Abstract
60S ribosomes undergo initial assembly in the nucleolus before export to the cytoplasm and recent analyses have identified several nucleolar pre-60S particles. To unravel the steps in the pathway of ribosome formation, we have purified the pre-60S ribosomes associated with proteins predicted to act at different stages as the pre-ribosomes transit from the nucleolus through the nucleoplasm and are then exported to the cytoplasm for final maturation. About 50 non-ribosomal proteins are associated with the early nucleolar pre-60S ribosomes. During subsequent maturation and transport to the nucleoplasm, many of these factors are removed, while others remain attached and additional factors transiently associate. When the 60S precursor particles are close to exit from the nucleus they associate with at least two export factors, Nmd3 and Mtr2. As the 60S pre-ribosome reaches the cytoplasm, almost all of the factors are dissociated. These data provide an initial biochemical map of 60S ribosomal subunit formation on its path from the nucleolus to the cytoplasm.
Keywords: Nmd3/nuclear export/nucleolus/rRNA processing/60S subunit
Introduction
Ribosomal biogenesis is a complicated process starting with the transcription of rDNA repeats by RNA polymerase I (25S, 18S and 5.8S rRNA) and polymerase III (5S rRNA) (Kressler et al., 1999; Venema and Tollervey, 1999). Polymerase I transcription generates the 35S pre-rRNA, which then undergoes rapid processing to the mature rRNAs by endonucleases and exonucleases, with concomitant modification of the rRNA by pseudouridylation and methylation (Venema and Tollervey, 1999). Ribosomal RNA biogenesis has been extensively studied, and is fairly well understood. During these processing reactions, a large number of non-ribosomal proteins associate with the pre-rRNAs, and many of the ∼80 ribosomal proteins are assembled onto the rRNA (Warner, 1989, 1999; Woolford, 1991). The process of ribosome assembly is, in contrast, just beginning to be understood (Warner, 2001). Early analyses reported that the ribosomal proteins and the 35S pre-rRNA primary transcript initially form a 90S particle (Trapman et al., 1975; Kruiswijk et al., 1978), which also includes many non-ribosomal proteins (Dragon et al., 2002; Grandi et al., 2002). Within the 90S pre-ribosomes the 35S pre-rRNA undergoes three rapid cleavage steps, which separate the precursors to the large and small subunits (Trapman et al., 1975; Venema and Tollervey, 1999). The precursor to the large subunit was initially described as the 66S particle, but is now known to consist of a family of pre-60S particles that carry many accessory proteins that show dynamic changes during maturation. Within the pre-60S particles the 27S pre-rRNAs are matured to the 25S and 5.8S rRNAs (Trapman et al., 1975), and the 5S rRNA associates with the complex. The early pre-60S particles are restricted to the nucleolus, but later particles are released into the nucleoplasm (Milkereit et al., 2001) and then exported to the cytoplasm. Nuclear export of the 60S pre-ribosome to the cytoplasm is facilitated by Nmd3, which is an NES-containing adapter protein that requires the export receptor Xpo1/Crm1/exportin-1 (Ho et al., 2000b; Gadal et al., 2001b), but may also require the function of other pre-60S associated proteins. The 40S subunit undergoes a comparatively simpler maturation (Venema and Tollervey, 1999) but its mechanism of export is still unknown (Moy and Silver, 1999).
Despite the speed with which the cell modifies and processes the pre-rRNA, several groups succeeded in isolating and characterizing stable pre-60S particles (Baßler et al., 2001; Harnpicharnchai et al., 2001; Saveanu et al., 2001; Fatica et al., 2002). These particles differed in protein and RNA composition and appeared to reflect the presence of a series of distinct nucleolar and/or nucleoplasmic pre-60S particles. To better define the pathway of 60S subunit synthesis we purified a series of particles to obtain ‘snapshots’ of the pre-60S ribosomes as they move from the nucleolus to the cytoplasm. Here, we report a detailed analysis of the composition of seven distinct 60S pre-ribosomes and describe their major location in the cell.
Results
Sedimentation of 60S pre-ribosomal particles
We previously isolated a pre-60S ribosomal particle by tandem-affinity purification of an associated component, Nug1, which is a putative GTPase (Baßler et al., 2001). This precursor contained ∼20 non-ribosomal proteins, including both characterized factors and the products of many previously uncharacterized open reading frames. To unravel the spatio-temporal changes that occur during ribosome formation inside the nucleus, from early assembly in the nucleolus until final ribosomal export, we employed a ‘reverse tagging’ methodology. The rationale behind this approach was that some of the associated proteins in the Nug1 pre-ribosomal complex could also be present in other precursor particles and thus allow purification of pre-ribosomes from different stages of maturation. We have selected the non-ribosomal proteins Nsa3, Nop7, Sda1, Rix1 and Arx1, which are components of the Nug1-containing pre-ribosome (Baßler et al., 2001). Moreover, we also chose Kre35, which is a homolog of Nug1 and a putative GTPase (E.Hurt, unpublished data). These bait proteins were genomically tagged at the C-terminus with the TAP tag for tandem affinity purification (Puig et al., 2001) and sedimentation analysis. In addition, these factors were tagged with the green fluorescent protein (GFP) for subcellular localization studies.
