTGTAGCGC 3′ | 1636-1655 | |||
| 4 | S: 5′GGAGGGGGCTCTGCTTCTG 3′ | 1658-1676 | Exon 13 | 220 bp |
| R: 5′ATGTACTTGCCACAGGGTGAG 3′ | 1858-1877 | |||
| 5 | S: 5′CTCCTGCGCCGACTGCCT 3′ | 1879-1896 | Exon 14 | 220 bp |
| R: 5′CTCGGCTCTCATCCACATAG 3′ | 2079-2098 | |||
| 6 | S: 5′ACAGAGTGCATCCAGGAGCA 3′ | 1252-1271 | Exon 11 | 147 bp |
| R: 5′CTCCAAGAAGCCCTTGCCAT 3′ | 1378-1397 | |||
| 7 | S: 5′GAGTCCTTGCTCTGAAGATGACT 3′ | −834,−812 | CD18 | 1.1 kb |
| R: 5′TGCTCTTGGTGGCAGGCACT 3′ | 230-249 | 5′UTR | ||
1F 5′ CAAGGAGGAAGAGTGCCA3′(nct579-596)
2F 5′ TTCGACTACCCATCG3′(nct895-909)
3R 5′ ATGTTGTTTTCAGCCAGC3′(nct831-848)
4F 5′ GCTTACAATAAACTCTCC3′(nct1075-1092)
5R 5′ GACAAAGGACTGCTCCTG3′(nct1281-1264)
6F 5′ CAGGTGAAGGTCACGGCC3′(nct1234-1251)
7F 5′ TTGGAGTGCGGCATCTGC3′(nct1393-1410)
8R 5′ CTCAGTGGTCCTCTCGCA3′(nct1725-1742)
9F 5′ TTGAGGGCTCAGCGTGCC3′(nct1703-1720)
10R 5′ ACAGGGTGAGGGGCAGCC3′(nct1848-1866)
11F 5′ CCGGCTGCCCCTCACCCTGT3′(nct1847-1867)
Sequence-tagged site
Genomic DNA prepared from each of the patients, their parents and the available family members as well as 30 non-affected Tunisian controls (randomly selected), was subjected to amplification by using primers 6S and 6R flanking the biallelic AvaII polymorphism, as described by Mastuura and Kishi [13]. Primers 7S and 7R (see Table 1) flanking the Xba1 site located at the 5′ end of the ITGB2 locus were designed and used to amplify a CD18 specific 1.12 Kb genomic fragment. PCR was performed using 500 ng of DNA, 25 pmoles of each primer and 1.5 U Taq polymerase in a 50 μl total volume of 10 mM Tris, 50 mM KC1, 1.0 M MgCl2, and 200 μM dNTP. The reaction mixture was subjected to 95°C for 2 min followed by 30 cycles of 94°C for 1 min, 62°C for 50 s and 72°C for 1 min 30 s. An aliquot of 25 μl of the PCR product was digested with Xba1 and size separated on a 0.8% agarose gel, stained with ethidium bromide and visualized under UV illumination.
Genomic DNA amplification
Exon 7, Exon 12, Exon 13, and Exon 14
Exon 7 and Exon 12 were amplified using, respectively, Oligonucleotides 2S and 2R and 3S and 3R designed according to Weitzman et al. [14]. PCR was performed using 500 ng of DNA, 25 pmoles of each primer and 1.5 U Taq polymerase in a 50 μl total volume of 1 OmM Tris, 5 OmM KC1, 1.0 M MgC12, and 200 μM dNTP. The reaction mixture was subjected to 95°C for 2 min followed by 30 cycles of 94°C for 1 min, 62°C for 30 s, and 72°C for 1 min. Exons 13 and 14 were amplified in a single fragment using oligos n°6 and n°7 at the same conditions described above. Thermocycling was as follows: 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min 30 s.
