Abstract
LimC and LimD are two novel LIM proteins of Dictyostelium, which are comprised of double and single LIM domains, respectively. Green fluorescent protein-fused LimC and LimD proteins preferentially accumulate at areas of the cell cortex where they co-localize with actin and associate transiently with cytoskeleton-dependent dynamic structures like phagosomes, macropinosomes and pseudopods. Furthermore, both LimC and LimD interact directly with F-actin in vitro. Mutant cells that lack either LimC or LimD, or both, exhibit normal growth. They are, however, significantly impaired in growth under stress conditions and are highly sensitive to osmotic shock, suggesting that LimC and LimD contribute towards the maintenance of cortical strength. Moreover, we noted an altered morphology and F-actin distribution in LimD– and LimC–/D– mutants, and changes in chemotactic motility associated with an increased pseudopod formation. Our results reveal both unique and overlapping roles for LimC and LimD, and suggest that both act directly on the actin cytoskeleton and provide rigidity to the cortex.
Keywords: actin binding protein/cytoskeleton/mutants/pseudopod formation
Introduction
Many cellular proteins are comprised of a variety of conserved protein-binding modules facilitating protein– protein interactions and thereby playing key roles in diverse cellular processes. One such conserved protein-binding module is the LIM domain, which is present in a wide variety of proteins in organisms representing an extensive range of evolution. The LIM domain displays a consensus amino acid sequence [CX2CX16–23HX2C]-X2-[CX2CX16–21CX2–3(C/H/D)] (Freyd et al., 1990). Structural studies have revealed that it is comprised of two zinc finger-like modules that co-ordinate the binding of two zinc atoms via the conserved cysteine, histidine and aspartate residues of the LIM consensus (Perez-Alvarado et al., 1994). Despite their ability to bind zinc ions and their structural similarity to zinc fingers, evidence available to date implicates LIM domains in protein–protein interactions rather than DNA binding (Schmeichel and Beckerle, 1994). Interaction of LIM domains with specific protein partners is now known to influence its subcellular distribution and activity; however, no single binding motif has been identified as a common target for LIM domains. LIM domains can dimerize with other LIM domains to form homo- or heterodimers, and interact with structural motifs found in transcription factors, signaling and cytoskeletal proteins, such as tyrosine-containing tight turn, Src homology 3 domain, helix–loop–helix domain, ankyrin repeat, PDZ domain and spectrin repeat (Tu et al., 1999; Flick and Konieczny, 2000).
LIM proteins have been classified based on the sequence relationships among LIM domains, number and position of LIM domains, and the presence of other functional domains besides the LIM domain, such as homeodomain, protein kinase domain and motifs involved in protein–protein interactions. Most group 1 proteins [e.g. LIM homeodomain (LHX) proteins and LIM only (LMO) proteins] are known to function in transcriptional regulation in the nucleus, whereas most group 2 (e.g. members of the cysteine-rich protein family, CRP1, CRP2 and CRP3) and group 3 (e.g. zyxin and paxillin) proteins are associated with the cytoskeleton (Khurana et al., 2002). Members of the CRP family are comprised primarily of only two LIM domains and have been shown to be associated with elements of the actin cytoskeleton, as all are capable of direct interaction with α-actinin and zyxin (Schmeichel and Beckerle, 1994; Pomies et al., 1997). CRP3 (also known as muscle LIM protein, MLP) specifically interacts through its C-terminal LIM domain with the cytoskeletal protein βI-spectrin (Flick and Konieczny, 2000). Gene disruption studies in the mouse have confirmed the requirement for CRP3/MLP in cytoarchitectural organization of both cardiac and skeletal muscle (Arber et al., 1997). In Dictyostelium, the LIM proteins DdLim and LIM2 have been observed to localize at the newly formed membrane extensions in motile cells. While DdLim is involved in the proper protrusion of lamellipodia during chemotaxis (Prassler et al., 1998), Dictyostelium cells lacking LIM2 are unable to properly organize their actin cytoskeleton in a chemotactic gradient and arrest at the mound stage of development (Chien et al., 2000).
Here we present the characterization of two novel Dictyostelium LIM proteins: LimC, which is comprised primarily of two LIM domains, and LimD, which harbors a single LIM domain. We have studied the dynamics of their subcellular redistribution using a green fluorescent protein (GFP)-tagged version of the two proteins and have generated mutant cells that lack either LimC or LimD, or both. Characterization of their phenotypes suggests that LimD affects cell polarity, pseudopod formation and chemotaxis, and that both LimC and LimD are required for maintaining the cell cortex under stress conditions. This might be due to direct interaction with the actin cytoskeleton.
Results
LimC and LimD are two novel Lim proteins in Dictyostelium
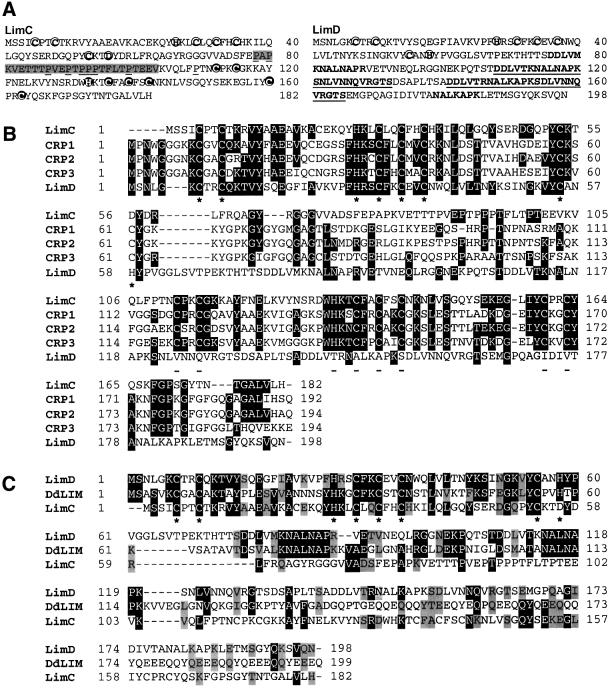
LimC and LimD are two novel LIM domain-containing proteins of 182 and 198 amino acids with calculated mol. wts of 20 000 and 21 500, respectively. LimC contains two LIM domains, one each at its N- and C-terminus, spanning amino acid residues 5–56 and 112–163, respectively, whereas LimD harbors a single LIM domain at its N-terminus, spanning amino acid residues 7–58 (Figure 1A). Based on the sequence relationship among the LIM domains and their overall structure, LimC and LimD belong to the group 2 LIM proteins (Dawid et al., 1998). Both LimC and LimD exhibit unique structural characteristics. In LimC, the sequence between the two LIM domains is proline rich (Figure 1A); LimD contains two nearly perfect repeat motifs of 25 amino acid resi dues DDLVT(K/R)NAL(N/K)APKS(N/D)LVNNQVRGTS (amino acid residues 108–132 and 141–165) at its C-terminus. Two shorter forms of this 25 amino acid repeat motif, DDLVMKNALNAPR (amino acid residues 76–88) and NALKAPK (amino acid residues 179–185), also appear once at the C-terminus of LimD (Figure 1A).
