Abstract
Candida albicans is a major fungal pathogen of humans. It regulates its morphology in response to various environmental signals, but many of these signals are poorly defined. We show that amino acid starvation induces filamentous growth in C.albicans. Also, starvation for a single amino acid (histidine) induces CaHIS4, CaHIS7, CaARO4, CaLYS1 and CaLYS2 gene expression in a manner reminiscent of the GCN response in Saccharomyces cerevisiae. These morphogenetic and GCN-like responses are both dependent upon CaGcn4, which is a functional homologue of S.cerevisiae Gcn4. Like ScGcn4, CaGcn4 activates the transcription of amino acid biosynthetic genes via the GCRE element, and CaGcn4 confers resistance to the histidine analogue, 3-aminotriazole. CaGcn4 interacts with the Ras-cAMP pathway to promote filamentous growth, but the GCN-like response is not dependent upon morphogenetic signalling. CaGcn4 acts as a global regulator in C.albicans, co-ordinating both metabolic and morphogenetic responses to amino acid starvation.
Keywords: amino acid starvation/Candida albicans/GCN4/morphogenesis/transcription
Introduction
The growth and survival of all microbes is dependent upon their ability to detect and respond to environmental change. This is particularly true for pathogenic microbes, which must counteract host responses and adapt to changing microenvironments during the establishment and progression of infections.
Candida albicans is a major fungal pathogen of humans. It causes frequent and recurrent oral and vaginal infections, and life-threatening systemic infections in immunocompromised patients (Odds, 1988). Candida albicans virulence is enhanced by several factors, one of which is its ability to undergo reversible morphological transitions between yeast, pseudohyphal and hyphal growth forms (Odds, 1994; Lo et al., 1997). Numerous conditions promote yeast–hypha morphogenesis in vitro, including growth of C.albicans at ambient temperatures >35°C, serum, neutral pH and nutrient starvation (Odds, 1988). These presumably reflect signals that control morphogenesis in vivo (Brown and Gow, 1999). However, the exact nature of many morphogenetic signals is poorly understood, particularly those detected by this fungus in serum or following nutrient starvation.
Several signalling pathways control morphogenesis in C.albicans. These include MAPK and Ras-cAMP signalling pathways that are thought to activate filamentous growth in response to starvation and/or serum signals (Ernst, 2000; Whiteway, 2000; Brown, 2001). The MAPK pathway is dependent upon Cph1, a homologue of Saccharomyces cerevisiae Ste12 (Liu et al., 1994). The Ras-cAMP pathway requires functional Efg1, which is a member of the APSES family of basic helix–loop– helix (bHLH) transcription factors (Stoldt et al., 1997; Bockmuhl and Ernst, 2001). Under most experimental conditions, yeast–hypha morphogenesis is blocked in a C.albicans cph1/cph1, efg1/efg1 double mutant (Lo et al., 1997), indicating that the transduction of most environmental signals is dependent upon Ras-cAMP or MAPK signalling.
In addition, a Prr2/CaRim101-dependent pathway activates morphogenesis in response to ambient pH (Ramon et al., 1999; Davis et al., 2000), a Czf1 pathway promotes hyphal development when C.albicans cells are embedded in a solid matrix (Brown et al., 1999), and a Cph2 pathway whose signals are poorly defined (Lane et al., 2001a). The Prr2/CaRim101 and Cph2 pathways appear to be integrated with the Ras-cAMP pathway, and Czf1 with the MAPK pathway (Brown et al., 1999; El Barkani et al., 2000; Lane et al., 2001b). In addition, a CaTup1–CaNrg1 pathway represses filamentous growth (Braun and Johnson, 1997; Braun et al., 2001; Murad et al., 2001).
In S.cerevisiae, starvation for a single amino acid stimulates the expression of genes on all amino acid biosynthetic pathways in a phenomenon termed general amino acid control (or the GCN response) (Hinnebusch, 1988). This response is dependent upon the bZIP transcriptional activator, ScGcn4, which interacts with general control response elements (GCREs) in the promoters of target genes (Arndt and Fink, 1986; Hinnebusch, 1988). ScGCN4 expression is regulated at both translational and transcriptional levels (Hinnebusch, 1988; Albrecht et al., 1998).
We noted that some conditions which promote morphogenesis impose amino acid starvation upon C.albicans (Odds, 1988), and that CaARO3 mRNA levels increase following amino acid starvation, suggesting that a GCN-like response might exist in C.albicans (Pereira and Livi, 1995). Here we report that amino acid starvation stimulates morphogenesis in C.albicans, and confirm that a GCN-like response does exist in this fungus. We show that both responses are dependent upon CaGcn4, a functional homologue of S.cerevisiae Gcn4. Hence, CaGcn4 plays a central role in co-ordinating morphogenetic and metabolic responses to amino acid starvation in this major pathogen of humans.
Results
Amino acid starvation stimulates morphogenesis in C.albicans
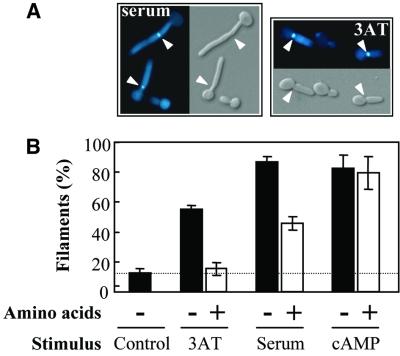
First, we tested whether amino acid starvation stimulates filamentous growth in C.albicans. Treatment with the histidine analogue, 3-aminotriazole (3AT), imposes histidine starvation in S.cerevisiae (Marton et al., 1998). Therefore, we exposed C.albicans cells to 3AT and examined their morphology. This treatment stimulated the growth of filamentous cells (Figure 1A).
Fig. 1. Amino acid starvation induces filamentous growth in C.albicans. (A) CAF-2 cells grown in YPD containing 10 mM 3AT or 10% serum for 2 h, stained with Calcofluor white, and visualized by light and fluorescence microscopy. Arrows highlight the positions of septa. (B) Percentage filament formation by C.albicans CAF2-1 (Table I) following 2 h growth at 37°C in YPD with the following additions: ± 20 mg/ml amino acids (open and closed bars, respectively); H2O control; 10 mM 3AT; 10% fetal calf serum; 10 mM dibutyryl cAMP.
The septin ring forms at the bud neck in pseudohyphal cells, but within the germ tube of a developing hypha (Sudbery, 2001). Therefore, to distinguish whether 3AT stimulated the development of pseudohyphae or true hyphae, we stained the cells with Calcofluor white, which preferentially stains septa and bud scars due to their relatively high chitin contents. Septa formed at the bud necks of filamentous cells induced by 3AT treatment, indicating that they were pseudohyphal (Figure 1A).
Many drugs exert unexpected side effects. Therefore, we tested whether the addition of amino acids to the growth medium suppresses the morphogenetic effects of 3AT. Histidine did not suppress the effect, possibly because addition of a single amino acid can induce amino acid starvation by inhibiting the biosynthesis of other amino acids (Hinnebusch, 1988). However, add ition of an amino acid mix did suppress 3AT-induced morphogenesis (Figure 1B), indicating that amino acid starvation, imposed by 3AT treatment, stimulates the formation of pseudohyphae. This confirms amino acid starvation as a morphogenetic signal in C.albicans.
