Abstract
The growth factor-activated AGC protein kinases RSK, S6K, PKB, MSK and SGK are activated by serine/threonine phosphorylation in the activation loop and in the hydrophobic motif, C-terminal to the kinase domain. In some of these kinases, phosphorylation of the hydrophobic motif creates a specific docking site that recruits and activates PDK1, which then phosphorylates the activation loop. Here, we discover a pocket in the kinase domain of PDK1 that recognizes the phosphoserine/phosphothreonine in the hydrophobic motif by identifying two oppositely positioned arginine and lysine residues that bind the phosphate. Moreover, we demonstrate that RSK2, S6K1, PKBα, MSK1 and SGK1 contain a similar phosphate-binding pocket, which they use for intramolecular interaction with their own phosphorylated hydrophobic motif. Molecular modelling and experimental data provide evidence for a common activation mechanism in which the phosphorylated hydrophobic motif and activation loop act on the αC-helix of the kinase structure to induce synergistic stimulation of catalytic activity. Sequence conservation suggests that this mechanism is a key feature in activation of >40 human AGC kinases.
Keywords: AGC kinase/docking site/PDK1/PKB/RSK
Introduction
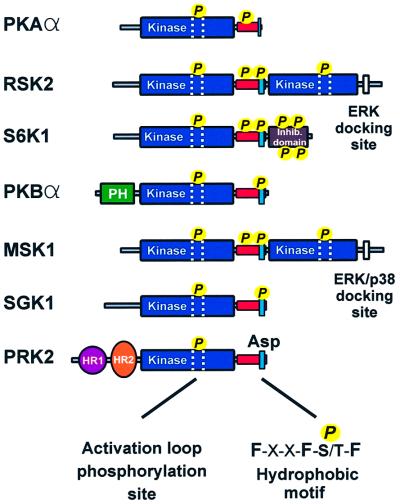
A significant part of growth factor signal transduction is mediated by a structurally related group of protein kinases belonging to the AGC kinase family. The group includes p90 ribosomal S6 kinase (RSK), p70 ribosomal S6 kinase (S6K), protein kinase B (PKB), mitogen- and stress-activated protein kinase (MSK), serum- and glucocorticoid-inducible kinase (SGK) and protein kinase C-related kinase (PRK). Collectively, these kinases phosphorylate a large array of cellular proteins and thereby regulate cellular division, survival, metabolism, transmembrane ion flux, migrative behaviour and differentiation. The kinases are activated by partly distinct signalling pathways. PKB, S6K and SGK are downstream mediators of phosphoinositide 3-kinase (PI3-K; Ming et al., 1994; Franke et al., 1995; Kobayashi and Cohen, 1999; Park et al., 1999), RSK and MSK are effectors of ERK and ERK/p38 mitogen-activated protein (MAP) kinases, respectively (Sturgill et al., 1988; Deak et al., 1998), and PRK2 is subject to control by Rho GTPase (Flynn et al., 2000). The differential responsiveness to upstream pathways is due, in part, to the fact that the kinases contain different signalling modules flanking the kinase domain, e.g. a PH domain in PKB, a Rho-binding domain in PRK, a unique inhibitory domain in S6K and a MAP kinase-activated kinase domain in RSK and MSK.
Despite the divergent regulation, these kinases have two regulatory features in common that are critical for activation (Figure 1). First, they all require phosphorylation of a serine or threonine residue in the activation loop within the kinase domain. The site is phosphorylated by 3-phosphoinositide-dependent kinase-1 (PDK1) in PKB (Alessi et al., 1997; Stokoe et al., 1997), S6K (Alessi et al., 1998; Pullen et al., 1998), RSK (Jensen et al., 1999; Richards et al., 1999), SGK (Kobayashi and Cohen, 1999; Park et al., 1999) and PRK (Flynn et al., 2000), whereas MSK may autophosphorylate at this site (Williams et al., 2000). Phosphorylation of the activation loop augments catalytic activity from absent to typically 10% of maximal activity. Secondly, this group of kinases all require phosphorylation of a serine or threonine residue in a so-called hydrophobic motif located in a conserved tail region C-terminally to the kinase domain. The motif is characterized by three aromatic amino acids surrounding the serine/threonine residue that becomes phosphorylated: Phe-X-X-Phe-Ser/Thr-Phe/Tyr. However, in PRK and atypical PKCs, which also belong to this kinase family, the hydrophobic motif contains a negatively charged amino acid (aspartic acid or glutamic acid) that mimicks the phosphoserine/threonine (Parekh et al., 2000). In RSK (Vik and Ryder, 1997) and MSK (Deak et al., 1998), the hydrophobic motif is phosphorylated by their respective C-terminal kinase domains. For PKB, S6K and SGK, the identity of the hydrophobic motif kinase has not yet been firmly established.