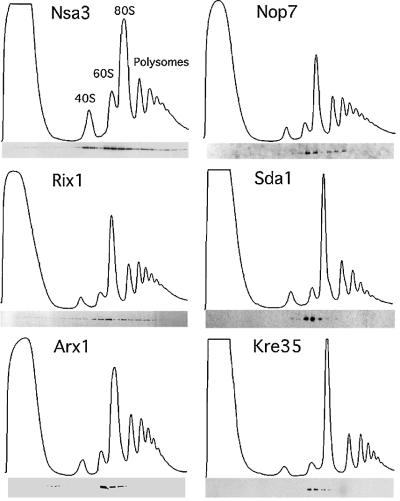
We first determined the size of the pre-ribosomal particles that purify with the different tagged proteins. Previous analyses showed that Nug1 associated with pre-60S particles on sucrose gradients (Baßler et al., 2001). Similar sucrose gradient centrifugation showed that each of the bait proteins associated with large particles, but with markedly different sedimentation profiles. Nsa3 displays a complicated sedimentation pattern with one peak at ∼40S, a second broader peak from 60S to 90S and some partitioning into fractions below 90S (Figure 1). In contrast, Sda1, Arx1 and Kre35 showed a distinct and confined peak at ∼60S (Figure 1). Nop7 and Rix1 reveal an ‘intermediate’ sedimentation pattern, with a pronounced peak at 60S and a second broader peak below 80S (Figure 1). We conclude from these studies that the bait proteins analyzed are associated with different 60S pre-ribosomal particles, and in some cases are associated with more than one particle (see Discussion).
Fig. 1. Reverse-tagged protein baits of the Nug1-containing 60S pre- ribosome are associated with pre-ribosomal particles of different sizes. The sedimentation behavior of the indicated TAP-tagged proteins was analyzed on sucrose density gradients, and ribosomal fractions (40S, 60S, 80S and polysomes) were determined by OD254 measurement of the gradient fractions (upper graph). Western blot analysis of these gradient fractions using anti-ProtA antibodies reveals the position of the indicated TAP-tagged baits (lower panel). Note that some baits (e.g. Nsa3) exhibit a broad distribution on the sucrose gradient, whereas other baits (e.g. Kre35) exhibit a distinct peak at ∼60S (see text).
Intracellular location of 60S pre-ribosomes
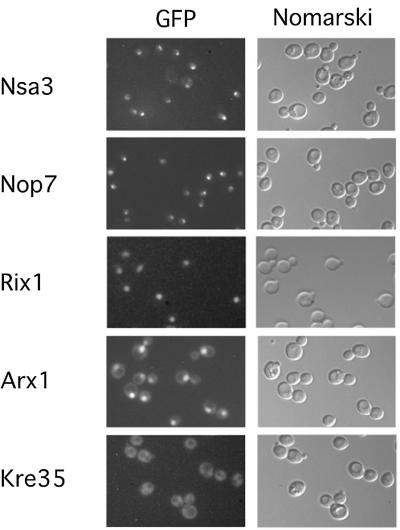
To determine the steady state location of these pre-ribosomal particles in living cells, yeast strains expressing GFP-tagged Nsa3, Nop7, Rix1, Arx1 or Kre35 were examined by fluorescence microscopy (Figure 2). Our previous data showed Nug1–GFP to be located in both the nucleolus and nucleoplasm (Baßler et al., 2001). The studies shown here suggest that Nsa3 and Nop7 are present in earlier particles, since they are predominantly concentrated in the nucleolus (Figure 2; see also Harnpicharnchai et al., 2001; Adams et al., 2002). In contrast, Rix1–GFP localized throughout the nucleus (Figure 2; see also Baßler et al., 2001). Sda1–GFP could not be tested, since it was expressed very inefficiently (data not shown), but myc-tagged Sda1 co-localized with DAPI staining, showing a nucleoplasmic localization (Buscemi et al., 2000). Arx1–GFP accumulates in the nucleoplasm but is also present in cytoplasm (Figure 2), suggesting that it accompanies the pre-60S particles to the cytoplasm. Finally, Kre35–GFP exhibits an exclusive cytoplasmic distribution with nuclear exclusion (Figure 2). This analysis allows us to propose a pathway for pre- 60S maturation, from the predominant nucleolar particles associated with Nsa3 and Nop7, to nucleolar/ nucleoplasmic particles (Nug1), nucleoplasmic particles (Rix1 and Sda1), nucleoplasmic/cytoplasmic particles (Arx1) and finally the cytoplasmic Kre35-associated particles.
Fig. 2. Subcellular location of GFP-tagged protein baits in yeast cells. The in vivo location of the indicated tagged proteins, associated with different 60S pre-ribosomes, was analyzed by fluorescence microscopy. For microscopic inspection, cells were grown to mid-log phase, mounted on a microscope slide and photographed with identical exposure times.
Composition of 60S pre-ribosomes
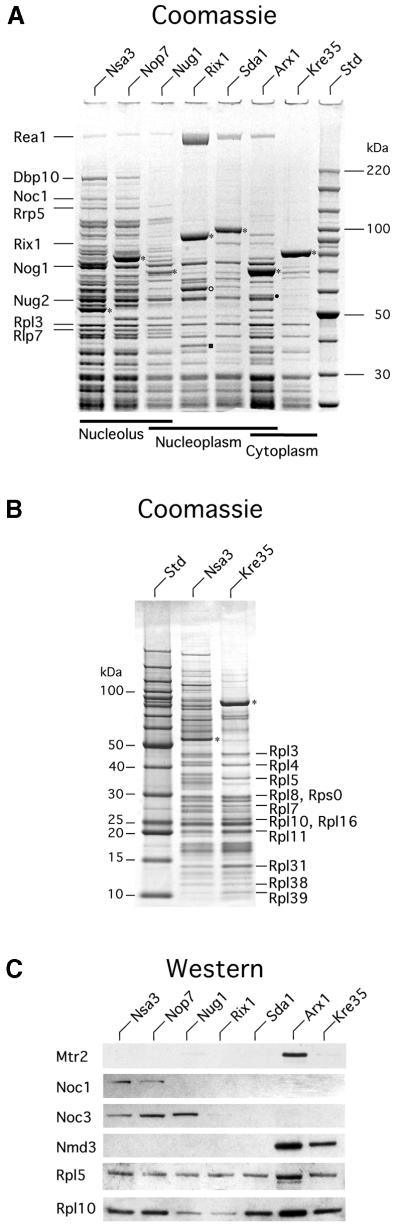
To obtain information about the protein and RNA composition of the different 60S pre-ribosomes, we TAP-purified each of the six tagged proteins under standardized conditions of cell growth and cell lysis, and compared them with the previously purified Nug1 particle (Baßler et al., 2001). Co-precipitated proteins were compared on the same SDS–polyacrylamide gel stained with Coomassie Blue (Figure 3A). The loading of the various particles can be estimated from the recovery of ribosomal L-proteins, which correspond to prominent bands below 50 kDa (Figure 3A; e.g. Rpl3) and is approximately equal, with the exception of Arx1, which is slightly overloaded. The samples were loaded (from left to right) in the order corresponding to their position in the predicted maturation pathway. This showed a striking decrease during non-ribosomal protein complexity in the pre-60S ribosome’s maturation.