Serum is considered to be a medically relevant morphogenetic stimulus, but the nature of this stimulus has not been defined (Ernst, 2000). Serum addition stimulated filamentous growth more strongly than 3AT treatment (Figure 1B). Furthermore, serum stimulated the formation of true hyphae, whereas pseudohyphae were formed in response to 3AT treatment (Figure 1A). However, amino acids partially suppressed the serum stimulus, and did so in a reproducible manner (Figure 1B), suggesting that amino acid starvation is one of several morphogenetic signals present in serum.
Candida albicans GCN4 is a functional homologue of S.cerevisiae GCN4
ScGcn4 mediates responses to amino acid starvation in S.cerevisiae (Hinnebusch, 1988). We reasoned, therefore, that a homologue of ScGcn4 might mediate amino acid starvation-induced morphogenesis in C.albicans. Hence, we isolated the C.albicans GCN4 gene.
Saccharomyces cerevisiae gcn4 mutants are unable to grow in the presence of 3AT. Therefore, we screened for CaGCN4 cDNA clones based on their ability to suppress the 3AT sensitivity of S.cerevisiae gcn4 cells. A cDNA clone was then used to isolate the complete CaGCN4 locus from a C.albicans genomic library. Both genomic and cDNA clones were sequenced. The sequence of the CaGCN4 locus revealed a major open reading frame (ORF) capable of encoding a protein of 322 amino acids. The longest cDNA sequences revealed that the CaGCN4 mRNA contains an unusually long 5′ leader sequence of at least 625 nucleotides with three upstream ORFs.
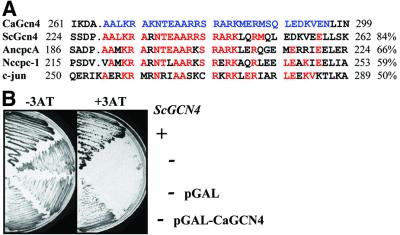
The C-terminal region of CaGcn4 displayed significant amino acid sequence similarity to the DNA-binding domains of ScGcn4 and Gcn4-like proteins from Aspergillus niger, Neurospora crassa and humans (Figure 2A). Significant sequence similarity amongst these proteins was limited to their DNA-binding regions. This is consistent with other transcription factors in C.albicans and S.cerevisiae, whose sequence conservation is restricted to their DNA-binding domains: for example, CaRfg1/ScRox1 and CaNrg1/ScNrg1 (Braun et al., 2001; Kadosh and Johnson, 2001; Murad et al., 2001). Hence, CaGcn4 belongs to a family of bZIP transcription factors that includes the global regulators ScGcn4, cpcAp, CPC1 and c-jun (Figure 2A).
Fig. 2. CaGcn4 is a functional homologue of S.cerevisiae Gcn4. (A) Alignment of the DNA-binding regions of the Gcn4-like proteins from C.albicans (CaGcn4; AAF18140), S.cerevisiae (ScGcn4; K02205), A.niger (cpcAp; X99215), N.crassa (CPC1; J03262) and humans (c-jun; J04111). The DNA-binding region in CaGcn4 is highlighted in blue, and identical residues in the other proteins are shown in red. Percentage identities to this region of CaGcn4 are shown. (B) Growth of S.cerevisiae strains on galactose plates ± 30 mM 3AT: +, GCN4 W303–1B; –, gcn4 H2036; pGAL, empty expression vector pRS-GAL1; pGAL-CaGCN4.
To test whether CaGcn4 is a functional homologue of ScGcn4, CaGCN4 was expressed in S.cerevisiae gcn4 cells under the control of the S.cerevisiae GAL1 promoter. This promoter is repressed by glucose and induced by galactose. The GAL1-CaGCN4 fusion suppressed the 3AT sensitivity of the gcn4 cells (Figure 2B). As expected, this suppression was galactose dependent (not shown), and was not observed in control cells containing the empty GAL1 expression plasmid. Therefore, CaGcn4 is a functional homologue of ScGcn4.
Inactivation of CaGcn4 blocks amino acid starvation-induced morphogenesis
Candida albicans gcn4/gcn4 mutants were constructed using the strain CAI4. The two CaGCN4 alleles in this diploid fungus were disrupted sequentially using a cassette that deleted codons 100–248 of the 322 codon ORF, thereby inactivating the DNA-binding domain. Disruption of the CaGCN4 locus was confirmed by Southern blotting. CaGCN4 mRNA was undetectable in gcn4/gcn4 cells by northern blot analysis or RT–PCR.
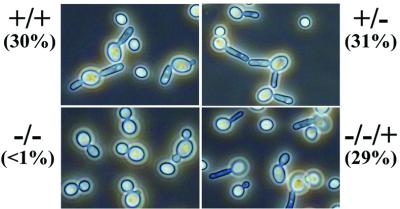
The role of CaGcn4 in amino acid starvation-induced morphogenesis was then investigated using these mutants (Table I). The morphology of cells grown in YPD and exposed to 10 mM 3AT for 2 h was examined. During the course of this experiment, both wild-type and mutant cells underwent one cell division. About 30% of cells containing a functional CaGCN4 locus formed filamentous projections, whereas the gcn4/gcn4 cells only formed buds (Figure 3). As a control, CaGCN4 was disrupted in a second C.albicans strain (CAI8; Table I). Again, inactivation of CaGcn4 blocked amino acid starvation-induced morphogenesis. To control for transformation-induced mutations, the CaGCN4 gene was reintroduced into both gcn4/gcn4 mutants. In each case, the reintroduction of CaGCN4 restored the wild-type phenotype. Clearly, CaGcn4 is required for amino acid starvation-induced filamentous growth in C.albicans.
Table I. Strains analysed in this study.