Fig. 1. Structural alignment of PKA and major growth factor-activated AGC kinases. The alignment illustrates that the growth factor-activated AGC kinases share two regulatory features: phosphorylation of the activation loop (stippled area) and phosphorylation of a hydrophobic motif (blue box), located in a tail region (red box) C-terminal to the kinase domain. Note that PRK2 contains a phosphate-mimicking aspartic acid residue and that PKA lacks a phosphorylation site in the hydrophobic motif.
Recent studies have shown that the hydrophobic motif can function as a docking site for PDK1 in PRK2 and PKCζ (Balendran et al., 2000), RSK2 (Frödin et al., 2000), S6K1 and SGK1 (Biondi et al., 2001). Furthermore, interaction of PDK1 with the hydrophobic motif of RSK2 (Frödin et al., 2000) or PRK2 (Biondi et al., 2000) increases the catalytic activity of PDK1 several fold, indicating that the motif functions to both recruit and activate PDK1. The binding site in PDK1 for the hydrophobic motif was identified through analysis of the PKA crystal structure (Biondi et al., 2000). PKA is also an AGC kinase, but it terminates in a partial hydrophobic motif (Phe-X-X-Phe-COOH) with no regulatory phosphorylation site. In the PKA crystal structure, the two phenylalanine residues in this motif interact with a hydrophobic pocket in the small lobe of the kinase domain (Knighton et al., 1991), and mutation of the phenylalanine residues nearly abolishes kinase activity (Etchebehere et al., 1997). Modelling of PDK1 based on the PKA crystal structure revealed that this pocket is conserved in PDK1 and constitutes the binding site for the hydrophobic motif of the PDK1 target kinases (Biondi et al., 2000, 2001).
An important feature of the interaction between PDK1 and the hydrophobic motif is that the motif must be phosphorylated (or contain a phosphate-mimicking acidic residue) for efficient interaction to occur (Balendran et al., 2000; Frödin et al., 2000; Biondi et al., 2001). Whether the hydrophobic pocket of PDK1 is endowed with the ability to recognize phosphoserine/phosphothreonine or whether the phosphate alters the conformation of the hydrophobic motif so that it can bind the hydrophobic pocket is not known. Moreover, the docking mechanism does not explain why, for example, PKB and MSK require phosphorylation in their hydrophobic motif, since PKB does not appear to use the motif for PDK1 docking (Biondi et al., 2001) and MSK is not a target of PDK1 (Williams et al., 2000).
In this study, we identify a phosphate-binding site next to the hydrophobic pocket of PDK1 that recognizes the phosphoserine/phosphothreonine in the hydrophobic motif. We extend these findings by showing that RSK2, S6K1, PKBα, MSK1 and SGK1 contain a similar phosphate-binding pocket in their kinase domain. They use the phosphate-binding pocket to interact with their own phosphorylated hydrophobic motif, resulting in large stimulation of kinase activity in synergy with activation loop phosphorylation. These findings define a critical function of the hydrophobic motif phosphorylation site and suggest that the phosphate-binding pocket is a key regulatory feature of the >40 human AGC kinases in which it is conserved. The phosphate-binding pocket may provide an attractive target for drugs aimed at activating or inhibiting these AGC kinases as an alternative to the commonly targeted ATP-binding site.
