Abstract
Previous studies have shown that the Schizo saccharomyces pombe Orc4 subunit is solely responsible for in vitro binding of origin recognition complex (ORC) to specific AT-rich sites within S.pombe replication origins. Using ARS3001, a S.pombe replication origin consisting of four genetically required sites, we show that, in situ as well as in vitro, Orc4 binds strongly to the Δ3 site, weakly to the Δ6 site and not at all to the remaining sequences. In situ, the footprint over Δ3 is extended during G1 phase, but only when Cdc18 is present and Mcm proteins are bound to chromatin. Moreover, this footprint extends into the adjacent Δ2 site, where leading strand DNA synthesis begins. Therefore, we conclude that ARS3001 consists of a single primary ORC binding site that assembles a pre-replication complex and initiates DNA synthesis, plus an additional novel origin element (Δ9) that neither binds ORC nor functions as a centromere, but does bind an as yet unidentified protein throughout the cell cycle. Schizosaccharomyces pombe may be an appropriate paradigm for the complex origins found in the metazoa.
Keywords: ARS3001/cell cycle/DNA replication origin/origin recognition complex/replication initiation point
Introduction
Eukaryotic DNA replication is a highly conserved process that begins with the assembly of a six subunit origin recognition complex (ORC) at specific DNA sites (replication origins) distributed throughout the genome (Bogan et al., 2000; Bell and Dutta, 2002). Nevertheless, ORC properties can differ markedly between species. For example, site-specific binding by the budding yeast Saccharomyces cerevisiae ORC requires five of its six subunits and ATP (Bell, 2002). In contrast, site-specific binding to replication origins by the fission yeast Schizosaccharomyces pombe ORC requires only its Orc4 subunit, and this binding does not require ATP (Kong and DePamphilis, 2001; Lee et al., 2001). Schizo saccharomyces pombe ORC is unique among eukaryotes in that the N-terminus of its Orc4 subunit contains nine AT-hook motifs that bind AT-rich sequences (Chuang and Kelly, 1999; Moon et al., 1999). Another example is mammalian ORC. While the six subunits of both S.pombe (Moon et al., 1999; Chuang et al., 2002) and S.cerevisiae (Bell, 2002) ORC form a stable complex in vitro, only the mammalian Orc2, 3, 4 and 5 proteins form a stable core complex in vitro, to which Orc1 and Orc6 are only weakly bound (Dhar et al., 2001; Vashee et al., 2001). During cell proliferation, mammalian Orc1 is selectively released during the S to M phase transition, ubiquitylated, and then rebound during the M to G1 phase transition to form a functional ORC (Natale et al., 2000; Kreitz et al., 2001; Li and DePamphilis, 2002; Mendez et al., 2002). Finally, the ORC subunits in Xenopus eggs, like those in yeast, exist as a stable complex (Rowles et al., 1996; Tugal et al., 1998), but unlike yeast, the entire Xenopus ORC is released from somatic cell chromatin following assembly of pre-replication complexes (pre-RCs) and prior to initiation of DNA synthesis (Sun et al., 2002).
Not surprisingly, replication origins from S.cerevisiae and S.pombe that have been shown to contain genetically required DNA regions and to bind ORC at specific sites also appear to differ markedly. Schizosaccharomyces pombe replication origins (0.5–1 kb) are five to 10 times larger than those in S.cerevisiae. Moreover, they lack a consensus sequence analogous to the S.cerevisiae A element (essential for ORC-specific binding), and they are not interchangeable with S.cerevisiae origins. Nevertheless, each of the four S.pombe origins analyzed so far contains two or more regions that are required for full ARS activity (Clyne and Kelly, 1995; Dubey et al., 1996; Kim and Huberman, 1998, 1999; Okuno et al., 1999). These required regions consist of AT-rich asymmetric sequences in which A residues are clustered on one strand and T residues on the other. However, while some segments of these regions are critical, others are redundant; and while some required regions are interchangeable, some are not. Furthermore, S.pombe Orc4, either alone or in combination with other ORC subunits, can bind to required regions (Kong and DePamphilis, 2001; Lee et al., 2001; Takahashi and Masukata, 2001) as well as to non-required AT-rich sequences (Lee et al., 2001; Chuang et al., 2002). Thus, the functions of the various genetically required regions in S.pombe replication origins are not clear.
These results raise the possibility that ORCs from different organisms interact with their cognate replication origins in markedly different ways. To evaluate this concept, we identified the sites for ORC binding, pre-replication complex assembly and DNA synthesis at the S.pombe replication origin, ARS3001. These results revealed that S.pombe ORC binds to the same sites in vivo as it does in vitro, but that only the strongest ORC binding site is used to assemble a pre-RC and initiate leading strand DNA synthesis. Schizosaccharomyces pombe ORC was bound preferentially to only one (Δ3) of the four required regions in ARS3001, and this region, together with an adjacent required region (Δ2), was the primary, if not exclusive, site for assembly of a pre-RC and initiation of DNA synthesis. Therefore, complex replication origins such as those in fission yeast are comprised of simpler elements, some of which are functionally analogous to those in S.cerevisiae replication origins, suggesting that the basic mechanism by which replication origins function is conserved throughout the eukaryotic kingdom. However, one of the four required regions (Δ9) appears to be a novel origin element that may be unique to the complex origins found in fission yeast and metazoa.
Results
Orc4 binds to two sites within ARS3001
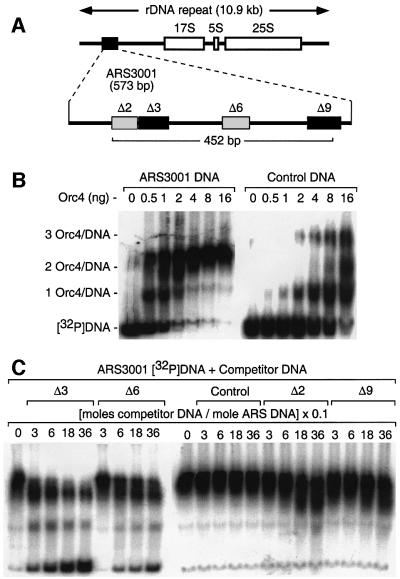
Chromosome III in S.pombe contains ∼100–150 copies of the 10.9 kb rDNA gene cluster (17S, 5.8S and 25S rRNA). Previous studies have shown that each non-transcribed spacer region contains one copy of ARS3001 (Figure 1A), a comparatively small S.pombe replication origin of ∼570 bp that contains four genetically required DNA regions (Kim and Huberman, 1998). Deletion of either the Δ3 or Δ9 region reduces ARS activity >30-fold, while deletion of either the Δ2 or Δ6 region reduces ARS activity ∼5-fold. Previous studies have also shown that the S.pombe Orc4 protein is solely responsible for binding the S.pombe ORC to specific AT-rich sites within S.pombe replication origins (Kong and DePamphilis, 2001; Lee et al., 2001). Therefore, to determine both the number and sequence specificity of S.pombe ORC binding sites in ARS3001, DNA band shift assays were used to quantify the interaction of purified Orc4 protein with a 452 bp ARS3001 [32P]DNA fragment that contains full ARS activity (Table I).