| Strain | Genotype | Source |
|---|---|---|
| S.cerevisiae | ||
| H2036 | MATa, ura3, leu2, trp1, gcn4 | Drysdale et al. (1995) |
| W303-1B | MATa, ura3, leu2, trp1, ade2, his3 | Thomas and Rothstein (1987) |
| C.albicans | ||
| SC5314 | Wild-type | Gillum et al. (1984) |
| CAF2-1 | URA3/ura3::λ imm434 | Fonzi and Irwin (1993) |
| CAI4 | ura3::λ imm434/ura3::λ imm434, | Fonzi and Irwin (1993) |
| GTC41 | ura3::λ imm434/ura3::λ imm434, GCN4/gcn4::hisG-URA3-hisG | This study |
| GTC42 | ura3::λ imm434/ura3::λ imm434, GCN4/gcn4::hisG | This study |
| GTC43 | ura3::λ imm434/ura3::λ imm434, gcn4::hisG-URA3-hisG/gcn4::hisG | This study |
| GTC44 | ura3::λ imm434/ura3::λ imm434, gcn4::hisG/gcn4::hisG | This study |
| GTC45 | ura3::λ imm434/ura3::λ imm434, gcn4::hisG/gcn4::hisG, CIp10-GCN4 | This study |
| GTC46 | ura3::λ imm434/ura3::λ imm434, gcn4::hisG/gcn4::hisG, CIp10-MET3p | This study |
| GTC47 | ura3::λ imm434/ura3::λ imm434, gcn4::hisG/gcn4::hisG, CIp10-MET3p-GCN4 | This study |
| GTC48 | ura3::λ imm434/ura3::λ imm434, gcn4::hisG/gcn4::hisG, CIp10-ACT1p | This study |
| GTC49 | ura3::λ imm434/ura3::λ imm434, gcn4::hisG/gcn4::hisG, CIp10-ACT1p-GCN4 | This study |
| CAI8 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG | Fonzi and Irwin (1993) |
| GTC81 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, GCN4/gcn4::hisG-URA3-hisG | This study |
| GTC82 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, GCN4/gcn4::hisG | This study |
| GTC83 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, gcn4::hisG-URA3-hisG/gcn4::hisG | This study |
| GTC84 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, gcn4::hisG/gcn4::hisG | This study |
| GTC85 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, gcn4::hisG/gcn4::hisG, CIp10-GCN4 | This study |
| GTC86 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, gcn4::hisG/gcn4::hisG, CIp10-MET3p | This study |
| GTC87 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, gcn4::hisG/gcn4::hisG, CIp10-MET3p-GCN4 | This study |
| JKC19 | ura3::λ imm434/ura3::λ imm434, cph1::hisG/cph1::hisG-URA3-hisG | Liu et al. (1994) |
| HLC52 | ura3::λ imm434/ura3::λ imm434, efg1::hisG /efg1::hisG-URA3-hisG | Lo et al. (1997) |
| HLC54 | ura3::λ imm434/ura3::λ imm434, cph1::hisG/cph1::hisG, efg1::hisG /efg1::hisG-URA3-hisG | Lo et al. (1997) |
| ras1-2/ras1-3 | ura3::λ imm434/ura3::λ imm434, ras1Δ::hisG/ ras1Δ::hph-URA3-hph | Feng et al. (1999) |
| RAS1/RAS1/RAS1V13 | RAS1/RAS1/ade2::Mal-RAS1V13-URA3 | Feng et al. (1999) |
| RAS1/RAS1 | RAS1/RAS1/ade2::Mal-URA3 | Feng et al. (1999) |
Fig. 3. Inactivation of CaGcn4 blocks amino acid starvation-induced morphogenesis in C.albicans. Phase contrast microscopy of C.albicans cells after 2 h growth at 37°C in YPD containing 10 mM 3AT: +/+, CAF2-1 (GCN4/GCN4); +/–, GTC41 (GCN4/gcn4); –/–, GTC43 (gcn4/gcn4); –/–/+, GTC45 (gcn4/gcn4, CIp10-GCN4) (Table I). Percentage filament formation is given in parentheses (errors <10%).
As described above, amino acid starvation is one of several morphogenetic signals present in serum. There fore, one would not expect the disruption of CaGCN4 to block serum-induced morphogenesis in C.albicans. As predicted, gcn4/gcn4 cells formed normal hyphae following serum stimulation (data not shown).
CaGcn4 activates transcription via the GCRE
ScGcn4 interacts with the GCRE (TGACTC) to activate transcription in S.cerevisiae (Arndt and Fink, 1986). CaGCN4 complements a S.cerevisiae gcn4 mutation (Figure 2B), suggesting that CaGcn4 might have a similar mode of action. Therefore, we tested whether CaGcn4 can activate transcription via the GCRE element. We used the Renilla reniformis luciferase (RrLUC) reporter, which functions in C.albicans despite the CUG codon reassignment in this fungus (Srikantha et al., 1996).
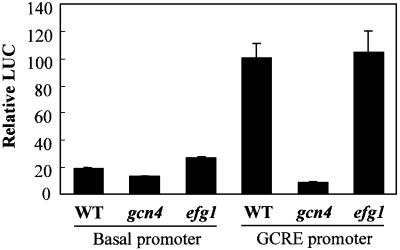
The activities of two RrLUC constructs were compared in 3AT-treated C.albicans cells. One construct contained a basal promoter upstream of the RrLUC reporter. This basal promoter contained the TATA box, RNA initiation site and 5′ leader region from the CaADH1 promoter (Bertram et al., 1996). The second promoter contained a (GCRE)5 sequence inserted upstream of this basal promoter. A single copy of each RrLUC fusion was integrated at the CaADE2 locus. Low luciferase activities were observed for the basal promoter, and these activities increased ∼5-fold following the introduction of the GCRE element. This increase was not observed in gcn4/gcn4 cells (Figure 4). Therefore, CaGcn4 activates transcription via the GCRE following amino acid starvation in C.albicans.
Fig. 4. CaGcn4 activates transcription in C.albicans via the GCRE and in an Efg1-independent fashion. Candida albicans strains were transformed with RrLUC reporter plasmids carrying either a basal promoter or a GCRE5 promoter: WT (wild type, CAI8); gcn4 null (GTC43); efg1 null (HLC52) (Table I). Relative luciferase activities following 4 h growth in SD with 20 mM 3AT.
Candida albicans displays a GCN-like response that is dependent upon CaGcn4
We tested whether C.albicans displays a GCN-like response. The 3AT sensitivity of S.cerevisiae gcn4 cells represents a classic reflection of the GCN response in budding yeast. Therefore, we examined the influence of CaGcn4 upon the 3AT sensitivity of C.albicans. All gcn4/gcn4 strains were 3AT sensitive, and the reintroduction of CaGCN4 into these strains restored 3AT resistance (Figure 5A). CaGCN4 was also reintroduced under the control of the CaMET3 promoter, which is repressed by methionine and cysteine (Care et al., 1999). As expected, the 3AT sensitivity of these gcn4/gcn4/MET3-GCN4 cells was suppressed in a methionine- and cysteine-dependent fashion (Figure 5A). Clearly, 3AT resistance in C.albicans is dependent upon CaGcn4.
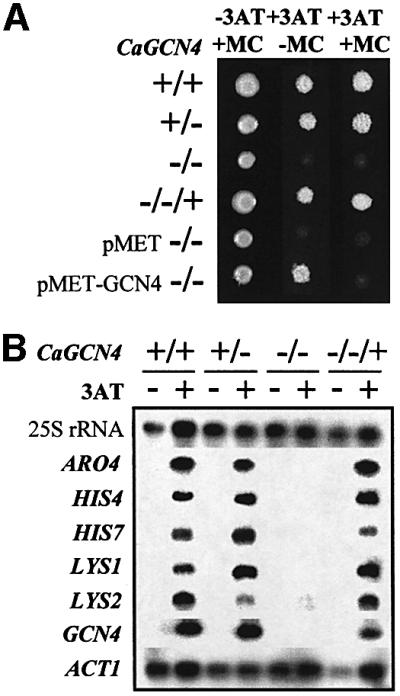
Fig. 5. Candida albicans displays a GCN-like response that is dependent upon CaGcn4. (A) Disruption of CaGCN4 confers 3AT sensitivity in C.albicans. Isogenic C.albicans strains were grown on SC plates ± 20 mM 3AT, and ± 10 mM methionine and 10 mM cysteine (+MC, –MC). Their CaGCN4 genotypes were as follows: +/+, CAF2-1; +/–, GTC41; –/–, GTC43; –/–/+, GTC45; –/– pMET, GTC46; –/– pMET-GCN4, GTC47 (Table I). (B) CaGcn4-dependent activation of amino acid biosynthetic mRNA levels in C.albicans in response to amino acid starvation. Northern blot analyses of amino acid biosynthetic mRNAs and the GCN4 mRNA were performed following 2 h of growth in YPD ± 10 mM 3AT, using 25S rRNA and ACT1 mRNA as loading controls: +/+, CAF2-1; +/–, GTC41; –/–, GTC43; –/–/+, GTC45 (Table I).