Fig. 3. SDS–PAGE analysis of the protein composition of the different pre-60S ribosomes. (A) The indicated TAP-tagged protein baits were isolated from yeast lysates by a two-step affinity purification (TAP method), and TAP-purified bait proteins were separated on an 4–12% SDS–polyacrylamide gradient gel and stained with colloidal Coomassie Blue. Co-purifiying proteins were identified by mass spectrometry (MALDI-TOF) and prominent co-migrating bands are indicated. The positions of tagged proteins are indicated by asterisks, Nmd3 in the Arx1 preparation by a filled circle, and Ynl182p and Yhr085p in the Rix1 preparation by an open circle and closed square, respectively. The molecular weight marker is indicated on the right. Note the relative higher loading of the Arx1 particle in comparison with the Rix1, Sda1 and Kre35 particle loading. (B) Lower molecular weight proteins in earliest (Nsa3) and latest (Kre35) particles as visualized with 10–20% SDS–PAGE. All bands, both in the high (Table I) and low molecular weight range (Table II), were identified by MS. Each bait protein is indicated with an asterisk. In addition, selected ribosomal proteins are indicated. (C) Western blot analysis of the purified bait proteins, analyzed by SDS–PAGE as shown in (A) and listed in Supplementary table SI. The western blot was incubated with the indicated antibodies to Mtr2, Noc1, Noc3, Nmd3, Rpl5 and Rpl10. Not shown are Sqt1, which was identified in all purifications, and Tif6, which was present in all except Kre35.
The earliest nucleolar particles that co-precipitated with tagged Nsa3 and Nop7 have similar protein profiles when the major bands are inspected following Coomassie staining (Figure 3A). Mass spectrometry (MALDI-TOF) identified additional non-ribosomal proteins associated with Nsa3 (∼50 species were identified in total; Table I; Supplementary table SI available at The EMBO Journal Online), including several factors known to act in early pre-rRNA processing and ribosome assembly steps. Among these are the methyltransferase Spb1 and components of both the box C/D and H/ACA classes of snoRNPs (Nop1, Nop56, Nop58, Spb1, Rrp9 and Cbf5), which direct rRNA methylation and pseudouridinylation and are required for early pre-rRNA cleavage. Moreover, some of the associated factors have also been implicated in 40S biogenesis (Nop1, Nop58, Rrp5, Rrp8, Rrp9 and Nop14). However, many of the Nsa3-associated components were also found in the Nug1 particle, including the helicases Dbp2, Dbp10 and Has1, the methyltransferase Spb1, and the GTPases Nug1, Nug2 and Nog1 (Table I), all of which are implicated in 60S subunit synthesis. These results suggested that Nsa3 is predominately associated with pre-60S particles and has lower association with earlier 90S pre-ribosomes, consistent with the results of sucrose gradient analyses (Figure 1) and RNA co-precipitation (see below).
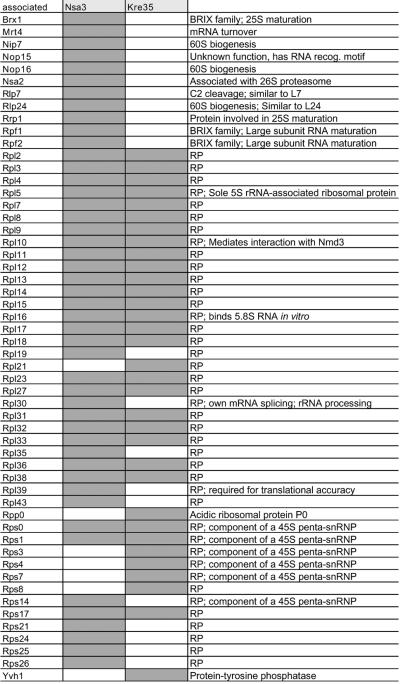
Table I. Non-ribosomal proteins associated with the different 60S pre-ribosomal particles.
TAP-tagged proteins are listed in the top row (bait, light gray). Co-purifying proteins larger than L3 (44 kDa) identified by mass spectrometry (row ‘associated’) are indicated vertically as gray rectangles in the columns below each bait protein. If known, the function of the associated proteins present in the purified 60S pre-ribosomes are indicated in the most right vertical column. The asterisk indicates proteins in the Nug1 preparation that have been newly identified in this study. Not indicated in the table are additional proteins associated with the bait proteins, which could be considered contaminants: Act1, Cdc10, Ded1, Gfa1, Mis1, Ssa1, Ssa2, Ssa4, Sbp1 and Tef1. Furthermore, selected proteins smaller than L3 (44 kDa) identified in the Arx1 particle using MS are Asc1, Nsa2, Pup2, YJL122w and Yvh1.
During transition from the nucleolar Nsa3- or Nop7-containing pre-ribosomes to the nucleolar/nucleoplasmic located Nug1-containing particles, many factors are dissociated causing a significant simplification of the protein composition (Figure 3A; Table I; Supplementary table SI). Among these are factors involved in processing in ITS1 (Rrp5, Rrp8, Rrp9, Ssf1 and Ssf2), the box C/D and H/ACA components, helicases and Noc1. Consistent with this, Noc1 was previously shown to be present in early nucleolar pre-ribosomes together with Noc2, but to be replaced by Noc3 upon release into the nucleoplasm (Milkereit et al., 2001). Western blot analysis (Figure 3C) confirms the exchange of Noc1 for Noc3. In the earliest particle (Nsa3), Noc1 is significantly enriched and Noc3 is present in lower amounts. As the particle matures (Nop7), Noc1 becomes less prevalent and Noc3 becomes more prominent. Subsequently, the Nug1 particle has largely lost Noc1 and is enriched for Noc3 (Figure 3C).
