Abstract
Hmo1 is one of seven HMG-box proteins of Saccharo myces cerevisiae. Null mutants have a limited effect on growth. Hmo1 overexpression suppresses rpa49-Δ mutants lacking Rpa49, a non-essential but conserved subunit of RNA polymerase I corresponding to the animal RNA polymerase I factor PAF53. This overexpression strongly increases de novo rRNA synthesis. rpa49-Δ hmo1-Δ double mutants are lethal, and this lethality is bypassed when RNA polymerase II synthesizes rRNA. Hmo1 co-localizes with Fob1, a known rDNA-binding protein, defining a narrow territory adjacent to the nucleoplasm that could delineate the rDNA nucleolar domain. These data identify Hmo1 as a genuine RNA polymerase I factor acting synergistically with Rpa49. As an HMG-box protein, Hmo1 is remotely related to animal UBF factors. hmo1-Δ and rpa49-Δ are lethal with top3-Δ DNA topoisomerase (type I) mutants and are suppressed in mutants lacking the Sgs1 DNA helicase. They are not affected by top1-Δ defective in Top1, the other eukaryotic type I topoisomerase. Conversely, rpa34-Δ mutants lacking Rpa34, a non-essential subunit associated with Rpa49, are lethal in top1-Δ but not in top3-Δ.
Keywords: nucleolus/RNA polymerase I/SGS1/topoisomerases/UBF
Introduction
Ribosomal RNAs (25S, 18S, 5.8S and 5S rRNA) are by far the most abundant RNA species of living cells. They are synthesized by RNA polymerase I (Pol I) for the three largest rRNAs and by RNA polymerase III (Pol III) for the 5S rRNA. Pol III also determines the synthesis of all tRNAs and of at least three non-translated RNAs (the U6 snRNA, the RNA component of RNase P and the 7S RNA required for co-translational secretion) in all eukaryotes examined so far including Saccharomyces cerevisiae (Paule and White, 2000; Briand et al., 2001b). RNA polymerase II (Pol II) produces all mRNAs and also makes a large number of non-translated RNA species.
To initiate transcription, eukaryotic Pols first have to recognize promoter-bound complexes organized around the TATA box-binding protein TBP. Pol II pre-initiation complexes contain TBP and the Pol II-specific factor TFIIB, whilst Pol III complexes contain TBP and the heterodimeric factor TFIIIB, where subunit Brf1 is akin to TFIIB. In both cases, the pre-initiation complexes are widely conserved among eukaryotes (Chédin et al., 1998; Hampsey, 1998). The Pol I enzyme and two of its associated factors, Rrn3 and TFIIH, are also conserved from yeast to human (Gadal et al., 1997; Carles and Riva, 1998; Moorefield et al., 2000; Iben et al., 2002). However, the yeast and human rDNA promoter-binding complexes are largely unrelated. In S.cerevisiae, this complex is made of two components. A three-subunit core factor (Keys et al., 1994; Lalo et al., 1996) directly binds the core promoter domain and is strictly essential for transcription. An upstream activating factor (UAF) containing histone H3 and H4 (Keener et al., 1997) and four other polypeptides (Keys et al., 1996) strongly enhances the binding of the core factor to the promoter via TBP.
In human cells, the Pol I pre-initiation complex is made of the SL1/TIF-IB factor (Comai et al., 1992; Eberhard et al., 1993) that combines TBP and three associated TAFs (TAFI48, TAFI63 and TAFI110). It also contains the important but auxiliary factor UBF (Jantzen et al., 1990; Smith et al., 1993; Moss and Stefanovsky, 2002 and references therein). UBF is a homodimeric protein bearing six HMG domains that interacts with rDNA in a sequence-independent way. Neither SLI nor UBF have sequence similarity with the yeast UAF or core factor. We show here that the yeast HMG-box protein Hmo1 (Lu et al., 1996a) also belongs to the rRNA transcription apparatus and may thus be functionally equivalent to UBF, which adds to the mounting evidence that Pol I transcription systems are conserved from yeast to human.
Results
General organization of Hmo1
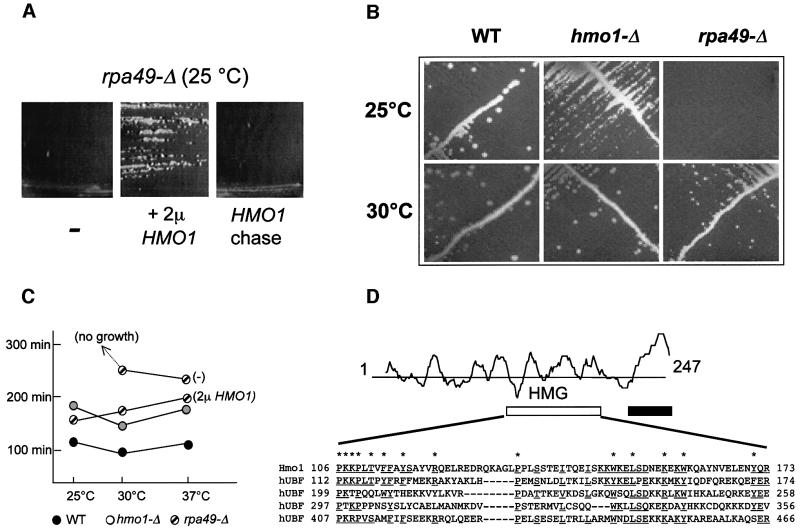
In the present study, we found that rpa49-Δ mutants lacking the conserved Rpa49/PAF53 Pol I subunit (Liljelund et al., 1992; Hanada et al., 1996) are suppressed in vivo by the overexpression of Hmo1 (Figure 1A). Hmo1, one of the seven HMG-box proteins encoded by the genome of S.cerevisiae, was first identified by its co-purification with an unidentified DNA helicase (Lu et al., 1996a). This protein has strong chromatin-binding properties (Freeman et al., 2000; Mitsouras et al., 2002) but its biological function so far was unknown. The original null mutants (Lu et al., 1996a) were viable, with a strong growth defect. The hmo1-Δ mutants of this study were prepared in a somewhat different genetic background (YPH500; Sikorski and Hieter, 1989). As shown Figure 1B and C, they have a <2-fold effect on growth rate between 25 and 37°C. hmo1 mutants were also isolated by their synthetic lethal effect on fpr1-Δ mutants lacking a non-essential prolyl isomerase (Dolinski and Heitman, 1999).
Fig. 1. General properties of Hmo1. (A) Suppression of rpa49-Δ::TRP1 by HMO1. Strain 49-ΔA (rpa49-Δ) was transformed with pFL44L without an insert (–), and with the multicopy plasmid pD10-ΔSph bearing HMO1 (2µ HMO1). A control strain was obtained by chasing pD10-ΔSph on FOA medium (HMO1 chase). Individual clones were selected and re-isolated on uracil omission plates, streaked on YPD and incubated for 4 days at 25°C. (B) Growth pattern of null mutants. Strains YPH499 (WT), SL13-1A (hmo1-Δ) and 49-ΔA (rpa49-Δ) were streaked on YPD and incubated for 4 days at 25 and 30°C. These three strains are isogenic and their complete genotype is given in Table I. (C) Doubling times of mutants. Strains YPH499 (WT), SL16-2C (hmo1-Δ) and 49-ΔA (rpa49-Δ) were grown in liquid YPD at the indicated temperatures. Doubling times were determined by nephelometry using a Hatch nephelometer. 49-ΔA (rpa49-Δ) was tested with or without the pD10-ΔSph plasmid bearing HMO1 (2µ HMO1). (D) Properties of the amino acid sequence of Hmo1. The hydrophilicity pattern of Hmo1 and the localization of its HMG (white box) and basic (black box) domains are indicated. The HMG domain is compared with the first four N-terminal domains of human UBF. Identical or strongly similar amino acids (F = W = Y, E = D, N = Q, E = Q, I = L) are underlined. Asterisks denote the general HMG consensus.