Fig. 1. (A) Deduced amino acid sequence of LimC and LimD. The conserved cysteine, histidine or aspartate residues of the N-terminal LIM domains of LimC and LimD (bold, circled letters) and the C-terminal LIM domain of LimC (bold, shaded and circled letters) are shown. The intervening sequence between the two LIM domains of LimC exhibiting low compositional complexity is shaded, the proline residues are underlined. The two nearly perfect repeat motifs of 25 amino acid residues (shown in bold, underlined letters) and two shorter versions of this repeat motif (shown in bold letters) in the C-terminal region of LimD are indicated. (B) Alignment of the LimC and LimD amino acid sequences with members of the CRP family, CRP1, CRP2 and CRP3. Residues that are identical between LimC and/or LimD and any of the other members of the CRP family are shadowed in dark gray. Asterisks and dashes below the aligned sequences indicate the N-terminal LIM consensus (in all sequences) and C-terminal LIM consensus (in all sequences except LimD), respectively. (C) Alignment of the LimC and LimD amino acid sequences with Dictyostelium DdLim. Residues that are identical between any two sequences are shadowed in dark gray, and residues that are similar are shadowed in gray. Asterisks below the aligned sequences indicate the N-terminal LIM consensus. Sequences were aligned using the Clustal_W program (Wisconsin package version 9.0). Dashes indicate gaps introduced in the sequence for optimal alignment. DDBJ/EMBL/GenBank accession Nos: LimC, AF348466; LimD, AF348467; DdLim, U97699.1; CRP1, P32965; CRP2, P50460; CRP3, P50463.
LimC exhibits the highest degree of homology (30–31% identity) to members of the mammalian CRP family, CRP1, CRP2 and CRP3 (Figure 1B), while LimD exhibits a very low homology (16–20% identity) to the three CRPs. LimD, however, is more closely related to Dictyostelium DdLim (27% identity and 40% similarity), comprised of a single LIM domain at its N-terminus, a glycine-rich region that follows the LIM domain and a coiled-coil region at its C-terminus (Prassler et al., 1998). The LIM domains of LimD and DdLim are 42% identical. Interestingly, one of the LimD shorter repeat motifs, NALNAP(K/R), also occurs twice at similar locations in DdLim (Figure 1C).
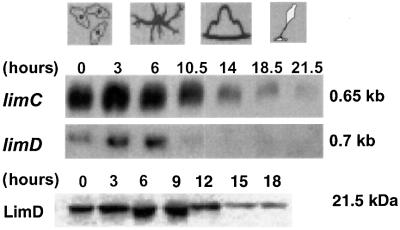
Both limC and limD are present as a single copy gene in the Dictyostelium genome. The coding regions of limC and limD genes are interrupted by three introns (of 130, 168 and 118 bp) and one intron (of 113 bp), respectively (not shown). limC and limD mRNA exists as a single species of 650 and 700 bp, respectively. The limC transcripts are abundant during the growth phase (0 h) and early development (3–6 h), and decline strongly once tight aggregates have formed (10.5 h). Likewise, limD mRNA is also present during the growth phase. The level of limD transcripts increases at 3 h of development, reaches a maximum at 6 h and then decreases strongly (10.5 h) (Figure 2). We also analyzed LimD protein levels with a monoclonal antibody and found high amounts during growth and early development. During mound formation (t12) and the following developmental stages, the amount decreased; however, the protein persisted throughout development (Figure 2). For LimC there is no antibody available with which we can detect the protein in western blots.
Fig. 2. Presence of limC and limD mRNAs and protein during development. The same amount (30 µg) of total RNA isolated from cells developed on phosphate-buffered agar plates at different hours of development as indicated was loaded in each lane. Hybridization was with 32P-labeled limC or limD cDNA. mRNA sizes are given in kb. For detection of LimD protein, total cellular extracts from AX2 cells (4 × 105 cells per lane) harvested at different time points were separated by SDS–PAGE (15% acrylamide). The resulting western blot was probed with LimD-specific monoclonal antibody K4-353-6.
Subcellular localization of LimC and LimD
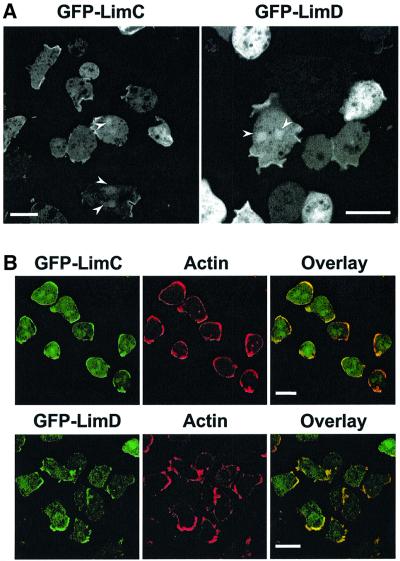
We used GFP-tagged versions of LimC and LimD, which complemented the null phenotype (see below), to investigate the subcellular localization of the two proteins in Dictyostelium. Immunoblot analyses performed with an anti-GFP antibody as well as LimC- and LimD-specific monoclonal antibodies indicated that both the GFP fusion proteins are stably produced (data not shown). GFP–LimC and GFP–LimD are present in the cytosol and associate with distinct areas of the cell cortex during various activities (see below). Occasional localization of GFP–LimC and GFP–LimD in the nucleus was also observed (Figure 3A). When GFP–LimC- and GFP– LimD-expressing cells were immunolabeled with anti-actin monoclonal antibody, we found that the pattern of GFP–LimC and GFP–LimD fluorescence coincided with that of the actin staining (Figure 3B). Furthermore, the GFP fusion proteins were recognized by Lim-specific monoclonal antibodies (data not shown).
Fig. 3. Distribution of GFP–LimC and GFP–LimD fusion proteins. (A) Both the fusion proteins accumulate to high levels at distinct areas of the cell cortex and are distributed throughout the cytoplasm. Occasional localization of both the fusion proteins in the nucleus was also observed (arrowheads). Bars, 10 µm. (B) GFP–LimC (upper panels) and GFP–LimD (lower panels) localization coincides with that of actin in the cell cortex. The cells were fixed with cold methanol and immunolabeled with anti-actin monoclonal antibody followed by Cy3-conjugated anti-mouse secondary antibody. Overlay images show the co-localization of GFP–LimC or GFP–LimD fluorescence with the actin staining in the cell cortex. Bars, 10 µm.
Localization of GFP–LimC and GFP–LimD during fluid- and solid-phase endocytosis
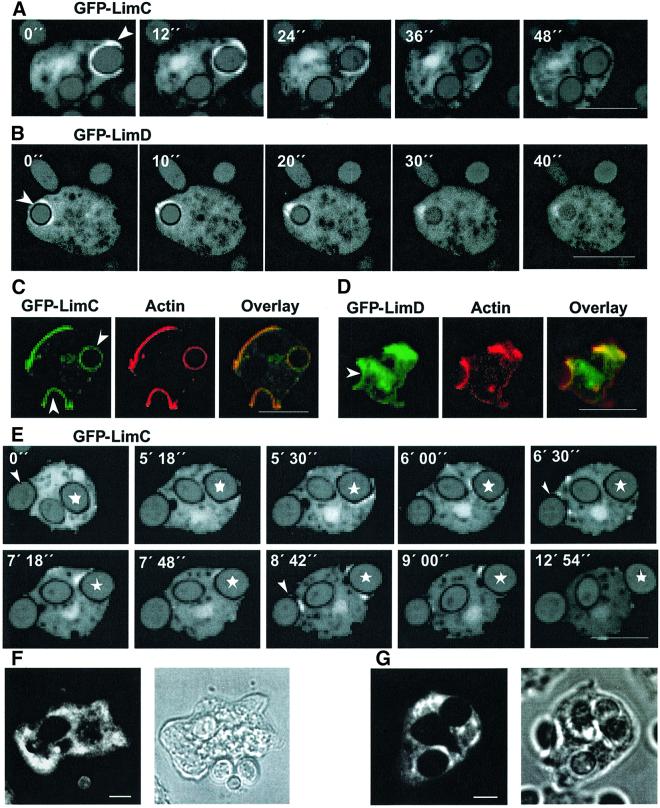
In Dictyostelium cells, macropinocytosis accounts for most of the fluid-phase uptake and depends on the integrity of the actin cytoskeleton (Hacker et al., 1997). We studied the dynamics of GFP–LimC and GFP–LimD distribution in Dictyostelium cells during uptake of TRITC–dextran. At the beginning of the sequence shown in Figure 4A, a GFP–LimC-rich membrane invaginates (0 s). The protrusion of the GFP–LimC-rich membrane progresses (12 and 24 s) until the edges of the protrusion fuse to form a macropinosome containing a portion of the surrounding liquid that is enclosed by a coat containing GFP–LimC (36 s). Thereafter, GFP–LimC gradually dissociates from the macropinosome (48 s) to liberate the macropinosome into the cytoplasm. We have observed essentially a similar pattern of redistribution of GFP–LimD in Dictyostelium cells during fluid-phase endocytosis (Figure 4B). This process is highly suggestive of an involvement of both LimC and LimD at early stages of fluid-phase endocytosis.
