Abstract
serpent (srp) encodes a GATA transcription factor essential for haematopoiesis in Drosophila. Previously, Srp was shown to contain a single GATA zinc finger of C-terminal type. Here we show that srp encodes different isoforms, generated by alternative splicing, that contain either only a C-finger (SrpC) or both a C- and an N-finger (SrpNC). The presence of the N-finger stabilizes the interaction of Srp with palindromic GATA sites and allows interaction with the Friend of GATA factor U-shaped (Ush). We have examined the respective functions of SrpC and SrpNC during embryonic haematopoiesis. Both isoforms individually rescue blood cell formation that is lacking in an srp null mutation. Interestingly, while SrpC and SrpNC activate some genes in a similar manner, they regulate others differently. Interaction between SrpNC and Ush is responsible for some but not all aspects of the distinct activities of SrpC and SrpNC. Our results suggest that the inclusion or exclusion of the N-finger in the naturally occurring isoforms of Srp can provide an effective means of extending the versatility of srp function during development.
Keywords: Drosophila/FOG/GATA/haematopoiesis/serpent
Introduction
Members of the GATA family are zinc finger transcription factors, conserved from yeast to vertebrates, that regulate a variety of developmental processes (Patient and McGhee, 2002). These factors bind to the consensus WGATAR DNA site (Martin and Orkin, 1990; Merika and Orkin, 1993) and contain one or two conserved zinc fingers with the characteristic Cys-X2-Cys-X17-Cys-X2-Cys spacing. In vertebrates, all GATA factors contain two distinctive zinc fingers separated by 29 amino acids and referred to as the N-finger (for N-terminal zinc finger) and C-finger (for C-terminal zinc finger), respectively. However, a number of invertebrate GATA factors, including the Drosophila GATA factor Serpent/dGATAb (Srp) (Abel et al., 1993; Rehorn et al., 1996), contain only a C-finger (Lowry and Atchley, 2000). The C-finger is responsible for DNA binding (Martin and Orkin, 1990) and for interaction with other transcription factors (Merika and Orkin, 1995; Rekhtman et al., 1999). In vertebrates, the GATA N-finger can modulate the binding of the C-finger to specific GATA sites (Trainor et al., 1996, 2000). Moreover, studies in mice have revealed a specific requirement for the GATA-1 N-finger for erythroid differentiation (Weiss et al., 1997) and for full rescue of GATA-1 loss of function (Shimizu et al., 2001). Similarly, specific mutations in the N-finger of the Drosophila GATA factor Pannier/dGATAa (Pnr) produce dominant effects on the formation of sensory bristles (Ramain et al., 1993). Among others, these data suggested that the N-finger could act as a binding domain for a cofactor regulating GATA activity. Most notably, it was shown in vertebrates and in flies that GATA N-finger mediates the interaction with transcriptional coregulators of the Friend of GATA (FOG)/U-shaped (Ush) family (Haenlin et al., 1997; Tsang et al., 1997).
FOG genes, described in mammals, Xenopus and Drosophila, code for structurally related proteins with multiple C2H2 and C2HC zinc fingers (Cubadda et al., 1997; Tsang et al., 1997; Tevosian et al., 1999; Deconinck et al., 2000). They specifically recognize the GATA N-finger through some of their C2HC fingers (Fox et al., 1998, 1999). Initially, FOG-1 was identified in mammals by a two-hybrid screen for factors that interact with the GATA-1 N-finger domain (Tsang et al., 1997). The interaction between GATA-1 and FOG-1 is necessary for erythroid and megakaryocytic differentiation (Tsang et al., 1998). Recently, human familial dyserythropoietic anaemia and thrombocytopenia has been associated with mis-sense mutations in the GATA-1 N-finger that diminish or abrogate GATA-1–FOG-1 interaction, thereby highlighting the importance of this interaction in vivo (Nichols et al., 2000; Mehaffey et al., 2001). Concomitantly with the cloning of FOG-1, we identified the Drosophila FOG homologue Ush by virtue of its antagonism of Pnr during adult neurogenesis (Cubadda et al., 1997; Haenlin et al., 1997). In addition, Drosophila Ush and vertebrate FOG-2 were shown to participate in cardiogenesis in flies and in vertebrates, inhibiting Pnr and GATA-4 functions, respectively (Fossett et al., 2000; Tevosian et al., 2000). However, the precise mode of action of the GATA–FOG complex remains unclear. In fact, FOG can either repress or enhance GATA-mediated transactivation, depending on the cell and promoter context (Tsang et al., 1997; Fox et al., 1999; Holmes et al., 1999).
Recent evidence suggests that blood cell differentiation in vertebrates and in Drosophila shares a common molecular basis (Fossett and Schulz, 2001). During Drosophila embryogenesis, blood cells (haemocytes) originate from the procephalic mesoderm and differentiate into two known lineages: plasmatocytes and crystal cells (Tepass et al., 1994; Lebestky et al., 2000). The plasmatocytes migrate throughout the embryo along several invariant paths and act as macrophages (Cho et al., 2002). They contribute to host defence by phagocyting microbes, and they play a crucial role in normal development by eliminating apoptotic bodies. This activity is largely dependent on the expression of croquemort, a member of the CD36 receptor family (Franc et al., 1999). They also participate in the synthesis of extracellular matrix components such as peroxidasin (Nelson et al., 1994). Crystal cells remain located around the proventriculus during embryogenesis and play a role in melanization, a defence-related process, during larval stages (Rizki et al., 1980).
Several genes that control blood cell formation and differentiation in Drosophila have been identified. Expression of the GATA transcription factor Srp in the procephalic mesoderm is required for the formation and differentiation of both classes of haemocytes (Rehorn et al., 1996). The transcription factor encoded by glial cell missing (gcm) is involved in plasmatocyte formation (Bernardoni et al., 1997), whereas the Runt factor Lozenge (Lz) is absolutely required for crystal cell formation (Lebestky et al., 2000). Finally, the Drosophila FOG protein, Ush, appears to repress crystal cell production (Fossett et al., 2001). All these three genes require the activity of srp since in its absence none of them is expressed in the haematopoietic anlage. Yet it is still not understood how these genes control blood cell formation at the molecular level. Of particular interest is the case of Srp and Ush. So far, all known functions of FOG proteins seem to be mediated by GATA factors (see for example Chang et al., 2002). Since Srp contains only a C-finger, it should be unable to interact with Ush. This suggests either that Ush has a GATA-independent function in haematopoiesis or that Ush acts via an uncharacterized GATA protein containing an N-finger.
In order to gain insight into the molecular mechanisms controlling blood cell formation, we sought new potential GATA protein-encoding genes in the Drosophila genome. We found that the srp locus contains an N-finger-coding exon that is alternatively spliced to give rise to proteins that contain either a C-finger (SrpC) only or both an N-finger and a C-finger (SrpNC). We have characterized these two isoforms in vitro and in vivo during haematopoieisis. Interestingly, we show that SrpC and SrpNC have both common and distinct activities. Finally, we provide evidence that Ush interacts with SrpNC and regulates its activity with respect to specific target genes. We propose that the co-expression of Srp proteins containing either one or two zinc fingers provides an extension of the regulatory properties of srp, consistent with its broad range of functions during development.