Hmo1 contains a single HMG domain matching the minimal consensus proposed for HMG domains and closely related to four of the six domains of the UBF Pol I factor associated with human Pol I (Figure 1D). The existence of a second HMG domain (Lu et al., 1996a) is not supported by our sequence alignments. A basic C-terminal extension (amino acids 210–247) can be deleted with no detectable phenotype (Lu et al., 1996a). The N-terminal half of the protein (positions 1–105) lacks homology to any known eukaryotic gene product. Indeed, Hmo1 has only been identified so far in yeasts closely related to S.cerevisiae such as Saccharomyces kluyveri (Neuvéglise et al., 2000).
HMO1 specifically suppresses rpa49-Δ mutants lacking the Pol I subunit Rpa49/PAF53
rpa49-Δ mutants grow slowly at 30°C and are lethal at 25°C (see Figure 1B and C). We exploited the latter property to isolate dosage-dependent suppressors of rpa49-Δ (strain 49-ΔA), using a genomic library borne on the URA3 multicopy vector pFL44L (Stettler et al., 1993). This yielded four different RPA49 clones and one extragenic suppressor clone containing HMO1. Subclones bearing HMO1 only restored a doubling of 180 min in rpa49-Δ cells, not very different from the 125 min of an isogenic wild-type (Figure 1C). Suppression was due to a gene dosage effect since a low-copy plasmid (pASD10-HMO1) hardly improves growth (data not shown). No other suppressor of rpa49-Δ was isolated in a second screen, using less stringent conditions. A reciprocal multicopy suppression effect of RPA49 on the slow growth of hmo1-Δ mutants was not observed.
Three other conditional Pol I mutants (rpa190-G728D, rpa12-Δ and rpa43-24) have been examined for dosage-dependent suppression. They do not respond to HMO1 but each of them is suppressed specifically by the overexpression of one particular protein of the Pol I transcription system. rpa190-G728D responds to Rpb6, a common subunit shared by Pol I, II and III (Briand et al., 2001a), and rpa12-Δ is suppressed by Rpa190, the largest subunit of Pol I (Nogi et al., 1993). rpa43-24 is suppressed by an increased gene dosage in the Pol I-specific initiation factor Rrn3 (Peyroche et al., 2000). Thus, rpa49-Δ, rpa12-Δ, rpa190-G728D and rpa43-24 each respond to a different suppressor, consistent with the fact that they affect distinct aspects of Pol I activity.
Hmo1 is synthetic lethal with several PolI-defective mutants
hmo1-Δrpa49-Δ double mutants are lethal (Figure 2A). In contrast, viable double mutants (e.g. SL11-1B, see Table I) are recovered readily in the presence of a complementing plasmid bearing either RPA49 (pSLA49 = 2µ URA3 RPA49) or HMO1 (pASD10 = CEN6 ADE2 HMO1). In both cases, the spontaneous loss of the RPA49 or HMO1 complementing plasmid can be monitored easily by the formation of 5-fluoro-orotic acid (FOA)-resistant colonies (loss of URA3) or of red sectors on YPD plates (loss of ADE2). Unlike their single mutant parents, hmo1-Δ and rpa49-Δ double mutants were unable to lose these plasmids, further demonstrating that the combination of these two mutations is lethal.
Fig. 2. Synthetic lethality between hmo1-Δ and RNA polymerase I mutants. (A) Synthetic lethality of hmo1-Δ rpa49-Δ double mutants. Strain SL6-10A (hmo1-Δ) was crossed to A49ΔA (rpa49-Δ). The resulting diploid (SL8) was submitted to tetrad analysis (20 asci). Replica plating on appropriate omission media identified the genotype of individual segregants. The growth pattern of one tetratype and one parental ditype ascus is shown after 6 days on YPD at 30°C (the position of the non-growing double-mutant spore is denoted ΔΔ). (B) Effect of hmo1-Δ on other Pol I mutants. Strain SL13-1A (hmo1-Δ) was crossed to T4-1D (rpa34-Δ) and GPY11-24 (rpa43-24). SL6-10b (hmo1-Δ) was crossed to SL9-6B (rpa12-Δ), D191-7C (rpa49-Δ), RY260 (rpb1-1) and D360-1A (rpa14-Δ). Genetic crosses were as described above, with at least 10 tetrads analysed in each case. The complete genotypes of the parental strains are given in Table I. Non-lethal growth patterns were assessed at 30°C and are indicated as follows: ++, +, (+): no, slight or intermediate growth defect, respectively. (C) Suppression of rpa49-Δ hmo1-Δ lethality by Pol II-dependent transcription. OG14-1a with plasmid pNOY103 (rpa49-Δ) and SL13-1a (hmo1-Δ) were crossed to give the SL14 diploid strain. Spores were germinated and grown on YPGal. In contrast to the SL8 cross described above, viable rpa49-Δ hmo1-Δ double mutants were obtained but invariably harboured the pNOY103 plasmid. The growth patterns of SL14-6A (hmo1-Δ) and of the SL14-5C (rpa49-Δ hmo1-Δ) double mutant, both harbouring pNOY103, are shown on YPD (repression) and YPGal (induction), after 4 days at 30°C. The complete genotype of these strains is given in Table I.