We then examined whether starvation for a single amino acid exerts a general effect upon amino acid biosynthesis in C.albicans. We selected five amino acid biosynthetic genes with GCRE sequences from the C.albicans genome sequence data generated at the Stanford DNA Sequencing and Technology Center (Tzung et al., 2001). CaARO4, CaHIS4, CaHIS7, CaLYS1 and CaLYS2 were PCR amplified and used as probes in northern blots to examine the effects of histidine starvation upon the levels of these mRNAs (Figure 5B). All five mRNAs were induced by 3AT, showing that genes on at least three different amino acid biosynthetic pathways are activated by histidine starvation. This effect was blocked in gcn4/gcn4 cells, and recovered following reintroduction of CaGCN4. There fore, histidine starvation stimulates a GCN-like response that is dependent upon CaGcn4.
Interestingly, CaGCN4 mRNA levels were elevated following exposure to 3AT, suggesting that CaGCN4 transcription is induced in response to amino acid starvation (Figure 5B). RT–PCR revealed low basal levels of CaGCN4 mRNA in non-starved cells carrying an active CaGCN4 gene (data not shown).
CaGCN4 stimulates filamentous growth in an Efg1-dependent fashion
Next we tested the effects of ectopic CaGCN4 expression upon C.albicans morphology. The CaGCN4 ORF was fused to the CaACT1 promoter and a single copy of this fusion was integrated into the C.albicans genome. The CaACT1-CaGCN4 fusion caused C.albicans to form wrinkly colonies in the absence of a morphogenetic stimulus, during growth on synthetic complete medium (Figure 6A). Microscopic analyses of cells from these colonies confirmed that this wrinkly phenotype correlated with filamentous growth. This confirmed the positive effect of CaGcn4 upon morphogenesis.
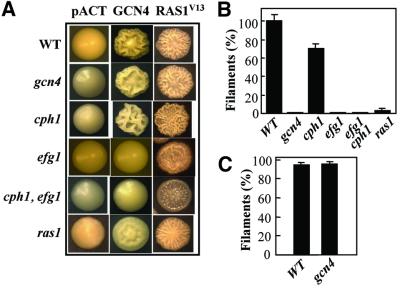
Fig. 6. Ras1–Efg1 signalling influences CaGcn4-dependent activation of morphogenesis in C.albicans. (A) Colony morphology of C.albicans strains following growth on synthetic complete medium containing amino acids and uridine at 30°C for 5 days: WT, CAF2-1; gcn4, GTC43; cph1, JKC19; efg1, HLC52; cph1, egf1, HLC54; ras1, ras1-2/ras1-3 (Table I). Cells were transformed with the following plasmids: pACT, control plasmid with the CaACT1 promoter; pACT-GCN4, plasmid carrying a CaACT1p-CaGCN4 fusion; RAS1V13, pQF145.2. (B) 3AT-induced filamentation in C.albicans signalling mutants. Filament formation was monitored after 2 h growth at 37°C in YPD containing 10 mM 3AT, and expressed as a percentage of filamentation in the wild-type strain. Strains are listed in (A). (C) CaGcn4 inactivation does not block RAS1V13-induced filamentation. Filament formation was monitored in pQF145.2 transformants after 2 h growth at 37°C in YPMal: WT, CAF2-1; gcn4, GTC43.
We then used this phenotype to test the relationship between CaGcn4 and the MAPK and Ras-cAMP pathways that activate hyphal growth in C.albicans (Lo et al., 1997; Brown and Gow, 1999). The wrinkly colonial phenotype was retained in the cph1/cph1 mutant (Figure 6A), and cph1/cph1 cells still formed filaments in response to amino acid starvation in liquid medium (Figure 6B), indicating that CaGcn4 does not act via the MAPK pathway. In contrast, efg1/efg pACT1-GCN4 cells formed smooth colonies, and 3AT-induced filamentous growth was blocked in an efg1/efg1 mutant (Figure 6). Hence, the morphological effects of CaGcn4 are dependent upon Efg1. EFG1 mRNA levels were not affected significantly by the gcn4/gcn4 mutation or by 3AT treatment (data not shown). Therefore, EFG1 expression is not regulated by CaGcn4.
The influence of CaRas1 upon CaGcn4-mediated morphogenesis was more complex. Ras1/ras1, pACT1- GCN4 colonies were less wrinkly than control colonies (RAS1/RAS1, pACT1-GCN4) (Figure 6A). However, they did contain filamentous cells, indicating that CaGcn4-mediated morphogenesis is not entirely dependent upon Ras1, at least after 5 days growth on solid medium. However, no filamentation was observed in ras1/ras1 cells after 2 h of 3AT induction in liquid medium (Figure 6B). Hence, CaRas1 inactivation might delay CaGcn4-mediated morphogenesis. Alternatively, the growth conditions might affect the degree to which CaGcn4-mediated morphogenesis depends upon CaRas1 signalling. The latter idea is consistent with previous reports indicating that ras1/ras1 cells display morphological defects that are medium dependent (Feng et al., 1999; Leberer et al., 2001). Whatever the explanation, clearly CaGcn4-mediated morphogenesis is partially dependent upon CaRas1.
Several observations indicated that CaGcn4 does not lie directly on the Ras-cAMP signalling pathway. First, in contrast to ras1/ras1 and efg1/efg1 cells (Lo et al., 1997; Feng et al., 1999), gcn4/gcn4 cells did not display morphological defects when exposed to a serum stimulus. Secondly, the addition of amino acids to the growth medium did not suppress cAMP-stimulated filament formation (Figure 1B). Thirdly, the ability of the constitutively active CaRAS1V13 allele to stimulate filamentous growth was retained in gcn4/gcn4 cells (Figure 6A and C), indicating that CaGcn4 is not required for Ras1 signalling. Nevertheless, CaGcn4 mediates its morphological effects by interacting with the Ras-cAMP pathway.
Activation of amino acid biosynthetic gene expression is not dependent upon Efg1
CaGcn4 influenced cellular morphogenesis in an Efg1-dependent fashion (Figure 6). Hence, we examined whether the GCN-like response was dependent upon Efg1.
Ectopic expression of CaGCN4, using the CaACT1 promoter fusion, increased the resistance of wild-type C.albicans cells to 3AT (Figure 7A), presumably by increasing CaGCN4 expression levels. This phenotype was not dependent upon Efg1, Ras1 or Cph1. Furthermore, the CaGcn4-dependent activation of the GCRE-RrLUC reporter was not blocked in efg1/efg1 cells (Figure 4). Therefore, the GCN-like response is not dependent upon Efg1, Ras1 or Cph1 signalling.
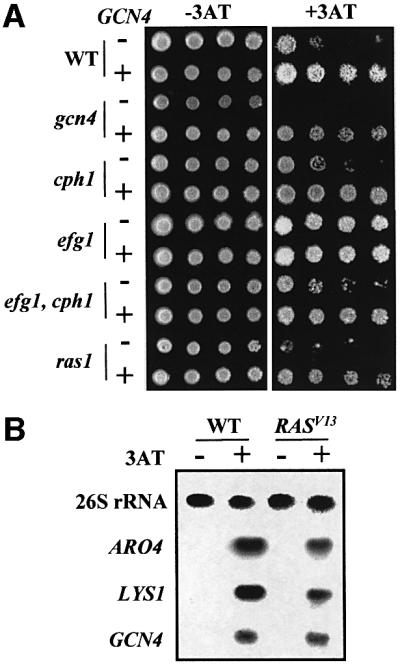
Fig. 7. The GCN-like response in C.albicans is not dependent upon Efg1, Cph1 or Ras1. (A) Constitutive activation of the GCN-like response is not blocked by inactivation of Efg1, Cph1 or Ras1. Ten-fold dilutions of C.albicans cells, transformed with pACT1 (–) or pACT-GCN4 (+), were plated on synthetic complete medium ± 10 mM 3AT: WT, CAF2-1; gcn4, GTC43; cph1, JKC19; efg1, HLC52; cph1, egf1, HLC54; ras1, ras1-2/ras1-3 (Table I). (B) Constitutive activation of Ras signalling does not activate the GCN response in C.albicans. Northern blot analysis of CaARO4, CaLYS1 and CaGCN4 mRNA levels, relative to the 26S rRNA loading control, during the growth of RAS1/RAS1 (WT) or RAS1/RAS1/RAS1V13 cells (RASV13; Table I) on synthetic complete medium containing maltose ± 10 mM 3AT.