The next set of particles, which are associated with Rix1 and Sda1, showed a further reduction in complexity (Figure 3A). These nucleoplasmic particles (see Figure 2) are still associated with the putative GTPases Nug1, Nug2 and Nog1, but other factors, including the ribosomal-like protein Rlp7 (Dunbar et al., 2000; Gadal et al., 2002) and the putative helicases Dbp2, Dbp10 and Has1, are substantially reduced (Figure 3A; Table I). Moreover, western analysis revealed that Noc3 is largely absent from these latter particles (Figure 3C). Concomitant with the removal of certain factors, other components join these particles. In particular, the Rix1-containing pre-ribosome is associated with large amounts of Rea1, a putative AAA-type ATPase (for ribosome export/assembly; see Baßler et al., 2001), and Ynl182p, an essential unknown protein that exhibits homology to Rrp9 and Pwp2 (Figure 3A; Table I). A large amount of the uncharacterized protein Yhr085p is associated with Rix1 but has not been found in any other purified pre-ribosomal particle. The Sda1-containing particle exhibits decreased association of Rea1 and Ynl182p, and is somewhat simpler than the Rix1-associated particle (Figure 3A).
A next major stage in the biogenesis of 60S subunits is represented by the Arx1-containing pre-ribosome, which has a dual location in both the nucleoplasm and cytoplasm. Biochemical and western analysis of the TAP-purified Arx1 show that the NES-containing export factor Nmd3 joins the 60S pre-ribosome at this stage (Figure 3A and C). Nmd3 binds to the general nuclear export receptor Xpo1/Crm1/exportin-1, which is crucial for nuclear export of 60S ribosomal subunits (Ho et al., 2000b; Gadal et al., 2001b). In contrast, the ribosomal protein Rpl10, which interacts with Nmd3 both in vivo (Karl et al., 1999) and in vitro (Gadal et al., 2001b) and is also required for nuclear export of 60S subunits (Gadal et al., 2001b), is present in all particles, from early nucleolar (Nsa3) until late cytoplasmic 60S pre-ribosomes (Kre35) (Figure 3C). In addition to Nmd3, Ybr267p, which has a C2H2 zinc finger motif, and the putative GTPase Kre35, which is homologous to Nug1 and Nug2, were found to be associated with the Arx1-containing 60S pre-ribosome (Figure 3A; Table I). Western blot analysis also demonstrated that the nuclear export factor Mtr2 specifically associates with the Arx1 particle (Figure 3C). Mtr2 forms a complex with the conserved mRNA export factor Mex67 but the mtr2-33 allele was previously observed to specifically inhibit pre-60S export (Baßler et al., 2001; see Discussion).
The final particle characterized is associated with the putative GTPase Kre35. This late, cytoplasmic pre-60S ribosome was associated with many ribosomal proteins but few non-ribosomal proteins (Figure 3A; Tables I and II). These include Nmd3, which is speculated to also function in the cytoplasm (Ho et al., 2000a), and Arx1, consistent with the proposal that it accompanies the pre-60S particle to the cytoplasm (see above). In addition, an uncharacterized zinc finger protein Ybr267p was identified in the particle. Notably absent were the nucleolar and nucleoplasmic GTPases, Nug1, Nug2 and Nog1, as well as the AAA ATPase Rea1.
Table II. Identification of low molecular weight proteins in the Nsa3 and Kre35 preparations.
TAP-tagged proteins are listed in the top row (bait, light gray). Co-purifying proteins smaller than L3 (44 kDa) identified by mass spectrometry (row ‘associated’) are indicated vertically as gray rectangles in the columns below each bait protein. If known, the function of the associated proteins present in the purified 60S pre-ribosomes are indicated in the most right vertical column.
Ribosomal proteins were compared in detail for the early Nsa3-associated and late Kre35-associated particles. The pattern of bands in the low molecular weight range of the SDS–polyacrylamide gel was strikingly similar (Figure 3B). Mass spectrometry of these bands revealed that both particles contain predominantly the same L-proteins, suggesting that most of the L-proteins are already assembled onto early pre-60S particles (Table II; Supplementary table SI). Moreover, we also found a low number of S-proteins in the Nsa3 and Kre35 preparations. Nsa3 is expected to co-precipitate with some small subunit proteins due to its association with 90S pre-ribosomes. The recovery of small subunit proteins with Kre35 could reflect some association with 40S subunits in the cytoplasm. However TAP purifications are frequently contaminated by ribosomal proteins (Gavin et al., 2002) and therefore this could be non-specific.