Fig. 4. GFP–LimC and GFP–LimD redistribution during fluid-phase endocytosis. Cells expressing GFP–LimC (A) or GFP–LimD (B) were allowed to adhere to glass coverslips and the supernatant was replaced by phosphate buffer containing 1 mg/ml TRITC–dextran. Confocal sections were taken at the times indicated. Arrowheads in the 0 s panels indicate the site of formation of pinocytic vesicles and arrows in subsequent images indicate newly formed pinosomes. Bars, 10 µm.
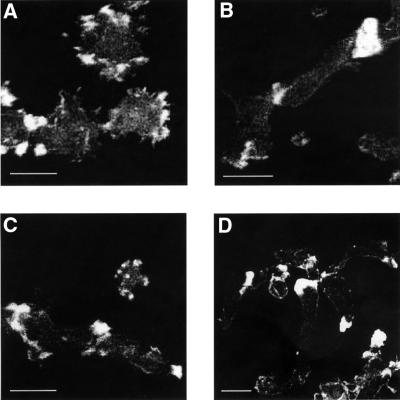
Phagocytosis is initiated by adhesion of a particle to the surface of a Dictyostelium cell. A phagocytic cup is formed at the cell surface around the particle, which, like fluid-phase endocytosis, involves active rearrangement of the actin cytoskeleton (Maniak et al., 1995). The rim of the cup extends around the particle, resulting in the internalization of the particle. GFP–LimC- and GFP–LimD-expressing Dictyostelium cells were challenged with TRITC-labeled yeast cells and observed under a confocal laser scan microscope. The GFP–LimC fusion protein accumulated to high levels at the phagocytic cup (0 s). Cup progression led to the formation of a coat around the yeast cell that is enriched with GFP–LimC (12 s), resulting in engulfment of the yeast cell. Immediately after the yeast cell has been successfully ingested, GFP–LimC is gradually removed from the phagosome (24–48 s) (Figure 5A). The GFP–LimD fusion protein exhibited an identical distribution pattern (Figure 5B). GFP–LimD, like GFP–LimC, gradually dissociates from the phagosome (10–40 s) after the yeast cell has been engulfed. Immunolabeling of phagocytosing cells with anti-actin monoclonal antibody showed that the fluorescence of the two GFP fusion proteins on the phagocytic cups coincides with the actin staining (Figure 5C and D). The endogenous proteins, as detected with monoclonal antibodies, were present throughout the cells and in areas close to the plasma membrane and around phagocytic cups. This was more pronounced for LimD (Figure 5F and G). LimD protein was also present in isolated phagosomes (data not shown).
Fig. 5. LimC and LimD in phagocytosis and exocytosis. (A and B) Dynamics of GFP–LimC and GFP–LimD redistribution during phagocytosis. GFP–LimC (A) or GFP–LimD (B) expressing cells were incubated with TRITC-labeled, heat-killed yeast cells. Confocal sections were taken at the times indicated. Arrowheads in the 0 s panel (A and B) mark the site of formation of the phagosomes. Bars, 10 µm. (C and D) Co-localization of GFP–LimC and GFP–LimD with actin at the phagocytic cups. GFP–LimC (C) or GFP–LimD (D) expressing cells were incubated for 10 min with heat-killed yeast cells and labeled after methanol fixation with anti-actin monoclonal antibody. Fluorescence patterns of the GFP–LimC (C) and the GFP–LimD (D) fusion proteins at the phagocytic cups as well as the cell cortex coincide with actin staining (overlay). The arrowheads in the GFP panels mark the position of the yeast cell being phagocytosed. Bars, 10 µm. (E) Dynamics of GFP–LimC redistribution during exocytosis. GFP–LimC cells were incubated with TRITC-labeled, heat-killed yeast cells for ∼1 h. Confocal sections were obtained after every 6 s. Selected images are shown. Asterisks (in all the frames) mark the yeast cell of interest and the arrowhead (in 0 s panel) marks the yeast that the cell is trying to phagocytose. Note the accumulation of GFP–LimC around the exocytotic vacuole as the cell prepares for exocytosis of the phagocytosed yeast (marked with an asterisk). Arrowheads in 6 min 30 s and 8 min 42 s panels show the simultaneous accumulation of GFP–LimC fusion protein on the phagocytic cup. Bar, 10 µm. Localization of endogenous LimC (F) and LimD (G) during uptake of yeast particles. Fixation and immunofluorescence detection with the respective monoclonal antibodies was carried out as for Figure 3. The corresponding phase-contrast images are shown. Bars, 5 µm.
Occasionally we observed exocytosis events and found that the GFP–LimC fusion protein reassembles on the endocytic vesicle as it returns to the cell cortex for exocytosis of the yeast particle. The series of images shown in Figure 5E suggests that LimC plays a role in exocytosis and might possibly facilitate association of the late vacuole with the cell cortex.
Localization of GFP–LimC and GFP–LimD in motile cells
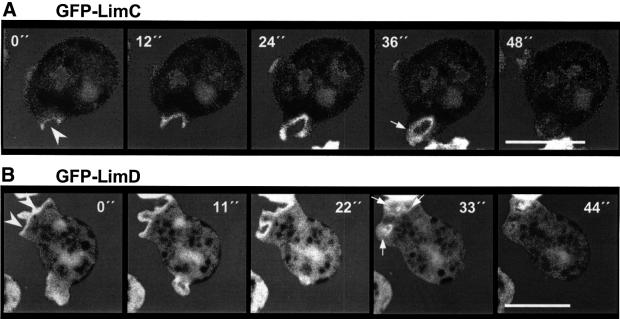
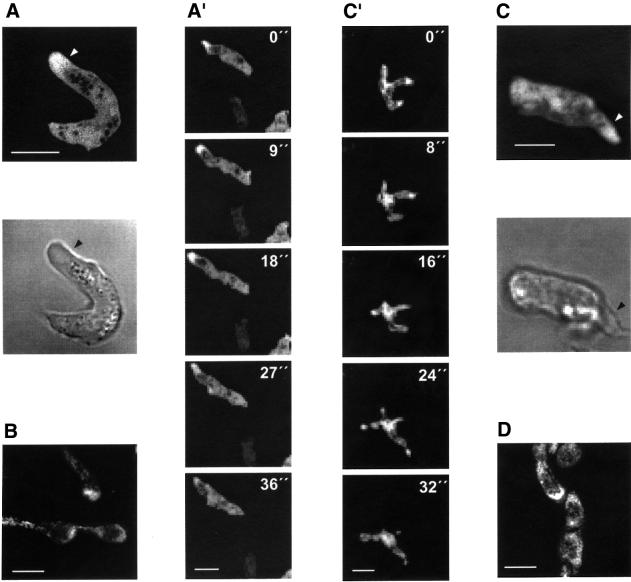
Dictyostelium cells are highly motile and move by extension of pseudopods. Extensive remodeling of the actin network accompanies the extension of pseudopods. Figure 6A and C shows the fluorescence images of aggregation-competent GFP–LimC- and GFP–LimD-expressing cells, respectively, which are extending a pseudopod. The presence and location of the pseudopod can be easily distinguished in the respective phase-contrast images (Figure 6A and C). GFP–LimC as well as GFP–LimD fusion protein were found to accumulate to high levels in the pseudopods (Figure 6A′ and C′). For the endogenous proteins we detected a similar enrichment in extending pseudopods of aggregating cells (Figure 6B and D).
