Abstract
Hepatitis E virus (HEV) is the causative agent of hepatitis E, an acute form of viral hepatitis. The open reading frame 2 (ORF2) of HEV encodes the viral capsid protein, which can self-oligomerize into virus-like particles. To understand the domains within this protein important for capsid biogenesis, we have carried out in vitro analyses of association and folding patterns of wild type and mutant ORF2 proteins. When expressed in vitro or in transfected cells, the ORF2 protein assembled as dimers, trimers and higher order forms. While N-terminal deletions up to 111 amino acids had no effect, the deletion of amino acids 585–610 led to reduced homo-oligomerization. This deletion also resulted in aberrant folding of the protein, as determined by its sensitivity to trypsin. This study suggests that a C-terminal hydrophobic region encompassing amino acids 585–610 of the ORF2 protein might be critical for capsid biogenesis.
INTRODUCTION
The hepatitis E virus (HEV) is endemic in many resource-poor regions of the world, where it is responsible for large epidemics and rampant sporadic cases of acute viral hepatitis [1, 2, 3, 4]. In developed countries, this disease is seen primarily in travellers to areas of HEV endemicity. Though largely a self-limited infection, it results in significant morbidity and mortality, especially among pregnant women [5], in whom the disease is exacerbated by the development of fulminant liver disease. In sporadic acute hepatitis E, outside of pregnancy as well, a fraction of patients develop fulminant disease with high mortality [6]. The transmission of HEV is feco-oral, with only human-to-human transfer recognized so far [7]. However, the recent discovery of a novel virus closely related to HEV in domestic swine [8] suggests possible zoonotic reservoirs as well.
In the absence of an in vitro system for virus propagation, the biology of HEV remains poorly studied. The viral genome has been cloned from multiple geographically distinct isolates and shows a high degree of sequence conservation [9, 10, 11, 12 13, 14, 15]. The genome of HEV is a positive-stranded RNA of about 7.5 kb with short 5' and 3' noncoding regions spanning a coding region that includes three open reading frames (ORFs) [9]. The ORF1 encodes a putative nonstructural protein with domains for a viral methyltransferase, papain-like cysteine protease, RNA helicase, and an RNA-dependent RNA polymerase [16]. The ORF2 encodes the viral capsid protein (pORF2), and ORF3 expresses a small protein of unknown function (pORF3). Earlier we have shown that pORF3 is a cytoskeleton-associated phosphoprotein, which appears to be phosphorylated by the cellular mitogen-activated protein kinase [17].
The ORF2 of HEV has been expressed using various systems, including E. coli [14, 18], insect cells using baculoviruses [19], and in animal cells using transfection [20], vaccinia virus [21] and alphaviruses [22]. The expression studies in insect cells have shown that pORF2 can form virus-like particles (VLPs) that are secreted from infected cells [23]. Multiple immunodominant B-cell epitopes have been identified on pORF2 and the protein contains a highly basic N-terminal half with about 10% arginine residues, presumably to neutralize the negative charge on the RNA genome backbone. These observations support the premise that ORF2 encodes the HEV capsid protein. In earlier studies, we have observed ORF2 to express approximately 74–88 kDa protein, one form being N-glycosylated [20]. The glycosylation has been mapped to asparagine residues at positions 137, 310, and 562 [24]. We have further shown that pORF2 carries an N-terminal signal sequence that translocates it across the endoplasmic reticulum (ER) membrane; the ER also appears to be the major site of pORF2 glycosylation and accumulation [24].
The structural protein of a simple virus such as HEV should have the ability to self-assemble into a capsid structure. In this work, we have explored the homo-oligomerization potential of pORF2 using cell transfection, in vitro expression and cross-linking experiments. The results reveal that homo-oligomerization of pORF2 depends largely upon a hydrophobic region towards the C-terminus of the protein.
MATERIALS AND METHODS
Vectors and mutagenesis
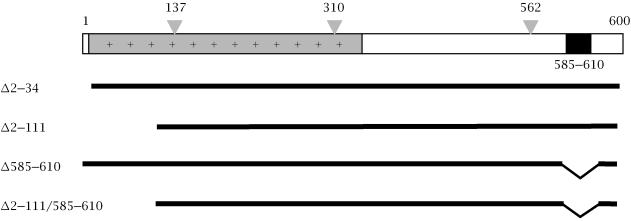
The expression vectors pSG-ORF2, pSG-ORF2[Δ2–34] and pSG-ORF2[137/310/562], expressing the wild type, signal sequence-deleted and glycosylation-null ORF2 proteins, respectively, have been described earlier [9]. The ORF2[Δ2–111] mutant was generated by digesting ORF2 at a SalI site (nucloetide 381), followed by oligonucleotide-based reconstruction. The expression vectors pSG-ORF2[Δ585–610] and pSG-ORF2[Δ2–111/Δ585–610] were generated by deleting a C-terminal hydrophobic region encompassing amino acids 585–610 by site-directed mutgenesis Figure 1.