RT/PCR and DNA Sequencing
Reverse transcription (RT) was performed on 1 μg of total RNA or 0.5 μg mRNA prepared by affinity oligodT column (Qiagen mRNA extraction kit) by first, a 5 min incubation at 65°C then transfer to ice followed by one hour incubation at 62°C in a 20 μl reaction containing 0.5 pM of OligodT, 0.2 mM of each dNTP, 50 mM Tris-HCl pH 8, 50 mM KC1, 5 mM MgCl2, RNase inhibitor (Rnasin, Boehringer Mannheim) and 1 unit of AMV reverse transcriptase. The reaction was stopped by a 5 min heat pulse at 90°C.
CD18 specific DNA fragments were amplified by PCR using 10 μl aliquot of the RT reaction, 1 μM of each primer in a 50 μl reaction containing 2 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl, and 2.5 units of Taq Polymerase (Amersham, France). Cycling was as follows: 35 cycles of 1 min at 94°C, 1 min at 62°C, and 2 min at 72°C to amplify the 1.6 Kb fragment using primers 1S and 1R. The Venta DNA polymerase (New England Biolabs) was alternatively used to generate blunt end PCR fragments that were ligated into the pCDNA3 vector. To rule out PCR artifacts and mutations introduced by the Taq polymerase. A minimum of four independent PCR reactions were performed using three different batches of enzyme and the fragments were controlled by hybridization to an ITGB2 specific cDNA probe and/or by restriction mapping.
Direct sequencing of PCR products was performed using the sequenase Kit (Amersham) and the cloned PCR products were sequenced as double stranded DNA using the same Kit as recommended by the manufacturer (Amersham).
Parentage assessment
Paternity was established in family KH by DNA Fingerprinting using VNTR Probes 33.6 and 33.15 (kindly provided by Prof A. Jeffreys, Leicester, UK) as described elsewhere [15].
RESULTS
β2 integrins expression level in LAD patients
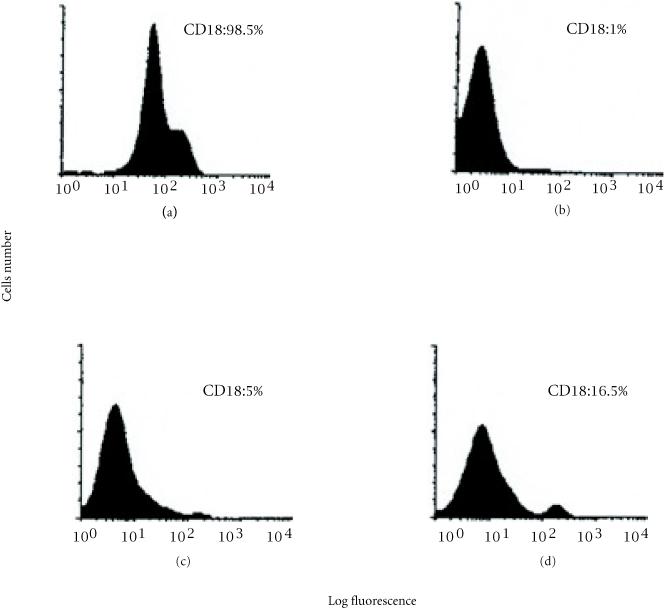
We used immunofluorescence flow cytometry to analyze the level of the CD18 subunit expression on the surface of PBMCs from five Tunisian LAD patients and their parents. As shown in Figure 1, low levels of CD18 expression (1% to 5%) were observed in the patients with the severe form of the disease. Patient K had a 16.5% CD18 expression level as compared to the normal control, which is consistent with the moderate form of LAD. All patients exhibited a normal cell surface expression of the leukocyte markers CD3, CD4, CD8, CD19, and HLA DR antigens (data not shown).
Figure 1.
FACS analysis of the β2 integrin CD18 expression at the surface of PBMCs from Tunisian patients with leukocyte adhesion deficiency (LAD). (a) control from a non-affected individual. (b), (c) and (d) represent respectively, the CD18 expression pattern obtained in patients (I, S, R), M, and K.