Results
serpent encodes isoforms including N and C zinc fingers
In a systematic search for GATA zinc finger-coding sequences in the Drosophila genome, we found five genes (see Materials and methods for details): dGATA-E (CG10278), dGATA-D (CG5034), pnr, grain and srp. dGATA-E and dGATA-D appear to include only a C-finger, while Pnr and Grain have already been shown to contain both an N- and a C-finger (Ramain et al., 1993; Lin et al., 1995). Interestingly, while Srp was reported previously to contain a single C-finger, our search revealed the presence of a putative exon (E4A) coding for an N-finger motif in srp. Using RT–PCR assays with various combinations of oligonucleotides (see Materials and methods and Figure 1A), we showed that E4A is expressed and that E4A and E4B are alternatively spliced to exon 5 (data not shown). In the course of these experiments, we also identified an additional splice acceptor site within E7 (Figure 1A). This downstream acceptor site in E7 is out-of-frame and leads to the deletion of the Srp glutamine-rich C-terminal region. Our data indicate that four alternatively spliced mRNAs are transcribed from srp, two encoding products with a single C-finger (SrpC and SrpCδ) and two encoding products with both N- and C-fingers (SrpNC and SrpNCδ) (Figure 1A). Interestingly, in SrpNC and SrpNCδ, the two fingers present the same conserved organization as in other GATA factors (Figure 1B). Notably, they are separated by 29 amino acids, as in all vertebrate GATA. In the following experiments, we used the two isoforms that contain the full-length exon 7, i.e. srpC and srpNC, to address the functional consequences of the alternative splicing of E4A and E4B.
Fig. 1. (A) Schematic representation of the srp locus and alternatively spliced transcripts. The location and orientation of the primers used for RT–PCR analysis are indicated. The non-coding regions in srp transcripts are indicated as open boxes. Exons 4A and 5, coding for the N- and the C-finger, respectively, are indicated as grey boxes. Exon 4A starts at position 121 031 and ends at 121 202 with reference to Drosophila scaffold region AE003711 (Flybase). Similarly, the internal splice acceptor site in exon 7 is located at position 123 288. (B) Alignment of the GATA zinc finger domains of SerpentNC with Pannier, Grain, mouse GATA-1, -2, -3 and -4 and Caenorhabditis elegans Elt-1. Arrows above the SrpNC sequence delineate the region coded by exon 4A. Conserved residues in each column are coloured according to the consensus character assigned to that column; brown, R and K; green, N, T, Q and S; purple, E and D; blue, I, F, L, A, V and M; red, H and Y; orange, G; light green, P; and yellow, C. (C) Semi-quantitative RT–PCR analysis of srp transcripts containing either exon 4A or exon 4B. RT–PCR was performed with E3 and E5 primers on RNA extracted from stage 5–14 embryos. Aliquots of the PCR were taken after different numbers of cycles. Products containing exon 4A or 4B were resolved on acrylamide gel, and their relative amount was quantified.
SrpNC shows specific features of a two-fingered GATA factor
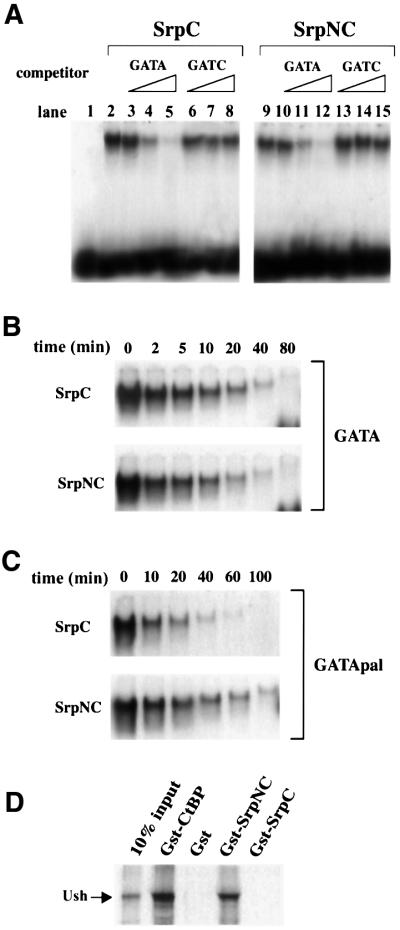
We first determined whether SrpC and SrpNC displayed different properties in vitro. While the C-finger is necessary and sufficient for specific DNA binding, it has also been shown in vertebrates that the N-finger can stabilize the binding to particular double GATA sites (Trainor et al., 1996). We tested, by electophoretic mobility shift assays (EMSAs), whether SrpNC and SrpC had similar DNA-binding properties. As shown in Figure 2A (lanes 2 and 9), both in vitro translated SrpC and SrpNC proteins bound to an oligonucleotide containing a consensus GATA site. The binding was specific, since it could be competed out efficiently by an excess of cold GATA oligonucleotide (Figure 2A, lanes 3–5 and 10–12), but not by an excess of the GATC oligonucleotide (Figure 2A, lanes 6–8 and 13–15). The stability of the SrpN and SrpNC complex on a single or on a palindromic GATA site was assessed by dissociation experiments. While the rate of dissociation was similar for SrpC and SrpNC on a single GATA probe (Figure 2B), SrpNC bound more stably than SrpC to the palindromic GATA sites (Figure 2C).
Fig. 2. (A–C) EMSAs using in vitro translated SrpC or SrpNC proteins. (A) SrpC and SrpNC both specifically bind a consensus GATA probe. Increasing concentrations (5- to 500-fold excess) of unlabelled wild-type (GATA) or mutant (GATC) competitors were added to the reaction as indicated in the upper part of the panel. No GATA-binding activity was observed with unprogrammed reticulocyte lysate (lane 1). (B and C) SrpC and SrpNC have distinct site-dependent binding properties as revealed by dissociation rate assays. After formation of the complexes between SrpC (upper panel) or SrpNC (lower panel) with either a single GATA site (B) or a palindromic double GATA site (C), an excess of the corresponding unlabelled oligonucleotides was added to the reaction mix and samples were loaded on the gel at various times, as indicated. (D) Only SrpNC interacts with Ush in vitro. Equivalent molar amounts of the GST fusion proteins were tested for their interaction with in vitro translated 35S-labelled Ush as indicated in the upper part of the panel.
The GATA N-finger allows interaction with cofactors of the FOG family (Fox et al., 1998). Key residues that are required for the interaction between GATA and FOG are conserved in the Srp N-finger. In order to test the binding between Ush and srp products, we performed pull-down assays in vitro. We found that in vitro translated [35S]methionine-labelled Ush bound to GST–SrpNC, but not to GST alone nor to GST–SrpC (Figure 2D). Thus, Ush specifically interacts with Srp isoforms that contain the N-finger. In addition, like its vertebrate homologues (Fox et al., 1999), Ush interacted with the transcriptional corepressor dCtBP in this assay (Figure 2D).