Fig. 1. Orc4 bound to two sites, Δ3 and Δ6, in ARS3001. (A) ARS3001 is located in the non-transcribed spacer of the rDNA repeats and contains two strongly required DNA regions (Δ3, Δ9) and two moderately required DNA regions (Δ2, Δ6) (Kim and Huberman, 1998). (B) DNA band shift assays were carried out with a 452 bp [32P]DNA fragment (5 ng) (panel A) radiolabeled at both 5′-ends, incubated with the indicated amount of Orc4, and then fractionated by gel electrophoresis. This sequence exhibited full ARS3001 activity (Table I). Control DNA consisted of 688 bp of average sequence taken from pBluescript KSII. (C) Competitive DNA band shift analysis was carried out with 5 ng of ARS3001 [5′-32P]DNA plus a 52–68 bp DNA fragment containing either Δ2, Δ3, Δ6, Δ9 or control DNA, at the indicated molar ratio. This DNA was then incubated with 8 ng Orc4. Competitor DNA sequences were: (control DNA) 5′-TAAATTTTTCAGGG TCGGTAGAGTCAGAGATGGGTGTGGGAAGGGGTAGTTGTAGG TAGG-3′; (Δ2 DNA) 5′-TTATGGGAAGGTGGAGAGAAAAAATG AAAAAAACAAGGTAATTTGTAGGATT-3′; (Δ3 DNA) 5′-AAT TTGTAGGATTTTTACAAAATAAATAAATACATTTTATATAATT TAACCAAAAGTAATGT-3′; (Δ6 DNA) 5′-AACAAAAAAAGTG CAAACAAATAAAAGAAAAAATAAGAAAACAAAAAAACAACT ACAAAGGTA-3′; and (Δ9 DNA) 5′-ATGAAAAAATAAAGAA AAATTTAATTTATAATTTAACAAAACAATATTTATTGAAAAGCCAATTTTAA-3′.
Table I. ARS3001 activity.
| Origin elements | Size (bp) | Transformation frequency (%) |
|---|---|---|
| Δ2-Δ3-Δ6-Δ9 | 906 | 100 |
| Δ2-Δ3-Δ6-Δ9 | 665 | 100 |
| Δ2-Δ3-Δ6-Δ9 | 452 | 100 |
| Δ2-Δ3-Δ6 | 320 | 1 |
| Δ2-Δ3-Δ6-1.8 kb-Δ9 | 320 | 1 |
| Δ2-Δ3-Δ6-1.8 kb-Δ9(<-) | 320 | 1 |
Plasmids containing strong ARS elements generate transformants of large, uniform size that do not rearrange (Kim and Huberman, 1999); therefore, small colonies were not scored. Δ9(<-) orientation was opposite to Δ9 wild type.
DNA and protein were briefly incubated at room temperature and then fractionated by neutral agarose gel electrophoresis (Figure 1B). Two Orc4–[32P]DNA complexes were detected at low ratios of Orc4 to DNA. As the ratio of protein to DNA reached saturation, only the larger complex was observed. A non-ARS DNA fragment consisting of average sequence composition did not bind Orc4 at low protein:DNA ratios, although it did bind up to three Orc4 proteins at high ratios of protein to DNA. Comparison of ARS3001–Orc4 complexes with control DNA–Orc4 complexes suggested that ARS3001 DNA contains only two Orc4 binding sites.
Competitive DNA band shift assays both confirmed and extended this conclusion. The 452 bp ARS3001 [32P]DNA fragment was mixed with increasing amounts of a 52–68 bp DNA fragment spanning one of the four required regions before incubating it with Orc4 and then fractionating the mixture by gel electrophoresis (Figure 1C). As the amount of either the Δ3 or Δ6 competitor was increased, two protein–DNA complexes became evident, and unbound [32P]DNA appeared at the bottom of the gel. Therefore, Orc4 bound to the Δ3 and Δ6 regions, but not to either the Δ2 or Δ9 regions. Moreover, the affinity of Orc4 for Δ3 was about six times greater than for Δ6, because similar amounts of unbound [32P]DNA were present at a molar ratio of Δ3:ARS3001 of 60 and a ratio of Δ6:ARS3001 of 360 (Figure 1C, compare lanes 3 and 9). Since competition between either the Δ2 or Δ9 fragment and ARS3001 DNA for Orc4 binding was similar to competition with control DNA, the Δ2 and Δ9 regions did not bind Orc4 under these conditions. Nevertheless, Orc4 binding to DNA was clearly facilitated by longer DNA lengths, because release of ∼50% of the [32P]DNA required ∼180-fold excess of the 63 bp Δ3 DNA fragment over the 452 bp ARS3001 DNA fragment. Similar results were observed by Lee et al. (2001). Therefore, binding of Orc4 to DNA appears to involve cooperativity between different sequences, consistent with the large size of replication origins in S.pombe.
Orc4 binds to specific ARS3001 DNA sequences in vitro
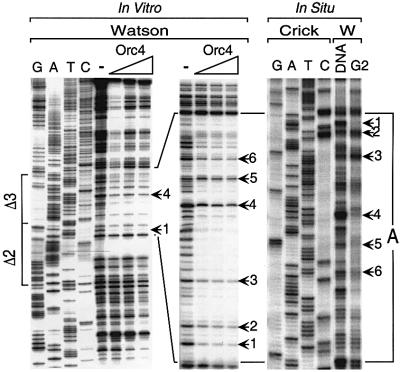
The results from DNA band shift assays suggested the presence of only two Orc4 binding sites in the 452 bp ARS3001 fragment, a strong site in the Δ3 region and a weaker one in the Δ6 region. To test this conclusion, purified Orc4 protein was incubated with the same ARS3001 [32P]DNA and then subjected to DNase I footprinting analysis throughout its entire length. Only two Orc4 DNA binding sites were detected in vitro. The clearest footprints were routinely observed in the Δ3 region where Orc4 protected ∼80 bases on the Watson strand (Figure 2, site A) and 22 bases on the Crick strand (Figure 3, site A). A footprint also was observed in Δ6 where Orc4 protected ∼36 bases on the Crick strand (Figure 4A, site B), but protection of the Watson strand was not detected (data not shown). No footprint was detected on either strand of Δ2 (Figure 2) or Δ9 (Figure 4C). Therefore, Orc4 preferentially bound specific DNA sites in the Δ3 and Δ6 regions of ARS3001.
Fig. 2. Orc4 produced a footprint on the Watson strand of the Δ3 region in vitro that matched a genomic footprint detected in situ. In vitro footprinting was carried out with a 665 bp ARS3001 DNA fragment incubated with either 0, 10, 40 or 80 ng of Orc4 (lanes indicated by right triangle). The 5′-end of the Watson strand (top of gel) was labeled with 32P. The sequence of the Watson strand (lanes G, A, T and C), beginning at the same 5′-nucleotide, was run in parallel using an appropriate DNA primer. To display the footprint (site A) in the Δ3 region more clearly, the same experiment was carried out with a 452 bp ARS3001 DNA fragment. Both fragments exhibited full ARS activity. Sequences of Δ3 and site A are given in Figure 8. In situ genomic footprinting was carried out on nuclei isolated from cells arrested in G2 phase. Nuclei (G2 lane) and genomic DNA (DNA) were isolated and subjected to DNase I digestion in parallel. Samples in which the extent of digestion was similar were subjected to primer extension using a primer annealed to the Crick strand. The same primer used to locate the 3′-ends of the Watson strand that was cut by DNase I was also used to display the sequence. Therefore, the sequence shown is the Crick strand, while the DNase I digestion pattern is from the Watson strand. Arrows indicate critical nucleotides that appear in both in vitro and in situ footprints. Sequences and footprinting data for Δ2 and Δ3 are given in Figure 8.
Fig. 3. Orc4 produced a footprint on the Crick strand of the Δ3 region in vitro that matched a genomic footprint detected in situ. The same footprinting analyses described in Figure 2 were carried out on the Crick strand of the Δ2–Δ3 region using a 320 bp ARS3001 DNA fragment that lacks the Δ9 region. Only the pyrimidine sequencing lanes are shown for simplicity. Sequences and footprinting data for Δ2 and Δ3 are given in Figure 8.