Identification of a novel polymorphic biallelic marker in the ITGB2 locus
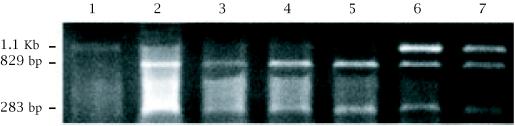
An STS assay was designed to amplify a 1.1 Kb genomic DNA a fragment that contains an Xba1 site. Digestion of this PCR product with Xba I permitted the identification of two alleles: 1.1 kb (allele A1 or Xba−) and 829 bp + 283 bp (allele A2 or Xba+) (see Figure 2. The Xba I alleles frequency was 0.5 for each allele within the same sample of non-affected Tunisian individuals.
Figure 2.
Polymorphism of the Xba1 site in a 1.1 Kb fragment generated from the 5′ region of the human ITGB2 (CD18) gene. 1.2% agarose gel electrophoresis of amplified DNA from Tunisian individuals following Xba1 restriction enzyme digestion. Lane 1: individual homozygous for allele A1 (absence of the polymorphic Xba1 site). Lanes 2, 3, 4, and 5: individuals homozygous for allele A2 (829 and 283 bp). Lanes 6 and 7: individuals heterozygous A1/A2.
Analysis of the LAD patients CD18 cDNA fragments
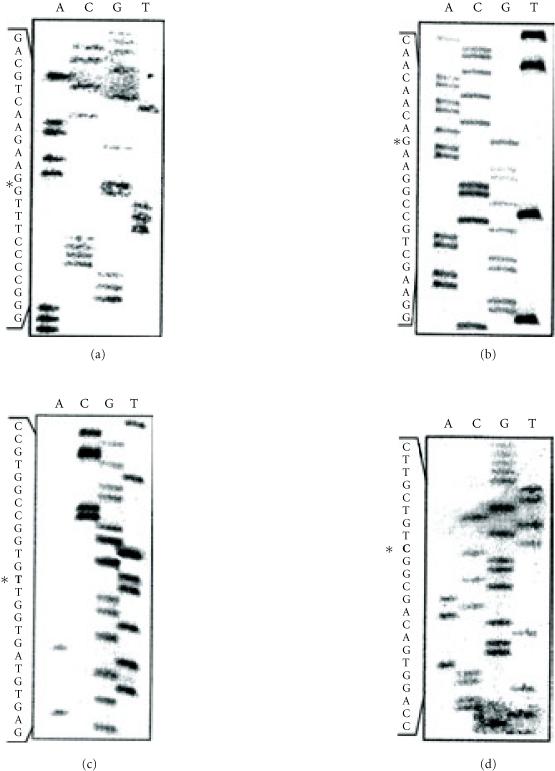
Northern blot showed that all the patients exhibited a normal sized CD18 specific mRNA (data not shown). We used the RT/PCR strategy to generate CD18 specific cDNA fragments. We have first made a 1.6 kb fragment corresponding to most of the region coding for the extracellular domain of the CD18 subunit. The majority of the mutations reported to date mapped to this region of the molecule. Sequencing of several independently generated PCR products from each patient revealed (see Figure 3) that all the patients have a deletion of the G at position 1497 (1497delG) causing a frame shift and yielding a premature stop codon, 28 residues downstream of the deletion. Patient R was homozygous for this mutation. PCR products from both parents subcloned into the pCDNA3 vector exhibited clones missing the G at position 1497 as well as normal clones. This finding is in agreement with patient R's parents being first cousins. The second mutation, identified in patient M, was another single-bp deletion: 1920delG that results in a frame shift and the occurrence of a stop codon 16 residues down stream of the deletion. The third mutation was a G to A transition affecting codon 284 (GGC/AGC) leading to the substitution of a conserved glycine by a serine (G284S). Patients S and I were compound heterozygous 1497delG/G284S. The G284S mutation was reported independently in at least one unrelated patient [16]. The fourth mutation was a single nucleotide transition in codon 593 (CGT/TGT) that leads to the replacement of the arginine normally encoded at this position by a cysteine (R593C). This mutation found in patient K, was reported as an inherited mutation in two other patients, one of whom being with certainty of different ethnic origin [16, 17]. Sequencing of multiple RT/PCR products generated from the patient's parents showed that no one of them carried this mutation. Patient K was a compound heterozygous 1497delG/R593C. The latter occurred on a different allele than the one carrying the 1497delG. It has previously been shown using CD18 transfection studies that the G284S and R593C mutations prevent β2 integrin heterodimer formation and therefore account for LAD phenotype [16, 17]. The mutations were confirmed in all the individuals studied by amplification and sequencing from the genomic DNA of the corresponding coding exon: G284S (Exon 7) 1497delG (Exon 12), 1920delG, and R593C within Exon 13 and Exon 14, respectively.