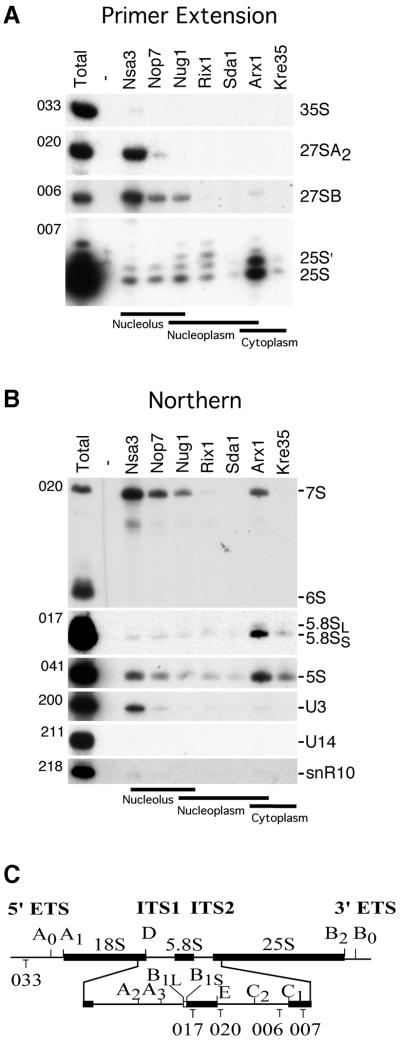
The RNA composition of the pre-ribosomal particles was also compared by primer extension (Figure 4A) and northern blot hybridization (Figure 4B). The Nsa3 particle predominantly contained the 27SA2 and 27SB pre-rRNAs, as shown by primer extension (Figure 4A), and by northern hybridization, the 7S rRNA (Figure 4B). The Nsa3 particle is unique in the presence of the U3 snoRNA, but in contrast no U14 or snR10 snoRNA could be observed (Figure 4B). Furthermore, none of the particles contained the 18S rRNA (data not shown). Taken together, these data mark Nsa3 as the earliest ribosomal pre-60S particle.
Fig. 4. RNA analysis of the different pre-60S particles. (A) Primer extension and (B) northern hybridization analyses for 35S, 27S and 25S rRNA were performed on RNA extracted from whole cells (Total) and affinity-purified tagged (Nsa3, Nop7, Nug1, Rix1, Sda1, Arx1 and Kre35) and non-tagged (Negative) wild-type strain. RNA was loaded proportionally to a Coomassie stained protein SDS–PAGE from the same purifications. The oligonucleotide used for each panel is indicated on the left side of each gel (see Materials and methods). The annealing location of oligonucleotides used for primer extension and northern hybridization analysis are indicated (C).
Similar to Nsa3, both Nop7 and Nug1 contained mainly 27SB, 7S pre-rRNA and 5S rRNA, but less of the 27SA2 precursor RNA and a higher ratio of matured 25S rRNA. These results are similar to previous results (Baßler et al., 2001; Harnpicharnchai et al., 2001) with only small variations in relative abundance, probably due to alternative purification procedures or strain background differences (see Harnpicharnchai et al., 2001).
For the Rix1, Arx1 and Kre35 particles, the most abundant ribosomal RNA species were mature 25S, 5.8S and 5S with smaller quantities of 27S and 7S precursors (Figure 4A and B). These data strongly suggest that rRNA maturation of the 60S ribosomes is largely completed at the nucleoplasmic stage of the Rix1 particle. This is supported by the finding that the rix1-1 mutant very efficiently accumulates the Rpl25–GFP reporter inside the nucleus, but does not show any rRNA processing defect (Baßler et al., 2001). In several independent preparations Sda1 did not detectably co-precipitate any pre-rRNAs, despite good protein precipitation, leading to the conclusion that it is predominately associated with the mature rRNAs.
These RNA analyses further confirm the ordering of these particles suggested by sedimentation, localization and protein composition analysis. Overall, this ordering showed that the particles represent five stages: (i) highly complex nucleolar pre-60S particles (Nsa3, Nop7); (ii) complex nucleolar/nuclear pre-60S particles (Nug1); (iii) intermediate complex nucleoplasmic particles (Rix1, Sda1); (iv) intermediate complex nuclear–cytoplasmic 60S pre-ribosome (Arx1); and (v) simple cytoplasmic 60S pre-ribosome (Kre35). Finally, it is important to note that the 5S rRNA (Figure 4B) and its binding partner the L5 protein (Figure 3C) were detected in each of the particles, indicating that the 5S RNP joins an early pre-60S particle. Together these results strongly indicate that maturation of the 25S rRNA is largely or fully completed prior to release of the particle from the nucleolus, whereas the final maturation of 7S to 5.8S rRNA occurs in the nucleoplasm.
Discussion
We have adopted a proteomic approach to follow the 60S pre-ribosomal particles from its assembly in the nucleolus until appearance in the cytoplasm. To this end, we isolated and compared the pre-60S ribosomes that were associated with seven proteins. Notably, a given bait protein can occur in different pre-ribosomal particles (for example Arx1), yet when purified is preferentially associated with a class (early, medium or late) of 60S pre-ribosomes. An explanation for this could be that the concentration of a given bait protein differs in the various precursor particles at steady state. Accordingly, Arx1 may be at its highest concentration in late nuclear/cytoplasmic particles, and occur in lower amounts in earlier particles. We have succeeded in ordering the purified complexes from early to late, consistent with the protein sedimentation and localization as well as the rRNA content of these particles. Thus, these analyses provide sequential ‘snapshots’ of the biochemical composition of these particles along the maturation and export pathway from nucleolar processing through nuclear export to cytoplasmic maturation. The particles characterized here contain many non-ribosomal proteins that were not isolated with previous 60S pre-ribosome purifications. The TAP purifications of Nug1, Sda1, Kre35 and Nop7 have been reported previously and the results are generally similar to our findings (Baßler et al., 2001; Harnpicharnchai et al., 2001; Gavin et al., 2002). Here we purified all seven bait proteins under identical conditions and further analyzed them for localization, sedimentation and RNA content.