Figure 1.
The 660 amino acid protein (pORF2) encoded by the HEV ORF2 is shown schematically, with three major regions: the extreme N-terminus, containing the putative signal sequence (open box), the highly basic N-terminal half of the protein (+) and a C-terminal hydrophobic stretch (filled box). The positions of three potential N-linked glycosylation sites are shown at amino acid residues 137, 310, 562. The deletion mutants used in this study are also shown schematically.
ORF2 expression
The transfection and labeling of cultured cells was carried out essentially as described earlier [24]. For in vitro expression, a coupled transcription-translation system (TNT; Promega, USA) was used. The reactions were quenched with 200 μg/ml cycloheximide and 200 μM methionine and the mixture incubated further at 30°C for 60 min to permit oligomerization.
Cross-linking and gradient analysis
Cell lysates were prepared in 10 mM sodium phosphate, pH 7.4 containing 100 mM NaCl, 1% Triton X-100 and a cocktail of protease inhibitors (Invitrogen, USA). To 500 μL of cell lysate, 50 μL of 10× cross-linking buffer (500 mM Tris-HCl, pH 8.0, 1 M NaCl) and 13 μL of 2% glutaraldehyde (final 0.05% ) were added. Following incubation at room temperature for 1 h, the cross-linking reactions were quenched with 50 μL of 10% SDS. For cross-linking of in vitro translation reactions, 13 μL of the reaction mixture was diluted to 500 μL with PBS, pH 7.4 and treated as above. The lysates, either directly or after cross-linking, were layered on a 3.5 mL linear gradient of 5%–25% (w/v) sucrose in PBS, pH 7.4 containing 0.1% Triton X-100. Following ultracentrifugation in a Beckman SW60 rotor for 20 h at 41,000 rpm and 4°C, 0.5 mL fractions were collected by puncturing the bottom of the tubes. Each fraction was immunoprecipitated with anti-pORF2 antibodies, subjected to electrophoresis on 3.5% or 7.5% nonreducing polyacrylamide gels and the proteins detected by fluorography. The 3.5% gels were prepared and run as described [25].
Translocation assays and proteolytic analysis
The in vitro expression of wild type and mutant ORF2 proteins was carried out as described above, in the absence or presence of microsomal membranes (Promega, USA), according to the supplier's protocol. Following protein synthesis, the reactions were chilled on ice and membrane vesicles were stabilized with 3 mM tetracaine and 10 mM CaCl2. The reaction mixtures were each divided into three aliquots, which were then treated on ice for 1 h as follows: (1) no addition, (2) 250 μg/mL trypsin, and (3) 250 μg/mL trypsin and 0.7% Triton X-100. The proteolysis was terminated by adding an equal volume of boiling SDS-PAGE loading dye buffer to the reaction mixture. The samples were boiled for 5 min and analyzed on reducing SDS-7.5% polyacrylamide gels followed by fluorography.
RESULTS
The ORF2 protein undergoes homo-oligomerization
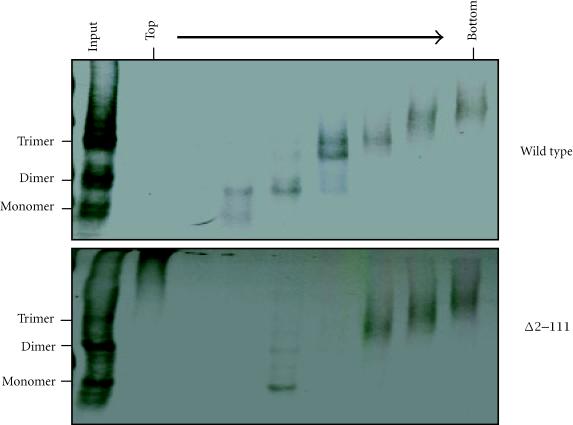
To determine if pORF2 can oligomerize, COS-1 cells were transiently transfected with vectors expressing wild type ORF2 or the N-terminal deletion mutant ORF2[Δ2–111]. After cross-linking and sucrose gradient sedimentation, the distribution of ORF2 proteins across the gradient was determined by immunoprecipitation of each fraction. Multiple species ranging from monomers to trimers were evident (Figure 2), showing that pORF2 has the ability to homo-oligomerize. For wild type pORF2, very little monomeric and some dimeric forms were evident; most of the protein was seen as trimers or higher oligomers. On deletion of the N-terminal 111 amino acids, the dimeric forms were significantly reduced. However, the trimers and higher oligomers were still observed towards the bottom of the gradient. The ORF2 protein oligomers were also analyzed after a 3 h chase following the 30 min labeling period. Though the signals were weaker, there was no change in the oligomer patterns following the chase period. This suggested that pORF2 turnover in cells was rapid and that oligomer formation took place either cotranslationally or very soon following translation. These results also indicated that up to 111 amino acids at the N-terminus were dispensable for the homo-oligomerization of pORF2.