Fig. 6. LimC and LimD during cell motility. Cells starved for 6 h aggregate and, in general, acquire an elongated shape with a well-defined front and tail. Fluorescence images of aggregation-competent GFP–LimC (A) and GFP–LimD (C) cells and their respective phase-contrast images are shown. The arrows point to an extending pseudopod. Time series of aggregation-competent GFP–LimC (A′) and GFP–LimD (C′) expressing cells, which are rapidly extending pseudopods. Confocal images were taken at the times indicated. Both GFP–LimC and GFP–LimD accumulate to high levels in the pseudopods, as do endogenous LimC (B) and LimD (D) in aggregating AX2 cells. Detection was as described in Figure 5. Bars, 10 µm.
All LimC domains co-localize with the actin cytoskeleton
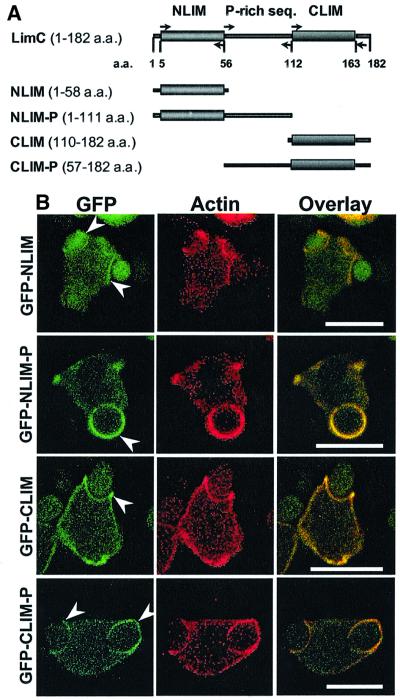
Since individual LIM domains are capable of docking with unique protein partners and the LimC protein is comprised primarily of two LIM domains, we were interested in the possibility that the site on LimC that is responsible for its co-localization with actin and its involvement in the dynamic processes might be contained within a single LIM domain. Confocal studies performed with the cells expressing GFP–NLIM, GFP–NLIM-P, GFP–CLIM and GFP–CLIM-P revealed that all four GFP fusion proteins exhibit a fluorescence pattern identical to the parent GFP–LimC, with preferential localization at the cell cortex, leading pseudopods of the motile cells, phagocytic cups and macropinosomes. Immunolabeling of phagocytosing cells with anti-actin monoclonal antibody showed that the fluorescence of all four GFP-fused LimC deletion products coincides with the actin staining (Figure 7). Our results suggest that either LIM domain of LimC is sufficient for its co-localization with actin and its involvement in the dynamic processes.
Fig. 7. Localization of N- and C-terminal LimC deletion products. (A) Schematic representation of terminally deleted LimC proteins used for localization studies. Domain organization of LimC is shown. NLIM and CLIM represent the N- and C-terminal LIM domain, respectively. ‘P-rich seq.’ represents the intervening region between the two LIM domains that is rich in proline residues. Arrows indicate the position and orientation of oligonucleotide primers used for amplification. (B) Distribution of GFP–NLIM, GFP–NLIM-P, GFP–CLIM and GFP–CLIM-P during phagocytosis. Cells expressing these proteins exhibit a fluorescence pattern that is identical to that of the parent GFP–LimC fusion protein (see Figure 5C). All the truncated GFP fusion proteins accumulate at the phagocytic cup (arrowheads) during uptake of the yeast cell and their fluorescence pattern coincides with actin staining (overlay). Bars, 10 µm.
LimC and LimD bind to F-actin in vitro
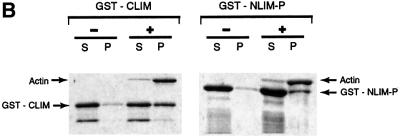
The finding that LimC and LimD co-localize with F-actin prompted us to study their interaction directly in actin sedimentation assays. Glutathione S-transferase (GST) fusions of LimC or LimD were used in this assay. Both proteins partially co-sedimented with filaments of α-actin purified from rabbit skeletal muscle as well as Dictyostelium actin, even in the presence of high salt concentration (100 mM KCl) (Figure 8), whereas GST–LimC and GST–LimD remained almost completely in the supernatant in the absence of actin. The GST protein itself did not sediment with F-actin (data not shown), thus suggesting a role for LimC and LimD in binding F-actin. Degradation products of GST–LimC and GST–LimD as well as a high molecular weight protein that is co-eluted with GST–LimD remain entirely in the supernatant, indicating the specificity of GST–LimC and GST–LimD binding to actin (Figure 8A).
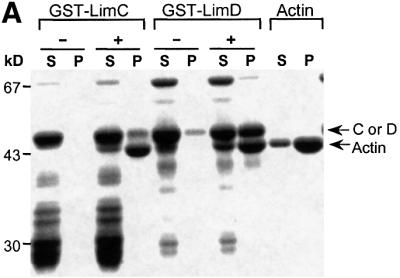
Fig. 8. Binding of LimC and LimD to F-actin. Co-sedimentation of (A) GST–LimC, GST–LimD and (B) GST–LimC deletion constructs GST–CLIM and GST–NLIM-P with F-actin. G-actin (5 µM) from D.discoideum was incubated with the GST fusion proteins in polymerization buffer containing 2 mM MgCl2, 100 mM KCl and 1 mM EGTA. Pellets (P) and supernatants (S) were separated by high-speed centrifugation, and the proteins in these fractions were analyzed by SDS–PAGE (12% acrylamide) and staining with Coomassie Blue. Controls for the fusion proteins alone were performed without the addition of G-actin in the reaction mixture. The presence (+) or absence (–) of G-actin in the reaction mixture is indicated on the top of the lanes. Arrows marked with C and D indicate GST–LimC and GST–LimD, respectively. Varying amounts of polymerized (P) and unpolymerized actin (S) in GST–LimC and GST–LimD panels might be due to different actin preparations.
The affinity of LimC and LimD for F-actin was measured at varying molar ratios of actin to LimC or LimD by densitometric analysis of supernatants and pellets of Coomassie Blue-stained SDS–polyacryl amide gels. The affinity constants obtained were 48.08 ± 8.92 µM for LimC and 11.85 ± 5.33 µM for LimD (data not shown). LimD had a binding capacity of ∼1 mol of GST–LimD per mol of actin. These numbers are in the range observed for other actin-binding proteins (Jung et al., 1996). When we tested the individual LimC domains we found that the GST–CLIM construct encompassing the C-terminal Lim domain was able to bind to F-actin as well as GST–NLIM-P, whereas GST–NLIM was not significantly enriched in the F-actin pellet in this assay, despite the fact that GFP fusions of this domain co-localize with F-actin (Figure 8B). This might, however, also be due to improper folding of the recombinant polypeptide.
Generation of LimC– and LimD– single mutants and LimC–/LimD– double mutant
To gain insight into the functions of LimC and LimD in vivo, we generated LimC– and LimD– single mutants lacking limC and limD genes, respectively, as well as a LimC–/LimD– double mutant. The gene replacement events were confirmed using Southern blot analyses. At the RNA level, no limC message could be detected in the LimC– single mutant and LimC–/LimD– double mutant strains. Although negligible amounts of truncated transcripts for limD were detected in the LimD– single mutant and LimC–/LimD– double mutant strains, the LimD protein was found to be absent in both LimD– and LimC–/LimD– mutants (data not shown). In all subsequent analyses, AX2 (wild-type parent strain), LimC– and LimD– single mutants, and LimC–/LimD– double mutant strains were assayed.