Figure 3.
Detection of mutations in Tunisian LAD patients CD18 and cDNA. The DNA sequences shown were performed on cloned PCR products obtained by RT/PCR of the patients PBMC total RNA. (a) Nucleotide sequence showing the deletion of a single G (1920delG) in the glycine codon at position 640 causing a frame shift introducing a stop codon 16 residues downstream. The mutated CD18 protein ends as following: GPFGFS RTAARRVRPCSCRTTP• (b) Detection of the G (1497delG) in the aspartic acid at position 500, causing a frame shift and a stop codon 28 residues downstream. The CD18 protein is truncated as following: SCRKFS TTTPSS AQGWRTVSAG SACATPATSPAS• (c) The C to T transition causing the substitution Arg 593 by Cyst detected in patient K is indicated by an asterisk. (d) Nucleotide sequence showing the missense mutation that affects residue 284 of the Cd18 in patients S and S changing a C into a G. FS = frame shift.
For further confirmation of the de novo character of the R593C mutation, an eventual mismatching between patient K and his biological parents was ruled out by paternity testing using DNA fingerprinting (data not shown).
Allelic association analysis
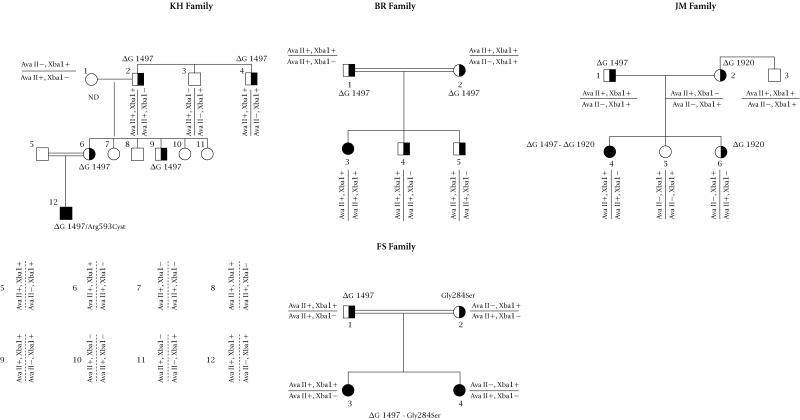
Segregation of two CD18 intragenic polymorphic markers Ava II and Xba1 with the mutations underlying LAD in our patients was studied using site tagged sequence analysis (STS). The Ava II polymorphism was studied by an STS assay described elsewhere [13]. Familial studies have shown (see Figure 4) that the 1497delG mutation, present in all the patients, segregates with the (Ava II+, Xba I+) markers. This strong linkage desequilibrium was confirmed by the study of the patients close relatives; Chi square = 41.5, p = 0.001 as calculated using the Four Fold Table method described by Feiss [18]. This allelic association was present in all the chromosomes exhibiting the 1497delG mutation and in 18 out of the 54 chromosomes studied. Interestingly, the allele frequency of the Ava II marker in the non-affected Tunisian population was identical to the one reported in the Japanese population [13] (i.e., 0.7 for the Ava II+ allele and 0.3 for the Ava II-allele).