Figure 2.
COS-1 cells transfected with wild type or Δ2–111 ORF2 expression plasmids were lysed 48 hours post-transfection, the lysates cross-linked with glutaraldehyde and separated on a sucrose gradient as described in materials and methods. The gradient fractions were immunoprecipitated with anti-pORF2 antibodies and analyzed on nonreducing 3.5% polyacrylamide gels.
In vitro homo-oligomerization of pORF2
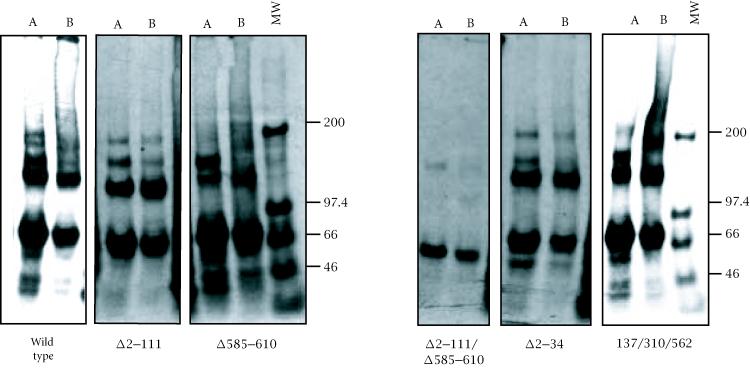
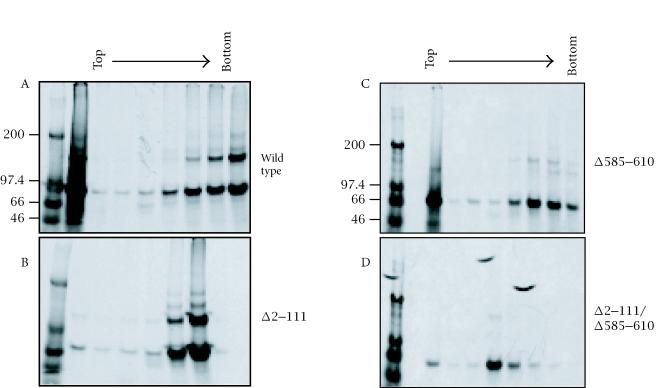
To determine if the in vitro expressed pORF2 can also oligomerize, the proteins were immunoprecipitated before or after cross-linking and analyzed on a nonreducing 3.5% gel (Figure 3). About 50% of the wild type ORF2 protein was found as dimers, with a small fraction as higher oligomers as well. The deletion of 111 N-terminal amino acids again showed no effect on this distribution. However, the deletion of amino acids 585–610 resulted in a drastic loss of pORF2 dimers. This effect was even more pronounced in the Δ2–111/Δ585–610 double mutant. The Δ2–34 and 137/310/562 mutants showed wild type patterns. To further evaluate the complexes formed, sucrose gradient sedimentation of in vitro expressed wild type and mutant ORF2 proteins was carried out (Figure 4). Following immunoprecipitation and nonreducing SDS-PAGE, two predominant forms, corresponding to monomers and dimers, were again observed. As expected, the proportion of dimers was higher in fractions towards the bottom of the gradient. When compared to the wild type protein, the Δ2–111 mutant being significantly smaller, sedimented to a lower density in the gradient. In agreement with earlier results, while the deletion of 111 N-terminal residues showed no effect on the monomer-to-dimer ratio, the deletion of amino acids 585–610 resulted in loss of the dimeric species. Taken together, these results suggested that residues 585 to 610 were required for pORF2 oligomerization.