Growth and morphology of mutant strains
Loss of LimC and LimD does not affect growth under optimal conditions, and phagocytosis and endocytosis were unimpaired in all mutant strains. Qualitative examination of cultures of the LimC–, LimD– and LimC–/LimD– mutants grown in axenic medium suggested that cells of the mutant strains were bigger in size compared with the wild-type AX2 cells. In a quantitative analysis, the cells of the mutant strains were found to be comparatively larger, with an average diameter of 13 µm for LimD– single mutants and 15 µm for LimC–/LimD– double mutants, compared with an average diameter of 11 µm for AX2 cells. LimC– mutants showed a slightly increased size (data not shown). DAPI labeling revealed that the large-sized mutant cells were multinucleated. When we quantified the number of nuclei per cell in the AX2 and mutant strains grown under shaking conditions we found that the majority of AX2 wild-type cells were mononucleated and binucleated, whereas many LimC–, LimD– and LimC–/LimD– mutant cells possessed four or more nuclei, suggesting that cytokinesis is not normal in all mutant strains (data not shown). We also determined the number of nuclei in cells attached to a substratum. Under these conditions, cells can undergo traction-mediated cyto fission (Neujahr et al., 1997). Again, more bi- and multinucleate cells were observed in the cultures of the Lim mutants than of the AX2 wild-type cells, confirming the mild cytokinesis defect.
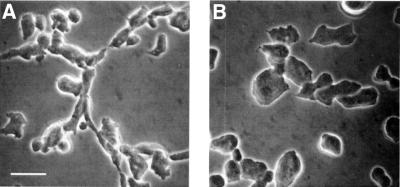
Dictyostelium wild-type cells after 6 h of starvation have an altered cell shape and are typically elongated. LimC– cells were also elongated, whereas LimD– cells did not show a change in cell morphology and had a more irregular cell shape. This was not due to a delay in development, since the aggregation stage-specific marker contact site A was present in comparable amounts and showed the same pattern of expression during development. The F-actin distributed primarily in a cortical pattern and concentrated at the leading edges in AX2 and in LimC–. In LimD– cells, F-actin was found in patches (Figure 9). The F-actin polymerization responses upon cAMP stimulation, however, were not impaired in all mutant cells (data not shown). Introduction of a GFP-tagged LimD led to a complete reversal of the abnormal cell shape and actin distribution in the LimD– strain. During the aggregation stage, the cells were no longer rounded, but were highly polarized and elongated and were indistinguishable from wild type (Figure 10). These results suggest that LimD regulates cell morphology and organization of the actin cytoskeleton.
Fig. 9. F-actin distribution and cell polarization in mutant cells. (A) LimD– cells have a polarization defect and have F-actin acumulated in patches at the cortex. (B) LimC– cells are polarized and show F-actin accumulation at cell–cell contacts comparable to AX2 (D). (C) LimC–/LimD– cells resemble LimD– single mutants. Aggregation stage cells (t6) were fixed with picric acid/paraformaldehyde and stained with TRITC–phalloidin. Bar, 10 µm.
Fig. 10. Expression of GFP–LimD rescues the polarity defect in LimD– cells. Phase-contrast images of (A) aggregating (t6) LimD– cells expressing GFP–LimD and (B) aggregating LimD– cells. Bar, 20 µm.
Chemotactic motility in mutant strains
Chemotaxis of starving cells towards cAMP was analyzed in the capillary assay, where cells are stimulated with a micropipette filled with cAMP (10–3 M), and the velocity was determined. LimC– cells migrated with an average velocity of 10.19 ± 1.63 µm/min towards the pipette, which is similar to wild-type velocity (11.32 ± 1.23 µm/min). For LimD– we observed a slightly lower velocity (8.07 ± 2.93 µm/min) and the double mutant was impaired even more (7.65 ± 1.86 µm/min). To analyze motile cells in more detail we used a chemotaxis assay and followed the cells with time-lapse video microscopy. Wild-type as well as LimC– cells were polarized and migrated in a rapid and directed way towards the micropipette filled with cAMP (Table I). Furthermore, they extended their pseudopods mainly in the direction of the micropipette and formed very few lateral pseudopods (Table II). LimD– cells also moved to the cAMP source. However, they did not migrate as fast and formed lateral pseudopods in addition to the main pseudopod extended in the direction of cAMP, whereas persistence and directionality were only slightly impaired (Tables I and II; data not shown). LimD– cells re-expressing LimD protein as a GFP fusion behaved like wild type (Table I).
Table I. Motility and chemotaxis parameters in a spatial gradient of cAMP.
| Cell type | Cell number | Velocity (µm/min) | Persistence (µm/min-deg) | Roundness (%) |
|---|---|---|---|---|
| AX2 | 47 | 11.32 ± 1.23 | 4.40 ± 0.80 | 75.04 ± 3.43 |
| LimC– | 14 | 10.19 ± 1.63 | 4.22 ± 1.28 | 74.02 ± 0.31 |
| LimD– | 27 | 8.07 ± 2.47 | 2.93 ± 1.05 | 80.22 ± 3.97 |
| LimC–/LimD– | 66 | 7.65 ± 1.86 | 2.73 ± 0.94 | 79.51 ± 4.19 |
| Rescue | 12 | 8.96 ± 1.06 | 3.11 ± 0.77 | 72.83 ± 5.57 |
Images were taken at 10× magnification every 30 s. In all cases cells were analyzed for at least 10 min. Persistence is essentially speed divided by the directional change. If an object is not turning, its persistence is the same as its speed. The roundness parameter indicates polarization. Generally, a more polarized cell shape produces less roundness.
Table II. Lateral pseudopod formation by AX2 wild-type and LimD– cells crawling in buffer or in a spatial gradient of cAMP.
| Cell type | Number of cells | 0–2 lateral pseudopods per 10 min (%) | 3–5 lateral pseudopods per 10 min (%) | >5 lateral pseudopods per 10 min (%) | Average frequency of lateral pseudopods per cell per 10 min | |
|---|---|---|---|---|---|---|
| Buffer | AX2 | 28 | 15 | 53 | 30 | 5.3 |
| LimD– | 23 | 5 | 60 | 34 | 4.9 | |
| Gradient | AX2 | 22 | 81 | 18 | – | 1.45 |
| LimD– | 25 | 40 | 60 | – | 2.7 |
Images were taken at 20× magnification every 30 s. In all cases cells were analyzed for 10 min. A χ2 test was performed between AX2 and LimD– cells on the combined data of the three categories of lateral pseudopods formed. The difference between AX2 and LimD– in the cAMP gradient was found to be highly significant (1 × 10–3).
Growth under stress conditions
Since Dictyostelium discoideum lives as a natural phagocyte in soil and feeds on yeast and bacteria, fluctuations in the environmental temperature, humidity and osmolarity pose physiological challenges to growth and survival. It is, therefore, believed that certain proteins that are not essential under optimal laboratory conditions might play a role under stress conditions.
Cultures of AX2 cells and mutant strains grown at a lower temperature (15°C) in axenic medium exhibited similar growth patterns with regard to maximum cell density and doubling time. However, growth of all the mutant strains was significantly impaired when the cultures were grown at a higher temperature (27°C) in axenic medium. Under this condition, AX2 cells reached a cell density of 5.4 × 106 cells/ml with a doubling time of 13 h. The LimC– and LimD– mutants were clearly impaired, showing a prolonged doubling time of 19 h and reduced cell density at saturation. The growth of LimC–/LimD– double mutant was nearly completely abolished at 27°C, as it exhibited a very low saturation cell density of 1.8 × 106 cells/ml.