Figure 4.
Segregation analysis of the CD18 specific markers Xba1 and Ava II with the mutations causing leukocyte adhesion deficiency in four Tunisian families. Family trees were drawn using the Pedraw software. Individuals within each nuclear family are numbered using Arabic numbers. The double bars indicate consanguineous mating. ND = not determined.
DISCUSSION
We have analyzed the molecular basis of LAD in five patients from a highly inbred population of North Africa (Tunisia) [9]. One patient had the moderate LAD phenotype and the four others had the severe one. Extensive DNA sequencing of the patients ITGB2 specific cDNA allowed the characterization of two novel mutations consisting of deletions of the nucleotides G at position 1497 and 1920 (1497delG and 1920delG). Each mutation resulted in a premature stop codon predicting the synthesis of a truncated protein.
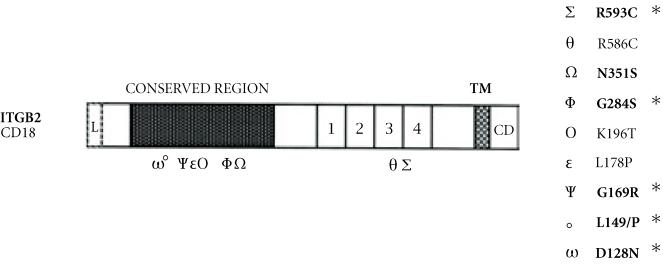
The spectrum of mutations revealed in our patients sample is peculiar in that respect that all the patients exhibited a unique mutation: 1497delG in one allele with only one patient being homozygous for this mutation. The observation of a preferential association between the 1497delG mutation and Ava II+, Xba I+ alleles and the fact that all the patients were born from consanguineous marriages, strongly suggests the presence of a founder effect at the basis of this mutation in our group of patients. It also suggests that this mutation was introduced in this population on an ancestral chromosome, tightly linked to the Ava II+, Xba+ markers. This is the first observation of an association between mutation and polymorphic markers in LAD patients. A previously reported haplotype analysis in five patients showed that LAD is not associated with a particular haplotype [19]. However, no informations on the patients ethnic origin or their degree of relatedness were available. The four other patients were compound heterozygous exhibiting two different molecular abnormalities: the 1497delG in one allele and either one of the 1920delG, G284S, or R593C mutations in their second allele. The R593C mutation was already reported in a patient of different ethnic origin as a parental allele transmitted by descent [17]. However this mutation seems to have arisen de novo in our patient. This mutation occurs at a CPG dinucleotide known to be a highly mutable dinucleotide. Another instance of a de novo in the CD18 gene associated with LAD phenotype was reported [20]. It concerned an A to G transition resulting in the substitution of the asparagine at position 351 by a serine (AAT/AGT). These observations suggest that there might be a predisposition for this type of DNA alteration in the CD18 gene. The report of a patient homozygous for the G284S mutation resulting from a G to A transition (GGC/AGC) [16] suggests that this mutation occurred independently in our patient who is a compound heterozygous G284S-1497delG. The GG dinucleotide affected by this mutation is considered to be a highly mutable dinucleotide according to the classification of the dinucleotides by virtue of their derived relative mutability established by Cooper and Krowczack [21]. Also of interest, is the observation that the mutation causing LAD in human (Japanese patients) and in bovine (Holstein Cattle) [22, 23] affects the same codon: GAC128 coding for a conserved Asparagine suggesting that the codon at this position might be more susceptible to mutagenesis. Other instances of probably recurrent mutations in the CD18 (see Figure 5) are observed and affect residues L149 (L149P: CTA/CCA) (see [16, 24, 25]), G169 (G169R: GGG/AGG) [24, 26], and R586 (R586W: CGT/TGT) [16, 20]. The high propensity to mutate of some of the ITGB2 gene codons, where de novo and recurrent mutations occurred, argues in favor of these sites being potential mutational “Hot spots.” Interestingly, most of these hot spots are clustered in a 250 amino-acid domain of the CD18 molecule that is highly conserved among the β (β1–8) integrins [12]. This region of the CD18 molecule is sought to be involved in the association of the α and β subunits. Furthermore, the elevated number and the nature of the mutations (de novo, recurrent) affecting the ITGB2 gene as well as their distribution suggest that this gene might be prone to spontaneous mutations.