Results
Phosphoserine/phosphothreonine recognition by the hydrophobic pocket of PDK1
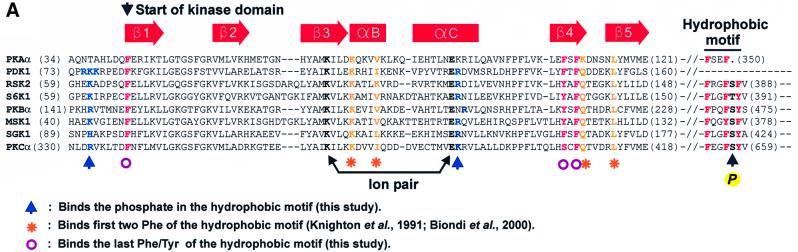
Previous studies provided a partial characterization of the hydrophobic pocket of PDK1 by showing that residues Lys115, Ile119, Gln150 and Leu155 in the small lobe of the kinase domain form a hydrophobic pocket that probably binds the first two phenylalanines of the hydrophobic motif of PDK1 target kinases (Biondi et al., 2000, 2001). Here, we wished to identify amino acids near the pocket that could account for recognition of phosphoserine/threonine in the hydrophobic motif. The program Swiss-Pdb Viewer (Guex and Peitsch, 1997) was used to model the sequence of PDK1 over the crystal structure coordinates of PKA (Engh et al., 1996), the only AGC kinase whose structure had been solved. The model of PDK1 visualized that the above-mentioned pocket residues cluster in one end of a larger groove (data not shown). Further into the groove and topographically close to this cluster, we noticed an arginine residue (131) located in the so-called αC-helix of the conserved kinase fold. Moreover, we noticed four consecutive basic residues, 75Arg-Lys-Lys-Arg78, N-terminal to β-strand 1. Although the basic cluster is outside the conserved kinase fold, and its position therefore less predictable, the start of β-strand 1 is topographically close to the hydrophobic pocket. We decided to investigate whether these positively charged amino acids could be involved in phosphate binding by performing more advanced molecular modelling and biochemical characterization.
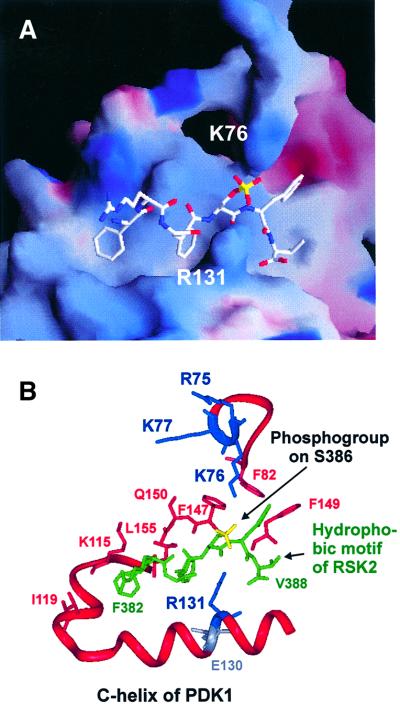
Residues 82–351 of PDK1, encoding the kinase domain, were used to model PDK1 based on the coordinates of a PKA structure in the closed, active conformation with a phosphorylated activation loop (Engh et al., 1996). The sequence of the hydrophobic motif of RSK2 phosphorylated at the regulatory site (Ser386) was then modelled into the pocket by first fixing the first two phenylalanines of the motif in the position held by the corresponding phenylalanines in the PKA crystal structure. The remaining residues of the motif were then modelled without template (ab initio modelling) into the pocket. Finally, residues 75–81 of PDK1 (encompassing the basic cluster) were modelled ab initio onto the PDK1 kinase domain. The resulting model of the docking interaction showed juxtaposition of the phosphogroup of Ser386 and Arg131 of PDK1 with the possibility of hydrogen bond formation between the two (Figure 2). Intriguingly, Arg131 is next to a glutamic acid residue (130 in PDK1) that is one of very few residues conserved in all kinases and that has a regulatory role. Modelling also suggested binding of the phosphogroup of Ser386 to Arg75, Lys76 and Lys77 of the basic cluster in PDK1, but suggested that only one of these residues may bind the phosphate at a time. Moreover, the side chains of Phe82, Phe147 and Phe149 of PDK1 appear to form a hydrophobic site that accommodates the last aromatic residue of the hydrophobic motif. Finally, the valine after the motif may form hydrophobic interactions with Leu145 of PDK1. In conclusion, the model suggests that the hydrophobic motif completely occupies the groove in the small lobe of the PDK1 kinase domain and that the phosphate in the motif is grabbed by two oppositely positioned basic residues.
Fig. 2. Model of the docking interaction between PDK1 and the phosphorylated hydrophobic motif of RSK2. (A) Electrostatic surface potential model of the hydrophobic pocket of PDK1 with positive potential in blue and negative potential in red. The RSK2 hydrophobic motif peptide (FRGFpSFV) is shown in white with the phosphogroup on Ser386 in yellow. (B) Ribbon representation of the pocket with side chains of the residues discussed in the text. PDK1 residues are red, blue or grey, whereas the RSK2 hydrophobic motif peptide is shown in green with the phosphogroup in yellow.