Taken together, our results indicate that SrpNC displays features characteristic of two-fingered GATA factors. The two types of naturally occurring isoforms encoded by srp (with or without the N-finger) have different DNA-binding properties, and only the isoforms including an N-finger can interact with Ush.
srpC and srpNC transcripts have identical expression patterns
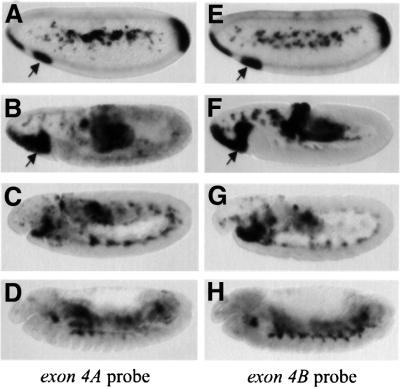
In order to determine whether a spatial regulation of the alternative splicing leading to SrpC and SrpNC occurs during embryonic development, we assessed the distribution of the corresponding srp transcripts by in situ hybridization using specific probes for exon 4A or 4B. At the blastoderm stage and during gastrulation, srpC and srpNC show the same expression pattern (Figure 3A, B, E and F). They are expressed in the procephalic mesoderm, the haemocyte primordium (arrows in Figure 3), at the anterior and posterior pole, in the primordium of the anterior and posterior midgut as well as in the amnioserosa and in the yolk cells. Later, during germ band extension, and after germ band retraction, srpC and srpNC are expressed identically in the developing fat body (Figure 3C, D, G and H) (for a full description of srp expression see Rehorn et al., 1996). Thus, srpC and srpNC transcripts are not differentially regulated spatially during embryonic development. However, the level of the transcripts is not identical. Indeed, by means of semi-quantitative RT–PCR, we determined that exon 4B-containing mRNA is five times more abundant than exon 4A-containing mRNA (Figure 1C), suggesting that two-fingered isoforms of Srp are less abundant than single-fingered isoforms.
Fig. 3. The transcripts containing either exon 4A (srpNC and srpNCδ) or exon 4B (srpC and srpCδ) have similar expression patterns during embryogenesis. Side views of stage 5 (A and E), stage 8 (B and F), stage 11 (C and G) or stage 14 (D and H) embryos hybridized with an RNA probe directed against either exon 4A (A–D) or exon 4B (E–H). Arrows indicate the expression of srp in the haemocyte primordium.
SrpC and SrpNC differ in their capacity to activate certain target genes in vivo
In order to analyse SrpC and SrpNC activities, we tested their capacities to activate gene expression in vivo during Drosophila embryonic haematopoiesis. Using the UAS-GAL4 system, we ectopically expressed them in the mesoderm and we then assessed the expression pattern of various haematopoietic markers.
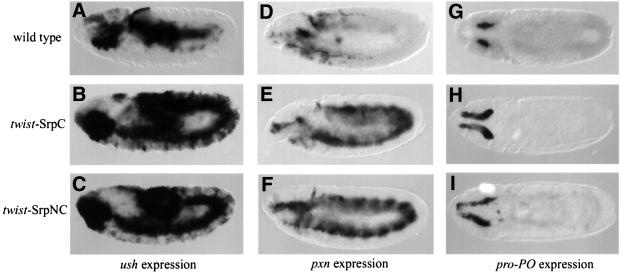
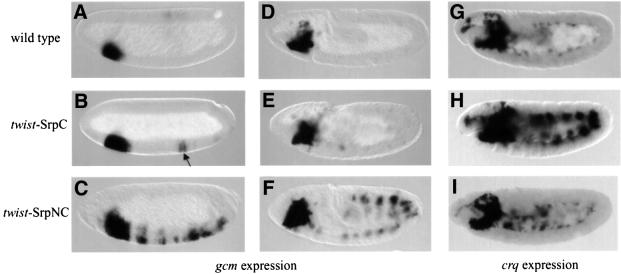
The two genes ush and gcm play critical roles in embryonic haematopoiesis. Their expression in the haematopoietic primordium occurs early and appears to depend on srp activity (Bernardoni et al., 1997; Fossett et al., 2001). Therefore, we decided to determine whether they are transcriptional targets of SrpC and/or SrpNC. We found that, whereas in a wild-type early embryo, ush expression is restricted to the anterior mesoderm, twist-driven expression of SrpC (twist-SrpC) or SrpNC (twist-SrpNC) induced strong expression of ush throughout the mesoderm (Figure 4, compare A with B and C). In contrast, twist-SrpC induced gcm expression poorly and in a limited number of mesodermal cells of stage 5 embryos (arrow in Figure 5B), whereas twist-SrpNC strongly activated gcm expression segmentally from stage 5 to 9 (Figure 5C and F).
Fig. 4. SrpC and SrpNC have a similar capacity to activate ectopically the expression of ush, pxn and pro-PO. (A–C) Side views of ush mRNA expression in stage 10 embryos. (D–F) Side views of pxn mRNA expression in stage 11 embryos. (G–I) Dorsal views of pro-PO mRNA expression in stage 11 embryos. (A, D and G) Wild type, (B, E and H) twist-Gal4; UAS-SrpC and (C, F and I) twist-Gal4; UAS-SrpNC.
Fig. 5. SrpC and SrpNC differentially activate gcm and crq expression. (A–F) Side views of gcm mRNA expression in stage 5 (A–C) or stage 9 (D–F) embryos. (G–I) Side views of crq mRNA expression in stage 10 embryos. (A, D and G) Wild type, (B, E and H) twist-Gal4; UAS-SrpC and (C, F and I) twist-Gal4; UAS-SrpNC. The arrow in (B) points to the few cells that express gcm ectopically in response to SrpC.
Next we looked at the expression of haematopoietic lineage-specific markers. As plasmatocyte markers, we used peroxidasin (pxn) and croquemort (crq). Since, Rizki et al. (1980) suggested that crystal cells are the only source of prophenoloxidase (pro-PO) in Drosophila, we used expression of this gene to monitor crystal cell formation. pro-PO transcripts were indeed detected in these cells from early stage 11 to the end of embryogenesis. We confirmed that pro-PO expression is specific to the crystal cells since it was not detected in lz mutant embryos (data not shown). Analysing these markers, we again observed two situations. On the one hand, twist-SrpC and twist- SrpNC had similar abilities to induce expression of the plasmatocyte marker pxn and of the crystal cell marker pro-PO (Figure 4E and F, and H and I, respectively). On the other hand, expression of crq was induced by twist-SrpC but not by twist-SrpNC (Figure 5, compare H with I). Note that pxn and crq were induced through most of the mesoderm, while pro-PO activation was restricted to the head region.
Taken together, our data show that SrpC and SrpNC have both common and different activities during haematopoiesis. Indeed, both isoforms activated the expression of ush, pxn and pro-PO in a similar manner. However, SrpC and SrpNC differentially stimulate the expression of crq and gcm, respectively, in the mesoderm.