Table I. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| 49-ΔA | MATa rpa49-Δ::TRP1 ura3-52 his3-Δ200 trp1Δ-1 lys2-801 ade2-1 | Liljelund et al. (1992) |
| D191-7C | MATα rpa49-Δ::TRP1 ura3-52 his3-Δ200 trp1 leu2-Δ1 lys2-801 ade2-1 | This work |
| SL6-10B | MATα ade2-1 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 hmo1-Δ::URA3 | This work |
| SL16-2C | MATα ade2-1 ura3-52 lys2-801 his3-Δ200 leu2-Δ1 hmo1-Δ::HIS3 | This work |
| SL13-1A | MATa ade2-1 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 hmo1-Δ::HIS3 | This work |
| SL6-10A | MATα ade2-1 ura3-52 lys2-801 his3-Δ200 leu2-Δ1 hmo1-Δ::URA3 | This work |
| BMA64-1A | MATa leu2-3 112 his3-11,15 trp1Δ2 CAN1-100 ade2-1 ura3-52 | Galy et al. (2000) |
| BMA64-1B | MATα leu2-3 112 his3-11,15 trp1Δ2 CAN1-100 ade2-1 ura3-52 | Galy et al. (2000) |
| BMA-Hmo1–GFP | MATa leu2-3 112 his3-11,15 trp1Δ2 CAN1-100 ade2-1 ura3-52 HMO1::GFP-TRP1 | This work |
| BMA-Fob1–GFP | MATa leu2-3 112 his3-11,15 trp1Δ2 CAN1-100 ade2-1 ura3-52 FOB1::GFP-TRP1 | This work |
| BMA-Hmo1–YFP | MATa leu2-3 112 his3-11,15 trp1Δ2 CAN1-100 ade2-1 ura3-52 HMO1::YFP-HIS3MX6 | This work |
| BMA-Fob1–CFP | MATα leu2-3 112 his3-11,15 trp1-Δ2 CAN1-100 ade2-1 ura3-52 FOB1::CFP-TRP1 | This work |
| D432-6D | MATa rpa190-G728D trp1 leu2 lys2-801 ura3-52 | This work |
| SL9-6B | MATα ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpa12-Δ::LEU2 | Gadal et al. (1997) |
| RY260 | MATa ura3-52 rpb1-1 | Nonet et al. (1987) |
| GPY11-24 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpa43-Δ::LEU2 (CEN TRP1 rpa43-24) | Peyroche et al. (2000) |
| SL11-1B | MATα ade2-1 ura3-52 lys2-801 trp1 his3-Δ200 leu2-Δ1 hmo1Δ::URA3 rpa49-Δ::TRP1 (pASD10 = CEN6 ADE2 HMO1) | This work (SL6-10A × 49-ΔA, transformed with pASD10) |
| SL14-5C | MATa leu2-D1 hmo1-D::HIS3 rpa49-D::TRP1 ura3-52 his3-Δ200 trp1 lys2-801 ade2-1 (pNOY103 = pGAL7::rDNA ADE3 URA3 2µ) | This work (SL13-1A × OG14-1A) |
| SL14-6A | MATa leu2-D1 rpa49-D::TRP1 hmo1-D::HIS3 ura352 his3-Δ200 trp1 lys2-801 ade2-1 (pNOY103 = pGAL7::rDNA ADE3 URA3 2µ) | This work (SL13-1A × OG14-1A) |
| OG14-1A | MATa rpa49-D::TRP1 ura3-52 his3-Δ200 trp1Δ-1 lys2-801 ade2-1 leu2-D1 (pNOY103 = pGAL7::rDNA ADE3 URA3 2µ) | This work (see Gadal et al., 1997) |
| SL7-4A | MATα rpa34-Δ::HIS3 ura3 ade2-1 lys2-801 his3-Δ200 trp1-Δ lhmo1-Δ::URA3 leu2 | This work (T4-1C × SL6-10B) |
| T4-1C | MATα rpa34-Δ::HIS3 ura3-52 ade2-1 lys2-801 his3-Δ200 trp1-Δ1 | Gadal et al. (1997) |
| D313-3D | MATα his3 trp1 ura3-1 ade2-1 lys2-801 top3-4Δ::URA3 sgs1-Δ::TRP1 | D64-10B × YPH52 (Sikorski and Hieter, 1989) |
| D64-10B | MATa CAN1-100 his3-11,15 leu2-3 112 trp1-1 ura3-1 ade2-1 top3-Δ::URA3 sgs1-Δ::TRP1 | Gangloff et al. (1994) |
| A14-U | MATa ade2-1 ura3-52 lys2-801 trp1-Δ63 rpa14::URA3 his3-Δ200 leu2-Δ1 | Smid et al. (1995) |
| D308-6B | MATα his3 trp1 ura3-1 ade2-1 leu2-3 112 top3-Δ::URA3 | This work |
| D211-3B | MATα ade2-1 his3-11,15 leu2-3 112 trp1-1 ura3-1 top3-Δ::HIS3 lys2 | This work |
| D360-1A | MATα ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpa14-Δ::HIS3 | This work |
| D360-1A | MATα ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpa14-Δ::HIS3 | This work |
| D386-9B | MATa lys2-Δ201 ura3-52 trp1-Δ63 his3-Δ200 rpb9-Δ::HIS3 | Van Mullem et al. (2002b) |
As shown in Figure 2B, lethality with hmo1-Δ extends to at least two other Pol I-defective mutants (rpa12-Δ and rpa43-24) but not to the rpa14-Δ and rpa34-Δ mutants of the non-essential subunits Rpa14 and Rpa34. Moreover, hmo1-Δ is viable when combined with the temperature-sensitive Pol II mutant rpb1-1 (Nonet et al., 1987). The synthetic lethality of hmo1-Δ with rpa12-Δ is consistent with the fact that Rpa49 dissociates from the immunopurified rpa12-Δ mutant enzyme (Van Mullem et al., 2002a). The lethality of rpa43-24 hmo1-Δ is consistent with recent data showing that Rpa43 and Rpa49 have synergic effects in vivo (S.Labarre and P.Thuriaux, unpublished results). The fact that hmo1-Δ exacerbates the growth defect of several Pol I mutants supports the idea that Hmo1 contributes to the Pol I-dependent synthesis of rRNA (see below). The suppression effect associated with its overexpression, however, seems specific for rpa49-Δ. It implies that the absence of Rpa49 is somehow compensated by a mere increase in the cellular concentration in Hmo1, therefore suggesting that both proteins affect the same aspect of Pol I-dependent transcription.
Hmo1 stimulates rRNA synthesis in rpa49-Δ cells
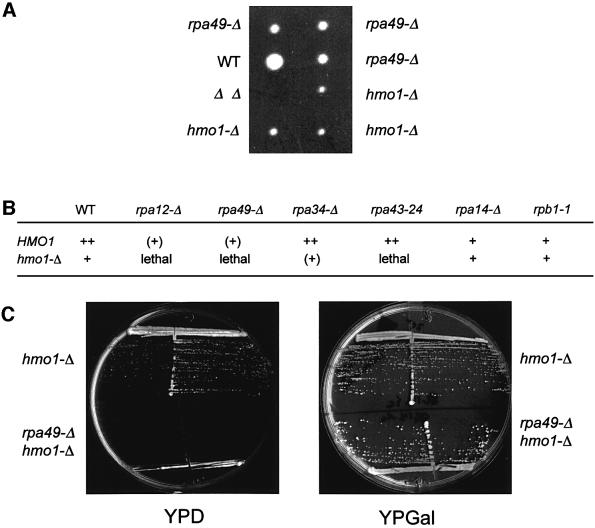
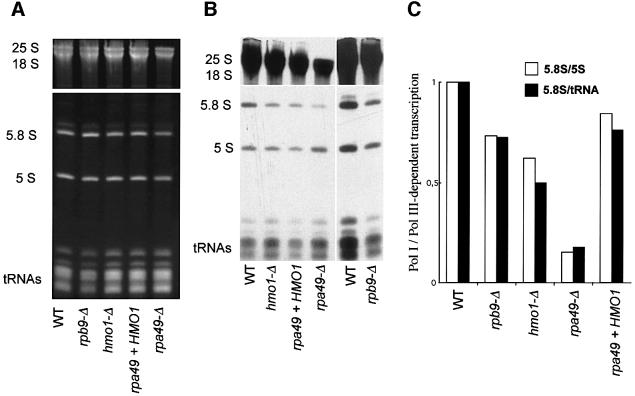
Figure 3 compares isogenic hmo1-Δ, rpa49-Δ (with or without HMO1 overexpression), rpb9-Δ and wild-type cells for their de novo synthesis of rRNAs and tRNAs by [3H]uracil pulse labelling. The steady-state levels of rRNAs and tRNAs were estimated by ethidium bromide staining. rpb9-Δ has a partial Pol II defect, due to the inactivation of the non-essential Pol II subunit Rpb9 (Woychik et al., 1991), and serves here as a slow-growing control. Since rRNAs contribute ∼80% of the total RNA in wild-type cells (Waldron and Lacroute, 1975), partial Pol I defects are already apparent when comparing steady-state rRNA and tRNA content. As shown in Figure 3A, the wild-type and rpb9-Δ controls have a similar rRNA/tRNA ratio. Hmo1-Δ and, even more so, rpa49-Δ are characterized by a shortage of rRNA compared with the tRNA level.