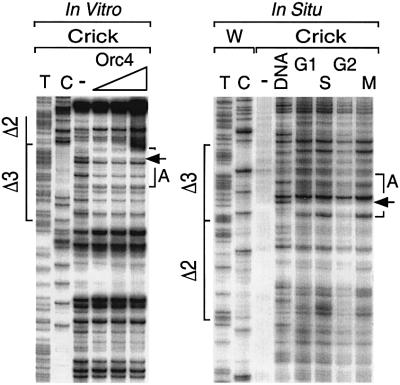
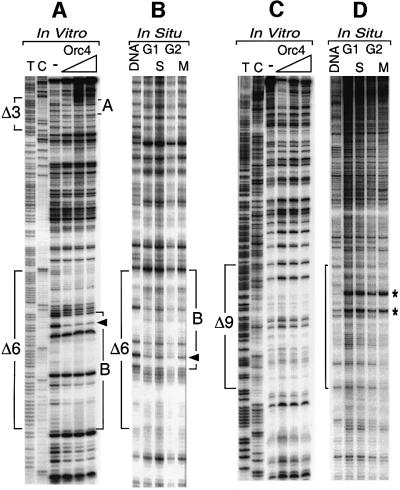
Fig. 4. Orc4 produced a footprint on the Crick strand of the Δ6 region in vitro that matched a genomic footprint detected in situ. The same footprinting analyses described in Figure 3 were carried out on the Crick strand of regions Δ3 and Δ6 using ARS3001-320. Genomic footprinting of the Δ6 region was analyzed using ARS3001-2 (Table II). A footprint (site B) was observed both in vitro (A) and in situ (B). No footprint was detected in the Δ9 region in vitro (C), although two DNase I-hypersensitive sites (*) were detected in situ (D). Region Δ9 was analyzed using the Crick strand of a 665 bp ARS3001 DNA fragment. In situ genomic footprinting of the Δ9 region was analyzed using ARS3001-3 (Table II). Sequences and footprinting data for Δ6 and Δ9 are given in Figure 8.
Orc4 binds preferentially to the T-rich strand in replication origins
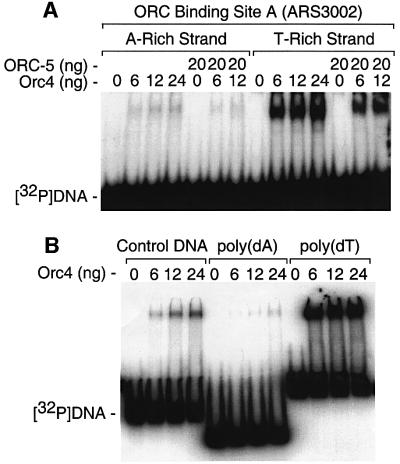
In both ARS3001 (Figures 2–4) and ARS3002 (Kong and DePamphilis, 2001), Orc4 bound to AT-rich asymmetric sequences in which one strand was rich in A residues and the other strand in T residues. To determine whether or not Orc4 had a preference for one of the two complementary strands, DNA band shift assays were carried out with two complementary single-stranded oligonucleotides from binding site A in ARS3002 (Kong and DePamphilis, 2001). Both oligonucleotides contained 46 residues that were 93% AT, but one oligonucleotide contained 20 consecutive A residues while the other contained 20 consecutive T residues. The results revealed that Orc4 strongly preferred the T-rich strand over the A-rich strand (Figure 5A). Moreover, purified ORC-5 complex alone (consisting of Orc1, 2, 3, 5 and 6; Kong and DePamphilis, 2001) did not bind to these oligonucleotides or alter the specificity of Orc4 for the T-rich strand (Figure 5A), consistent with previous studies which showed binding of S.pombe ORC to DNA to be attributed exclusively to Orc4.
Fig. 5. Orc4 preferentially bound the T-rich strand at ORC DNA binding sites. (A) DNA band shift assays containing 1 ng [5′-32P]oligonucleotide plus the indicated amounts of Orc4 and ORC-5 complex were incubated at room temperature for 10 min before fractionating the material by gel electrophoresis. The A-rich strand of Orc4 binding site A in ARS3002 DNA (Kong and DePamphilis, 2001) was represented by 5′-TAATACTATTTTTTATATTAATTAAAAA AAAAAAAAAAAAAAACCT-3′. The T-rich strand was represented by 5′-AGGTTTTTTTTTTTTTTTTTTTTAATTAATATAAAAAATA GTATTA-3′. (B) A non-ARS ssDNA sequence from the S.pombe Orc3 gene was used as a control DNA [5′-CCGGCCTCGAGATGC ATCACCATCACCATCACTCAGCAATACTACAATATGATTC-3′]. Poly(dA) and poly(dT) sequences were each 46 residues long.
To determine whether or not Orc4’s preference for the T-rich strand of ARS3002 was due to the presence of oligo(T)20, the same experiment was repeated using oligo(A)46, oligo(T)46 and a non-ARS control sequence of 55 nucleotides with 53% AT. Again, Orc4 strongly preferred the T-oligomers (Figure 5B). The strong preference of Orc4 for oligo(T) residues in one strand helps to account for the sequence specificity exhibited by Orc4 and indicates that Orc4 binds only in one orientation.
Orc4 binding sites in vitro are occupied in vivo
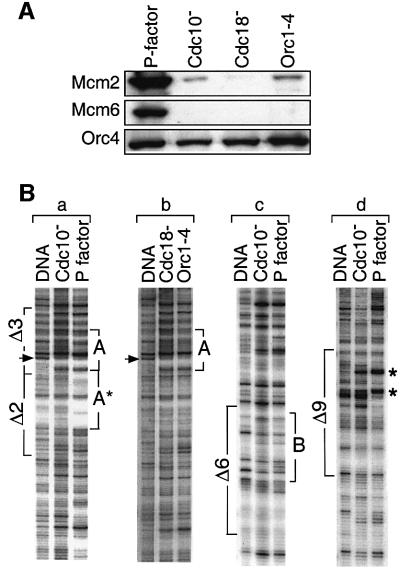
To determine whether or not the Orc4 DNA binding sites identified in vitro were also utilized in vivo, DNase I genomic footprinting was carried out in situ using nuclei isolated from S.pombe cells arrested at specific points in its cell cycle. G1 phase cells were prepared either by addition of P-factor or by inactivating Cdc10 protein. P-factor arrests cell division in S.pombe by inhibiting Cdc2 (Stern and Nurse, 1997), a cyclin-dependent protein kinase that is required for initiating S phase by activating pre-RCs (Bell and Dutta, 2002). Therefore, P-factor-arrested cells contained chromatin-bound Orc4 (Figure 6A), Cdc18 (Stern and Nurse, 1997), and Mcm2 and Mcm6 (Figure 6A), the components of a pre-RC. In contrast, cells lacking Cdc10 contained neither Cdc18 (Kelly et al., 1993) nor chromatin-bound Mcm proteins (Figure 6A; Ogawa et al., 1999), because Cdc10 is required for expression of Cdc18 (Kelly et al., 1993) and Cdc18 is required for loading Mcm proteins at ORC–chromatin sites (Bell and Dutta, 2002). FACS analysis confirmed that Cdc18– cells were arrested in G1 phase and contained chromatin-bound Orc4 but not Mcm proteins (Figure 6A). Schizosaccharomyces pombe was arrested in S phase by inactivating ribonucleotide reductase (Cdc22 gene; Fernandez Sarabia et al., 1993), an enzyme required for dNTP synthesis, and in G2 phase by inactivating the Cdc25 protein kinase, which is required to activate Cdk1–Cyclin B (Russell and Nurse, 1987). Cells were arrested in metaphase by addition of benomyl, an inhibitor of microtubule assembly (Svoboda et al., 1995).