SrpC and SrpNC both induce the crystal cell and plasmatocyte lineages in an srp null embryo
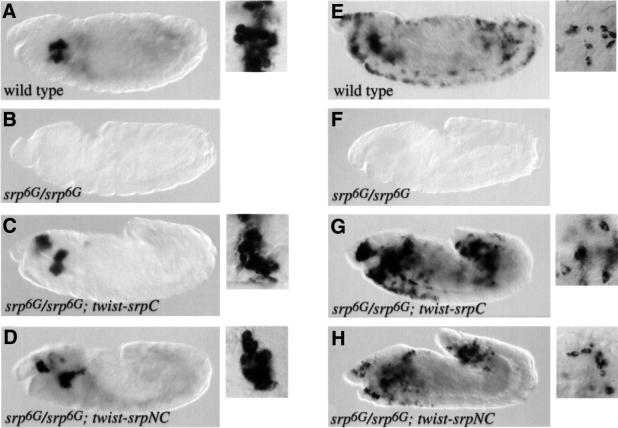
srp is absolutely required for determination of all haematopoietic lineages (Rehorn et al., 1996; Lebestky et al., 2000). However, the relative contributions of SrpC and SrpNC to this process are unknown, since there is no known mutation in srp that affects only one of the two classes of isoforms. To address this question, we asked whether SrpC and SrpNC individually could rescue the plasmatocyte and/or crystal cell lineages in an srp mutant background. During haematopoiesis, srp expression is first detected at the blastoderm stage in a patch of cells within the mesoderm. Therefore, we used the twist-Gal4 driver to express UAS-SrpC or UAS-SrpNC in the mesoderm of srp mutant embryos. To monitor crystal cell formation and plasmatocyte formation, we assessed the expression of pro-PO and pxn, respectively. Whereas no pro-PO expression was detected in srp mutant embryos, pro-PO expression was restored around the proventriculus and in an additional patch of cells located above the pharynx upon expression of twist-SrpC or twist-SrpNC (Figure 6, compare B with C and D). This phenotype is equivalent to that which we observed previously in wild-type embryos expressing twist-SrpC or twist-SrpNC. Higher magnification views showed that the pro-PO-expressing cells had the typical morphology of crystal cells. Similarly, no expression of pxn was detected in srp mutant embryos, while scattered pxn-positive cells were observed in srp– embryos expressing twist-SrpC or twist-SrpNC (Figure 6, compare F with G and H). Morphological analysis confirmed that these cells were genuine plasmatocytes (Tepass et al., 1994). Thus our results suggest that SrpC and SrpNC are each able to induce the formation of both lineages.
Fig. 6. SrpC and SrpNC individually can rescue the lack of crystal cells and plasmatocytes due to an srp null mutation. In situ hybridization revealing pro-PO (A–D) or pxn (E–H) expression in stage 13–14 embryos. (A and E) Wild type, (B and F) srp6G/srp6G, (C and G) twist-Gal4/+; UAS-SrpC; srp6G/srp6G and (D and H) twist-Gal4/UAS-SrpNC; srp6G/srp6G. Ten-fold higher magnifications of labelled cells are shown to the right of the wild-type and rescued embryo panels.
Misexpression of SrpC or SrpNC controls crystal cell formation independently of Ush
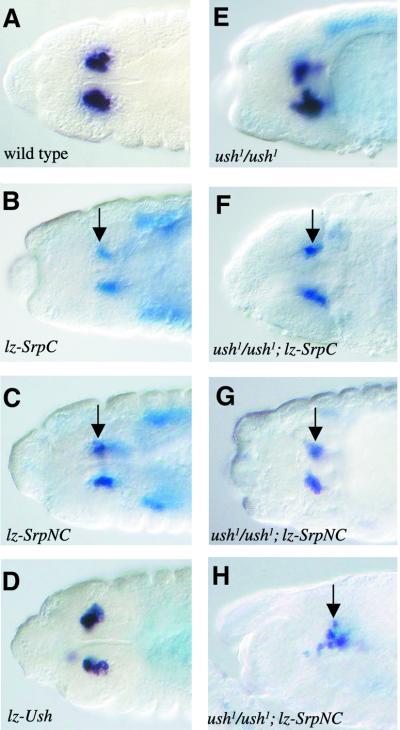
Previous studies have shown that ush acts to repress crystal cell formation. Notably, Fossett et al. (2001) showed that lz-Gal4-driven expression of UAS-Ush led to a variable decrease in the number of crystal cells, as monitored by the expression of a Uas-lacZ reporter gene driven by lz-Gal4. However, the molecular mechanism of action of ush in this process remained largely unknown. The expression of SrpNC during haematopoiesis suggested that it might act with Ush. Thus we decided to analyse the consequences of misexpressing either SrpC or SrpNC in the crystal cells. Because only SrpNC can interact with Ush, we surmised that SrpNC, but not SrpC, would repress crystal cell formation. We used the lz-Gal4 driver to express UAS-SrpC, UAS-SrpNC or UAS-Ush in the crystal cells. Misexpression of each transgene was visualized by in situ hybridization with a probe against srp or ush, respectively, and differentiation of the crystal cells was monitored by analysing pro-PO expression. As expected, UAS-Ush induced a reduction in the number of crystal cells in stage 13–16 embryos (Figure 7, compare A with D). Surprisingly, both UAS-SrpC and UAS-SrpNC also reduced the number of crystal cells (Figure 7B and C). Note that the reduction induced by SrpC or SrpNC was greater than that observed with Ush, although, as for Ush, we observed considerable variations of phenotype within the population of embryos. Most interestingly, we observed strong expression of UAS-SrpC or UAS-SrpNC driven by lz-Gal4 and no expression of pro-PO in the same cells in stage 13–16 embryos (arrows in Figure 7B and C). In contrast, lz-Gal4-driven expression of UAS-Ush did not prevent pro-PO expression in these cells (Figure 7D). This suggests that SrpC and SrpNC can control both the number and the differentiation of the crystal cells, while Ush only affects their number.
Fig. 7. Forced expression of SrpC or SrpNC in the crystal cells represses their formation independently of ush. Side views (A–G) or dorsal view (H) of stage 13 embryos processed to reveal pro-PO mRNA expression (black staining) and either srp mRNA expression (blue staining in B, C, F, G and H) or ush mRNA expression (blue staining in D). (A) Wild type, (B) lz-Gal4/+; UAS-SrpC, (C) lz-Gal4/+; UAS-SrpNC, (D) lz-Gal4/+; UAS-Ush, (E) ush1/ush1, (F) lz-Gal4/+; ush1/ush1; UAS-SrpNC and (G and H) lz-Gal4/+; ush1/ush1; UAS-SrpNC. Arrows indicate crystal cells expressing srp but not pro-PO.
However, it still remained possible that SrpNC was acting in a complex with Ush to prevent crystal cell formation and differentiation. Additionally, the effect of SrpC and SrpNC on crystal cell number could be related to Ush activity, since SrpC and SrpNC can induce its expression (at least in the mesoderm, see above). In order to test this hypothesis, we misexpressed UAS-SrpC or UAS-SrpNC under the control of lz-Gal4 in an ush null mutant background. As shown in Figure 7F, G and H, even under these conditions, SrpC and SrpNC reduced the number of crystal cells and repressed pro-PO expression. Therefore, SrpC and SrpNC can both inhibit crystal cell formation and differentiation independently of ush activity.