The ability of the mutant strains to grow in the presence of increased osmolarity was tested by supplementing the axenic medium with either 30 mM NaCl or 115 mM sorbitol. In the presence of 30 mM NaCl, AX2 cells grew to a density of 4.3 × 106 cells/ml at saturation with a doubling time of 18 h. LimC– cells exhibited a similar growth pattern, with a similar cell density at saturation and comparable doubling times. Similar cell density at saturation but a slightly longer doubling time of 22.5 h were observed for LimD–. The LimC–/LimD– cells showed a prolonged doubling time of 30 h and reduced cell density at saturation. The presence of 115 mM sorbitol led to an increased doubling time of 20 h and a reduced maximum cell density for wild-type and mutant cells alike.
Since the role of a cytoskeletal protein in the cytoskeletal rearrangements elicited by adaptation to sustained altered osmolarity could differ from that of the acute osmotic shock, the response of the mutant strains to acute osmotic shock was analyzed. To this end, a viability assay was performed by exposing the cells to 0.4 M sorbitol for 2 h and then diluting into a solution of low osmolarity. Both the single mutants as well as the double mutant cells exhibited an increased sensitivity to acute osmotic shock. While the AX2 cells showed a high viability of 81 ± 23.7%, the viability of the LimC–, LimD– and LimC–/LimD– mutant cells after osmotic shock was drastically reduced to 22.8 ± 10.6, 14.8 ± 6.2 and 22.7 ± 8.4%, respectively. This indicates that the mutant cells are less tolerant to osmotic shock, which might reflect a reduced strength of the cortical cytoskeleton in these mutants.
Development of mutant strains
Dictyostelium cells can aggregate in starvation buffer under submerged conditions, while post-aggregation development and fruiting require a solid substratum. Under these conditions, all mutant strains developed normally, expressed developmental markers at the appropriate time points and formed fruiting bodies with viable spores. Only when we placed the cells in monolayer under starvation buffer did we note differences. Under these conditions, wild-type AX2 cells form large aggregates by 13 h of development, whereas the mutant strains still display streams of moving cells at this time point, which persist even after 16 h (LimC– and LimD–) and 18 h (LimC–/LimD–) of development. Moreover, rather than coalescing to a single center as in case of the AX2 cells, individual streams of all the mutant strains often fractured along their length, forming smaller aggregates (data not shown).
Discussion
LimC and D directly associate with F-actin
Members of the CRP family (CRP1, CRP2 and CRP3/MLP) have been observed to play a regulatory or structural role in the actin cytoskeleton owing to their interaction with cytoskeletal proteins and their localization at focal contacts and stress fibers in mammalian cells (Schmeichel and Beckerle, 1994). Several other cytoplasmic LIM domain-containing proteins control cytoskeletal rearrangements in mammalian cells, such as paxillin, LIM kinase and zyxin (Beckerle, 1997). The Dictyostelium DdLim protein localizes in the cell cortex, where it might be involved in a receptor-mediated Rac-1 signaling pathway that leads to actin polymerization in lamellipodia and ultimately cell motility (Prassler et al., 1998). Likewise, the Dictyostelium LIM2 protein is enriched in the cell cortex, although it does not co-localize with F-actin, and is required for cell motility and chemotaxis (Chien et al., 2000). In contrast, LimC and LimD co-localize with F-actin in vivo and interact with F-actin directly in vitro. Based on their homology to CRPs and DdLim, respectively, they are classified as members of the group 2 LIM proteins.
LimC and LimD not only co-localize with F-actin, they also bind to F-actin directly, and GST fusion proteins co-sedimented with actin filaments even in the presence of high salt concentration (100 mM KCl), a property that has not previously been attributed to any other LIM domain only protein. In all the cases reported, the association with the actin cytoskeleton was mediated by proteins like α-actinin or zyxin.
LimC– and LimD– cells have defects associated with the actin cytoskeleton
LimC and LimD contribute to the maintenance of the strength of the cortical actin cytoskeleton, as is reflected by the inability of the LimC–, LimD– and LimC–/LimD– mutant strains to grow under conditions of high temperature as well as the increased sensitivity of all the mutant strains to osmotic shock. In addition, growth of LimD– and LimC–/LimD– cells is impaired under conditions of increased osmolarity (30 mM NaCl). Slow growth under conditions of reduced temperature and increased osmolarity has been reported in mutants lacking actin cross-linking proteins, a 34 kDa actin-bundling protein and α-actinin, respectively (Rivero et al., 1999b). These and other reports implicate the actin cytoskeleton in adaptation to conditions of altered tonicity. Dictyostelium cells respond to hyperosmotic stress by shrinking spontaneously, followed by rearrangement of cytoskeletal proteins. The two main components of the cytoskeleton, actin and myosin II, are phosphorylated as a consequence of osmotic shock (Zischka et al., 1999). Disassembly of myosin II filaments is an essential part of the hyperosmotic stress reponse in Dictyostelium, which allows the cell to adopt a spherical shape and provides the mechanical strength necessary to resist extensive shrinkage (Kuwayama et al., 1996). The Dictyostelium LIM protein, DdLim, decreases in amount in the cytoskeletal fraction isolated from osmotically shocked cells (Zischka et al., 1999). In addition, increased sensitivity to osmotic shock has been observed in Dictyostelium cells that lack actin cross-linking proteins α-actinin and gelation factor (Rivero et al., 1996b).
Both GFP–LimC and GFP–LimD preferentially accumulate in the pseudopods of the cells, where they remain until the pseudopod retracts. DdLim has also been observed to accumulate at the extreme membrane rims of newly formed protrusions in aggregation-competent cells and is involved in cell motility, and Lim2– cells have a motility as well as a cell polarity defect (Prassler et al., 1998; Chien et al., 2000). Since they do not directly interact with actin, it was suggested that LIM2 controls the actin cytoskeleton by an as yet unknown mechanism, whereas Dlim is supposed to act in a receptor-mediated Rac-1 signaling pathway, since it interacts with Rac-1. LimC and LimD, due to their F-actin binding activity, might act differently at the molecular level, although Lim2 and LimD mutants have similar characteristics.
Loss of LimD causes a dramatic polarization defect. In chemotaxing cells, this is manifested by the occurrence of multiple pseudopods that are extended in addition to the leading pseudopod that has formed in the direction of the cAMP source. Overall, lateral pseudopod formation did not strongly affect orientation and directionality during chemotaxis; the reduced speed, however, could result from this. Normally, pseudopod formation is restricted to the leading edge. This appears to be ensured by the cortical actin cytoskeleton, which upon chemotactic stimulation allows locally and in restricted areas a decrease in rigidity with subsequent pseudopod formation. Myosin II and unconventional myosins have been implicated in this process, based on the observation that mutations in the corresponding genes led to a loss of polarity and increased lateral pseudopod formation (Wessels et al., 1988, 1991; Titus et al., 1993; Zhang et al., 2002). Through their direct interaction with F-actin, LimC and LimD might play a similar role.
Our studies reveal that LimC and LimD share similar and overlapping functions, as is apparent in the LimC–/LimD– double mutant cells, which exhibit more delay in aggregation, increased sensitivity to high temperature and large average cell size in comparison with LimC– and LimD– single mutants. The LimD– cells show a specific defect in salt sensitivity, cell polarity and F-actin distribution, thus suggesting a unique role for LimD, whereas LimC– cells are least impaired in the cellular functions we have analyzed. Taken together, we have described Lim proteins that couple the cortical actin cytoskeleton to intracellular signaling pathways by directly interacting with F-actin, and thereby modulating the chemotactic response during early development and contributing towards the maintenance of the strength of the actin cytoskeleton.