Fig. 3. Effect of hmo1-Δ and rpa49-Δ on rRNA. (A) Steady-state level of rRNA and tRNA. Strains YPH499 (WT), D386-9B (rpb9-Δ), SL16-2C (hmo1-Δ) and 49-ΔA (rpa49-Δ) with or without the pD10-ΔSph plasmid bearing HMO1 (2µ HMO1) were grown on YPD at 30°C and harvested in mid-log phase (OD600 = 0.5). Total rRNA was extracted in hot phenol (Hermann-Le Denmat et al., 1994) and quantified by spectrophotometry at 260 and 280 nm. A 10 µg aliquot of total RNA was separated by gel electrophoresis on polyacrylamide and revealed by ethidium bromide staining. (B) In vivo labelling of rRNA and tRNA. The same strains as above were transformed with the URA3 centromeric plasmid YCp50 to complement their ura3-52 mutation. The resulting transformants were grown in uracil omission medium and exposed to 150 µCi of [3H]uracil for 20 min at 30°C (Hermann-Le Denmat et al., 1994). Total RNA was extracted and separated as described above. [3H]Uracil incorporation was revealed by overnight autoradiography (see Material and methods). The last two lanes correspond to a 4 day exposure. (C) Ratio of Pol I (5.8S) and Pol III (5S, tRNA) −signals. Non-saturated autoradiograms were scanned as described in Materials and methods. The two ratios (5.8S/5S) and (5.8S/tRNA) were normalized relatively to the wild-type control.
To measure the de novo synthesis of rRNA, cells were exposed to a 20 min pulse with [3H]uracil. At 30°C, rpa49-Δ has an ∼5-fold effect on Pol I-dependent synthesis of rRNA (e.g. 5.8S rRNA) relative to Pol III transcripts such as 5S rRNA and tRNAs. The presence of HMO1 largely reverses this effect (Figure 3B and C), confirming that the gene dosage of HMO1 is important for Pol I-dependent transcription in cells lacking the Rpa49 subunit of Pol I. hmo1-Δ cells also partly compromise the de novo synthesis of rRNA, with no detectable accumulation of pre-rRNA intermediates. Along with their low steady-state level in rRNA, this is consistent with a partial Pol I defect. However, these data should be considered with some caution, since slowing down growth also has some effect on rRNA synthesis (Waldron and Lacroute, 1975), as can be seen in our rpb9-Δ control (Figure 3B and C).
rpa49-Δ hmo1-Δ is rescued by the Pol II-dependent synthesis of rRNA
Nogi et al. (1991) have shown that Pol I-specific growth defects are bypassed by high copy number plasmids where rDNA transcription is under the control of a Pol II promoter such as the GAL7 promoter. We thus repeated the hmo1-Δ × rpa49-Δ crosses mentioned above in diploid cells harbouring pNOY103, a URA3 pGAL7::rDNA vector. Tetrad analysis was done on YPGal medium at 25°C. Under these conditions, viable hmo1-Δ rpa49-Δ double mutants were readily recovered (e.g. SL14-5C, Table I). They invariably harboured pNOY103 and were unable to lose it in the presence of FOA. Moreover, they failed to grow when turning off the GAL7 promoter on glucose-containing medium (Figure 2C), demonstrating that the synthetic lethality of hmo1-Δ rpa49-Δ is due solely to a Pol I-dependent defect.
Nucleolar localization of Hmo1
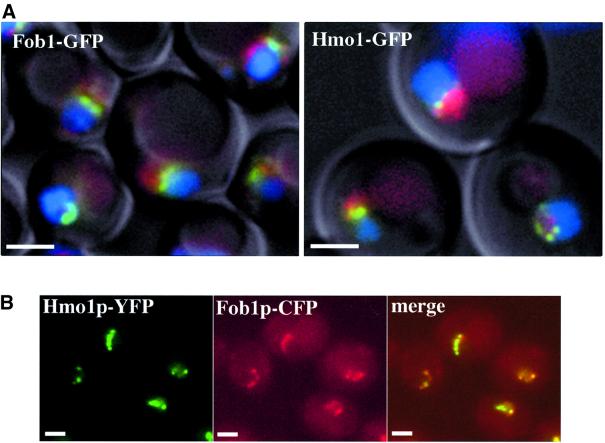
C-terminal fusions of GFP to Hmo1 and to Fob1, a proven rDNA-binding protein, were expressed from their chromosomal loci [strains BMA-Hmo1–green fluorescent protein (GFP) and BMA-Fob1–GFP, see Table I]. Their localization was compared with that of a DsRed–Nop1 fusion protein, where Nop1 is a part of the nucleolar dense fibrillar component (Léger-Silvestre et al., 1999), and with DNA staining (Hoechst 33352), which preferentially reveals the nucleoplasmic DNA with nucleolar staining exclusion. As shown in Figure 4A, Fob1 stains a structure located at the nucleolar–nucleoplasmic interface (compare with DNA and Nop1 staining). Hmo1 has a very similar localization pattern. Fob1 is a bona fide rDNA-binding protein (Defossez et al., 1999) and we are therefore inclined to believe that this staining pattern defines the localization of rDNA in living cells. In a second experiment (Figure 4B), Fob1 and Hmo1 were tagged with different GFP spectral variants, cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP), in a diploid strain prepared by crossing strains BMA-Fob1–CFP and BMA-Hmo1–YFP (Table I). Staining the resulting diploid entirely confirmed the co-localization of these two proteins, supporting our conclusion that Hmo1 is an rDNA-binding component of the yeast Pol I transcription system.