Figure 3.
In vitro synthesized pORF2 and mutant proteins were subjected to immunoprecipitation before (A) or after (B) cross-linking. The mixtures were analyzed on a nonreducing 7.5% polyacrylamide gel. Molecular size markers are shown in kilodaltons (MW).
Figure 4.
Coupled transcription-translation reactions were carried out for the in vitro synthesis of wild type or mutant ORF2 proteins. The reaction mixtures were separated on a sucrose gradient, the fractions immunoprecipitated and analyzed on nonreducing 7.5% polyacrylamide gels. Molecular size markers are shown in kilodaltons.
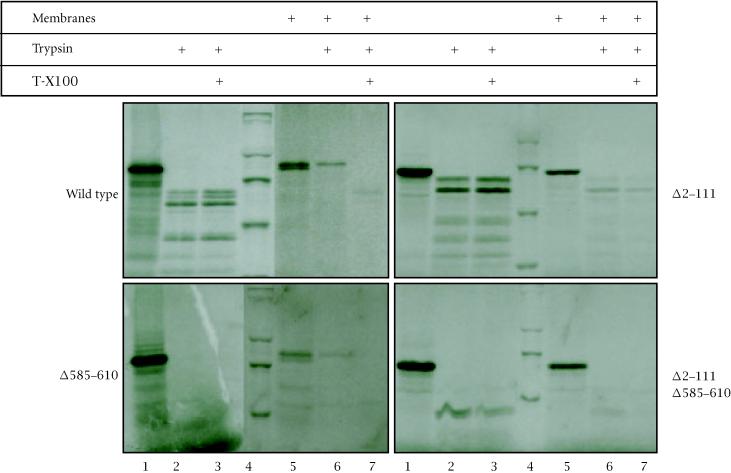
Sensitivity of pORF2 to proteolysis
Wild type and mutant ORF2 proteins were synthesized in vitro, in the absence or presence of added microsomal membranes and then subjected to trypsin digestion (Figure 5). The wild type and Δ585–610 proteins, with an intact N-terminus, were found to be protected from trypsin digestion when synthesized in the presence of membranes (lanes 5–7). The Δ2–111 and Δ2–111/Δ585–610 mutant proteins, lacking the N-terminus, were not protected under identical conditions. Further, the trypsin protection was lost when the membrane vesicles were lysed with detergent. These results were in agreement with our earlier findings of a functional membrane-translocating signal sequence at the N-terminus of pORF2. In the absence of membranes, the wild type as well as mutant proteins were susceptible to trypsin, but the polypeptide patterns generated were vastly different (lanes 1–3). The wild type protein showed at least four prominent polypeptides ranging in size from about 40–60 kDa. The Δ2–111 mutant showed two major polypeptides in the 50–60 kDa size range. The Δ585–610 mutant protein, however, was completely sensitive to trypsin, an effect seen with the Δ2–111/Δ585–610 protein as well. These results suggested that the region encompassing amino acids 585–610 also influenced the folding of pORF2.
Figure 5.
Wild type or mutant ORF2 proteins were synthesized in vitro in the absence (lanes 1–3) or presence (lanes 5–7) of canine pancreatic membranes. Subsequently, each reaction mix was divided into three parts, treated with trypsin and Triton-X100 as shown and analyzed on reducing SDS-7.5% polyacrylamide gels. Molecular size markers (lane 4) are, from top to bottom, 200 kDa, 97.4 kDa, 66 kDa, and 46 kDa.
DISCUSSION
Here we have analyzed the HEV capsid protein, pORF2, by chemical cross-linking and sucrose gradient sedimentation to show that the protein has the ability to homo-oligomerize. These oligomers form shortly after synthesis of the polypeptides and their presence in specific fractions of the gradient, indicated that they were of discrete sizes. The assembly of oligomers generally takes place post-translationally after folding of the individual subunits, which require one or more folded domains to have structurally defined surfaces for specific contacts. We have shown earlier that pORF2 is translocated across the ER membrane by virtue of an N-terminal signal sequence [24]. The in vitro translation, translocation, and trypsin sensitivity experiments reported here also supported those observations. Further, analysis of pORF2 glycosylation patterns in the presence of protein translocation inhibitors such as Brefeldin A and Monensin suggested that ER was also the major site of pORF2 accumulation [24].