Materials and methods
Growth and development of Dictyostelium
Growth and development of D.discoideum wild-type AX2 and the derived mutant strains were analyzed as described previously (Rivero et al., 1996a, 1999a). Development under submerged conditions on plastic surfaces in a monolayer was carried out at 21°C in starvation buffer (10 mM MES pH 6.5, 10 mM NaCl, 10 mM KCl, 1 mM CaCl2, 1 mM MgSO4).
Cloning of limC and limD cDNAs
Sequence information for two novel LIM domain-containing proteins of Dictyostelium was obtained from the Dictyostelium cDNA sequencing project, University of Tsukuba, Japan (accession numbers SSC 504 and FC-AE02). clone FC-AE02 (referred to as limD) was a full-length clone; a full-length limC cDNA was isolated from a λgt11 cDNA library derived from growth phase cells. N- and C-terminal deletion constructs of limC, NLIM (amino acids 1–58), NLIM-P (amino acids 1–111), CLIM (amino acids 110–182) and CLIM-P (amino acids 57–182), were generated by PCR. All the amplified PCR products were confirmed by sequencing.
Protein expression and purification and actin sedimentation assay
LimC, LimD and deletion constructs of LimC corresponding to NLIM, NLIMP-P, CLIM and CLIM-P were expressed as GST fusion proteins using a pGEX-2T expression vector (Amersham Pharmacia Biotech, Freiburg, Germany). The GST fusion proteins were purified as described by Vithalani et al. (1998). Binding of GST fusion proteins to F-actin was carried out as described by Jung et al. (1996). Dictyostelium actin was purified according to Haugwitz et al. (1991).
Expression of GFP fusion proteins
The full-length limC and limD cDNAs as well as deletion constructs of limC were fused to the C-terminus of the gene encoding the red-shifted S65T mutant of GFP cloned into the expression vector pDEXRH (Westphal et al., 1997). Transformation and selection of transformants were carried out as described previously (Witke et al., 1987).
Gene replacement mutants of Dictyostelium
For disruption of limC and limD genes in strain AX2, gene replacement vectors were constructed. limC- and limD-containing genomic DNA fragments were isolated from AX2 (DDBJ/EMBL/GenBank accession Nos: limC, AF348466; limD, AF348467). For construction of a limC gene replacement vector, the 1.4 kb blasticidin resistance cassette (Adachi et al., 1994) was inserted at the HincII site located 145 bp downstream of the LimC translation start codon. A limD gene replacement vector was constructed by cloning the 1.4 kb Bsr cassette at the ScaI site located 315 bp downstream of the LimD translation start codon. AX2 cells were transformed with limC or limD gene replacement vector. The clones with gene replacement events were identified by Southern and northern blot analyses. To generate a double mutant strain lacking both LimC and LimD (LimC–/LimD–), another limC gene replacement vector was constructed by inserting the 2.0 kb G418 resistance cassette from pDNeoII (Witke et al., 1987) at the HincII site in the coding region of the limC gene. The resulting vector was introduced into LimD– cells. LimC–/LimD– clones were verified by Southern and northern blot hybridization.
Generation of monoclonal antibodies
For generation of monoclonal antibodies specific to LimC and LimD, BALB/c mice were immunized with GST–LimC and GST–LimD fusion proteins. PAIB3Ag81 myeloma cells were used for fusion. Monoclonal antibody K4-353-6, specific for LimD, detected the endogenous protein in immunoblots of whole-cell homogenates; K5-253-3, specific for LimC, detected only the GFP fusion protein. Detection of immunolabeled bands was by chemiluminescence using horseradish peroxidase-coupled anti-mouse IgG. Both monoclonal antibodies were used in immunofluorescence studies.
Mutant analyses
The cell size of exponentially growing AX2 and derived mutant strains was determined after treating the cells with 20 mM EDTA for 1 h. Phagocytosis was assayed by challenging the Dictyostelium cells with TRITC-labeled, heat-killed yeast cells (Maniak et al., 1995). Resistance of cells against osmotic shock was determined according to Rivero et al. (1996b). The F-actin content of cells after stimulation with cAMP was determined according to Haugwitz et al. (1994).
Chemotaxis
Vegetative cells were resuspended at 1 × 107 cells/ml in Soerensen phosphate buffer and starved for 6 h. Cells were then diluted to 1–3 × 105 cells/ml and transferred onto a 5 cm glass coverslip with a plastic ring placed on an Olympus IX70 inverse microscope equipped with a 10× or 20× UplanFl 0.3 objective. Cells were stimulated with a glass capillary micropipette (Eppendorf Femtotip) filled with 1 × 10–3 M cAMP (Gerisch and Keller, 1981). Time-lapse image series were captured and stored on a computer hard drive at 30 s intervals with a JAI CV-M10 CCD camera and an Imagenation PX610 frame grabber (Imagenation Corp., Beaverton, OR) controlled through Optimas software (Optimas Corp., Bothell, WA). The instantaneous velocity and chemotaxis parameters were analyzed by the Dynamic Image Analysis System (DIAS). Lateral pseudopods were counted manually from the images taken by a 20× objective (Wessels et al., 1998; Zhang et al., 2002).
Fluorescence microscopy
The distribution of GFP fusion proteins during uptake of TRITC–dextran or TRITC-labeled yeast cells was followed as described previously (Maniak et al., 1995; Hacker et al., 1997). The distribution of GFP fusion proteins during motility was assayed in aggregation-competent cells that had been starved for 6 h in suspension. For immunolabeling studies, cells were fixed either in cold methanol (–20°C) or at room temperature with picric acid/paraformaldehyde followed by 70% ethanol. The distribution of actin was investigated by immunolabeling cells with anti-actin monoclonal antibody (Simpson et al., 1984), followed by incubation with Cy3-conjugated anti-mouse IgG (Sigma) or with TRITC–phalloidin (Sigma). Nuclei were stained with DAPI (Sigma). Confocal microscopy was carried out as described previously (Rivero et al., 1999a).
Acknowledgments
Acknowledgements
We gratefully acknowledge the Tsukuba cDNA sequencing project, University of Tsukuba, Japan, for providing sequence information, Drs M.Schleicher, F.Rivero and A.Hofmann for critical discussion and helpful suggestions, Dr E.Korenbaum for rabbit actin, B.Gassen and M.Stumpf for excellent technical assistance, and R.Müller for providing Dictyostelium actin and for help with experiments. We are especially grateful to R.Blau-Wasser for immunofluorescence data and for help in finalizing the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft and Köln Fortune to A.A.N. B.K. was a recipient of a fellowship from the Graduate College, Institute of Genetics, Cologne.