Fig. 4. In vivo localization of Hmo1–GFP and Fob1–GFP fusion proteins. (A) Fob1–GFP and Hmo1–GFP are concentrated at the interface between the nucleoplasm and the nucleolus. Strains BMA-Fob1–GFP and BMA-Hmo1–GFP bearing the pUN100-DsRed-NOP1 plasmid (Gadal et al., 2001) were grown at 30°C to mid-log phase (OD600 = 0.5) in YPD. Cells were washed with water. Nucleoplasm and nucleolus were visualized using DNA staining and Nop1 staining, respectively. GFP (green), DsRed (red) and Hoechst 33352 (blue) signals were monitored by fluorescence microscopy as described in Materials and methods. Cells were examined by Nomarski imaging. (B) Fob1 and Hmo1 co-localize. Fob1 and Hmo1 were tagged with different GFP spectral variants, CFP and YFP, and visualized in a diploid strain prepared by crossing strains BMA-Fob1–CFP and BMA-HMO1–YFP as described in Materials and methods (scale bars correspond to 2 µm).
hmo1-Δ and rpa49-Δ are synthetic lethal with top3-Δ but not with top1-Δ mutants
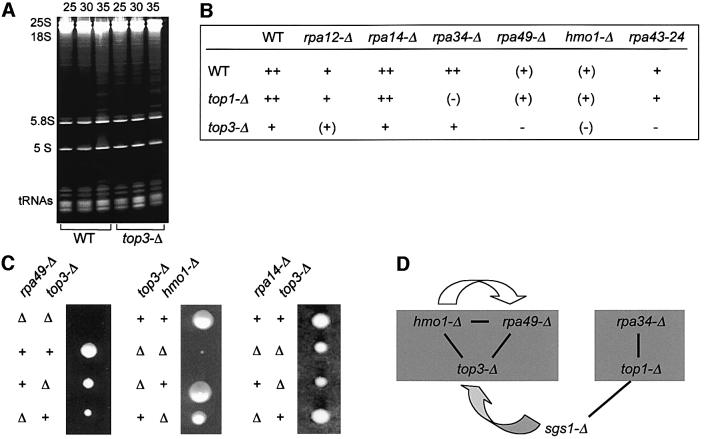
Like other eukaryotes, yeast has two unrelated type I topoisomerases, Top1 and Top3. Their separate or simultaneous inactivation has little (Top1) or moderate (Top3) effect on growth (Thrash et al., 1985; Gangloff et al., 1994). top1-Δ mutants strongly affect the topology of rDNA and increase mitotic recombination within the rDNA cluster (Christman et al., 1993), but have little or no effect on rRNA formation (Thrash et al., 1985). top3-Δ mutants also strongly increase mitotic recombination at the rDNA repeat (Gangloff et al., 1994). They grow poorly above 35°C, but their effect on rRNA transcription had not been examined. We show here that top3-Δ mutants have a wild-type steady-state level of tRNA and rRNA at 35°C (Figure 5A). Hence, their adverse effect at high temperature probably is not due to a preferential defect in rRNA synthesis.
Fig. 5. Synthetic lethality and extragenic suppression between RNA polymerase I, type 1 topoisoimerases and Hmo1 mutants. (A) Steady-state levels in rRNA and tRNA. Strains YPH499 (WT) and D211-3B (top3-Δ) were grown on YPD. Growth temperatures (°C) are given above each lane. Cells were harvested in mid-log phase (OD600 = 0.5). Note that top3-Δ has a severe growth defect at 35°C (Gangloff et al., 1994). Total RNA was extracted in hot phenol (Hermann-Le Denmat et al., 1994) and quantified by spectrophotometry at 260 and 280 nm. For each lane, 10 µg of total RNA were separated by polyacrylamide gel electrophoresis and revealed by ethidium bromide staining. (B) Synthetic lethality pattern. D308-6B (top3-Δ) was crossed to strains SL13-1A (hmo1-Δ), T4-1D (rpa34-Δ), 49-ΔA (rpa49-Δ) and GPY11-24 (rpa43-24). Strain D211-3B (top3-Δ) was used in the genetic cross to A14::URA3 (rpa14-Δ). At least 10 tetrads were analysed in each cross. Since top3-Δ mutants tend to accumulate sgs1 suppressors (Gangloff et al., 1994), the parental top3-Δ SGS1+ mutants were re-isolated freshly before each cross by meiotic out-crossing from top3-Δ sgs1-Δ double mutants. Growth patterns are as follows ++, +, (+): growth ranging from wild-type to intermediate level after 5 days at 30°C. –, (–): no or barely detectable growth, respectively. (C) Tetratype asci corresponding to top3-Δ crossed with rpa49-Δ, hmo1-Δ and rpa14-Δ. The wild-type (++), double mutant (ΔΔ) and parental single mutant genotype are indicated on the left of each panel. Spores were germinated on YPD and incubated at 30°C for 5 days. (D) Genetic interactions between Hmo1, Rpa49, Top3, Sgs1, Rpa34 and Top1. The two shaded grey boxes correspond to epistatic null mutants. Epistasis is defined here by the fact that double mutants between these two groups are viable (with a growth pattern similar to that of the slowest growing parent), whilst double mutants within each group are lethal or barely grow. Dark lines denote synthetic lethality. The white arrow indicates the dosage-dependent suppression of rpa49-Δ by HMO1. The grey arrow stands for the extragenic suppression of top3-Δ by sgs1-Δ. Data based on Gangloff et al. (1994), Lu et al. (1996a), Gadal et al. (1997) and this study.
As shown in Figure 5B and C, top3-Δ interferes with a subset of Pol I mutants, namely hmo1-Δ, rpa49-Δ and the conditional rpa43-24 mutant. rpa12-Δ, rpa34-Δ and rpa14-Δ are not or slightly (rpa12-Δ) impaired by top3-Δ. Moreover, this effect is specific for the type I topoisomerase considered, since hmo1-Δ, rpa49-Δ and rpa43-24 are not affected by top1-Δ mutants. Conversely, rpa34-Δ is nearly synthetic lethal with top1-Δ (Gadal et al., 1997). In vitro, human Top1 stimulates Pol II-dependent transcription (Kretzschmar et al., 1993) in a way that does not depend on catalytic activity (Shykind et al., 1997). In contrast, the Pol I-specific effect associated with the lethality of rpa34-Δ top1-Δ strictly depends on the catalytic activity of Top1, since it is not reversed by a plasmid bearing the top1-Y727W mutant at the catalytic tyrosine (data not shown).
Figure 5D summarizes the genetic interactions relating null mutants of Hmo1, Top1, Top3 and the Pol I-specific subunits Rpa49 and Rpa34 (Gadal et al., 1997). Hmo1/Rpa49/Top3 and Top1/Rpa34 clearly define two groups of null mutants. Crosses within a group invariably yield lethal double mutants. Crosses between groups produce double mutants comparable with the slowest-growing single mutant. In addition, null mutants of the Sgs1 DNA helicase discriminate between the two topoisomerases since sgs1-Δ suppresses the adverse effects of top3-Δ (Gangloff et al., 1994) and its lethality with hmo1-Δ and rpa49-Δ (data not shown), but are synthetic lethal with top1-Δ (Lu et al., 1996b). sgs1-Δ has many pleiotropic properties including a strong accumulation of the 3µ rDNA episome (Sinclair and Guarente, 1997). The latter phenotype tentatively suggests that the Pol I-specific effects associated with the loss of Hmo1, Rpa49 or Top3 may not concern the episomal form of rDNA.
Discussion
Pol I-specific subunits and transcription factors that are specialized in yeast rRNA synthesis were discovered by a genetic screen based on the Pol II-dependent rescue of lethal Pol I mutants (Nogi et al., 1991). However, factors like Hmo1 that only lead to a partial growth defect would be overlooked by this approach. Hmo1 was first identified by its co-purification with an unidentified DNA helicase (Lu et al., 1996a). Apart from their slow growth rate, hmo1-Δ null mutants may alter plasmid stability (Lu et al., 1996a) and are synthetic lethal with fpr1-Δ mutants lacking a non-essential prolyl isomerase (Dolinski and Heitman, 1999).