Fig. 6. The genomic footprint at Δ3 was extended to Δ2 in the presence of pre-RCs. (A) Schizosaccharomyces pombe cells were arrested in G1 phase either by addition of P-factor, or by inactivating Cdc10 or Orc1, or by depletion of Cdc18. Chromatin was isolated and its proteins were fractionated by SDS–PAGE and then analyzed with antibodies against Mcm2, Mcm6 and Orc4, as described previously (Kong and DePamphilis, 2001). (B) In situ genomic footprinting was performed on the Crick strand as in Figures 3 and 4. The footprint (A) in Δ3 was extended (A*) to Δ2 in P-factor arrested cells (a), but it was not extended in Cdc10–, Cdc18– or Orc1-4 arrested cells (a and b). Similar changes were not detected in the Δ6 (c) or Δ9 regions (d).
Conditions for DNase I digestion were adjusted so that both purified genomic DNA and chromatin were digested to similar extents. Note that the same [5′-32P]DNA primers used to map DNase I cleavage sites in the Watson and Crick strands were also used to reveal the sequence of these regions. Therefore, the sequence shown is the strand complementary to the sequence subjected to in situ footprinting. Note also that the in situ footprints are in the opposite direction to the in vitro footprints, because the in situ footprints were detected indirectly by primer extension of a [5′-32P]oligonucleotide into the region of interest, while the in vitro footprints were detected directly by radiolabeling the 5′-end of the footprinted strand.
Both the Watson (Figure 2) and Crick (Figure 3) strands of site A were protected in situ, and the pattern of protection was strikingly similar to the Orc4 footprint observed in vitro. The arrows in each panel indicate unique nucleotide positions that allow one to compare the in vitro and in situ footprints. Moreover, the same footprint was detected in S, G2 and M phases of the cell cycle, consistent with previous studies showing that S.pombe ORC is bound to chromatin throughout the cell cycle (Lygerou and Nurse, 1999; Kong and DePamphilis, 2001), and that Cdc18 and Mcm proteins are absent from chromatin during S, G2 and M phases (Bell and Dutta, 2002). The same genomic footprint was also detected in cells arrested in G1 phase when either Cdc10 (Figures 3 and 6Ba) or Orc1 (Figure 6Bb) was inactivated, or when Cdc18 (Figure 6Bb) was absent. These conditions prevented assembly of pre-RCs. Furthermore, a footprint was also detected in situ on the Crick strand of Δ6 (Figure 4B) that corresponded to Orc4 binding site B in vitro (Figure 4A), but not on the Watson strand where no footprint was detected in vitro (data not shown). It is equally important to note that in sequences such as Δ2 and Δ9, where no Orc4 footprint was detected in vitro, either no genomic footprint was observed in situ (Δ2; Figures 2 and 3) or the genomic footprint observed in situ was unrelated to binding of Orc4 protein (Δ9; discussed below).
These experiments revealed that the same Orc4 DNA binding sites that were identified in vitro were occupied in situ. Moreover, the DNase I footprints observed in situ were remarkably similar to the ones observed in vitro, did not appear at sites that did not bind Orc4 in vitro and were not dependent on Orc1, Cdc18 or Mcm proteins. Therefore, we conclude that these genomic footprints resulted from Orc4, suggesting that Orc4 is responsible for selection of S.pombe ORC-specific binding sites in vivo as well as in vitro. In support of this conclusion, only the footprint at the strongest Orc4 binding site (site A) was expanded when pre-RCs were present.
A pre-RC is assembled at Δ3 and Δ2
In S.cerevisiae, the presence of a pre-RC at a specific replication origin was recognized in situ by a Cdc6-dependent extension of the ORC footprint by ∼50 bp during G1 phase (Santocanale and Diffley, 1996). Following initiation of DNA replication, the extended footprint disappeared (Diffley et al., 1994), because Cdc6 and Mcm proteins were no longer present at the origin (Bell and Dutta, 2002). Therefore, to determine whether or not one or both of the S.pombe ORC binding sites identified above acted as a pre-RC assembly site, their G1 phase genomic footprints were compared in the presence and absence of Cdc18 (the S.pombe equivalent of Cdc6) and chromatin-bound Mcm proteins.
Cells arrested in G1 phase either by defective Cdc10 or Orc1, or the absence of Cdc18, contained chromatin bound Orc4, but not Mcm proteins. These cells all exhibited the same genomic footprint at site A in Δ3. In contrast, cells arrested in G1 phase by P-factor contained chromatin-bound Mcm proteins, and the genomic footprint at Δ3 in these cells was extended ∼45 bp into Δ2 (Figure 6Ba). No changes were detected in the genomic footprint at Δ6 or Δ9 between P-factor and Cdc10– arrested cells (Figure 6Bc and d). Therefore, regions Δ2 and Δ3 constitute a pre-RC assembly site.
DNA synthesis begins in the Δ2 region
The site where DNA replication begins [origin of bi-directional replication (OBR)] is defined by the transition between continuous and discontinuous DNA synthesis that must occur on each strand of the replication origin (DePamphilis, 1999). Using a new experimental protocol for mapping the 5′-ends of nascent DNA strands in replicating intermediates, this transition has identified, with nucleotide resolution, the start sites for leading strand DNA synthesis at replication origins in SV40 (Gerbi and Bielinsky, 1997), S.cerevisiae (Bielinsky and Gerbi, 1999), S.pombe (Gomez and Antequera, 1999) and human cells (Abdurashidova et al., 2000). Therefore, the same protocol was applied to ARS3001 in S.pombe in order to determine the relationship between ORC binding sites and the OBR.
RNA-primed DNA chains were purified from genomic DNA, annealed with a sequence-specific [5′-32P]DNA primer, and then the primer was extended to the 5′-terminal deoxyribonucleotide of the template strand by DNA polymerase. Thus, the relative amount of each [5′-32P]DNA chain was proportional to the number of such chains, and not to their length. The same [5′-32P]DNA primer was also used to generate a sequence ladder that was fractionated in parallel with the [5′-32P]DNA products from the primer extension reaction. Thus, the 5′-nucleotide map position of the nascent DNA strand was identified by direct comparison with its sequence.
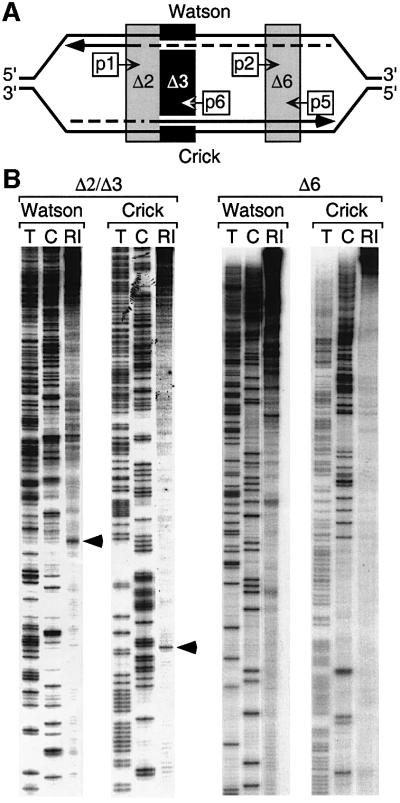
Three sets of primers were used that covered a 700 bp region containing ARS3001 (Figure 7A). Only one OBR was detected, located in the Δ2 region (Figure 7), the same region where a pre-RC was assembled. Arrowheads mark the position where a transition from discontinuous to continuous DNA synthesis was detected on both the Watson and Crick DNA templates (Figure 7B, Δ2/Δ3). These two transition points were separated by 10 bp. Additional initiation sites were detected upstream of these two transition points, on each DNA template, representing initiation events that resulted from synthesis, extension and eventually ligation of Okazaki fragments to the 5′-ends of long growing nascent DNA strands. These experiments exhibited a light background of polymerase pause sites, as seen on the leading strand template of Δ6 (Figure 7B, Crick strand, Δ6). These bands may result from failure to eliminate all DNA fragments, as well as a low level of initiation events at Δ6.