References
- Adachi H., Hasebe,T., Yoshinaga,K., Ohta,T. and Sutoh,K. (1994) Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun., 205, 1808–1814. [DOI] [PubMed] [Google Scholar]
- Arber S., Hunter,J.J., Ross,J.,Jr, Hongo,M., Sansig,G., Borg,J., Perriard,J.C., Chien,K.R. and Caroni,P. (1997) MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy and heart failure. Cell, 88, 393–403. [DOI] [PubMed] [Google Scholar]
- Beckerle M.C. (1997) Zyxin: zinc fingers at sites of cell adhesion. BioEssays, 19, 949–957. [DOI] [PubMed] [Google Scholar]
- Chien S., Chung,C.Y., Sukumaran,S., Osborne,N., Lee,S., Ellsworth,C., McNally,J.G. and Firtel,R.A. (2000) The Dictyostelium LIM domain-containing protein LIM2 is essential for proper chemotaxis and morphogenesis. Mol. Biol. Cell, 11, 1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I.B., Breen,J.J. and Toyama,R. (1998) LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet., 14, 156–162. [DOI] [PubMed] [Google Scholar]
- Flick M.J. and Konieczny,S.F. (2000) The muscle regulatory and structural protein MLP is a cytoskeletal binding partner of βI-spectrin. J. Cell Sci., 113, 1553–1564. [DOI] [PubMed] [Google Scholar]
- Freyd G., Kim,S.K. and Horvitz,H.R. (1990) Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature, 344, 876–879. [DOI] [PubMed] [Google Scholar]
- Gerisch G. and Keller,H.U. (1981) Chemotactic reorientation of granulocytes stimulated with micropipettes containing fMet-Leu-Phe. J. Cell Sci., 52, 1–10. [DOI] [PubMed] [Google Scholar]
- Hacker U., Albrecht,R. and Maniak,M. (1997) Fluid-phase uptake by macropinocytosis in Dictyostelium. J. Cell Sci., 110, 105–112. [DOI] [PubMed] [Google Scholar]
- Haugwitz M., Noegel,A.A., Rieger,D., Lottspeich,F. and Schleicher,M. (1991) Dictyostelium discoideum contains two profilin isoforms that differ in structure and function. J. Cell Sci., 100, 481–489. [DOI] [PubMed] [Google Scholar]
- Haugwitz M., Noegel,A.A., Karakesisoglou,J. and Schleicher,M. (1994) Dictyostelium amoebae that lack G-actin sequestering profilins show defects in F-actin content, cytokinesis and development. Cell, 79, 303–314. [DOI] [PubMed] [Google Scholar]
- Jung E., Fucini,P., Stewart,M., Noegel,A.A. and Schleicher,M. (1996) Linking microfilaments to intracellular membranes: the actin-binding and vesicle-associated protein comitin exhibits a mannose-specific lectin activity. EMBO J., 15, 1238–1246. [PMC free article] [PubMed] [Google Scholar]
- Khurana T., Khurana,B. and Noegel,A.A. (2002) LIM proteins: association with the actin cytoskeleton. Protoplasma, 219, 1–12. [DOI] [PubMed] [Google Scholar]
- Kuwayama H., Ecke,M., Gerisch,G. and Van Haastert,P.J. (1996) Protection against osmotic stress by cGMP-mediated myosin phosphorylation. Science, 271, 207–209. [DOI] [PubMed] [Google Scholar]
- Maniak M., Rauchenberger,R., Albrecht,R., Murphy,J. and Gerisch,G. (1995) Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein tag. Cell, 83, 915–924. [DOI] [PubMed] [Google Scholar]
- Neujahr R., Heizer,C. and Gerisch,G. (1997) Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J. Cell Sci., 110, 123–137. [DOI] [PubMed] [Google Scholar]
- Perez-Alvarado G.C., Miles,C., Michelsen,J.W., Louis,H.A., Winge,D.R., Beckerle,M.C. and Summers,M.F. (1994) Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat. Struct. Biol., 1, 388–398. [DOI] [PubMed] [Google Scholar]
- Pomies P., Louis,H.A. and Beckerle,M.C. (1997) CRP1, a LIM domain protein implicated in muscle differentiation, interacts with α-actinin. J. Cell Biol., 139, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prassler J., Murr,A., Stocker,S., Faix J., Murphy,J. and Marriott,G. (1998) DdLim is a cytoskeleton-associated protein involved in the protrusion of lamellipodia in Dictyostelium. Mol. Biol. Cell, 9, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F., Furukawa,R., Noegel,A.A. and Fechheimer,M. (1996a) Dictyostelium discoideum cells lacking the 34,000-dalton actin-binding protein can grow, locomote, and develop, but exhibit defects in regulation of cell structure and movement: a case of partial redundancy. J. Cell Biol., 135, 965–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F., Köppel,B., Peracino,B., Bozzaro,S., Siegert,F., Weijer,C.J., Schleicher,M., Albrecht,R. and Noegel,A.A. (1996b) The role of the cortical cytoskeleton: F-actin crosslinking proteins protect against osmotic stress, ensure cell size, cell shape and motility, and contribute to phagocytosis and development. J. Cell Sci., 109, 2679–2691. [DOI] [PubMed] [Google Scholar]
- Rivero F., Albrecht,R., Dislich,H., Bracco,E., Graciotti,L., Bozzaro,S. and Noegel,A.A. (1999a) RacF1, a novel member of the Rho protein family in Dictyostelium discoideum, associates transiently with cell contact areas, macropinosomes, and phagosomes. Mol. Biol. Cell, 10, 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F., Furukawa,R., Fechheimer,M. and Noegel,A.A. (1999b) Three actin cross-linking proteins, the 34 kDa actin-bundling protein, α-actinin and gelation factor (ABP-120), have both unique and redundant roles in the growth and development of Dictyostelium. J. Cell Sci., 112, 2737–2751. [DOI] [PubMed] [Google Scholar]
- Schmeichel K.L. and Beckerle,M.C. (1994) The LIM domain is a modular protein-binding interface. Cell, 79, 211–219. [DOI] [PubMed] [Google Scholar]
- Simpson P.A., Spudich,J.A. and Parham,P. (1984) Monoclonal antibodies prepared against Dictyostelium actin: characterization and interactions with actin. J. Cell Biol., 99, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus M.A., Wessels,D., Spudich,J.A. and Soll,D. (1993) The unconventional myosin encoded by the myoA gene plays a role in Dictyostelium motility. Mol. Biol. Cell, 4, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Li,F., Goicoechea,S. and Wu,C. (1999) The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol. Cell. Biol., 19, 2425–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithalani K.K., Parent,C.A., Thorn,E.M., Penn,M., Larochelle,D.A., Devreotes,P.N. and De Lozanne,A. (1998) Identification of darlin, a Dictyostelium protein with Armadillo-like repeats that binds to small GTPases and is important for the proper aggregation of developing cells. Mol. Biol. Cell, 9, 3095–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels D., Soll,D.R., Knecht,D., Loomis,W.F., De Lozanne,A. and Spudich,J. (1988) Cell motility and chemotaxis in Dictyostelium amoebae lacking myosin heavy chain. Dev. Biol., 128, 164–177. [DOI] [PubMed] [Google Scholar]
- Wessels D., Murray,J., Jung,G., Hammer,J.A.,III and Soll,D.R. (1991) Myosin IB null mutants of Dictyostelium exhibit abnormalities in motility. Cell Motil. Cytoskeleton, 20, 301–315. [DOI] [PubMed] [Google Scholar]
- Wessels D., Voss,E., Von Bergen,N., Burns,R., Stites,J. and Soll,D.R. (1998) A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil. Cytoskeleton, 41, 225–246. [DOI] [PubMed] [Google Scholar]
- Westphal M., Jungbluth,A., Heidecker,M., Mühlbauer,B., Heizer,C., Schwartz,J.M., Marriott,G. and Gerisch,G. (1997) Microfilament dynamics during cell movement and chemotaxis monitored using a GFP–actin fusion protein. Curr. Biol., 7, 176–183. [DOI] [PubMed] [Google Scholar]
- Witke W., Nellen,W. and Noegel,A. (1987) Homologous recombination in the Dictyostelium α-actinin gene leads to an altered mRNA and lack of the protein. EMBO J., 6, 4143–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wessels,D., Fey,P., Daniels,K., Chisholm,R.L. and Soll,D.R. (2002) Phosphorylation of the myosin regulatory light chain plays a role in motility and polarity during Dictyostelium chemotaxis. J. Cell Sci., 115, 1733–1747. [DOI] [PubMed] [Google Scholar]
- Zischka H., Oehme,F., Pintsch,T., Ott,A., Keller,H., Kellermann,J. and Schuster,S.C. (1999) Rearrangement of cortex proteins constitutes an osmoprotective mechanism in Dictyostelium. EMBO J., 18, 4241–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]