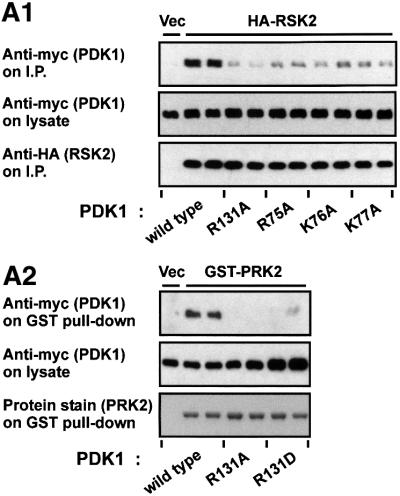
We next assessed the effect of mutating the predicted phosphate-binding residues in PDK1 on its ability to bind RSK2 in vivo using co-immunoprecipitation experiments. Prior to lysis, the cells were exposed to epidermal growth factor (EGF) in order to induce phosphorylation of the RSK2 hydrophobic motif and recruitment of PDK1 to RSK2 (Frödin et al., 2000). RSK2 co-precipitated wild-type PDK1, but much lower amounts of PDK1-R131A mutant (Figure 3A1, upper panel) or R131D, R131L and R131H mutants (data not shown). Furthermore, alanine mutation of either Arg75, Lys76 or Lys77 in PDK1 all resulted in much reduced precipitation of PDK1 by RSK2. PRK2, that contains aspartic acid in the position of phosphoserine/threonine in the hydrophobic motif, co-precipitated wild-type PDK1, but not PDK1-R131A or PDK1-R131D (Figure 3A2, upper panel), or R131L and R131H (data not shown). All PDK1 constructs expressed protein at a similar level (Figure 3A, middle panels).
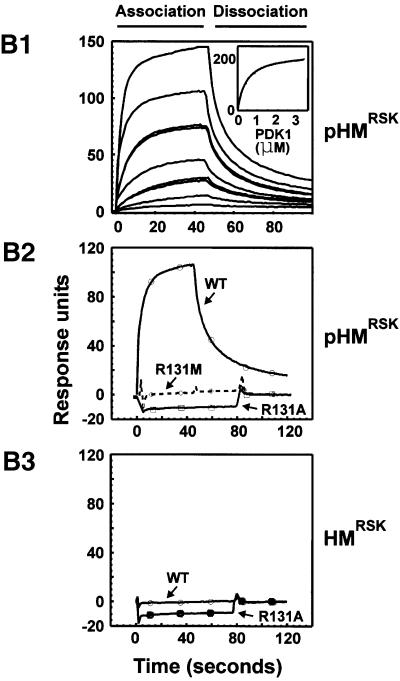
Fig. 3. Identification of arginine/lysine residues in PDK1 required for interaction with the phosphorylated hydrophobic motif. (A) COS7 cells were co-transfected with plasmids expressing wild-type or mutant myc-PDK1 together with HA-RSK2, GST–PRK2 or empty vector (Vec). After 48 h and a final 3 h serum starvation period, the cells were lysed subsequent to 35 min EGF treatment of RSK-expressing cells. HA-RSK2 and GST–PRK2 were precipitated from the cell lysates using anti-HA antibody or glutathione beads, respectively. The precipitates were subjected to SDS–PAGE and immunoblotting with anti-myc antibody to detect co-precipitated myc-PDK1 (upper panels) or to anti-HA immunostaining or protein staining to assess the amounts of HA-RSK2 and GST–PRK2 (lower panels). Pre-precipitation lysates were subjected to immunoblotting for the myc tag (middle panels). (B) Interaction of PDK1 with the hydrophobic motif of RSK2 was analysed using surface plasmon resonance measurements in a BiaCore3000 system. Biotinylated peptides of the motif phosphorylated (pHMRSK) or non-phosphorylated (HMRSK) at Ser386 were used to coat Sensor Chips SA (10 response units). (1) GST–PDK1 was injected at different concentrations (0.013–3.33 µM) onto pHM-coated chips. In the inset, the steady-state binding is plotted against the various concentrations of PDK1. Kinetic constants were obtained by fitting the data to a hyperbola using Kaleidagraph software. (2 and 3) GST–PDK1 wild-type, –R131A or –R131M were injected at 400 nM onto chips coated with pHMRSK and HMRSK, respectively. All experiments in (A) and (B) were performed three times with similar results.