Fig. 7. Leading strand DNA synthesis began in Δ2. The nucleotide locations of nascent strand start sites were mapped using replicating intermediates (RI) enriched for RNA-primed nascent DNA. Primers 1 and 6 (Table II) were annealed to the Crick and Watson strands, respectively, to identify the ends of nascent DNA strands in the Δ2–Δ3 region, primer set 2 and 5 were used for the Δ6 region, and primer set 3 and 4 examined the Δ9 region (data not shown). The same primers were used to display the sequence of these regions. Arrowheads indicate transition points from discontinuous to continuous DNA synthesis on each strand of Δ2 region (sequence given in Figure 8).
Analysis of Δ6 (Figure 7) and Δ9 (data not shown) revealed intense RNA-primed initiation sites on the nascent DNA strand complementary to the Watson template, but not on the nascent DNA strand complementary to the Crick template. Thus, DNA synthesis at replication forks in these regions was primarily discontinuous on the ‘lagging’ strand template and continuous on the ‘leading’ strand template, consistent with the conclusion that most, if not all, leading strand DNA synthesis began at the OBR identified in Δ2.
Δ9 is required specifically for origin function
Genomic footprinting revealed two DNase I hypersensitive sites in the Δ9 region, indicating that one or more protein(s) were bound to Δ9 throughout the cell cycle (Figures 4D and 6D). A similar result has been observed by J.Huberman (personal communication). However, since Orc4 did not bind to Δ9 in vitro (Figures 1 and 4C), some protein(s) other than ORC apparently bind to Δ9 in situ.
To confirm that Δ9 is required for ARS activity, the transformation efficiency of a 452 bp fragment containing all four required regions (Figure 1A) was compared with the transformation efficiency of 906 and 665 bp fragments that contained the 452 bp fragment extended at both ends. The transformation frequencies of these three fragments were indistinguishable, whereas the transformation frequency of a 320 bp fragment containing Δ2, Δ3 and Δ6, but missing Δ9, was reduced ∼100-fold (Table I), consistent with previous studies (Kim and Huberman, 1998).
In the yeast Yarrowia lipolytica, ARS elements of ∼1 kb contain an AT-rich centromeric sequence at one end and an origin of replication at the other end (Vernis et al., 1997), and only centromeres can supply the partition system required for ARS function (Vernis et al., 2001). In S.pombe, proteins that bind to centromeres also bind to AT-rich sequences in ARS elements (Lee et al., 1997; Sanchez et al., 1998), but these centromere binding proteins do not have AT-hook domains, and therefore they may exhibit different sequence specificity than Orc4 protein.
Therefore, to determine whether Δ9 is required for origin function or some other plasmid specific function such as stability, nuclear association or segregation during cell division, the distance between Δ6 and Δ9 was increased by 1.8 kb with Δ9 in either its normal or reverse orientation. If Δ9 is required specifically for origin function, then this change should decrease ARS3001 activity. If, however, Δ9 is required for a plasmid maintenance function, then this change should have no effect on ARS activity. In fact, increasing the distance between Δ9 and Δ6 reduced the transformation frequency of ARS3001 to the same extent as deleting Δ9, regardless of the orientation of Δ9 (Table I). Therefore, the four required regions identified by Kim and Huberman (1998) comprise a minimal replication origin with full ARS activity, and are each required specifically for origin function.
Discussion
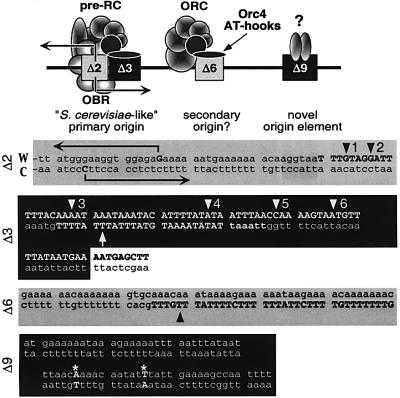
Previous studies have shown that, in vitro, S.pombe Orc4 is solely responsible for selecting ORC binding sites in S.pombe DNA replication origins, and that these sites consist of AT-rich asymmetric sequences (see Introduc tion). Here we confirm and extend these conclusions by showing that Orc4 binds to the same sites in vivo as it does in vitro, regardless of the presence or absence of the remaining Orc subunits. Furthermore, only one of the four required regions in ARS3001 strongly binds Orc4, and it is this region where a pre-RC is assembled and DNA synthesis is initiated (Figure 8). Thus, despite the fact that S.pombe DNA replication origins are larger and less well defined than S.cerevisiae origins, their mechanisms of action appear remarkably similar, with one exception. ARS3001 contains a DNA region that is essential for origin activity, but that is not associated either with ORC binding or with centromeric function (Figure 8). This region may represent a novel element unique to the complex origins found in fission yeast and the metazoa.
Fig. 8. Schizosaccharomyces pombe ARS3001 contains two strongly required regions (Δ3, Δ9) and two moderately required regions (Δ2, Δ6). ORC binds (through the AT-hook domain of its Orc4 subunit) strongly to Δ3 and moderately to Δ6 (capital letters), but not to either Δ2 or Δ9. Arrowheads in Figures 2–4 are reproduced here. Leading strand DNA synthesis begins at Δ2, marking this as the OBR. Δ9 is required for origin function, not for plasmid segregation, and binds a protein(s) of unknown function throughout the cell cycle.
Yeast replication origins fit a common paradigm
Both DNA band shift assays (Figure 1) and DNA footprinting assays (Figures 2–4) revealed that purified S.pombe Orc4 bound to only two sites in ARS3001, and these sites corresponded to the genetically required regions Δ3 and Δ6. However, the affinity of Orc4 for Δ3 was ∼6-fold greater than for Δ6. Since deletion of Δ3 effectively eliminates ARS3001 activity, while deletion of Δ6 reduces it only ∼5-fold (Kim and Huberman, 1998), Δ3 is the primary site for ORC binding, analogous to elements A and B1 in S.cerevisiae replication origins (Bell, 2002). This conclusion is consistent with the fact that neither Δ6, which binds Orc4 weakly, nor Δ9 which does not bind Orc4, can substitute for Δ3, but Δ3, which binds Orc4 strongly, can replace Δ6 (Kim and Huberman, 1998).