The SrpNC–Ush complex represses crq expression
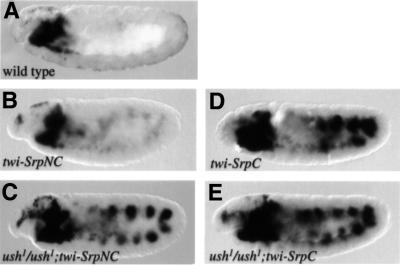
We have shown that the misexpression of SrpC, but not SrpNC, activates ectopic expression of crq. Conversely, SrpNC is a much stronger activator of gcm expression than SrpC. These differences in activity on particular target genes could be due to the capacity of SrpNC, unlike SrpC, to form a complex with Ush. Given that twist-SrpNC induces ush expression in the mesoderm, it is possible that Ush exerts a feedback action on SrpNC, preventing it from activating crq expression and/or enhancing its capacity to activate gcm. We decided to check this possibility by assaying the expression of crq and gcm in an ush mutant embryo expressing twist-SrpNC. The absence of ush had no detectable effect on the activation of gcm by twist-SrpNC, showing that ush is not involved in this specific activity of SrpNC (data not shown). On the contrary, we observed ectopic expression of crq by twist-SrpNC in the absence of ush function (Figure 8, compare A and B with C). Note that twist-SrpC-mediated activation of crq was similar in a wild-type and ush mutant embryo (Figure 8, compare D with E). These data strongly suggest that Ush can form a complex with SrpNC, thereby modulating the transactivation of crq.
Fig. 8. ush inhibits SrpNC-mediated activation of crq. Side views of crq mRNA expression in stage 10 embryos. (A) Wild type, (B) twist-Gal4/+; UAS-SrpNC, (C) twist-Gal4/+; ush1/ush1; UAS-SrpNC, (D) twist-Gal4/+; UAS-SrpC and (E) twist-Gal4/+; ush1/ush1; UAS-SrpC.
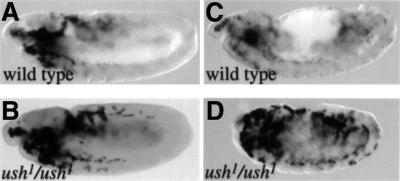
This also led us to consider that ush might repress the expression of crq in the wild-type embryo. We thus assayed crq expression in an ush mutant. In wild-type stage 11 embryos, the expression of crq was barely detectable in migrating plasmatocytes, especially those localized beyond the head (Figure 9A). However, the level of expression of crq was increased in ush mutants, and crq-positive plasmatocytes were clearly visible in the trunk region (Figure 9B). Moreover, in ush embryos, the expression of crq persisted longer than in wild type (Figure 9, compare C with D). Note that this effect was specific to crq, as the expression of pxn in the plasmatocytes appeared normal in ush mutant embryos (data not shown). These observations are consistent with a role for Ush in the control of crq expression in association with SrpNC. In conclusion, these results support the idea that some, but not all, of the differential activities of SrpC and SrpNC in vivo depend on a physical interaction between SrpNC and Ush.
Fig. 9. ush downregulates crq expression. Side views of crq mRNA expression in stage 11 (A and B) or 14 (C and D) embryos. (A and C) Wild type, (B and D) ush1/ush1.
Discussion
We have identified new isoforms, encoded by the previously characterized gene srp, which are produced by alternative splicing. The alternative use of exon 4A and 4B allows the production of GATA proteins containing either a single C-finger or both N- and C-fingers. Analysis of the expression pattern of these isoforms indicated that the splicing mechanism is not spatially regulated and thus that isoforms with one or two fingers most probably co-exist in the same cells. We also identified a new splice acceptor site within exon 7. Use of this internal site leads to the synthesis of Srp proteins with a shorter C-terminal region. The Srp C-terminal domain has no significant homology to other proteins suggestive of a possible function. Here, we have focused our study on the functional differences of GATA factors harbouring one or two zinc fingers and on the characterization of the two isoforms SrpC and SrpNC.
SrpC and SrpNC have common and different features
It is remarkable that srp encodes both single and dual zinc finger-containing products. Our results provide strong evidence that this alternative splicing allows production of transcription factors with specific activities. On the one hand, the two isoforms activated the expression of ush and pxn with similar efficiency, suggesting that SrpC and SrpNC have similar transactivating properties in vivo. On the other hand, SrpC, but not SrpNC, activated crq expression, while SrpNC was a much stronger activator of gcm expression than SrpC. The domain coded by exon 4B that is present only in SrpC has no known motif and we do not know if and how it participates in SrpC-specific function. However, the presence of the N-terminal zinc finger encoded by exon 4A may explain some of the distinct features of SrpNC as discussed below.
We show that, as in the case of vertebrate GATA-1, the presence of the N-finger in Srp stabilizes binding to double palindromic GATA sites. Although the N-finger of GATA-1 modulates the binding and the transactivating properties of GATA-1 on synthetic promoters (Trainor et al., 2000), the functional importance of these effects has remained elusive, particularly as no GATA-1 isoform contains only the C-finger. In the case of srp, these distinct binding properties may have direct functional consequences. For instance, the fact that SrpC and SrpNC activate a common target, ush, whereas only SrpNC strongly activates a specific target, gcm, could be related to the DNA-binding specificity of the two isoforms. A scan of the ush upstream regulatory region shows that it contains several GATA consensus sequences, nine of which are clustered in <1 kb and are organized as three repetitions of three sites. In contrast, GATA sites are far less frequent in gcm regulatory regions and are often organized in palindromes. Considering that ush and gcm are likely to be direct target genes for srp, the different organization of their regulatory regions may explain the differential effect we observed.
SrpC and SrpNC can both induce blood cell formation in Drosophila
We were able to rescue the lack of plasmatocyte and crystal cell formation due to an srp null mutation by expressing SrpC or SrpNC in the mesoderm. No difference between the two isoforms was seen in this assay, suggesting that the N-finger is not absolutely required for srp function in embryonic blood cell formation. However, in the absence of a functional test, we cannot determine to what extent the formation of embryonic blood cells is fully rescued. Interestingly, rescue experiments with the mouse GATA-1 mutant indicate that the GATA-1 N-finger is dispensable for primitive erythro poiesis but is required for definitive erythopoiesis (Shimizu et al., 2001). In Drosophila, a second wave of haematopoiesis, occurring at the larval stage, gives rise to four different lineages: plasmatocytes, crystal cells, secretory cells and lamellocytes (Lanot et al., 2001). srp is expressed in the dorsal lymph gland (i.e. the main larval haematopoietic organ) and it probably controls larval haematopoiesis (Lebestky et al., 2000). By analogy to vertebrate GATA-1, the Srp N-finger may provide an additional function for larval haematopoiesis, perhaps during formation of the new cell types.
In our assay, the expression of the transgene was limited to the mesoderm but it still rescued blood cell formation. This finding suggests that the early expression of srp in the haematopoietic primordium is sufficient to initiate the genetic programme that controls haemocyte formation and differentiation. Interestingly, in the wild-type embryo, srp transcripts are not expressed detectably in haemocytes after stage 11, but Srp protein is detected in plasmatocytes and crystal cells throughout most of embryogenesis (Sam et al., 1996; Lebestky et al., 2000). Persistence of srp products in haemocytes might be critical for srp function, and control of srp products at the post-translational level may play a crucial role in the correct regulation of blood cell differentiation. Rescue of crystal cell formation by mesodermal expression of SrpC and SrpNC contrasts with the observation that later expression driven by lz-Gal4 in crystal cells represses their development. Lebetsky et al. (2000) reported that Srp levels were reduced in crystal cells compared with surrounding plasmatocytes. Therefore, our results are consistent with a two-step model in which Srp expression is first necessary to induce lz expression and subsequently is downregulated to allow crystal cell differentiation.