Overexpressing Hmo1 strongly suppresses the cold-sensitive defect of rpa49-Δ mutants lacking the conserved Pol I-specific Rpa49/PAF53 subunit. Null mutants are lethal in rpa49-Δ cells. This defect is rescued by Pol II-dependent transcription of rDNA. It extends to other Pol I mutants, in a mutant-specific way. Finally, Hmo1 co-localizes with Fob1, a bona fide rDNA-binding protein (Defossez et al., 1999). By fluorescence microscopy, Hmo1 and Fob1 were shown to delineate a narrow segment located at the nucleolar–nucleoplasmic interface, that we propose to be the rDNA localization domain in living cells. These data strongly suggest that Hmo1 is a nucleolar factor specifically associated with rDNA, that acts synergistically with the conserved Rpa49/PAF53 subunit of Pol I during rDNA transcription.
Hmo1 is only found in S.cerevisiae and closely related yeast such as S.kluyveri, where the HMO1 gene is immediately near the rDNA cluster (Neuvéglise et al., 2000). No homologue protein is encoded by the Schizosaccharomyces pombe genome (Wood et al., 2002). This poor conservation may be a general property of HMG-box proteins since S.cerevisiae and S.pombe each encode seven HMG-box proteins that are largely unrelated to each other, except for Nhp6A and Nhp6B, two nearly identical proteins that control the Pol III-dependent transcription of U6 splicing snRNA (Kruppa et al., 2001; Lopez et al., 2001) in S.cerevisiae and are closely related to an S.pombe gene product. This poor conservation is not incompatible with a conserved function, as the Abf2 HMG-box protein of S.cerevisiae may be functionally equivalent to the human mitochondrial transcription factor A (Parisi et al., 1993). It is therefore tempting to speculate that Hmo1 is functionally related to the animal UBF factor acting in Pol I-dependent transcription (Jantzen et al., 1990; Moss and Stefanovsky, 2002). UBF is homodimeric and contains six HMG domains, but nothing is known of the number of Hmo1 molecules associated with rDNA.
UBF is present in mammals and amphibians (Xenopus laevis) but is probably restricted to vertebrates. Like Hmo1, UBF is important but partly dispensable for rDNA transcription (Smith et al., 1993). Its overexpression strongly increases rDNA transcription in human cardiomyocytes, in a way that is reminiscent of the dosage-dependent suppression phenotype found in the present work (Hannan et al., 1996). HMG domains have strong DNA-bending properties, and the corresponding proteins may act as architectural factors favouring the recruitment and co-operative interaction of transcription factors (Thomas and Travers, 2001; Mitsouras et al., 2002). Animal UBFs, for example, could form an rDNA ‘enhanceosome’ facilitating the recycling of Pol I between closely associated rDNA repeats (Moss and Stefanovsky, 2002). Likewise, our genetic suppression data suggest that Hmo1 alters the accessibility of individual rDNA to the Pol I transcription complex, in a way that compensates for the lack of Rpa49/PAF53. These data are based on the bypass of null mutants and thus need not imply a physical interaction between Hmo1 and Rpa49 itself. In animal cells, such an interaction is actually suggested by protein pull-down and far-western blotting data between UBF and PAF53 (Hanada et al., 1996).
The bending induced by HMG proteins may also strongly alter the topology of DNA. In the case of Hmo1, this would be consistent with its tight association with a DNA helicase (Lu et al., 1996a). Topological constraints on rDNA could explain the curious pattern of synthetic lethality that relates type 1A topoisomerases (Top1 and Top3) to Hmo1 and to non-essential subunits of Pol I. Briefly, cells lacking either Hmo1 or Rpa49 are lethal in a top3-Δ context but are not affected detectably by top1-Δ, whilst the converse pattern is observed on rpa34-Δ (Gadal et al., 1997). Another player in the game is Sgs1, a DNA helicase that can be inactivated genetically with no major effect on growth (Gangloff et al., 1994), but strongly interferes with genetic recombination (Gangloff et al., 2000). sgs1-Δ suppresses the adverse effects of top3-Δ (Gangloff et al., 1994), including its lethality with hmo1-Δ and rpa49-Δ, but is synthetic lethal with top1-Δ (Lu et al., 1996b). sgs1-Δ cells accumulate rDNA in its 3µ episomal form (Sinclair and Guarente, 1997). A simple way of explaining the genetic interactions just mentioned could be that episomal transcription does not require Hmo1, Rpa49 or Top3, but is strictly dependent on Top1.
Twelve of the 14 subunits of yeast Pol I are present in the human enzyme (Gadal et al., 1997; Carles and Riva, 1998). Three components of the Pol I transcription system, TBP (Comai et al., 1992; Schultz et al., 1992), TFIIH (Iben et al., 2002) and Rrn3/TIF-IA (Schnapp et al., 1990; Yamamoto et al., 1996; Moorefield et al., 2000) are also conserved from yeast to man. On the other hand, the TAFI48, TAFI63 and TAFI110 subunits of the mammalian SL1/TIF-IB factor (Eberhard et al., 1993; Zomerdijk et al., 1994) bear no detectable homology to any S.cerevisiae protein. Conversely, the Pol I-specific subunits of the yeast core factor and UAF (Rrn5, Rrn6, Rrn9, Rrn10 and Rrn11) are only found in S.cerevisiae and closely related species such as Kluyveromyces lactis (Bolotin-Fukuhara et al., 2000).
This lack of conservation evidently reflects the very fast evolution of the rDNA promoter (Grummt et al., 1982). It raises a curious paradox by imposing that a highly conserved enzyme (Pol I) must be able to recognize the highly variable platform defined by the yeast and human rDNA-binding complex. The initiation factor Rrn3/TIF-IA could be part of the answer, as the yeast and human forms can be exchanged in vivo despite a moderate sequence conservation (Moorefield et al., 2000). On the other hand, recent results suggest that Rrn3/TIF-IA may not determine Pol I recruitment, as initially proposed (Peyroche et al., 2000; Miller et al., 2001), but could instead be involved in a later step of Pol I initiation (Aprikian et al., 2001). Interestingly, Hmo1 stimulates the in vitro binding of a viral Pol II activator (the Rta factor of the Epstein–Barr virus) to its target gene by the simple virtue of its DNA-bending effect, apparently without any specific protein– protein interaction (Mitsouras et al., 2002). This supports the idea that Hmo1 and UBF could help in organizing yeast and animal rDNA-binding complexes into a similar target structure for Pol I-dependent transcription.
Materials and methods
Yeast media and genetic techniques were described previously (Stettler et al., 1993; Hermann-Le Denmat et al., 1994). Cells were grown on rich medium YPD or YPGal (with glucose or galactose as the main carbon source), and tested for auxotrophy on omission media derived from the synthetic complete medium (SC). URA3 plasmids were counter-selected on FOA medium (SC with 0.1% FOA). Dosage-dependent suppressors of rpa49-Δ were selected from a genomic library prepared in the multicopy vector pFL44L (2µ URA3), with a total number of clones (∼50 000) corresponding roughly to five yeast genomes (Stettler et al., 1993). This yielded four plasmids with different RPA49 inserts and a fifth plasmid (pFL44-HMO1) with a 7 kbp insert bearing several genes including HMO1. The latter was the only gene retained in pD10-ΔSph, a suppressor plasmid obtained by deleting the 6.5 kbp SphI fragment of pFL44-HMO1. Eliminating pD10-ΔSph or pFL44-HMO1 by selection on FOA restored the initial mutant growth defect.