The efg1/efg1 strain HLC52 was more resistant to 3AT than its isogenic parent CAF2-1 (Figure 7A). The basis for this was not clear, but Efg1 might exert indirect effects upon 3AT resistance by affecting cell wall integrity, and hence the uptake of the drug. Significantly, this effect was not observed in the efg1/efg1, cph1/cph1 double mutant, HLC54, indicating that the increase in 3AT resistance mediated by ectopic CaGCN4 expression is not dependent upon Efg1.
It has been reported that in S.cerevisiae, ScGCN4 can be activated by Ras signalling (Engelberg et al., 1994). Therefore, we tested whether this was the case in C.albicans. Northern blot analyses revealed that the CaRAS1V13 allele did not increase CaARO4, CaLYS1 or CaGCN4 mRNA levels in the absence of 3AT (Figure 7B). Instead, a normal GCN-like response was observed in CaRAS1V13 cells. Hence, constitutive Ras1 signalling did not stimulate the GCN-like response in C.albicans. Taken together, our data indicate that the GCN response is not mediated by MAPK or Ras-cAMP signalling.
Discussion
Effect of amino acid starvation upon C.albicans morphogenesis
Numerous treatments influence yeast–hypha morphogenesis in C.albicans (Odds, 1988) but, in most cases, the exact nature of each morphogenetic signal remains obscure. We have shown that amino acid starvation promotes pseudohyphal growth in C.albicans (Figure 1), and that this morphogenetic response is dependent upon CaGcn4 (Figures 3 and 6). Hence, amino acid starvation can now be added to the shortlist of well-defined morphogens in C.albicans.
Serum is a strong inducer of hyphal development which is commonly used to stimulate morphogenesis in vitro. However, the specific factor(s) in serum that mediates this effect has not been defined. Difficulties in purifying a specific morphogen from serum have led to the suggestion that a combination of several components mediates the strong serum stimulus (Feng et al., 1999). Our data are consistent with this idea. Amino acid starvation is not as strong a morphogenetic stimulus as serum, and it leads to the formation of pseudohyphae rather than true hyphae (Figure 1). Nevertheless, amino acids partially suppress the stimulatory effect of serum, and they do so in a reproducible manner, suggesting that amino acid starvation might comprise one component of the serum stimulus. Candida albicans gcn4/gcn4 mutants still formed hyphae following serum stimulation, showing that other serum signals are transduced by CaGcn4-independent signalling. Therefore, serum appears to contain several distinct morphogenetic stimuli, one of which might be amino acid starvation.
General amino acid control in C.albicans
We have confirmed a previous suggestion (Pereira and Livi, 1995) that C.albicans exhibits general amino acid control when challenged with amino acid starvation. Histidine starvation imposed by 3AT treatment stimulated not only histidine biosynthetic gene expression but also the expression of genes involved in lysine, tyrosine, phenylalanine and tryptophan biosynthesis (Figure 5). This GCN-like response is dependent upon CaGcn4 (Figure 5), a functional homologue of ScGcn4 (Figure 2).
Similar responses have been described in divergent fungal species. Starvation for a single amino acid leads to increased synthesis of many amino acid biosynthetic enzymes in S.cerevisiae, N.crassa and A.niger. These responses are dependent upon the related bZIP transcription factors, ScGcn4, CPC1 and cpcAp, respectively (Hinnebusch, 1988; Paluh et al., 1988; Ebbole et al., 1991; Wanke et al., 1997). Like CaGcn4 (Figure 4), these factors stimulate amino acid biosynthetic gene expression via GCRE-like promoter elements. Like CaGcn4, ScGcn4, CPC1 and cpcAp are all encoded by mRNAs with long 5′ leader sequences that contain upstream ORFs, suggesting that these mRNAs might be translationally regulated. This has been demonstrated for the ScGCN4 and cpc-1 mRNAs (Hinnebusch, 1988; Luo et al., 1995). Finally, like CaGCN4 (Figure 5), the ScGCN4, cpc-1 and cpcA genes are transcriptionally regulated in response to amino acid starvation.
Clearly, the GCN response has been retained not only in budding yeast and in saprophytic filamentous fungi, but also in an opportunistic pathogenic fungus of mammals. This might suggest that, at some point during disease establishment or progression in the host, C.albicans encounters niches where amino acids are in short supply.
CaGcn4 signalling
Our data show that C.albicans can respond to amino acid starvation in at least two ways: by stimulating cellular morphogenesis and a GCN-like response. Both are dependent upon CaGcn4. How does CaGcn4 mediate these effects?
Three main observations indicated that CaGcn4 mediates amino acid starvation responses via a pathway that is independent of Cph1, Efg1 and Ras1. First, CaGcn4 activated a GCRE-RrLUC reporter in an Efg1-dependent fashion (Figure 4). Secondly, ectopic CaGCN4 expression enhanced the 3AT resistance of C.albicans in a Cph1-, Efg1- and Ras1-independent manner (Figure 7A). Thirdly, constitutive RAS1V13 signalling did not activate the GCN-like response (Figure 7B). Therefore, neither MAPK nor Ras-cAMP signalling is implicated in the GCN response in C.albicans (Figure 8). Instead, by analogy with S.cerevisiae (Hinnebusch, 1988), the GCN-like response might be regulated by CaGcn2 signalling. A ScGcn2 homologue is present in the C.albicans genome.
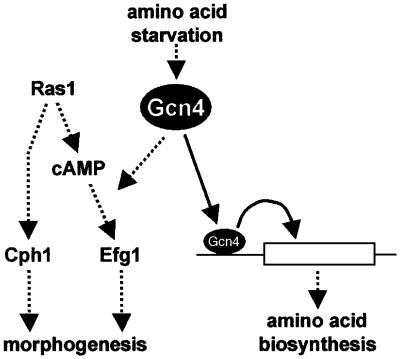
Fig. 8. Models describing the relationship between CaGcn4, morphogenetic signalling and general amino acid control in C.albicans. Amino acid starvation induces CaGcn4, which activates the transcription of amino acid biosynthetic genes in an Efg1-independent fashion via the GCRE element in their promoters. Ras1 stimulates morphogenesis via both Ras-cAMP (Efg1) and MAPK (Cph1) pathways, but does not activate amino acid biosynthetic gene expression. CaGcn4 stimulates morphogenesis by interacting with the Ras-cAMP pathway. CaGcn4 might act downstream in concert with a Ras1-dependent factor. Alternatively, CaGcn4 might act upstream of Ras1, activating the pathway specifically in response to amino acid starvation (see text). Hence, CaGcn4 coordinates morphogenetic and GCN-like responses following amino acid starvation in C.albicans.