It is tempting to speculate that oligomerization of pORF2 occurs inside the ER, in the intermediate compartment, or during transit to the cis-Golgi. However, when expressed in COS-1 cells, the Δ2–111 mutant, which lacks the ER translocating signal sequence, did not show any difference in its homo-oligomerization compared to the wild type protein. Further, pORF2 expressed in vitro was also found to homo-oligomerize. Though in both of these cases the ORF2 proteins displayed oligomerization, the type of interactions may be different. In the COS-1 cell-expressed protein, the major oligomeric species was a trimer, while in the protein expressed in vitro the predominant species was a dimer. Thus, while pORF2 being a capsid protein, can spontaneously oligomerize, the cellular compartment and its resident proteins are likely to play a role in the formation of productive capsids.
The analysis of pORF2 mutants clearly showed that while an N-terminal region as large as 111 amino acids was not required for its homo-oligomerization, a C-terminal hydrophobic stretch encompassing amino acids 585–610 was critical for it. When ORF2 was expressed in baculovirus-infected insect cells, multiple forms of the protein were found [19, 23, 26, 27]. In one study, a 50 kDa form secreted as virus-like particles [23], had its N-terminus at amino acid 112 of ORF2. Similar observations were made by other workers [26, 27], with one report placing the C-terminus of truncated forms at amino acid residues 578 and 607 [26]. The truncation of N-terminal 111 amino acids appeared to stabilize the oligomeric structure of pORF2. Our results on trypsin sensitivity, presented here, also support these observations. Recently, the C-terminal region encompassing amino acids 578–607 has been shown to be critical for generating neutralizing antibodies in experimentally infected chimpanzees [28]. Interestingly, this is the same region we find to be important for capsid assembly.
The sensivity of wild type and mutant ORF2 proteins has provided some clues about its folding. The ORF2 protein has 58 possible trypsin digestion sites and complete digestion with trypsin should theoretically release peptides in the range of 174 to 5556 Da. However, the generation of 40–60 kDa polypeptides on trypsin digestion of wild type or Δ2–111 mutant pORF2, suggested that a majority of trypsin sites were buried in the folded protein and were therefore not accessible to the protease. However, under identical reaction conditions, the Δ585–610 and Δ2–111/Δ585–610 mutants were completely sensitive to trypsin. This indicated that the 585–610 region, apart from its role in pORF2 oligomerization, was also critical for proper folding of the protein. It is likely that oligomerization depends upon proper folding of the protein.
We propose that the ORF2 protein has two distinct parts. The positively charged N-terminal part is important for RNA encapsidation, while the C-terminal part is required for oligomerization into a basic structural unit. While these in vitro studies do educate us about the nature of the HEV capsid protein, the details of pORF2 processing in an HEV-inefected cell and its role in capsid assembly remain to be determined. Such studies will have to wait for the availability of a viable in vitro culture system for HEV.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by internal grants from the ICGEB.
References
- Bradley DW. Enterically-transmitted non-A, non-B hepatitis. British Med Bull. 1990;46:442–461. doi: 10.1093/oxfordjournals.bmb.a072409. [DOI] [PubMed] [Google Scholar]
- Khuroo MS. Study of an epidemic of non-A, non-B hepatitis: possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am J Med. 1980;68:818–823. doi: 10.1016/0002-9343(80)90200-4. [DOI] [PubMed] [Google Scholar]
- Purcell RH, Ticehurst JR. Enterically-transmitted non-A, non-B Hepatitis: Epidemiology and Clinical Characteristics. In: Zuckerman, editor. Viral Hepatitis and Liver Disease. Alan R Inc; New York, NY: 1988. pp. 131–137. [Google Scholar]
- Wong DC, Purcell RH, Sreenivasan MA, Prasad SR, Pavri KM. Epidemic and endemic hepatitis in India: evidence for a non-A, non-B virus etiology. Lancet. 1980;2:876–879. doi: 10.1016/s0140-6736(80)92045-0. [DOI] [PubMed] [Google Scholar]
- Khuroo MS, Teli MR, Skidmore S, Sofi MA, Khuroo M. Incidence and severity of viral hepatitis in pregnancy. Am J Med. 1981;70:252–255. doi: 10.1016/0002-9343(81)90758-0. [DOI] [PubMed] [Google Scholar]
- Johnson YN, Sallie R, Fang JWS, et al. Detection of hepatitis E virus genome and gene products in two patients with fulminant hepatitis E. J Hepatol. 1995;20:605–610. doi: 10.1016/0168-8278(95)80215-0. [DOI] [PubMed] [Google Scholar]
- Krawczynski K. Hepatitis E. J Hepatol. 1993;17:932–941. [PubMed] [Google Scholar]