Since both Δ3 and Δ6 are required for full ARS activity, Δ6 either facilitates the activity of Δ3 or operates as an independent, secondary initiation site. In the first model, two or more ORCs in close proximity may facilitate recruitment of a single Cdc18 protein to a specific chromosomal site. Cdc18 is an extremely labile protein that is required for assembly of pre-RCs at ORC– chromatin sites, and its overexpression can result in reinitiation of DNA replication within a single S phase (Yanow et al., 2001). Thus, it is not surprising that Cdc18 expression is limited to just a few minutes prior to S phase, and then Cdc18 is rapidly destroyed once S phase begins (Muzi Falconi et al., 1996). In fact, the B2 element in S.cerevisiae ARS1 appears to function in just this manner. Sequences that substitute fully for B2 function were weak ORC binding sites in vitro, and mutations in B2 could be rescued by overexpression of Cdc6 (Wilmes and Bell, 2002). In the second model, S.pombe replication origins would be analogous to those S.cerevisiae origins that contain multiple ORC binding sites, all of which must be inactivated in order to abolish ARS activity (Hurst and Rivier, 1999; Theis and Newlon, 2001). The Δ2–Δ3 region may be the primary origin because it requires less energy to unwind its DNA than does the Δ6 region (Kim and Huberman, 1998).
The role of the Δ2 region in S.pombe ARS3001 is directly comparable to the B-region in S.cerevisiae replication origins: it is part of the site where a pre-RC is assembled and it is the site where leading strand DNA synthesis begins. The in situ footprint over Δ3 was extended into the adjacent Δ2 region during G1 phase, but only when Cdc18 was present and Mcm proteins were bound to chromatin. In S.cerevisiae, ORC alone protects 32 bp. During G1 phase, this footprint is extended to ∼80 bp, most of which occurs through the B1 and B2 elements (Santocanale and Diffley, 1996). This extension is dependent on the presence of Cdc6, the S.cerevisiae homolog of Cdc18. Mapping the sites where RNA-primed DNA synthesis began throughout ARS3001 revealed only one OBR (Figure 7), and this was located in Δ2, 25 bp from Δ3. The same experimental protocol identified a single OBR at S.cerevisiae ARS1 between elements B1 and B2, 28 bp from element A (Bielinsky and Gerbi, 1999). Thus, in both S.pombe ARS3001 and S.cerevisiae ARS1, the OBR lies adjacent to the ORC binding site in an easily unwound DNA region through which the ORC footprint is extended when Cdc6/Cdc18 is present and Mcm proteins are bound to chromatin.
The conclusion that Δ2 is the site where DNA replication begins is consistent with the fact that inverting the orientation of Δ3 has the same effect as deleting Δ3: both mutations reduce ARS activity to near background levels (Kim and Huberman, 1998). The N-terminal half of Orc4 consists of nine AT-hook motifs that bind to DNA, while the C-terminal half, which is homologous to Orc4 proteins in other eukaryotes, presumably interacts with one or more of the remaining Orc subunits. Therefore, ORC will be directed to one side of the Orc4 DNA binding site. The fact that Orc4 strongly prefers T-rich oligonucleotides (Figure 5) suggests that it will bind only in one orientation. Therefore, one would expect that the orientation of the Orc4 DNA binding site would be critical, because it will target assembly of the pre-RC to an adjacent DNA unwinding element, such as Δ2. Similar orientation effects have also been observed at S.cerevisiae replication origins (Bell, 2002).
Not only are S.pombe replication origins comprised of simpler elements, some of which are functionally analogous to those in S.cerevisiae replication origins, but S.cerevisiae replication origins are more complex than originally appreciated. First, the relative positions of the A element and the four known B elements are highly variable among S.cerevisiae origins. Secondly, the classical S.cerevisiae 11 bp ARS consensus sequence (ACS) (the A element) is actually a 17 bp AT-rich sequence that is much more diverse than initially thought (Theis and Newlon, 1997). This means that ORC can bind to a wider variety of sequences than previously realized. In fact, as many as one-third of the S.cerevisiae origins actually contain two to three ORC binding sites clustered within ∼40–800 bp (Hurst and Rivier, 1999; Theis and Newlon, 2001). Thus, replication origins in budding and fission yeast are clearly more similar than initially believed.
The Orc4 subunit targets ORC to specific AT-rich asymmetric sequences
The tight association of the S.pombe Orc4 subunit with chromatin (Moon et al., 1999) throughout the cell cycle (Kong and DePamphilis, 2001), the lack of any effect by the other five ORC subunits on Orc4 binding to DNA in vitro (Kong and DePamphilis, 2001; Figure 5) or in situ (Figure 6Bb), and the striking similarity between Orc4 footprints at the Δ3 and Δ6 regions of ARS3001 DNA in vitro and genomic footprints at the same sites in situ (Figures 2–4) support the conclusion that Orc4 is solely responsible for binding of S.pombe ORC to specific DNA sites in vivo as well as in vitro.
Inspection of the Orc4 protected regions of ARS3001 (Figures 2–4), ARS3002 (Kong and DePamphilis, 2001) and ARS1 (Kong and DePamphilis, 2001; Lee et al., 2001) reveals that Orc4 binds to consecutive runs of (T)3–7 or (T)3–4A motifs such as those found in the Crick strand of site A in Δ3 (Figure 8) that do not contain either alternating AT residues or interspersed G or C residues. Sequences containing either alternating AT or interspersed G and C residues did not bind Orc4, even when they contained (T)3–7 and (T)3–4A motifs (e.g. Δ9; Figure 8) or bound Orc4 weakly (e.g. Δ6; Figure 8). In support of this conclusion, asymmetric AT-rich sequences and the oligomer (AAAT/TTTA)10 compete strongly with ARS elements in binding S.pombe ORC, whereas sequences either with an average GC content (non-ARS control DNA in Figure 1; Kong and DePamphilis, 2001) or with oligomers of alternating AT do not (Lee et al., 2001; Takahashi and Masukata, 2001; Chuang et al., 2002). In fact, required regions in ARS2004 could be replaced by either (A:T)40 or (AAAT/TTTA)10, but not by (AT/TA)20 or (AAAC/TTTG)10 (Okuno et al., 1999).
The affinity of Orc4 for AT-rich asymmetric sequences can be accounted for by its preference for oligo(T)-rich deoxyribonucleotides. In both examples where an Orc4 binding site contained a large region of oligo(T) on one strand and oligo(A) on the other, it was the T-rich strand that was more strongly protected in the DNase I footprint assays [site B in ARS3001 (Figure 8); site A in ARS3002 (Kong and DePamphilis, 2001)]. Moreover, only the T-rich strand of ARS3002 bound Orc4, and this binding preference was reproduced with oligo(T) and oligo(A) (Figure 5). These data indicate that Orc4 binds DNA in only one orientation.
Δ9 appears to be a novel origin element
As discussed above, the Δ2–Δ3 region of ARS3001 represents an S.cerevisiae-like replication origin, and the Δ6 region is either a secondary origin that is used less frequently, or a weak ORC binding site that facilitates pre-RC assembly at Δ2–Δ3. However, the Δ9 region appears to be unique. It does not bind Orc4, but it does bind an as yet unidentified protein throughout the cell cycle in vivo. The function of this protein can be replaced by one or more of the ORC subunits, because Δ3 can replace Δ9 (Kim and Huberman, 1998), suggesting that Δ9 is required specifically for origin function and not for plasmid stability, binding to nuclear structure or segregation during cell division. In fact, while Δ9 still stimulates ARS3001 activity when it is substituted for Δ6 (Kim and Huberman, 1998) and thereby moved closer to the primary origin at Δ2–Δ3, Δ9 cannot be moved further away from the origin without loss of ARS activity (Table I). Therefore, the Δ9 region represents a novel element of the more complex replication origins found in S.pombe and perhaps the metazoa, as well. Its function in replication may be analogous to that of an enhancer in transcription.
Is S.pombe a model for the metazoa?