Plasmid pASD10-HMO1 contains the SacI–SphI fragment of pD10-ΔSph (with HMO1) cloned into the ADE2 centromeric vector pASZ11 (Stotz and Linder, 1990). pSLA49 (2µ URA3 RPA49) was constructed by cloning a 3.8 kbp RPA49 insert (from plasmid pFL44S-49; Liljelund et al., 1992) between the SalI and SacI sites of pFL44L. pUN100-DsRed-NOP1 was described previously (Gadal et al., 2001). pFA6-YFP-TRP1, pFA6-CFP-TRP1, pFA6-YFP-HIS3MX6 and pFA6-CFP-HIS3MX6 are derivatives of pFA6a-GFP(S65T)-HIS3MX6 and pFA6-GFP-TRP1 (Longtine et al., 1998) where the PacI–AscI fragment, bearing the GFP-coding sequence, was PCR exchanged with the corresponding eYFP and eCFP spectral variants of GFP, using vectors pECFP-C1 and pEYFP-C1 from Clontech.
Yeast strains are listed in Table I. hmo1-Δ::HIS3, hmo1-Δ::URA3 and hmo1Δ::TRP1 mutants were constructed in a YPH99 × YPH500 diploid (Sikorski and Hieter, 1989) by integrative transformation with a SmaI–SphI fragment where HMO1 has been interrupted by inserting the appropriate HIS3, URA3 or TRP1 cassette between the intragenic HpaI and KpnI sites. rpa14-Δ::HIS3 was constructed by direct in situ deletion in the wild-type strain YPH499 (Baudin et al., 1993). The genetic structure of the diploid transformants was checked by genomic hybridization, and the corresponding haploid segregants were obtained by tetrad analysis. Strains BMA-HMO1::GFP, BMA-FOB1::GFP, BMA-HMO1::CFP and BMA-FOB1::YFP were constructed in a BMA64-1A or BMA64-1B background by C-terminal fusions (see Table I) as described previously (Longtine et al., 1998).
Fluorescence microscopy was done on exponentially grown cells washed in water and stained with Hoechst 33352 (5 ng/µl) for 5 min. Samples were examined using a Leica DMRXA fluorescence microscope. Fluorescent signals were collected with single band pass filters for excitation of DsRed (XF137-2, Omega optical), GFP (GFP, Leica), YFP (XF104, Omega Optical), CFP (XF114-2, Omega optical) and Hoechst 33352 (A, Leica). Images were acquired with a Hamamatsu C4742-95 cooled CCD camera controlled by the Openlab® software (version 2.2.4, Improvision) and processed using Adobe Photoshop® software (version 5, Adobe).
Steady-state levels of rRNA and tRNAs were measured in exponentially growing cells by ethidium bromide staining, and their de novo synthesis was monitored after a 20 min pulse of [3H]uracil (Hermann- Le Denmat et al., 1994). The signal was quantified by scanning non-saturated autoradiogramms originating from two different assays with ImageQuant (Molecular Dynamic). The following ratios were measured: (18S + 25S) versus tRNAs (Pol I/Pol III), 5.8S versus 5S rRNA (Pol I/Pol III) and 5S versus tRNA (Pol III internal control).
Acknowledgments
Acknowledgements
We thank Ulf Nehrbass and André Sentenac for their kind support, Serge Gangloff for top3 and sgs1 strains, Rolf Sternglanz for the top1-Y827F mutant, Michel Werner for useful suggestions in the course of this work, and Frank Feuerbach-Fournier, Ivan Le Masson and Annette Boese for critical reading of the manuscript. O.G. received fellowships from the Institut de Formation Supérieure Bio-Médicale and from Institut Pasteur.
References
- Aprikian P., Moorefield,B. and Reeder,R.H. (2001) New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol., 21, 4847–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin-Fukuhara M. et al. (2000) Genomic exploration of the hemiascomycetous yeasts: 11. Kluyveromyces lactis. FEBS Lett., 487, 66–70. [DOI] [PubMed] [Google Scholar]
- Briand J.F., Navarro,F., Rematier,P., Boschiero,C., Labarre,S., Werner,M., Shpakovski,G.V. and Thuriaux,P. (2001a) Partners of Rpb8p, a small subunit shared by yeast RNA polymerases I, II and III. Mol. Cell. Biol., 21, 6056–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand J.F., Navarro,F., Gadal,O. and Thuriaux,P. (2001b) Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C. and Riva,M. (1998) Yeast RNA polymerase I subunits and genes. In Paule,M.R. (ed.), Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Springer Verlag, pp. 9–38.
- Chédin S., Ferri,M.L., Peyroche,G., Andrau,J.C., Jourdain,S., Lefebvre,O., Werner,M., Carles,C. and Sentenac,A. (1998) The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harbor Symp. Quant. Biol., 63, 381–389. [DOI] [PubMed] [Google Scholar]
- Christman M.F., Dietrich,F.S., Levin,N.A., Sadoff,B.U. and Fink,G.R. (1993) The rRNA-encoding DNA array has an altered structure in topoisomerase I mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 7637–7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tanese,N. and Tjian,R. (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell, 68, 965–976. [DOI] [PubMed] [Google Scholar]
- Defossez P.A., Prusty,R., Kaeberlein,M., Lin,S.J., Ferrigno,P., Silver,P.A., Keil,R.L. and Guarente,L. (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell, 3, 447–455. [DOI] [PubMed] [Google Scholar]
- Dolinski K.J. and Heitman,J. (1999) Hmo1p, a high mobility group 1/2 homolog, genetically and physically interacts with the yeast FKBP12 prolyl isomerase. Genetics, 151, 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D., Tora,L., Egly,J.M. and Grummt,I. (1993) A TBP-containing multiprotein complex (TIF-IB) mediates transcription specificity of murine RNA polymerase I. Nucleic Acids Res., 21, 4180–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide,L. and Strunnikov,A. (2000) The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol., 149, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Mariotte-Labarre,S., Chédin,S., Quemeneur,E., Carles,C., Sentenac,A. and Thuriaux,P. (1997) A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell. Biol., 17, 1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauss,D., Kessl,J., Trumpower,B., Tollervey,D. and Hurt,E. (2001) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol., 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin,J.C., Scherthan,H., Doye,V., Rascalou,N. and Nehrbass,U. (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature, 403, 108–112. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., Soustelle,C. and Fabre,F. (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet., 25, 192–194. [DOI] [PubMed] [Google Scholar]
- Grummt I., Roth,E. and Paule,M.R. (1982) Ribosomal RNA transcription in vitro is species specific. Nature, 296, 173–174. [DOI] [PubMed] [Google Scholar]
- Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev., 62, 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Song,C.Z., Yamamoto,K., Yano,K., Maeda,Y., Yamaguchi,K. and Muramatsu,M. (1996) RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J., 15, 2217–2226. [PMC free article] [PubMed] [Google Scholar]
- Hannan R.D., Stefanovsky,V., Taylor,L., Moss,T. and Rothblum,L.I. (1996) Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocytes: implications for cardiac hypertrophy. Proc. Natl Acad. Sci. USA, 93, 8750–8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann-Le Denmat S., Werner,M., Sentenac,A. and Thuriaux,P. (1994) Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol. Cell. Biol., 14, 2905–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iben S., Tschochner,H., Bier,M., Hoogstraten,D., Hozak,P., Egly,J.M. and Grummt,I. (2002) TFIIH plays an essential role in RNA polymerase I transcription. Cell, 109, 297–306. [DOI] [PubMed] [Google Scholar]