Ush regulates SrpNC activity on a specific target gene
One of the best characterized features of GATA N-fingers is their dimerization with cofactors of the FOG family. Consistent with this feature, we found that SrpNC interacts with the Drosophila FOG Ush, but SrpC does not. Previous analysis showed that ush regulates the number of crystal cells (Fossett et al., 2001). It was proposed that this function of ush could be mediated by a putative isoform of Srp containing an N-finger. Our findings strongly support this hypothesis. However, it was not possible to address this issue directly, since both SrpC and SrpNC display a strong Ush-independent repressive effect on crystal cell formation and differentiation.
A new function of ush revealed here is the regulation of the level of expression of the macrophage receptor crq, suggesting that ush displays a broader function in haematopoiesis than previously assumed. Notably, we provide evidence that Ush modulates SrpNC transactivation of crq. As Ush interacts with the corepressor dCtBP in vitro, the Ush–SrpNC complex could repress crq expression. However, we do not know whether crq is a direct target of srp, so we cannot rule out the possibility that the Ush–SrpNC complex activates a transcriptional repressor that regulates crq. Vertebrate FOGs can act as either a coactivator or a corepressor of GATA factors (Tsang et al., 1997; Crispino et al., 1999; Fox et al., 1999; Deconinck et al., 2000). In Drosophila, Ush was clearly shown to be a repressor of Pannier-induced activation in cell culture, and it probably also represses the expression of achaete in the dorso-central proneural cluster in vivo (Cubadda et al., 1997; Haenlin et al., 1997; Garcia-Garcia et al., 1999). Furthermore, in a heterologous assay in Drosophila, the CtBP-binding region of mFOG2 was shown to be required for repressing the formation of crystal cells but not cardiac cells (Fossett et al., 2000, 2001). Thus several mechanisms seem to regulate the function of the GATA–FOG complex.
Remarkably, some functions of SrpNC appear to be independent of Ush. Thus, gcm-specific activation by SrpNC is not affected in an ush mutant embryo. Moreover, SrpNC still represses crystal cell formation in the absence of ush. This is reminiscent of mouse erythropoiesis, where both FOG-dependent and FOG-independent regulation of gene expression by GATA-1 have been observed (Crispino et al., 1999). The molecular mechanisms underlying the regulation by Ush/FOG-1 of SrpNC/GATA-1 activity on some specific targets remain to be elucidated. It is tempting to speculate that the N-finger of SrpNC is involved in the recognition of promoter sequences, on gcm for example, and thus is not available to recruit Ush. Alternatively, other cofactors already localized to the promoter or bound to SrpNC might prevent Ush binding to the N-finger.
srp is a structural and functional homologue of vertebrate GATA genes
We have focused our study on haematopoiesis, but srp also participates in other developmental processes, such as germ band retraction (Frank and Rushlow, 1996), midgut differentiation (Reuter, 1994), fat body formation (Hayes et al., 2001), induction of the immune response (Petersen et al., 1999) and the ecdysone response (Brodu et al., 1999). It will be interesting to determine the respective roles of SrpC and SrpNC in these different phenomena. Phylogenetic analysis shows that SrpNC is closely related to vertebrate GATA factors. It has been suggested that srp is a functional homologue of the entire vertebrate GATA family, since srp is required in Drosophila for haematopoiesis, like GATA-1/2/3 in mice, and for endodermal development, like GATA-4/5/6 (Rehorn et al., 1996). Nevertheless, this hypothesis was at odds with the fact that Srp seemingly had a single zinc finger while all the vertebrate GATAs have two (Lowry and Atchley, 2000). The present identification of Srp isoforms with two fingers gives new force to this hypothesis. Further, the expression of isoforms of Srp with distinct activities helps to account for the broad range of functions ensured by this gene.
It is worth noting that alternative splicing eliminating the N-finger has also been described in Bombyx mori GATAβ (Drevet et al., 1995) and in chicken GATA-5 genes (MacNeill et al., 1997). Moreover, a BLAST search analysis revealed alternatively spliced human expressed sequence tags coding for two isoforms of a potential GATA factor with either one or two zinc fingers (L.Waltzer, unpublished results). This suggests that alternative splicing of GATA genes could be more general than previously thought, and as yet unnoticed splice variants of GATA vertebrate genes may generate proteins with only a C-finger.
In conclusion, our results shed further light on the molecular control of haematopoiesis by the GATA factor Srp. The alternative splicing of srp gives rise to different Ush-interacting and non-interacting Srp proteins with different target gene specificities, thereby contributing to the exquisite control of Drosophila blood cell formation. We speculate that alternative splicing of the GATA N-finger might be an important mechanism regulating the activity of other GATA genes from insects to man.
Materials and methods
Fly stocks
The twist-Gal4, lz-Gal4, srp6G, ush1 stocks were provided by the Drosophila Stock Center, Bloomington, IN. Several Uas-SrpC and Uas-SrpNC transgenic lines were generated by P-element-mediated germline transformation of the pUAST-srpC and pUAST-srpNC plasmids, respectively, into w1118 embryos according to standard protocols. Uas-Ush transgenic lines have already been described in Haenlin et al. (1997).
Embryos obtained from the mating of Uas-SrpC, Uas-SrpNC or Uas-Ush to twist-Gal4 or lz-Gal4 flies were collected at 25°C. To analyse the phenotype of the rescued srp mutant by SrpC or SrpNC, twist-Gal4; srp6G e/TM3, twist-lacZ females were crossed to Uas-SrpC; srp6G e/TM3, twist-lacZ or to Uas-SrpNC/Y, srp6G e/TM3, twist-lacZ males. lacZ staining was used to genotype the embryos. To analyse the effect of overexpression of SrpC or SrpNC on the production of crystal cells in an ush mutant background, ush1/CyO; Uas-SrpC or ush1/CyO; Uas-SrpNC females were crossed to lz-Gal4/Y, ush1/CyO males. In order to observe the effect of mesodermal expression of SrpNC in ush mutant embryos, twist-Gal4; ush1/Cyo females were crossed to ush1/CyO; Uas-SrpNC males. ush embryos were identified by their retraction phenotype. Embryos overexpressing SrpC or SrpNC were identified after in situ hybridization against srp.
Database search
In order to find all the potential GATA factor-encoding genes in the Drosophila genome, we used either the consensus GATA-type zinc finger sequence defined in PRODOM (reference PD000513), or the Drosophila GATA Pannier N-finger or C-finger sequences. These sequences were used as queries in three independent iterative PSI-BLAST searches against the database of predicted proteins encoded by the Drosophila genome (BLASTP). Pannier C-finger was also used as a query in a TBLASTN search against the whole Drosophila genomic sequence. Similar results were obtained in all searches.
RT–PCR
Total RNA was isolated from dechorionated embryos using Trizol™ according to the manufacturer’s instructions. A 2 µg aliquot of RNA was used as a template in a 20 µl reverse transcription reaction with 0.5 µM srp-specific oligonucleotide reverse primer or 1 µM oligo(dT) primer. Reverse transcription reaction mixture (0.4 µl) was then used in a standard PCR in the presence of 0.5 µM specific primers. The RT–PCR products were checked on agarose gels, subcloned into pGemT easy vector (Promega) and sequenced.