MAPK and Ras-cAMP signalling pathways have been implicated in morphogenetic signalling in C.albicans (Brown and Gow, 1999; Ernst, 2000; Whiteway, 2000). Our data showed that the MAPK pathway is not required for CaGcn4-mediated morphogenesis (Figure 6). Instead, the Ras-cAMP pathway is implicated, because CaGcn4-mediated morphogenesis was inhibited by inactivation of Efg1 or CaRas1 (Figure 6). However, RAS1V13-induced morphogenesis was not blocked in gcn4/gcn4 cells (Figure 6). These data suggest that CaGcn4 might lie above CaRas1, activating this pathway in response to amino acid starvation but not in response to other morphogenetic signals. This would explain why ras1/ras1 and gcn4/gcn4 mutants display different morphogenetic phenotypes (Figures 3 and 6; Feng et al., 1999). However, CaGcn4 is a transcription factor (Figures 2 and 4). Therefore, it is conceivable that CaGcn4 might activate morphogenesis by interacting with a downstream component of the Ras-cAMP pathway, the activity of which is largely dependent upon CaRas1. An obvious candidate is the transcription factor Efg1 (Stoldt et al., 1997) (Figure 8).
A similar picture is emerging for pH signalling in C.albicans. Prr2/CaRim101 is a pacC-like transcription factor that mediates pH-stimulated morphogenesis in C.albicans. Expression of a constitutively active version of Prr2/CaRim101 stimulates hyphal growth and, like pACT1-GCN4-mediated morphogenesis (Figure 6), this effect is dependent upon Efg1 (El Barkani et al., 2000). Furthermore, like CaGcn4, Prr2/CaRim101 activates other responses in C.albicans in an Efg1-independent manner. Therefore, several types of environmental signal, including ambient pH and amino acid starvation, appear to converge upon Efg1 to stimulate filamentous growth in C.albicans.
Divergence in the cellular roles of Gcn4
The biochemical activities of Gcn4-like proteins seem to be conserved. CaGcn4, ScGcn4, CPC1, cpcAp and c-jun are all bZIP transcriptional activators that interact with GCRE-like sequences in the promoters of their target genes (Figure 4; Arndt and Fink, 1986; Bohmann et al., 1987; Ebbole et al., 1991; Wanke et al., 1997). However, there has been divergence with respect to their cellular roles. The fungal proteins appear to act as global regulators, in that they activate other cellular responses in addition to general amino acid control. For example, ScGcn4 has been shown to regulate >1000 S.cerevisiae genes encoding biosynthetic and organellar functions, transporters, protein kinases and transcription factors, in addition to controlling amino acid biosynthetic gene expression (Natarajan et al., 2001). However, ScGcn4 has not been implicated in pseudohyphal development (Kron, 1997; Lengeler et al., 2000; Gancedo, 2001), and transcript profiling of the GCN response has not revealed any obvious links with pseudohyphal development in S.cerevisiae (Natarajan et al., 2001). In contrast, we have demonstrated that CaGcn4 does influence C.albicans morphogenesis. Therefore, there has been some divergence in the cellular roles of Gcn4 between S.cerevisiae and C.albicans. Hence, ScGcn4 and CaGcn4 represent significant examples of regulators in S.cerevisiae and C.albicans that display conservation of biochemical function, but divergence in their cellular roles. Other examples include ScTup1–CaTup1 (Braun and Johnson, 1997), ScRox1–CaRfg1 (Kadosh and Johnson, 2001) and ScNrg1–CaNrg1 (Murad et al., 2001). Presumably, this reflects the contrasting niches of these two fungi.
Materials and methods
Strains and growth conditions
Candida albicans strains (Table I) were grown in YPD or synthetic complete medium (Murad et al., 2001). Amino acid starvation was imposed using synthetic complete medium (SC) lacking histidine and containing 3AT at various concentrations, as specified. Morphogenesis was stimulated by adding 10 mM 3AT, 10 mM dibutyryl cAMP or 10% fetal calf serum to cells growing on YPD or synthetic complete medium at 37°C. The C.albicans cells were stained with 0.1% Calcofluor White, and cell and colony morphology was analysed as described before (Murad et al., 2001).
Isolation of CaGCN4
CaGCN4 cDNAs were isolated by transforming a C.albicans library in pRSGAL1 (Wiltshire et al., 1999) into S.cerevisiae H2036 (ura3, gcn4: Table I), and screening for transformants that displayed galactose-dependent 3AT resistance. One CaGCN4 cDNA was used to isolate the complete CaGCN4 locus from a C.albicans genomic library (Smith et al., 1992) by colony hybridization. cDNA and genomic clones were sequenced to completion (accession No. AF205716).
Construction of C.albicans strains
To disrupt the CaGCN4 locus, the CaGCN4 ORF (–106 to +995) was PCR amplified using the primers 5′-CTACTGCAGAGAGAAAGTCCTGCCTC and 5′-CGAGAGGCATGCATAGTAGTAAC (PstI and SphI sites underlined), cloned into pGEM-T Easy to make pGEM-GCN4, and resequenced. A deletion from +295 to +742 was made by reverse PCR on pGEM-GCN4 using the primers 5′-CTTTAGATCTGTCCATAATCAAATC and 5′-CATAGATCTCAACCTTTACAACCGA. This introduced a BglII site at the point of the deletion (underlined). The hisG-URA3-hisG sequence (Fonzi and Irwin, 1993) was then inserted at this BglII site to create the gcn4::hisG-URA3-hisG disruption cassette. This cassette was released from the pGEM-T backbone using NotI and transformed into C.albicans (Gietz and Woods, 1998). Both CaGCN4 alleles in the strains CAI4 and CAI8 (Table I) were disrupted using two rounds of ura-blasting (Fonzi and Irwin, 1993). Disruptions were confirmed by Southern blotting and PCR diagnosis.
To reintroduce CaGCN4 into gcn4/gcn4 mutants, the CaGCN4 locus (–106 to +995) was cloned into CIp10 (Murad et al., 2000) to generate pGCN4. This plasmid was linearized with BstXI, transformed into C.albicans, and single copy integration at the gcn4 locus confirmed by Southern blotting.
To create the MET3-GCN4 fusion, the CaGCN4 ORF (–106 to +995) was released from pGEM-GCN4 using PstI and SphI, and ligated into pCaEXPa (Care et al., 1999) to make pMET3-GCN4. This plasmid was linearized with StuI, transformed into C.albicans and integrated at single copy into the RP10 locus (Murad et al., 2000).
To create the ACT1-GCN4 fusion, a double-stranded oligonucleotide containing the S.cerevisiae CYC1 terminator region (Osborne and Guarente, 1989) (top strand 5′-TCGAGATCGATGGTTACCCGTACGACGCGTGTCGACCTGCAGAAGCTTGCATGCGCTAGCGTCCCTATTTATTTTTTTATAGTTATGTTAGTATTAAGAACGTTATTTATATTTCAAATT: ScCYC1 terminator emboldened; ClaI, BstEII, SunI, MluI, SalI and PstI sites underlined with alternate sites in italics) was cloned between the SalI and MluI sites in CIp10, thereby inactivating these sites, to make pCYC1t. yEGFP (Cormack et al., 1997) was cloned between the HindIII and NheI sites of pCYC1t. Then the CaACT1 promoter region (–2 to –1019) was PCR amplified using the primers 5′-ATCGCTCGAGCTATTAAGATCACCAGCCTC and 5′-CATACCACCAAGCTTTTTGAATGATTATATTTT (XhoI and HindIII sites underlined) and cloned between the XhoI and HindIII sites to make pACT1-GFP. The PstI–SphI CaGCN4 fragment from pMET3-GCN4 was then blunt-ended and cloned between the HindIII and NheI sites in pACT1-GFP to make pACT1-GCN4. To create the control plasmid, pACT1, the yEGFP sequence was removed from pACT1-GFP by digestion with HindIII and NheI, and the plasmid was end-filled and religated. pACT1 and pACT1-GCN4 were then linearized with StuI, transformed into C.albicans, and single copy integration at the RP10 locus confirmed by Southern blotting. To introduce RAS1V13, strains were transformed with pQF145.2 and analysed on media containing 2% maltose to activate the Mal-RAS1V13 fusion (Feng et al., 1999).