Initiation sites for DNA replication in mammals are determined by both genetic and epigenetic factors, but the size and composition of these sites has been diffi cult to define and consequently appear more complex than replication origins in S.cerevisiae (discussed in DePamphilis, 1999; Altman and Fanning, 2001; Aladjem et al., 2002). Metazoan replication origins may resemble ‘compound replication origins’ in S.cerevisiae that contain redundant ORC binding sites (Hurst and Rivier, 1999; Theis and Newlon, 2001), and ‘clustered origins’ in S.pombe (Dubey et al., 1994; Kim and Huberman, 1999) that contain multiple, closely spaced origins with one origin strongly preferred over the others. Mapping initiation events in these yeast origins using two-dimensional gel protocols, as in mammalian origins, failed to identify individual origins, detecting instead a broad zone of initiation events. Based on DNA sequence homology, metazoan ORCs may recognize a motif similar to the one recognized by the S.cerevisiae ORC (Bogan et al., 2000), but metazoan ORCs alone may bind weakly to such motifs, allowing them to change the number and locations of initiation sites during animal development. Site specificity may arise through association with other chromosomal proteins, such as HMG-I, that contain AT-hook motifs to direct ORC to specific sites. In fact, initiation loci in mammals (Aladjem et al., 1998; Abdurashidova et al., 2000; Altman and Fanning, 2001) and flies (Ina et al., 2001) all contain AT-rich asymmetric sequences, analogous to those in S.pombe replication origins.
Materials and methods
Reagents
ARS3001 DNA sequences (DDBJ/EMBL/GenBank accession No. AL512862) were isolated from the S.pombe genome by PCR and cloned into pBluescript II SK–-Ura4 (4.7 kb) at EcoRI and SpeI sites. pARS3001-906 contained a 906 bp fragment terminated by 5′-GAT CGACA···GGGAAAAGATACG-3′. pARS3001-665 contained a 665 bp fragment terminated by 5′-AGAGGAAA···TCAGAGATG-3′. pARS3001-452 contained a 452 bp fragment terminated by 5′-AGG TGGAGAG···TTGAAAGCC-3′. pARS3001-320 contained a 320 bp fragment terminated by 5′-AGGTGGAGAG···AATATGGGA-3′. In pARS3001-320/Δ9, Δ6 and (5′-TCGATAC··Δ9··GAGTAGG-3′) were separated by the ura4 gene (1.8 kb). P-factor was purchased from Research Genetics, Inc., and purified by HPLC to >90%. Schizosaccharomyces pombe Orc4 protein and ORC-5 complex were prepared as described previously (Kong and DePamphilis, 2001).
DNA band shift assay
dsDNA band shift assays were performed as described previously (Kong and DePamphilis, 2001). Reactions were incubated for 10 min at room temperature, adjusted to 0.02% NP-40, and then fractionated by electrophoresis at room temperature in 1.2% agarose gel (standard Tris–borate EDTA buffer pH 8.0) at 5 V/cm for 3 h. Conditions for ssDNA band shift assays were the same as for dsDNA band shift assays except that poly(dG–dC) was omitted, and products were resolved by electrophoresis in 5% polyacrylamide gels (standard Tris–borate EDTA buffer pH 8.0) at 10 V/cm.
Cell synchronization
Schizosaccharomyces pombe (1 l) JL197 ura4-D18 cdc10-129 (G1 phase arrest), Orc1-4 mutant (Grallert and Nurse, 1996) (G1 phase arrest), JL202 ura4-D18 cdc22-M45 (S phase arrest) and JL206 ura4-D18 cdc25-22 (G2 phase arrest) were grown in YE medium to OD590 0.4 at 26°C and then shifted to 36.5°C for 3.5 h. For M phase arrest, h– ura4-D18 Orp1-3HA was grown at 32°C to OD590 0.4, adjusted to 25 µg benomyl/ml and incubated for 3 h. For P-factor-induced G1 phase arrest, either h– cyr1Δ::Leu2 Sxa2Δ::ura4 Leu1-32 ura4-d18 or h– mat1-M Del-mat2,3::Leu2 cyr1::ura4+ ura4-D18 was incubated in EMM medium at 32°C to OD590 0.2, adjusted to 1.5 µg/ml P-factor and incubated for 6 h. Cdc18-deficient strain ura4-D18 leu1-32 ade6-M216 Δcdc18::ura4+ pREP81X-cdc18+ (Kelly et al., 1993) was grown to OD595 0.3 in EMM medium supplemented with adenine at 32°C, and then with 5 µg/ml thiamine for 4 h.
In vitro footprinting assay
DNase I footprinting assays were performed in vitro as described previously (Leblanc and Moss, 2001), with modifications (see Supplementary data, available at The EMBO Journal Online).
In situ genomic footprinting assay
DNase I genomic footprinting was performed as described previously (Huibregtse and Engelke, 1991), with modifications (see Supplementary data).
Mapping DNA synthesis initiation sites
Initiation sites for RNA-primed DNA synthesis were mapped as described for S.cerevisiae (Gerbi and Bielinsky, 1997), with adaptations for S.pombe (see Supplementary data).
Supplemenary data
Supplementary data are available at The EMBO Journal Online.
Table II. DNA primers used for analysis of ARS3001 DNA.
| Primer | Sequence |
|---|---|
| Primer 1 | 5′-GGGAGTAGAGGTAGTTGTATGGAGGAAG-3′ |
| Primer 6 | 5′-CAAAGTTGGTCAACCAAGCTCATTTTC-3′ |
| Primer 2 | 5′-TGAAAATGAGCTTGGTTGACCAACTTTG-3′ |
| Primer 5 | 5′-CTTTTGTTTTTCATGCTTTCCTCACATG-3′ |
| Primer 3 | 5′-CAACTACAAAAGGTATTGAAAATCGTGCG-3′ |
| Primer 4 | 5′-CTCCATTCCCTACCTACAACTACCCC-3′ |
Acknowledgments
Acknowledgements
We thank Paul Nurse, Richard Egel and Joon-Kyu Lee for providing yeast strains, and Joel Huberman for sharing unpublished observations.