To compare the levels of exon 4A- versus exon 4B-containing transcripts, reverse transcription was performed using the E5 primer. The PCR was performed in the presence of 0.5 µM E3 primer and a 0.5 µM 32P end-labelled E5 primer. From cycle 20 onward, aliquots were preserved for analysis every two cycles to ensure that amplification was in the logarithmic phase. To facilitate the separation of exon 4A- and exon 4B-specific bands, the RT–PCR products were digested by PstI before being run on a 4% acrylamide gel and quantified with a phosphoimager. RT–PCR experiments were repeated with three different preparations of RNA and gave similar results.
Forward primers used were the following: E2, 5′-TTATGCTGGC TCGTTGCTTACTC-3′; E3, 5′-ATACCTGGTTCGATCCGTTAAGC-3′, E4AS: 5-GTCAATGTGGTGCGATTTCAAC-3′; and E4BS, 5′-TG AATCAGGCGGGGATTTCTAT-3′. Reverse primers used were the following: E4AR, 5′-GGCTGTTTAATTAGGGGTCGATTC-3′; E4BR, 5-GGCGCGACTAACTGCTCGTCG-3′; E5, 5′-ATGGTGTCCTTTTT CATGGTCAGT-3′; and E7, 5′-CAGCGTGTCGCGCCTACTCC-3′.
Plasmids
pBS-KS Srp, containing the SrpC open reading frame (ORF) (Brodu et al., 1999), was a gift from C.Antoniewski. The full-length SrpNC ORF was cloned into pBS-KS by PCR. The resulting plasmid, pBS-SrpNC, was checked by sequencing. The SrpC or SrpNC ORF was subcloned into pUAST (for transgenesis) or into pGEX2TK (for GST fusion protein expression) by standard cloning techniques.
In situ hybridizations
In situ hybridizations were carried out as described previously using a Dig-UTP- or fluorescein-UTP-labelled antisense RNA probe (Peyrefitte et al., 2001). RNA probes for srp, ush, gcm and lacZ have been described previously. To generate RNA probes for srp exon 4A, srp exon 4B, crq exon 3 or pro-PO exon 3, the corresponding DNA sequences were cloned by PCR in pGemTeasy. The corresponding antisense RNAs were transcribed in vitro using T7 or SP6 RNA polymerase.
Pull-down assays
pGEX2TK-SrpC, pGEX2TK-SrpNC, pGEX-dCtBP (a generous gift from M.Levine) and pGEX2TK plasmids were used to produce GST–SrpC, GST–SrpNC, GST–dCtBP and GST proteins, respectively, in Escherichia coli BL21. pBS-Ush cDNA (Cubadda et al., 1997) was used as a template to produce full-length Ush protein in vitro using a coupled transcription/translation system (Promega) in the presence of [35S]methionine. Interaction assays were performed as described in Waltzer and Bienz (1999).
Electrophoretic mobility shift assays
The following double-stranded oligonucleotides were used in EMSAs: GATA (5′-CTCCGGCAACTGATAAGGACTCCC-3′), GATC (5′-CT CCGGCAACTGATCAGGACTCCC-3′) and GATApal (5′-CTCCGGC AACTATCAGATAAGGACTCCC-3′). EMSAs were performed by incubating in vitro translated SrpC or SrpNC with 5 × 104 c.p.m. of 5′-end 32P-labelled double-stranded GATA probe for 30 min at room temperature in 10 mM HEPES pH 7.9, 50 mM KCl, 1 mM MgCl2, 1 mM EDTA, 5% glycerol and 0.5 µg poly(dI–dC) in a final volume of 20 µl. The stability of the protein–DNA complexes was assessed by dissociation rate experiments as described in Trainor et al. (1996), using a 200-fold excess of unlabelled probe. The reactions were loaded on to a 6% poly acrylamide gel with 0.5× TBE and run at room temperature at 15 V/cm. The protein–DNA complexes were visualized by autoradiography.
Acknowledgments
Acknowledgements
We are grateful to Professor J.Smith and Dr B.G.Monster for critically reading the manuscript, and B.Augé for expert technical assistance. This work was supported by the Centre National de Recherche Scientifique (CNRS) and grants from the Association pour la Recherche sur le Cancer (ARC), from the Fondation pour la Recherche Médicale (FRM) and from the Ministère de l‘Education Supérieure et la Recherche (MESR) of France. S.P. was financed by graduate fellowships from the MESR and ARC.
References
- Abel T., Michelson,A.M. and Maniatis,T. (1993) A Drosophila GATA family member that binds to Adh regulatory sequences is expressed in the developing fat body. Development, 119, 623–633. [DOI] [PubMed] [Google Scholar]
- Bernardoni R., Vivancos,B. and Giangrande,A. (1997) glide/gcm is expressed and required in the scavenger cell lineage. Dev. Biol., 191, 118–130. [DOI] [PubMed] [Google Scholar]
- Brodu V., Mugat,B., Roignant,J.Y., Lepesant,J.A. and Antoniewski,C. (1999) Dual requirement for the EcR/USP nuclear receptor and the dGATAb factor in an ecdysone response in Drosophila melanogaster. Mol. Cell. Biol., 19, 5732–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A.N., Cantor,A.B., Fujiwara,Y., Lodish,M.B., Droho,S., Crispino,J.D. and Orkin,S.H. (2002) GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl Acad. Sci. USA, 99, 9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N.K., Keyes,L., Johnson,E., Heller,J., Ryner,L., Karim,F. and Krasnow,M.A. (2002) Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell, 108, 865–876. [DOI] [PubMed] [Google Scholar]
- Crispino J.D., Lodish,M.B., MacKay,J.P. and Orkin,S.H. (1999) Use of altered specificity mutants to probe a specific protein–protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell, 3, 219–228. [DOI] [PubMed] [Google Scholar]
- Cubadda Y., Heitzler,P., Ray,R.P., Bourouis,M., Ramain,P., Gelbart,W., Simpson,P. and Haenlin,M. (1997) u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev., 11, 3083–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deconinck A.E., Mead,P.E., Tevosian,S.G., Crispino,J.D., Katz,S.G., Zon,L.I. and Orkin,S.H. (2000) FOG acts as a repressor of red blood cell development in Xenopus. Development, 127, 2031–2040. [DOI] [PubMed] [Google Scholar]
- Drevet J.R., Swevers,L. and Iatrou,K. (1995) Developmental regulation of a silkworm gene encoding multiple GATA-type transcription factors by alternative splicing. J. Mol. Biol., 246, 43–53. [DOI] [PubMed] [Google Scholar]
- Fossett N. and Schulz,R.A. (2001) Functional conservation of hematopoietic factors in Drosophila and vertebrates. Differentiation, 69, 83–90. [DOI] [PubMed] [Google Scholar]
- Fossett N., Zhang,Q., Gajewski,K., Choi,C.Y., Kim,Y. and Schulz,R.A. (2000) The multitype zinc-finger protein U-shaped functions in heart cell specification in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 97, 7348–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossett N., Tevosian,S.G., Gajewski,K., Zhang,Q., Orkin,S.H. and Schulz,R.A. (2001) The Friend of GATA proteins U-shaped, FOG-1 and FOG-2 function as negative regulators of blood, heart and eye development in Drosophila. Proc. Natl Acad. Sci. USA, 98, 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A.H., Kowalski,K., King,G.F., Mackay,J.P. and Crossley,M. (1998) Key residues characteristic of GATA N-fingers are recognized by FOG. J. Biol. Chem., 273, 33595–33603. [DOI] [PubMed] [Google Scholar]