Surface plasmon resonance measurements showed that wild-type PDK1 bound with high affinity (Kd ≅ 400 ± 20 nM) to a synthetic peptide of the RSK2 hydrophobic motif phosphorylated at Ser386 (pHMRSK), whereas PDK1-R131M or PDK1-R131A had no detectable affinity for pHMRSK (Figure 3B1 and B2). Unphosphorylated RSK2 hydrophobic motif peptide (HMRSK) showed no binding to wild-type PDK1 or PDK1-R131A (Figure 3B3).
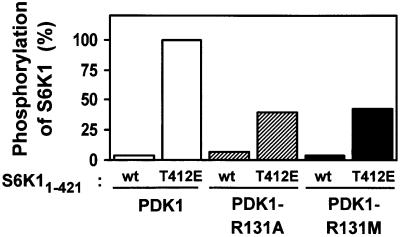
S6K11–421 becomes a much better substrate of PDK1 when Thr412, i.e. the phosphorylation site of the hydrophobic motif, is mutated to glutamic acid (Alessi et al., 1998; Dennis et al., 1998), which is due to enhanced docking of PDK1 to S6K1 via the hydrophobic pocket (Biondi et al., 2001). S6K11–421 has a deletion of the C-terminal inhibitory domain, which blocks PDK1 interaction with S6K1. We used this assay to evaluate the importance of Arg131 in PDK1 for the ability of PDK1 to phosphorylate S6K1. As shown in Figure 4, PDK1, PDK1-R131A and PDK1-R131M all phosphorylated S6K11–421 poorly. PDK1 phosphorylated S6K11–421T412E with high efficiency. In contrast, PDK1-R131A and PDK1-R131M showed reduced ability to phosphorylate S6K11–421T412E, suggesting that these mutants are impaired in recognizing the negative charge of E412. The reduced ability of PDK1-R131A or -R131M to phosphorylate S6K11–421T412E was not due to impairment of PDK1 catalytic activity by the mutations. In fact, these and other mutations in the hydrophobic pocket of PDK1 increased the kinase activity towards the PDK1 peptide substrate T308tide 2- to 3-fold (data not shown). This suggests that the unoccupied pocket can suppress PDK1 activity, as previously proposed (Biondi et al., 2000).
Fig. 4. R131 is important for the ability of PDK1 to phosphorylate S6K1. Wild-type or T412E His-S6K11–421 were incubated for 10 min with wild-type or mutant GST–PDK1 and Mg[γ-32P]ATP. Thereafter, the kinase reactions were subjected to SDS–PAGE. Radioactivity incorporated into the S6K1 protein band was quantitated and expressed as a percentage of the maximal value obtained. The results are representative of two independent experiments.
In contrast to alanine, aspartic acid, leucine, histidine or methionine (data above), the basic amino acid lysine could partially substitute for arginine at position 131 in PDK1. Against T308tide, PDK1-R131K showed basal activity similar to that of PDK1, but the dose–response curve for activation by pHMS6K and HMPRK2 was shifted to ∼10- and 100-fold higher concentrations, respectively (data not shown).
In conclusion, our data strongly suggest that PDK1 utilizes Arg131 in the C-helix and the N-terminal basic residues Arg75, Lys76 and Lys77 in order to recognize phosphoserine/phosphothreonine (or mimicking acidic residues) in the hydrophobic motif of its target kinases.