References
- Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. [DOI] [PubMed] [Google Scholar]
- Aladjem M.I., Rodewald,L.W., Kolman,J.L. and Wahl,G.M. (1998) Genetic dissection of a mammalian replicator in the human β-globin locus. Science, 281, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Aladjem M.I., Rodewald,L.W., Lin,C.M., Bowman,S., Cimbora,D.M., Brody,L.L., Epner,E.M., Groudine,M. and Wahl,G.M. (2002) Replication initiation patterns in the β-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Mol. Cell. Biol., 22, 442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman A.L. and Fanning,E. (2001) The Chinese hamster dihydrofolate reductase replication origin β is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol., 21, 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev., 16, 659–672. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- Bielinsky A.K. and Gerbi,S.A. (1999) Chromosomal ARS1 has a single leading strand start site. Mol. Cell, 3, 477–486. [DOI] [PubMed] [Google Scholar]
- Bogan J.A., Natale,D.A. and DePamphilis,M.L. (2000) Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell Physiol., 184, 139–150. [DOI] [PubMed] [Google Scholar]
- Chuang R.Y. and Kelly,T.J. (1999) The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl Acad. Sci. USA, 96, 2656–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang R.Y., Chretien,L., Dai,J. and Kelly,T.J. (2002) Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: interaction with origin DNA and Cdc18 protein. J. Biol. Chem., 277, 16920–16927. [DOI] [PubMed] [Google Scholar]
- Clyne R.K. and Kelly,T.J. (1995) Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J., 14, 6348–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M.L. (1999) Replication origins in metazoan chromosomes: fact or fiction? BioEssays, 21, 5–16. [DOI] [PubMed] [Google Scholar]
- Dhar S.K., Delmolino,L. and Dutta,A. (2001) Architecture of the human origin recognition complex. J. Biol. Chem., 276, 29067–29071. [DOI] [PubMed] [Google Scholar]
- Diffley J.F., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Dubey D.D., Zhu,J., Carlson,D.L., Sharma,K. and Huberman,J.A. (1994) Three ARS elements contribute to the ura4 replication origin region in the fission yeast, Schizosaccharomyces pombe. EMBO J., 13, 3638–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey D.D., Kim,S.M., Todorov,I.T. and Huberman,J.A. (1996) Large, complex modular structure of a fission yeast DNA replication origin. Curr. Biol., 6, 467–473. [DOI] [PubMed] [Google Scholar]
- Fernandez Sarabia M.J., McInerny,C., Harris,P., Gordon,C. and Fantes,P. (1993) The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol. Gen. Genet., 238, 241–251. [DOI] [PubMed] [Google Scholar]
- Gerbi S.A. and Bielinsky,A.K. (1997) Replication initiation point mapping. Methods, 13, 271–280. [DOI] [PubMed] [Google Scholar]
- Gomez M. and Antequera,F. (1999) Organization of DNA replication origins in the fission yeast genome. EMBO J., 18, 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert B. and Nurse,P. (1996) The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev., 10, 2644–2654. [DOI] [PubMed] [Google Scholar]
- Huibregtse J.M. and Engelke,D.R. (1991) Direct sequence and footprint analysis of yeast DNA by primer extension. Methods Enzymol., 194, 550–562. [DOI] [PubMed] [Google Scholar]
- Hurst S.T. and Rivier,D.H. (1999) Identification of a compound origin of replication at the HMR-E locus in Saccharomyces cerevisiae. J. Biol. Chem., 274, 4155–4159. [DOI] [PubMed] [Google Scholar]
- Ina S., Sasaki,T., Yokota,Y. and Shinomiya,T. (2001) A broad replication origin of Drosophila melanogaster, oriDα, consists of AT-rich multiple discrete initiation sites. Chromosoma, 109, 551–564. [DOI] [PubMed] [Google Scholar]
- Kelly T.J., Martin,G.S., Forsburg,S.L., Stephen,R.J., Russo,A. and Nurse,P. (1993) The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell, 74, 371–382. [DOI] [PubMed] [Google Scholar]
- Kim S.M. and Huberman,J.A. (1998) Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol. Cell. Biol., 18, 7294–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.M. and Huberman,J.A. (1999) Influence of a replication enhancer on the hierarchy of origin efficiencies within a cluster of DNA replication origins. J. Mol. Biol., 288, 867–882. [DOI] [PubMed] [Google Scholar]
- Kong D. and DePamphilis,M.L. (2001) Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol. Cell. Biol., 21, 8095–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S., Ritzi,M., Baack,M. and Knippers,R. (2001) The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem., 276, 6337–6342. [DOI] [PubMed] [Google Scholar]
- Leblanc B. and Moss,T. (2001) DNase I footprinting. Methods Mol. Biol., 148, 31–38. [DOI] [PubMed] [Google Scholar]
- Lee J.K., Huberman,J.A. and Hurwitz,J. (1997) Purification and characterization of a CENP-B homologue protein that binds to the centromeric K-type repeat DNA of Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 94, 8427–8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.K., Moon,K.Y., Jiang,Y. and Hurwitz,J. (2001) The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc. Natl Acad. Sci. USA, 98, 13589–13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.J. and DePamphilis,M.L. (2002) Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol., 22, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z. and Nurse,P. (1999) The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci., 112, 3703–3712. [DOI] [PubMed] [Google Scholar]
- Mendez J., Zou-Yang,X.H., Kim,S.Y., Hidaka,M., Tansey,W.P. and Stillman,B. (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell, 9, 481–491. [DOI] [PubMed] [Google Scholar]
- Moon K.Y., Kong,D., Lee,J.K., Raychaudhuri,S. and Hurwitz,J. (1999) Identification and reconstitution of the origin recognition complex from Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 96, 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzi Falconi M., Brown,G.W. and Kelly,T.J. (1996) cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 93, 1566–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K.A. (1977) Temperature-sensitive lethal mutants in the structural gene for DNA ligase in the yeast Schizosaccharomyces pombe. Cell, 12, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Natale D.A., Li,C.J., Sun,W.H. and DePamphilis,M.L. (2000) Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M–G1 transition in mammals. EMBO J., 19, 2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y., Takahashi,T. and Masukata,H. (1999) Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol., 19, 7228–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Satoh,H., Sekiguchi,M. and Masukata,H. (1999) Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol. Cell. Biol., 19, 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowles A., Chong,J.P., Brown,L., Howell,M., Evan,G.I. and Blow,J.J. (1996) Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell, 87, 287–296. [DOI] [PubMed] [Google Scholar]
- Russell P. and Nurse,P. (1987) Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell, 49, 559–567. [DOI] [PubMed] [Google Scholar]
- Sanchez J.P., Murakami,Y., Huberman,J.A. and Hurwitz,J. (1998) Isolation, characterization and molecular cloning of a protein (Abp2) that binds to a Schizosaccharomyces pombe origin of replication (ars3002). Mol. Cell. Biol., 18, 1670–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J.F. (1996) ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J., 15, 6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Stern B. and Nurse,P. (1997) Fission yeast pheromone blocks S-phase by inhibiting the G1 cyclin B–p34cdc2 kinase. EMBO J., 16, 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.H., Coleman,T.R. and DePamphilis,M.L. (2002) Cell cycle dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J., 21, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda A., Bahler,J. and Kohli,J. (1995) Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma, 104, 203–214. [DOI] [PubMed] [Google Scholar]
- Takahashi T. and Masukata,H. (2001) Interaction of fission yeast ORC with essential adenine/thymine stretches in replication origins. Genes Cells, 6, 837–849. [DOI] [PubMed] [Google Scholar]
- Theis J.F. and Newlon,C.S. (1997) The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl Acad. Sci. USA, 94, 10786–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J.F. and Newlon,C.S. (2001) Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites. Mol. Cell. Biol., 21, 2790–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugal T., Zou-Yang,X.H., Gavin,K., Pappin,D., Canas,B., Kobayashi,R., Hunt,T. and Stillman,B. (1998) The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. J. Biol. Chem., 273, 32421–32429. [DOI] [PubMed] [Google Scholar]
- Vashee S., Simancek,P., Challberg,M.D. and Kelly,T.J. (2001) Assembly of the human origin recognition complex. J. Biol. Chem., 276, 26666–26673. [DOI] [PubMed] [Google Scholar]
- Vernis L., Abbas,A., Chasles,M., Gaillardin,C.M., Brun,C., Huberman,J.A. and Fournier,P. (1997) An origin of replication and a centromere are both needed to establish a replicative plasmid in the yeast Yarrowia lipolytica. Mol. Cell. Biol., 17, 1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernis L., Poljak,L., Chasles,M., Uchida,K., Casaregola,S., Kas,E., Matsuoka,M., Gaillardin,C. and Fournier,P. (2001) Only centro meres can supply the partition system required for ARS function in the yeast Yarrowia lipolytica. J. Mol. Biol., 305, 203–217. [DOI] [PubMed] [Google Scholar]
- Wilmes G.M. and Bell,S.P. (2002) The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc. Natl Acad. Sci. USA, 99, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanow S.K., Lygerou,Z. and Nurse,P. (2001) Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J., 20, 4648–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]