Figure 5.
Localization of the missense mutations causing LAD, along the CD18 subunit: most of the mutations causing a single amino-acid substitution are clustered in the NH2 domain 250 residues that are highly conserved among the β integrins (β1–β8). A Greek letter indicates each point mutation. Asterisks (*) indicate potential hot spot mutations 1, 2, 3, 4 are the cystein rich repeats, TM = transmembrane domain, CD = cytoplasmic domain.
Under this assumption, the frequency of morbid alleles in highly inbred populations should be higher than the one observed, and screening for heterozygous carriers especially in affected families with extended pedigree should prove to be useful. Other children (five) with both the mild and the severe form of LAD were diagnosed during and after a genetic survey in the area from where the first patients originated (Fathallah and Bejaoui, unpublished data).
References
- Arnaout MA. Leukocyte adhesion molecules deficiency; its structural basis, pathophysiology, and implications for modulating the inflammatory response. Immunol Rev. 1990;114:145–180. doi: 10.1111/j.1600-065x.1990.tb00564.x. [DOI] [PubMed] [Google Scholar]
- Arnaout MA. Structure and function of the leukocyte adhesion molecules CD11/CD18. Blood. 1990;75:1037–1050. [PubMed] [Google Scholar]
- Wong EM, Davis M, Le Beau N, Springer TA. Cloning and chromosomal localization of a novel gene-encoding a human β2-integrin alpha subunit. Gene. 1996;171:291–294. doi: 10.1016/0378-1119(95)00869-1. [DOI] [PubMed] [Google Scholar]
- Anderson DC, Schmalsteig FC, Finegold MJ. The severe and moderate phenotypes of heritable Mac-1, LFA-1 deficiency: their quantitative definition and relation to leukocyte dysfunction and clinical features. J Infect Dis. 1985;152:668–688. doi: 10.1093/infdis/152.4.668. [DOI] [PubMed] [Google Scholar]
- Kishimoto TK, O'Connor K, Springer TA. Leukocyte adhesion deficiency: aberrant splicing of a conserved integrin sequence causes a moderate deficiency phenotype. Biol Chem. 1989;264(6):3588–3595. [PubMed] [Google Scholar]
- Sligh JE, Hurwitz MY, Zhu C, Anderson DC, Beaudet AL. An initiation codon mutation in CD18 in association with the moderate phenotype of leukocyte adhesion deficiency. Biol Chem. 1992;267(2):714–718. [PubMed] [Google Scholar]
- Lopez-Rodriguez C, Nueda A, Grospierre B, et al. Characterization of two new CD18 alleles causing severe leukocyte adhesion deficiency. Eur J Immunol. 1993;23:2792–2798. doi: 10.1002/eji.1830231111. [DOI] [PubMed] [Google Scholar]
- Arnaout MA, Michishta M. New Concepts in immunodeficiency diseases. CRC press; 1993. Genetic abnormalities in leukocyte adhesion molecules deficiency; pp. 191–202. [Google Scholar]
- Riou S, El Younsi C, Chaabouni H. Consanguinité dans la population du nord de la Tunisie. Tunisie Médicale. 1989;52:73–76. [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium yhiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsh EF, Maniatis T. Molecular cloning A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; New York, NY: 1989. [Google Scholar]
- Kishimoto TK, O′Connor K, Lee A, Roberts TM, Springer TA. Cloning of the β subunit of the leukocyte adhesion proteins: homology to an extracellular matrix receptor defines a novel supergene family. Cell. 1987;48:681–690. doi: 10.1016/0092-8674(87)90246-7. [DOI] [PubMed] [Google Scholar]