Reporter assays
Renilla reniformis LUC promoter fusions were made using a derivative of pCRW3 (Srikantha et al., 1996). A basal promoter was designed, based on CaADH1 (Bertram et al., 1996). Recognizable yeast enhancer elements (S.cerevisiae Promoter Database, http://cgsigma.cshl.org/jian/; MatInspecror V2.2, http://transfac.gbf.de/TRANSFAC/) were removed from the ADH1 RNA initiation region, and convenient restriction sites added to generate the sequence 5′-ACGCGTAGCAGGTGCCACCACGGCAAAGACATTGTCTGGAACCACTGCGATCGCTAAACTGTATAAAAGGACCTATGCATGCCTGGTCTTATCTACTCCAGAATTATTTTTTTTCTATCAGTTTAACAACAACAAACGTTATTGTCATACAACAACCTGCAG (MluI and PstI sites underlined; TATA and major RNA initiation site in bold and italics). This double-stranded oligonucleotide was inserted upstream of the RrLUC ORF between unique MluI and PstI sites in pCRW3N to generate the basal RrLUC construct. The oligonucleotide 5′-CTGACTCTGAGGTGACTCGGATCCTGACTCTACTGTGACTCTATAGTGACTCT (GCREs underlined) was then introduced between the BstEII and SpeI sites of this basal construct, upstream of the TATA element, to make the GCRE-RrLUC fusion. The RrLUC plasmids were linearized with HindIII, and transformed into C.albicans CAI8 (Gietz and Woods, 1998). The plasmids were also transformed into a ura3 derivative of HLC52 by co-transformation with the URA3 plasmid, CIp10 (Murad et al., 2000). Single copy integration of RrLUC plasmids at the ade2 locus was confirmed by PCR and Southern blotting. Luciferase activities were measured in quadruplicate (Murad et al., 2001) in C.albicans transformants after 4 h growth at 30°C in SC lacking histidine and containing 20 mM 3AT. Activities are expressed relative to those of wild-type cells carrying the GCRE-RrLUC fusion. Similar data were obtained in three experiments using independent transformants.
RNA analyses
To select genes for northern analysis, homologues of S.cerevisiae amino acid biosynthetic genes were identified in the C.albicans genome sequence data generated by the Stanford DNA Sequencing and Technology Center (http://www-sequence.stanford.edu/group/candida). Genes with ≥1 GCRE elements in their promoter regions were then identified using Regulatory Sequence Analysis Tools (http://www.ucmb. ulb.ac.be/bioinformatics/rsa-tools/; van Helden et al., 2000). From this list, CaARO4, CaHIS4, CaHIS7, CaLYS1 and CaLYS2 were selected for analysis. Their coding regions were PCR amplified, restriction mapped to confirm the authenticity of the products, radiolabelled by random prime labelling, and used to probe northern blots as described previously (Swoboda et al., 1994; Brown et al., 2001).
RT–PCR was performed using standard methods (Schaller et al., 1998) with the following primers: CaACT1, 5′-GATGAAGCCCAATCCAAAAG, 5′-GGAGTTGAAAGTGGTTTGGT (677 bp product); EFG1, 5′-ACTACCATGTGGGAAGATGA, 5′-CAGGAGCATTATACTGACCA (633 bp product); and CaGCN4, 5′-GTTGATACTGTTGCTACCAA, 5′-TTTTACGAGCTCTGGATCTT (763 bp product). The intron-containing EFB1 sequence was used to control for loading and contamination with genomic DNA (Schaller et al., 1998).
Acknowledgments
Acknowledgements
This paper is dedicated to the memory of Graham Gooday, the founder member of the Aberdeen Fungal Group. We thank N.Gow, F.Odds and J.Berman for stimulating discussions, and J.Ernst, A.Hinnebusch and G.R.Fink for providing strains. We are grateful to P.Carter for help with DNA sequencing, and the Stanford DNA Sequencing and Technology Center for access to their C.albicans genome sequence data (http://www-sequence.stanford.edu/group/candida). G.T. and S.M. are supported by the UK Biotechnology and Biological Sciences Research Council (1/P10256, 98/A1/P/04001). C.W. and A.B. were supported by the European Commission (BMH4-CT96-0310, QLK2CT-2000-00795). A.B. is supported by The Wellcome Trust (055015, 063204).
References
- Albrecht G., Mosch,H.-U., Hoffman,B., Reusser,U. and Braus,G.H. (1998) Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 273, 12696–12702. [DOI] [PubMed] [Google Scholar]
- Arndt K. and Fink,G.R. (1986) GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc. Natl Acad. Sci. USA, 83, 8516–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G., Swoboda,R.K., Gow,N.A.R., Gooday,G.W. and Brown,A.J.P. (1996) Structure and regulation of the Candida albicans ADH1 gene encoding an immunogenic alcohol dehydrogenase. Yeast, 12, 115–128. [DOI] [PubMed] [Google Scholar]
- Bockmuhl D.P. and Ernst,J.F. (2001) A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics, 157, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann D., Bos,T.J., Admon,A., Nishimura,T., Vogt,P.K. and Tjian,R. (1987) Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science, 238, 1386–1392. [DOI] [PubMed] [Google Scholar]
- Braun B.R. and Johnson,A.D. (1997) Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science, 277, 105–109. [DOI] [PubMed] [Google Scholar]
- Braun B.R., Kadosh,D. and Johnson,A.D. (2001) NRG1, a repressor of filamentous growth in Candida albicans, is down-regulated during filament induction. EMBO J., 20, 4753–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.J.P. (2001) Morphogenetic signalling pathways in Candida albicans. In Calderone,R. (ed.), Candida and Candidiasis. ASM Press, Washington, DC. pp. 95–106.