These data shed new light on the earliest and late intermediates and give insights into how pre-ribosomes gain export competence. The earlier pre-60S particles, which are associated with Nsa3, have many known 60S biogenesis factors. However, these particles also contain a few components that also play a role in 40S subunit biogenesis. Among these are core factors of both box C/D and box H/ACA snoRNAs (Maden, 1998), which are required for modification of the pre-rRNA. Most snoRNA-directed modification takes place on the 35S rRNA, but the presence of the snoRNP proteins in the pre-60S particle could also indicate that this is not fully completed prior to the early cleavages. A similar observation has been made for Xenopus pre-rRNA methylation (Yu et al., 1997). Also present is Rrp5, which is required for cleavage of the 27SA2 pre-rRNA at site A3, as well as for cleavage at earlier sites on the 40S synthesis pathway (Venema and Tollervey, 1996). Its presence in an early 60S pre-ribosome is not unexpected, but it was not identified in previous analyses. Less expected was the presence of the U3 snoRNP protein Rrp9p, which is required for cleavage at sites A0, A1 and A2 within the 35S pre-rRNA (Venema and Tollervey, 1996; Venema et al., 2000). Whether Rrp9 remains on the pre-rRNA after the other U3 snoRNP components have dissociated or has a separate function in 60S synthesis remains to be determined. Notably, Rrp9 was recently found to be associated with early 90S pre-ribosomes including the 35S pre-rRNA, the U3 snoRNP and 40S subunit processing factors, but predominantly lacking 60S synthesis factors (Grandi et al., 2002). However, purified Rrp9 also contains 27SA2 pre-rRNA, whereas the other purified 40S biogenesis factors do not contain this intermediate (Grandi et al., 2002). Thus, Rrp9 could be one among fewer factors that bridge between 60S and 40S biogenesis and are already present in 90S pre-ribosomes. Thus, it is possible that Rrp9, Nsa3 and a few other 60S biogenesis factors become associated with 35S pre-rRNA to trigger cleavage at A2 site as a prerequisite for generating 27S pre-rRNA and subsequent recruitment of the bulk 60S biogenesis machinery. Ribosomal RNA analysis (Figure 4) and sucrose gradient sedimentation (Figure 1) suggest that Nsa3 might bind to very early pre-60S particles and possibly already to 90S particles.
Previously purified 60S pre-ribosomes that were associated with Nop7, Nug1, Nog2/Nug2 and Ssf1 (Baßler et al., 2001; Harnpicharnchai et al., 2001; Saveanu et al., 2001; Fatica et al., 2002) represent nucleolar intermediates that are located prior to the pre-60S particles associated with Sda1, Arx1 and Kre35 in the 60S synthesis pathway (the protein profile of TAP-tagged purified Nug2 is similar to that of Nug1; T.A.Nissan and J.Baßler, unpublished data).
The pre-ribosomal particles isolated with Arx1 and Kre35 baits represent, respectively, the 60S pre-ribosome just prior and after export into the cytoplasm. Strikingly, these particles are relatively simple and have only a few non-ribosomal proteins bound. The nuclear export factor Nmd3 joins the 60S pre-ribosome at the level of Arx1 (Ydr101c, termed Arx1 for associated with ribosomal export complex), which could be the trigger for the acquisition of export competence. The adapter Nmd3 can interact with both the large subunit protein Rpl10 on the ribosome and the nuclear export receptor Xpo1 (Ho et al., 2000b; Gadal et al., 2001b). Xpo1 is thought to bind to nucleoplasmic NES-containing cargoes, such as Nmd3, in a RanGTP-dependent manner. However, Rpl10 was found to be present in all the purified 60S pre-ribosomes. It is thus conceivable that the Nmd3 binding site on Rpl10 is concealed in earlier 60S pre-ribosomes, but revealed in a later stages of maturation, e.g. by the action of ATPases or GTPases. In earlier studies, Rpl10p was reported to associate with 60S ribosomal subunits in the cytoplasm where it functions to mediate joining of 60S and 40S subunits (Eisinger et al., 1997). Our previous work suggested that Rpl10p may have a late nuclear function, as indicated by defects in ribosome export in rpl10 temperature-sensitive (ts) mutants (Gadal et al., 2001b).
Another nuclear export factor, Mtr2, was also found associated with the Arx1 particle. Most ts mutants of MTR2 show an mRNA export defect (Santos-Rosa et al., 1998). However, the mtr2-33 allele did not impair mRNA export, but showed nuclear accumulation of 60S pre-ribosomes at the restrictive temperature and a synthetic lethal relationship with the Nug1 GTPase, Ecm1 and Nmd3 (Baßler et al., 2001). Mtr2 was not identified in earlier or later 60S pre-ribosomal particles, suggesting a more direct role of Mtr2 in 60S subunit export. These data provide a biochemical link between the bona fide mRNA export factor Mtr2 and ribosomal export. We also found the importin Kap121/Pse1 associated with the Arx1-containing 60S pre-ribosomal particles (see Table I). The significance of this interaction is not clear. Kap121/Pse1 is known to be involved in the import of ribosomal proteins (Rout et al., 1997). However, a role of Kap121 in nuclear export of pre-ribosomes cannot be excluded. Notably, the karyopherin Kap142p/Msn5p mediates nuclear import and export of different cargo proteins (Yoshida and Blobel, 2001).
A striking feature of both early and late 60S pre-ribosomes is their association with several putative GTPases (Baßler et al., 2001; Harnpicharnchai et al., 2001; Saveanu et al., 2001; Fatica et al., 2002). The Nug1, Nug2 and Nog1 GTPases have nucleolar/nuclear locations (Baßler et al., 2001; Park et al., 2001; Saveanu et al., 2001) and co-purify with nuclear pre-ribosomal particles (from Nsa3 until Arx1), whereas the Kre35 GTPase is predominately localized in the cytoplasm and associates with the three later 60S pre-ribosomes (Sda1, Arx1 and Kre35). Thus, pre-60S associated GTPases could play regulatory roles, e.g. in the maturation and export of 60S subunits, or in signaling between ribosome synthesis and other cellular pathways.
Another conspicuous factor of the 60S pre-ribosome is the very large (559 kDa) protein Rea1. Rea1 is a member of the AAA family of ATPases (Baßler et al., 2001), which have a chaperone activity that has been implicated in dissociating protein–protein interactions (Vale, 2000). It is thus tempting to speculate that Rea1 functions in the late nucleoplasmic 60S pre-ribosome to dissociate non- ribosomal proteins at or prior to export. This class of ATPases may have another function as molecular motors (Vale, 2000), which could be relevant in the case of Rea1, as it has recently been shown to be related to dynein (Garbarino and Gibbons, 2002). Therefore, an alternative role of Rea1 may be in assisting transportation of the pre-ribosomal particle to the nuclear pore or through the nuclear pore complex.