- Fox A.H., Liew,C., Holmes,M., Kowalski,K., Mackay,J. and Crossley, M. (1999) Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J., 18, 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc N.C., Heitzler,P., Ezekowitz,R.A. and White,K. (1999) Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science, 284, 1991–1994. [DOI] [PubMed] [Google Scholar]
- Frank L.H. and Rushlow,C. (1996) A group of genes required for maintenance of the amnioserosa tissue in Drosophila. Development, 122, 1343–1352. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia M.J., Ramain,P., Simpson,P. and Modolell,J. (1999) Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development, 126, 3523–3532. [DOI] [PubMed] [Google Scholar]
- Haenlin M., Cubadda,Y., Blondeau,F., Heitzler,P., Lutz,Y., Simpson,P. and Ramain,P. (1997) Transcriptional activity of Pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev., 11, 3096–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S.A., Miller,J.M. and Hoshizaki,D.K. (2001) serpent, a GATA-like transcription factor gene, induces fat-cell development in Drosophila melanogaster. Development, 128, 1193–1200. [DOI] [PubMed] [Google Scholar]
- Holmes M., Turner,J., Fox,A., Chisholm,O., Crossley,M. and Chong,B. (1999) hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. J. Biol. Chem., 274, 23491–23498. [DOI] [PubMed] [Google Scholar]
- Lanot R., Zachary,D., Holder,F. and Meister,M. (2001) Postembryonic hematopoiesis in Drosophila. Dev. Biol., 230, 243–257. [DOI] [PubMed] [Google Scholar]
- Lebestky T., Chang,T., Hartenstein,V. and Banerjee,U. (2000) Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science, 288, 146–149. [DOI] [PubMed] [Google Scholar]
- Lin W.H., Huang,L.H., Yeh,J.Y., Hoheisel,J., Lehrach,H., Sun,Y.H. and Tsai,S.F. (1995) Expression of a Drosophila GATA transcription factor in multiple tissues in the developing embryos. Identification of homozygous lethal mutants with P-element insertion at the promoter region. J. Biol. Chem., 270, 25150–25158. [DOI] [PubMed] [Google Scholar]
- Lowry J.A. and Atchley,W.R. (2000) Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J. Mol. Evol., 50, 103–115. [DOI] [PubMed] [Google Scholar]
- MacNeill C., Ayres,B., Laverriere,A.C. and Burch,J.B. (1997) Transcripts for functionally distinct isoforms of chicken GATA-5 are differentially expressed from alternative first exons. J. Biol. Chem., 272, 8396–8401. [DOI] [PubMed] [Google Scholar]
- Martin D.I. and Orkin,S.H. (1990) Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev., 4, 1886–1898. [DOI] [PubMed] [Google Scholar]
- Mehaffey M.G., Newton,A.L., Gandhi,M.J., Crossley,M. and Drachman,J.G. (2001) X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood, 98, 2681–2688. [DOI] [PubMed] [Google Scholar]
- Merika M. and Orkin,S.H. (1993) DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol., 13, 3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M. and Orkin,S.H. (1995) Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol., 15, 2437–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R.E., Fessler,L.I., Takagi,Y., Blumberg,B., Keene,D.R., Olson,P.F., Parker,C.G. and Fessler,J.H. (1994) Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J., 13, 3438–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols K.E., Crispino,J.D., Poncz,M., White,J.G., Orkin,S.H., Maris, J.M. and Weiss,M.J. (2000) Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA-1. Nat. Genet., 24, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient R.K. and McGhee,J.D. (2002) The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev., 12, 416–422. [DOI] [PubMed] [Google Scholar]
- Petersen U.M., Kadalayil,L., Rehorn,K.P., Hoshizaki,D.K., Reuter,R. and Engstrom,Y. (1999) Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. EMBO J., 18, 4013–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrefitte S., Kahn,D. and Haenlin,M. (2001) New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix–loop–helix genes. Mech. Dev., 104, 99–104. [DOI] [PubMed] [Google Scholar]
- Ramain P., Heitzler,P., Haenlin,M. and Simpson,P. (1993) pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development, 119, 1277–1291. [DOI] [PubMed] [Google Scholar]
- Rehorn K.P., Thelen,H., Michelson,A.M. and Reuter,R. (1996) A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development, 122, 4023–4031. [DOI] [PubMed] [Google Scholar]
- Rekhtman N., Radparvar,F., Evans,T. and Skoultchi,A. (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev., 13, 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter R. (1994) The gene serpent has homeotic properties and specifies endoderm versus ectoderm within the Drosophila gut. Development, 120, 1123–1135. [DOI] [PubMed] [Google Scholar]
- Rizki T.M., Rizki,R.M. and Grell,E. (1980) A mutant affecting the crystal cells in Drosophila melanogaster. Wilhelm Roux’s Arch. Dev. Biol., 188, 91–99. [DOI] [PubMed] [Google Scholar]
- Sam S., Leise,W. and Hoshizaki,D.K. (1996) The serpent gene is necessary for progression through the early stages of fat-body development. Mech. Dev., 60, 197–205. [DOI] [PubMed] [Google Scholar]
- Shimizu R., Takahashi,S., Ohneda,K., Engel,J.D. and Yamamoto,M. (2001) In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis. EMBO J., 20, 5250–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U., Fessler,L.I., Aziz,A. and Hartenstein,V. (1994) Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development, 120, 1829–1837. [DOI] [PubMed] [Google Scholar]
- Tevosian S.G., Deconinck,A.E., Cantor,A.B., Rieff,H.I., Fujiwara,Y., Corfas,G. and Orkin,S.H. (1999) FOG-2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl Acad. Sci. USA, 96, 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian S.G., Deconinck,A.E., Tanaka,M., Schinke,M., Litovsky,S.H., Izumo,S., Fujiwara,Y. and Orkin,S.H. (2000) FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell, 101, 729–739. [DOI] [PubMed] [Google Scholar]
- Trainor C.D., Omichinski,J.G., Vandergon,T.L., Gronenborn,A.M., Clore,G.M. and Felsenfeld,G. (1996) A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high- affinity interaction. Mol. Cell. Biol., 16, 2238–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor C.D., Ghirlando,R. and Simpson,M.A. (2000) GATA zinc finger interactions modulate DNA binding and transactivation. J. Biol. Chem., 275, 28157–28166. [DOI] [PubMed] [Google Scholar]
- Tsang A.P., Visvader,J.E., Turner,C.A., Fujiwara,Y., Yu,C., Weiss,M.J., Crossley,M. and Orkin,S.H. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell, 90, 109–119. [DOI] [PubMed] [Google Scholar]
- Tsang A.P., Fujiwara,Y., Hom,D.B. and Orkin,S.H. (1998) Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev., 12, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1999) A function of CBP as a transcriptional co-activator during Dpp signalling. EMBO J., 18, 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M.J., Yu,C. and Orkin,S.H. (1997) Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol., 17, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]