- Jantzen H.M., Admon,A., Bell,S.P. and Tjian,R. (1990) Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature, 344, 830–836. [DOI] [PubMed] [Google Scholar]
- Keener J., Dodd,J.A., Lalo,D. and Nomura,M. (1997) Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl Acad. Sci. USA, 94, 13458–13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys D.A., Vu,L., Steffan,J.S., Dodd,J.A., Yamamoto,R.T., Nogi,Y. and Nomura,M. (1994) RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev., 8, 2349–2362. [DOI] [PubMed] [Google Scholar]
- Keys D.A., Lee,B.S., Dodd,J.A., Nguyen,T.T., Vu,L., Fantino,E., Burson,L.M., Nogi,Y. and Nomura,M. (1996) Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev., 10, 887–903. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M., Meisterernst,M. and Roeder,R.G. (1993) Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA, 90, 11508–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa M., Moir,R.D., Kolodrubetz,D. and Willis,I.M. (2001) Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA poly merase III in S.cerevisiae. Mol. Cell, 7, 309–318. [DOI] [PubMed] [Google Scholar]
- Lalo D., Steffan,J.S., Dodd,J.A. and Nomura,M. (1996) RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J. Biol. Chem., 271, 21062–21067. [DOI] [PubMed] [Google Scholar]
- Léger-Silvestre I., Trumtel,S., Noaillac-Depeyre,J. and Gas,N. (1999) Functional compartmentalization of the nucleus in the budding yeast Saccharomyces cerevisiae. Chromosoma, 108, 103–113. [DOI] [PubMed] [Google Scholar]
- Liljelund P., Mariotte,S., Buhler,J.M. and Sentenac,A. (1992) Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 89, 9302–9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lopez S., Livingstone-Zatchej,M., Jourdain,S., Thoma,F., Sentenac,A. and Marsolier,M.C. (2001) High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol. Cell. Biol., 21, 3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Kobayashi,R. and Brill,S.J. (1996a) Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem., 271, 33678–33685. [DOI] [PubMed] [Google Scholar]
- Lu J., Mullen,J.R., Brill,S.J., Kleff,S., Romeo,A.M. and Sternglanz,R. (1996b) Human homologues of yeast helicase. Nature, 383, 678–679. [DOI] [PubMed] [Google Scholar]
- Miller G., Panov,K.I., Friedrich,J.K., Trinkle-Mulcahy,L., Lamond,A.I. and Zomerdijk,J.C. (2001) hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J., 20, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsouras K., Wong,B., Arayata,C., Johnson,R.C. and Carey,M. (2002) The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol. Cell. Biol., 22, 4390–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorefield B., Greene,E.A. and Reeder,R.H. (2000) RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl Acad. Sci. USA, 97, 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. and Stefanovsky,V.Y. (2002) At the center of eukaryotic life. Cell, 109, 545–548. [DOI] [PubMed] [Google Scholar]
- Neuvéglise C., Bon,E., Lepingle,A., Wincker,P., Artiguenave,F., Gaillardin,C. and Casaregola,S. (2000) Genomic exploration of the hemiascomycetous yeasts: 9. Saccharomyces kluyveri. FEBS Lett., 487, 56–60. [DOI] [PubMed] [Google Scholar]
- Nogi Y., Vu,L. and Nomura,M. (1991) An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 88, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano,R., Dodd,J., Carles,C. and Nomura,M. (1993) Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol. Cell. Biol., 13, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe,C., Sexton,J. and Young,R. (1987) Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol., 7, 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M.A., Xu,B. and Clayton,D.A. (1993) A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol., 13, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule M.R. and White,R.J. (2000) Transcription by RNA polymerases I and III. Nucleic Acids Res., 28, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche G., Milkereit,P., Bischler,N., Tschochner,H., Schultz,P., Sentenac,A., Carles,C. and Riva,M. (2000) The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J., 19, 5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Pfleiderer,C., Rosenbauer,H. and Grummt,I. (1990) A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J., 9, 2857–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M.C., Reeder,R.H. and Hahn,S. (1992) Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II and III promoters. Cell, 69, 697–702. [DOI] [PubMed] [Google Scholar]
- Shykind B.M., Kim,J., Stewart,L., Champoux,J.J. and Sharp,P.A. (1997) Topoisomerase I enhances TFIID–TFIIA complex assembly during activation of transcription. Genes Dev., 11, 397–407. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Smid A., Riva,M., Bouet,F., Sentenac,A. and Carles,C. (1995) The association of three subunits with yeast RNA polymerase is stabilized by A14. J. Biol. Chem., 270, 13534–13540. [DOI] [PubMed] [Google Scholar]
- Smith S.D., O‘Mahony,D.J., Kinsella,B.T. and Rothblum,L.I. (1993) Transcription from the rat 45S ribosomal DNA promoter does not require the factor UBF. Gene Expr., 3, 229–236. [PMC free article] [PubMed] [Google Scholar]
- Stettler S., Chiannilkulchai,N., Hermann-Le Denmat,S., Lalo,D., Lacroute,F., Sentenac,A. and Thuriaux,P. (1993) A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet., 239, 169–176. [DOI] [PubMed] [Google Scholar]
- Stotz A. and Linder,P. (1990) The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene, 95, 91–98. [DOI] [PubMed] [Google Scholar]
- Thomas J.O. and Travers,A.A. (2001) HMG1 and 2 and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci., 26, 167–174. [DOI] [PubMed] [Google Scholar]
- Thrash C., Bankier,A.T., Barrell,B.G. and Sternglanz,R. (1985) Cloning, characterization and sequence of the yeast DNA topoisomerase I gene. Proc. Natl Acad. Sci. USA, 82, 4374–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mullem V., Landrieux,E., Vandenhaute,J. and Thuriaux,P. (2002a) Rpa12p, a conserved RNA polymerase I subunit with two functional domains. Mol. Microbiol., 43, 1105–1113. [DOI] [PubMed] [Google Scholar]
- Van Mullem V., Wery,M., Werner,M., Vandenhaute,J. and Thuriaux,P. (2002b) The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and elongator histone acetyltransferases. J. Biol. Chem., 277, 10220–10225. [DOI] [PubMed] [Google Scholar]
- Waldron C. and Lacroute,F. (1975) Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J. Bacteriol., 122, 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- Woychik N.A., Lane,W.S. and Young,R.A. (1991) Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J. Biol. Chem., 266, 19053–19055. [PubMed] [Google Scholar]
- Yamamoto R.T., Nogi,Y., Dodd,J.A. and Nomura,M. (1996) RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J., 15, 3964–3973. [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk J.C., Beckmann,H., Comai,L. and Tjian,R. (1994) Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science, 266, 2015–2018. [DOI] [PubMed] [Google Scholar]