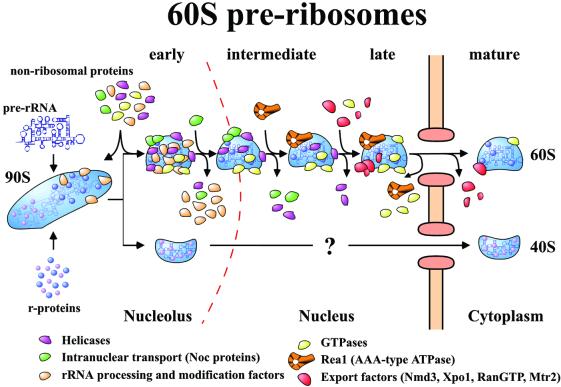
Taken together, our data allow us to draw a new model for maturation and export of the 60S ribosomal subunit (Figure 5). The initial ribosomal precursor particle is a 90S assembly, which after cleavage at A2 separates into 40S and 60S pre-subunits. The derived pre-60S ribosomes undergo a series of RNA processing reactions, which are likely to be exclusively nucleolar. These particles carry many associated factors that fulfill many different functions (see Fatica and Tollervey, 2002). Following rRNA maturation, the 60S pre-ribosomes move from the nucleolus to the nucleoplasm, accompanied by major changes in the protein composition of the particles. In the nucleoplasm, maturation and removal of factors from the particle continues. However, new components join the 60S pre-ribosome in a sequential manner, of which the huge AAA-type ATPase may catalyze the restructuring or the export of the 60S pre-ribosomes. Prior or concomitantly to export, most of the GTPases are removed from the 60S pre-ribosome and an Nmd3 binding site is revealed, which triggers recruitment of the general export receptor exportin-1. The final cytoplasmic pre-60S ribosomes have lost almost all of the non-ribosomal proteins and await the final structural rearrangements that will convert them into mature subunits competent for translation.
Fig. 5. Model of the pathway of 60S pre-ribosome maturation and export. The maturation of the pre-ribosome is depicted. The various classes of factors that are associated with the particle are identified by color. The factors coming on and off the particles represent actual identified proteins by mass spectrometry. For simplicity, the alterations in RNA composition are not depicted.
Materials and methods
Yeast strains and plasmids
Genomic integration of GFP (HIS3MX6-Marker) and TAP (TRP1-Marker) C-terminal tags resulting in fusion proteins of Nsa3 (YHR052w), Nop7 (YGR103w), Rix1 (YHR197w), Sda1 (YGR245c), Arx1 (YDR101c) and Kre35 (YGL099w) were performed as described previously (Longtine et al., 1998; Puig et al., 2001) into the yeast strain DS1-2b (MATa, ura3, trp1, his3, leu2) derived from cross of FY23 with FY86. The following yeast strains and plasmids used in this study were as described previously: Nug1-TAP (Baßler et al., 2001), FY23 and FY86 (Gadal et al., 2001b).
Sucrose density gradient centrifugation
Isolation of ribosomes under low salt conditions by sucrose gradient centrifugation was performed as described previously (Baßler et al., 2001). Briefly, cycloheximide was added to yeast grown to OD600 0.5, after a 15 min incubation, cells were washed and lysed. After ultracentrifugation, a gradient collector was used to record the UV profile and collect fractions for analysis.
RNA analysis
Northern hybridization and primer extension of RNA extracted from the reverse-tagged and purified protein baits, which were isolated using the complete TAP purification protocol with modifications to minimize RNA degradation. Briefly, the purification were performed using 1 mM ribonucleoside vanadyl complex (Sigma) as an RNase inhibitor, with time reduction in centrifugation and TEV cleavage steps to further reduce ribonuclease degradation. After purifications, samples were flash-frozen in liquid nitrogen. The northern hybridization and primer extention were performed as described previously (Beltrame and Tollervey, 1992; Tollervey et al., 1993). Oligonucleotides used were: 006, AGATTAGCCGCAGTTGG; 007, CTCCGCTTATTGATATGC; 017, GCGTTGTTCATCGATGC; 020, TGAGAAGGAAATGACGCT; 033, CGCTGC TCACCAATGG; 041, CTACTCGGTCAGGCTC; 070, CTCCGCTTATTGATATGC; 200, UUAUGGGACUUGUU (2′-O-methyl RNA); 211, TGCGAATGTTAAGGAACC; 218, CUIUUAAAUUUICIUU (2′-O-methyl RNA).
Mass spectrometry
Mass spectrometry using tryptic digest from Coomassie-stained SDS–PAGE was performed as described previously (Baßler et al., 2001). Proteins were identified using Mascot (Matrix Science) and the MSDB protein database. The MS/MS results were supported by peptide fingerprints (involving less than five peptides).
Miscellaneous
Affinity purification of TAP-tagged proteins was performed as described previously using 2–6 l of yeast culture (Puig et al., 2001). Protein was visualized using Novex 4–12% gradient or NuPAGE 10–20% gradient SDS–PAGE (Invitrogen) stained with colloidal Coomassie (Sigma). Western blot analysis were performed according to Siniossoglou et al. (1996). Fluorescence microscopy was carried out as described previously (Gadal et al., 2001a).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Dr Trumpower (Dartmouth Medical School, Hanover, NH) for providing antibodies against Rpl10 and Sqt1, Dr A.Johnston (University of Texas, Austin, TX) for Nmd3 antibodies, Dr F.Fasiolo (IBMC, Strasbourg, France) for Tif6 antibodies, and Dr H.Tschochner (BZH, Heidelberg, Germany) for the Noc antibodies. The excellent technical assistance of Petra Ihrig under the supervison of Dr J.Lechner (Mass Spectrometry Unit, BZH, Heidelberg, Germany) is acknowledged. E.H. is recipient of grants from the Deutsche Forschungs gemeinschaft (Schwerpunktprogramm ‘Funktionelle Architektur des Zellkerns’ and Gottfried Wilhelm Leibniz Program). E.P. and D.T. were supported by the Wellcome Trust.