- Mastuura S, Kishi F. Investigation of the polymorphic Ava II site by a PCR-based assay at the human CD18 gene locus. Hum Genet. 1994;93:721. doi: 10.1007/BF00201583. [DOI] [PubMed] [Google Scholar]
- Weitzman JB, Wells CE, Wright AH, Clark PA, Law SKA. The gene organization of the human β2 integrin subunit (CD18) FEBS Lett. 1991;294(1):97–103. doi: 10.1016/0014-5793(91)81351-8. [DOI] [PubMed] [Google Scholar]
- Helminen P, Ehnholm C, Lokki ML, Jeffreys A, Peltonen L. Application of DNA “Fingerprints” to paternity determinations. Lancet. 1988;12:574–576. doi: 10.1016/s0140-6736(88)91363-3. [DOI] [PubMed] [Google Scholar]
- Wright AH, Douglas WA, Taylor GM, et al. Molecular characterization of leukocyte adhesion deficiency in six patients. Eur J Immunol. 1995;25:717–722. doi: 10.1002/eji.1830250313. [DOI] [PubMed] [Google Scholar]
- Arnaout MA, Dana N, Gupta SK, Tenen DG, Fathallah DM. Point mutations impairing cell Surface expression of the common β subunit (CD18) in a patient with leukocyte adhesion molecule (Leu-CAM) deficiency. J Clin Invest. 1990;85:977–981. doi: 10.1172/JCI114529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss J. Statistical Methods for Rates and Proportions. 2nd ed. John Wiley & Sons; New York, NY: 1980. [Google Scholar]
- Law SKA, Taylor GM. Restriction fragment length polymorphism of the gene of the human leukocyte integrin β-subunit (CD18) Immunogenetics. 1991;34:341–345. doi: 10.1007/BF00211998. [DOI] [PubMed] [Google Scholar]
- Nelson C, Rabb H, Arnaout MA. Genetic cause of leukocyte adhesion molecule deficiency: abnormal splicing and a missense mutation in a conserved region of CD18 impair cell surface expression of β2 integrins. J Biol Chem. 1992;267(5):3351–3357. [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum Genet. 1990;85:55–74. doi: 10.1007/BF00276326. [DOI] [PubMed] [Google Scholar]
- Matsuura S, Kishi F, Tsukahara M, Nunoi H, Kobayashi K, Kajii T. Leukocyte adhesion deficiency: identification of novel mutations in two Japanese patients with a severe form. Biochem Biophys Res Commun. 1992;184:1460–1467. doi: 10.1016/s0006-291x(05)80047-6. [DOI] [PubMed] [Google Scholar]
- Shuster DE, Marcus EK Jr, Gilbert RO. Identification and prevalence of a genetic defect that causes leukocyte adhesion deficiency in Holstein cattle. Proc Natl Acad Sci USA. 1992;89:9225–9229. doi: 10.1073/pnas.89.19.9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw AJ, Hibbs ML, Stacker AS, Springer TA. Distinct mutations in two patients with leukocyte adhesion deficiency and their functional correlates. J Exp Med. 1990;172:335–345. doi: 10.1084/jem.172.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back AL, Kwok WW, Hickstein DD. Identification of two molecular defects in a child with leukocyte adherence deficiency. J Biol Chem. 1992;267:5482–5487. [PubMed] [Google Scholar]
- Corbi AL, Vara A, Ursa A, Garcia Rodriguez MC, Fontan G, Sanchez-Madrid F. Molecular basis for a severe case of leukocyte adhesion deficiency. Eur J Immun. 1992;22:1877–1881. doi: 10.1002/eji.1830220730. [DOI] [PubMed] [Google Scholar]