- Brown A.J.P. and Gow,N.A.R. (1999) Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol., 7, 333–338. [DOI] [PubMed] [Google Scholar]
- Brown A.J.P. et al. (2001) Transcript analysis of 1003 novel yeast genes using high-throughput northern hybridizations. EMBO J., 20, 3177–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.H., Giusani,A.D., Chen,X. and Kumamoto,C.A. (1999) Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol., 34, 651–662. [DOI] [PubMed] [Google Scholar]
- Care R.S., Trevethick,J., Binley,K.M. and Sudbery,P.E. (1999) The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol., 34, 792–798. [DOI] [PubMed] [Google Scholar]
- Cormack B., Bertram,G., Egerton,M., Gow,N.A.R., Falkow,S. and Brown,A.J.P. (1997) Yeast enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology, 143, 303–311. [DOI] [PubMed] [Google Scholar]
- Davis D., Wilson,R.B. and Mitchell,A.P. (2000) RIM101-dependent and -independent pathways govern pH responses in Candida albicans.Mol. Cell. Biol., 20, 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale C.M., Dueñas,E., Jackson,B.M., Reusser,U., Braus,G.H. and Hinnebusch,A.G. (1995) The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol. Cell. Biol., 15, 1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D.J., Paluh,J.L., Plamann,M., Sachs,M.S. and Yanofsky,C. (1991) cpc-1, the general regulatory gene for genes of amino acid biosynthesis in Neurospora crassa, is differentially expressed during the asexual life cycle. Mol. Cell. Biol., 11, 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Barkani A., Kurzai,O., Fonzi,W.A., Ramon,A., Porta,A., Frosch,M. and Muhlschlegel,F.A. (2000) Dominant active alleles of RIM101 (PRR2) bypass the pH restriction on filamentation of Candida albicans. Mol. Cell. Biol., 20, 4635–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg D., Klein,C., Martinetto,H., Struhl,K. and Karin,M. (1994) The UV response involving the Ras signaling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell, 77, 381–390. [DOI] [PubMed] [Google Scholar]
- Ernst J.F. (2000) Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology, 146, 1763–1774. [DOI] [PubMed] [Google Scholar]
- Feng Q., Summers,E., Guo,B. and Fink,G. (1999) Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol., 181, 6339–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi W.A. and Irwin,M.Y. (1993) Isogenic strain construction and gene mapping in Candida albicans.Genetics, 134, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J.M. (2001) Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev., 25, 107–123. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Woods,R.A. (1998) Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method. Methods Microbiol., 26, 53–66. [Google Scholar]
- Gillum A.M., Tsay,E.Y. and Kirsch,D.R. (1984) Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S.cerevisiae ura3 and E.coli pyrF mutations. Mol. Gen. Genet., 198, 179–182. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. (1988) Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev., 52, 248–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D. and Johnson,A.D. (2001) Rfg1, a protein related to the S.cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in C.albicans. Mol. Cell. Biol., 21, 2496–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C.A., Redd,M.J., Schultz,J., Carlson,M. and Johnson,A.D. (1992) Ssn6–Tup1 is a general repressor of transcription in yeast. Cell, 68, 709–719. [DOI] [PubMed] [Google Scholar]
- Kron S.J. (1997) Filamentous growth in budding yeast. Trends Microbiol., 5, 450–454. [DOI] [PubMed] [Google Scholar]
- Lane S., Zhou,S., Pan,T., Dai,Q. and Liu,H. (2001a) The basic helix–loop–helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via Tec1. Mol. Cell. Biol., 21, 6418–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S., Birse,C., Zhou,S., Matson,R. and Liu,H. (2001b) DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2 and Efg1 in Candida albicans. J. Biol. Chem., 276, 48988–48996. [DOI] [PubMed] [Google Scholar]
- Leberer E., Harcus,D., Dignard,D., Johnson,L., Ushinsky,S., Thomas,D.Y. and Schroeppel,K. (2001) Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol., 42, 673–687. [DOI] [PubMed] [Google Scholar]
- Lengeler K.B., Davidson,R.C., D’Souza,C., Harashima,T., Shen,W.-C., Wang,P., Pan,X., Waugh,M. and Heitman,J. (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev., 64, 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kohler,J.R. and Fink,G.R. (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science, 266, 1723–1726. [DOI] [PubMed] [Google Scholar]
- Lo H.J., Kohler,J.R., DiDomenico,B., Loebenberg,D., Cacciapuoti,A. and Fink,G.R. (1997) Nonfilamentous C.albicans mutants are avirulent. Cell, 90, 939–949. [DOI] [PubMed] [Google Scholar]
- Luo Z., Freitag,M. and Sachs,M.S. (1995) Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol. Cell. Biol., 15, 5235–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton M.J. et al. (1998) Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat. Med., 4, 1293–1301. [DOI] [PubMed] [Google Scholar]
- Murad A.M.A., Lee,P.R., Broadbent,I.D., Barelle,C.J. and Brown,A.J.P. (2000) CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast, 16, 325–327. [DOI] [PubMed] [Google Scholar]
- Murad A.M.A. et al. (2001) NRG1 represses yeast–hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J., 20, 4742–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K., Meyer,M.R., Jackson,B.M., Slade,D., Roberts,C., Hinnebusch,A.G. and Marton,M.J. (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol., 21, 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F.C. (1988) Candida and Candidosis, 2nd edn. Baillière Tindall, London, UK.
- Odds F.C. (1994) Candida species and virulence. ASM News, 60, 313–318. [Google Scholar]
- Osborne B.I. and Guarente,L. (1989) Mutational analysis of a yeast transcriptional terminator. Proc. Natl Acad. Sci. USA, 86, 4097–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh J.L., Orbach,M.J., Legerton,T.L. and Yanofsky,C. (1988) The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc. Natl Acad. Sci. USA, 85, 3728–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S.A. and Livi,G.P. (1995) A GCN-like response in Candida albicans. Cell Biol. Int., 19, 65–69. [DOI] [PubMed] [Google Scholar]
- Ramon A.M., Porta,A. and Fonzi,W.A. (1999) Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-regulated transcription factor encoded by PRR2. J. Bacteriol., 181, 7524–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M., Schafer,W., Korting,H.C. and Hube,B. (1998) Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol. Microbiol., 29, 605–615. [DOI] [PubMed] [Google Scholar]
- Smith D.J., Cooper,M., DeTiani,M., Losberger,C. and Payton,M.A. (1992) The Candida albicans PMM1 gene encoding phospho mannomutase complements a Saccharomyces cerevisiae sec53-6 mutation. Curr. Genet., 22, 501–503. [DOI] [PubMed] [Google Scholar]
- Srikantha T., Klapach,A., Lorenz,W.W., Tsai,L.K., Laughlin,L.A., Gorman,J.A. and Soll,D.R. (1996) The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol., 178, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt V.R., Sonneborn,A., Leuker,C.E. and Ernst,J. (1997) Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J., 16, 1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P.E. (2001) The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localisation. Mol. Microbiol., 41, 19–31. [DOI] [PubMed] [Google Scholar]
- Swoboda R.K., Bertram,G., Delbruck,S., Ernst,J.F., Gow,N.A.R., Gooday,G.W. and Brown,A.J.P. (1994) Fluctuations in glycolytic mRNA levels during the yeast-to-hyphal transition in Candida albicans reflect underlying changes in growth rather than a response to cellular dimorphism. Mol. Microbiol., 13, 663–672. [DOI] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein,R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell, 56, 619–630. [DOI] [PubMed] [Google Scholar]
- Tzung K.-W. et al. (2001) Genomic evidence for a complete sexual cycle in Candida albicans.Proc. Natl Acad. Sci. USA, 98, 3249–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J., André,B. and Collado-Vides,J. (2000) A web site for the computational analysis of yeast regulatory sequences. Yeast, 16, 177–187. [DOI] [PubMed] [Google Scholar]
- Wanke C., Eckert,S., Albrecht,G., van Hartingsveldt,W., Punt,P.J., van den Hondel,C.A. and Braus,G.H. (1997) The Aspergillus niger GCN4 homologue, cpcA, is transcriptionally regulated and encodes an unusual leucine zipper. Mol. Microbiol., 23, 23–33. [DOI] [PubMed] [Google Scholar]
- Whiteway M. (2000) Transcriptional control of cell type and morphogenesis in Candida albicans. Curr. Opin. Microbiol., 3, 582–588. [DOI] [PubMed] [Google Scholar]
- Wiltshire C., Black,S. and Brown,A.J.P. (1999) Overexpression of Candida albicans mitochondrial ribosomal protein S9 (MrpS9p) disturbs mitochondrial function in Saccharomyces cerevisiae. Yeast, 15, 139–143. [DOI] [PubMed] [Google Scholar]