References
- Adams C.C., Jakovljevic,J., Roman,J., Harnpicharnchai,P. and Woolford,J.L.,Jr (2002) Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA, 8, 150–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J., Grandi,P., Gadal,O., Leßmann,T., Tollervey,D., Lechner,J. and Hurt,E.C. (2001) Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell, 8, 517–529. [DOI] [PubMed] [Google Scholar]
- Beltrame M. and Tollervey,D. (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscemi G., Saracino,F., Masnada,D. and Carbone,M.L. (2000) The Saccharomyces cerevisiae SDA1 gene is required for actin cytoskeleton organization and cell cycle progression. J. Cell Sci., 113, 1199–1211. [DOI] [PubMed] [Google Scholar]
- Dragon F. et al. (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature, 417, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar D.A., Dragon,F., Lee,S.J. and Baserga,S.J. (2000) A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl Acad. Sci. USA, 97, 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger D.P., Dick,F.A. and Trumpower,B.L. (1997) Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol., 17, 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A. and Tollervey,D. (2002) Making ribosomes. Curr. Opin. Cell Biol., 14, 313–318. [DOI] [PubMed] [Google Scholar]
- Fatica A., Cronshaw,A.D., Dlakic,M. and Tollervey,D. (2002) Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell, 9, 341–351. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauß,D., Braspenning,J., Hoepfner,D., Petfalski,E., Philippsen,P., Tollervey,D. and Hurt,E.C. (2001a) A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J., 20, 3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauß,D., Kessl,J., Trumpower,B., Tollervey,D. and Hurt,E. (2001b) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a NES-containing factor Nmd3p that associates with the large subunit protein Rpl10p. Mol. Cell. Biol., 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauß,D., Petfalski,E., Gleizes,P.-E., Gas,N., Tollervey,D. and Hurt,E. (2002) Rlp7p is associated with 60S pre-ribosomes, restricted to the granular component of the nucleolus and required for pre-rRNA processing. J. Cell Biol., 157, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino J.E. and Gibbons,I.R. (2002) Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics, 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.-C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Grandi P. et al. (2002) 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP and 40S subunit processing factors but predominately lack 60S synthesis factors. Mol. Cell, 10, 105–115. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai P. et al. (2001) Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell, 8, 505–515. [DOI] [PubMed] [Google Scholar]
- Ho J.H.N., Kallstrom,G. and Johnson,A.W. (2000a) Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA, 6, 1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H.N., Kallstrom,G. and Johnson,A.W. (2000b) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol., 151, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl T. et al. (1999) GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr. Genet., 34, 419–429. [DOI] [PubMed] [Google Scholar]
- Kressler D., Linder,P. and De La Cruz,J. (1999) Protein trans-acting factors involved in ribsome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T., Planta,R.J. and Knop,J.M. (1978) The course of the assembly of ribosomal subunits. Biochim. Biophys. Acta, 517, 378–389. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 10, 953–961. [DOI] [PubMed] [Google Scholar]
- Maden B.E.H. (1998) Eukaryotic rRNA methylation: the calm before the Sno storm. Trends Biochem. Sci., 23, 447–450. [DOI] [PubMed] [Google Scholar]
- Milkereit P. et al. (2001) Maturation of pre-ribosomes requires Noc-proteins and is coupled to transport from the nucleolus to the nucleoplasm. Cell, 105, 499–509. [DOI] [PubMed] [Google Scholar]
- Moy T.I. and Silver,P.A. (1999) Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev., 13, 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Jensen,B.C., Kifer,C.T. and Parsons,M. (2001) A novel nucleolar G-protein conserved in eukaryotes. J. Cell Sci., 114, 173–185. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary,F., Rigaut,G., Rutz,B., Bouveret,E., Bragado-Nilsson,E., Wilm,M. and Seraphin,B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods, 24, 218–229. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Blobel,G. and Aitchison,J.D. (1997) A distinct nuclear import pathway used by ribosomal proteins. Cell, 89, 715–725. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno,H., Simos,G., Segref,A., Fahrenkrog,B., Panté,N. and Hurt,E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C., Bienvenu,D., Namane,A., Gleizes,P.E., Gas,N., Jacquier,A. and Fromont-Racine,M. (2001) Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J., 20, 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer,C., Rieger,M., Doye,V., Tekotte,H., Weise,C., Emig,S., Segref,A. and Hurt,E.C. (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell, 84, 265–275. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen,H., Jansen,R.P., Kern,H. and Hurt,E.C. (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation and ribosome assembly. Cell, 72, 443–457. [DOI] [PubMed] [Google Scholar]
- Trapman J., Retèl,J. and Planta,R.J. (1975) Ribosomal precursor particles from yeast. Exp. Cell Res., 90, 95–104. [DOI] [PubMed] [Google Scholar]
- Vale R.D. (2000) AAA proteins: lords of the ring. J. Cell Biol., 150, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1996) RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J., 15, 5701–5714. [PMC free article] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- Venema J., Vos,H.R., Faber,A.W., van Venrooij,W.J. and Raue,H.A. (2000) Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA, 6, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. (1989) Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev., 53, 256–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci., 24, 437–440. [DOI] [PubMed] [Google Scholar]
- Warner J.R. (2001) Nascent ribosomes. Cell, 107, 133–136. [DOI] [PubMed] [Google Scholar]
- Woolford J.L. Jr, (1991) The structure and biogenesis of yeast ribosomes. Adv. Genet., 29, 63–118. [DOI] [PubMed] [Google Scholar]
- Yoshida K. and Blobel,G. (2001) The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol., 152, 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.T., Shu,M.D. and Steitz,J.A. (1997) A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA, 3, 324–331. [PMC free article] [PubMed] [Google Scholar]