- Meng XJ, Purcell RH, Halbur PG, et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam AW, Smith MM, Guerra ME, et al. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:20–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Nguyen D, Fernandez J, et al. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV) Virology. 1992;191:550–558. doi: 10.1016/0042-6822(92)90230-m. [DOI] [PubMed] [Google Scholar]
- Tasarev SA, Emerson SU, Reyes GR, et al. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XZ, Lida F, Shikata T, Zhuang H, Win KM. Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986–1988) of China. Nucleic Acids Res. 1992;20:3512. doi: 10.1093/nar/20.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi SL, Purdy MA, McCaustland KA, Margolis HS, Bradley DW. The sequence of hepatitis E virus isolated directly from a single source during an outbreak in China. Virus Res. 1993;28:233–247. doi: 10.1016/0168-1702(93)90024-h. [DOI] [PubMed] [Google Scholar]
- Panda SK, Nanda SK, Zafrullah M, Ansari IH, Ozdener MH, Jameel S. An Indian strain of hepatitis E virus (HEV): Cloning, sequence, and expression of the structural region and antibody responses in sera from individuals from an area of high-level HEV endemicity. J Clin Microbiol. 1995;33:2653–2659. doi: 10.1128/jcm.33.10.2653-2659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlauder GG, Dawson GJ, Erker JC, et al. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J Gen Virol. 1998;79:447–456. doi: 10.1099/0022-1317-79-3-447. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. Computer-assisted assignment of functional domains in the non structural polyprotein of hepatitis E virus: delineation of an additional group of positive-stranded RNA plant and animal viruses. Proc Natl Acad Sci USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M, Ozdener MH, Panda SK, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Gen Virol. 1997;71:9045–9053. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Toressi J, Locarnini SA, et al. Amino-terminal epitopes are exposed when full length open reading frame 2 of hepatitis E virus is expressed in Eschericia coli, but carboxyl-terminal epitopes are masked. J Med Virol. 1997;52:289–300. doi: 10.1002/(sici)1096-9071(199707)52:3<289::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- McAtee PC, Zhang Y, Yarbough PO, Bird T, Fuerst TR. Purification of a soluble hepatitis E open reading frame 2-derived protein with unique antigenic properties. Protein Expr Purif. 1996;8:262–270. doi: 10.1006/prep.1996.0099. [DOI] [PubMed] [Google Scholar]
- Jameel S, Zafrullah M, Ozdener MH, Panda SK. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J Virol. 1996;70:207–216. doi: 10.1128/jvi.70.1.207-216.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl M, Isaacs SN, Kaur M, et al. Expression of hepatitis E virus putative structural proteins in recombinant vaccinia viruses. Clin Diagn Lab Immunol. 1994;1:253–256. doi: 10.1128/cdli.1.2.253-256.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torresi J, Meanger J, Lambert P, Li F, Locarnini SA, Anderson DA. High level expression of the capsid protein of hepatitis E virus in diverse eukaryotic cells using the Semliki Forest virus replicon. J Virol Methods. 1997;69:81–91. doi: 10.1016/s0166-0934(97)00142-0. [DOI] [PubMed] [Google Scholar]
- Li T, Yamakawa Y, Suzuki K, et al. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol. 1997;71:7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M, Ozdener MH, Kumar R, Panda SK, Jameel S. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J Virol. 1999;73:4074–4082. doi: 10.1128/jvi.73.5.4074-4082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M, McCammon K, Sambrook J. Protein folding and intracellular transport: evaluation of conformational changes in nascent exocytotic proteins. Meth Cell Biol. 1989;32:185–206. doi: 10.1016/s0091-679x(08)61171-1. [DOI] [PubMed] [Google Scholar]
- Robinson RA, Burgess WH, Emerson SU, et al. Structural characterization of recombinant hepatitis E virus ORF2 protein in baculovirus-infected insect cells. Protein Expr Purif. 1998;12:75–84. doi: 10.1006/prep.1997.0817. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McAtee P, Yarbough PO, Tam AW, Fuerst T. Expression, characterization, and immunoreactivities of a soluble hepatitis E virus putative capsid protein species expressed in insect cells. Clin Diagn Lab Immunol. 1997;4:423–428. doi: 10.1128/cdli.4.4.423-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield DJ, Glamann J, Emerson SU, Purcell RH. Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J Virol. 2000;74:5548–5555. doi: 10.1128/jvi.74.12.5548-5555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]