A phosphate-binding pocket is conserved in the growth factor-activated AGC kinases
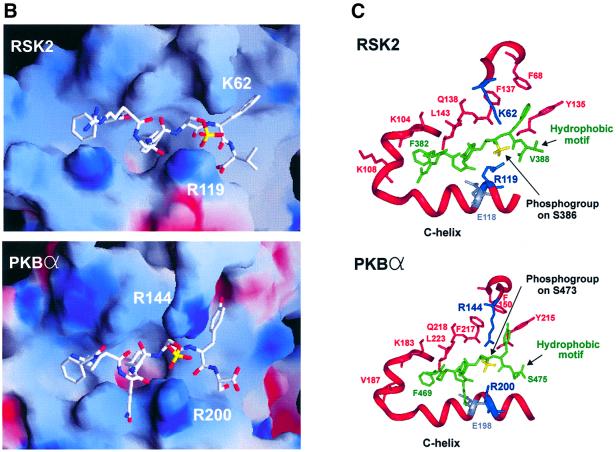
PDK1 itself belongs to the AGC kinase family, but is atypical since it does not contain a hydrophobic motif. Interestingly, sequence alignment suggests that a phosphate-binding pocket is also present in RSK2, S6K1, PKBα, MSK1, SGK1 and PKCα (Figure 5A). First, an arginine or lysine residue equivalent to Arg131 in the C-helix of PDK1 is conserved. Secondly, a basic residue equivalent to Lys76 of PDK1 is conserved. Finally, the residues for binding the three aromatic residues of the hydrophobic motif are conserved. This raised the possibility that the phosphorylated hydrophobic motif of these kinases may interact with a phosphate-binding pocket within the kinase domain, which could serve to stimulate their catalytic activity by an intramolecular mechanism. To address this question, we first modelled the sequence encompassing the kinase domains of RSK2 (the N-terminal kinase domain) or PKBα over the structure of PKA and then modelled their respective phosphorylated hydrophobic motif into the presumed pocket. As shown in Figure 5B and C, the hydrophobic motif fitted perfectly into the hydrophobic pocket of RSK2 and PKBα, respectively. Moreover, in the two kinases, the phosphogroup of the hydrophobic motif was juxtaposed to the conserved arginine in the C-helix, with the possibility of hydrogen bond formation between the two. Both models also allowed binding of the phosphate to the positively charged residue N-terminal to the kinase domain, i.e. K62 in RSK2 and R144 in PKBα. Very similar results were obtained through modelling of S6K1, MSK1 and SGK1 (data not shown).
Fig. 5. A phosphate-binding pocket is conserved in major growth factor-activated AGC kinases. (A) Amino acid sequence alignment of selected AGC kinases in the region of the kinase domain that contains the hydrophobic pocket as well as the region forming the hydrophobic motif. Conserved residues predicted to interact with the phosphogroup, the first two aromatic residues and the last aromatic residue of the hydrophobic motif are indicated by an arrow, asterisk and circle, respectively. The ion pair formation between the lysine and glutamic acid residues conserved in all kinases is indicated. (B and C) Model of the intramolecular interaction of the hydrophobic pocket of RSK2 and PKBα with their respective phosphorylated hydrophobic motifs, FRGFpSFV (RSK2) and FPQFpSYS (PKBα). (B) Electrostatic surface potential models of the pockets, with positive potential in blue and negative potential in red. The hydrophobic motif peptides are shown in white with the phosphogroup in yellow. (C) Ribbon representation of the pockets with side chains of the residues discussed in the text. The hydrophobic motif peptides are shown in green with the phosphogroup in yellow.
Evidence for intramolecular activation of AGC kinases by the hydrophobic motif via the phosphate-binding pocket
To test for an intramolecular function of the hydrophobic motif, a reconstitution assay was designed that could assess the contributions of hydrophobic motif phosphorylation, activation loop phosphorylation and their interaction in catalytic stimulation of RSK2, S6K1, MSK1, PKBα and SGK1. The assay was based on isolated kinase domains with deletion of the hydrophobic motif and other C-terminal domains, i.e. the C-terminal kinase domain of RSK2 and MSK1 and the inhibitory domain in S6K1. In this assay, hydrophobic motif phosphorylation was conferred by the addition of synthetic peptides of the motif phosphorylated at the regulatory site (pHM). Activation loop phosphorylation was conferred by pre-exposure to PDK1, which was then removed, except in the RSK2 assay. Although not a physiological substrate, MSK1 can be phosphorylated and activated by PDK1 (Frödin et al., 2000), which was exploited here.
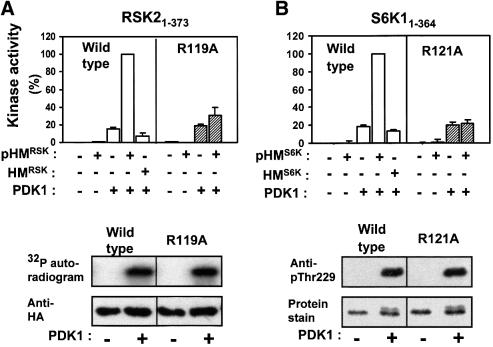
The kinase domain of RSK2 (RSK21–373) was completely inactive when incubated alone (Figure 6A). No activation was achieved by the addition of pHMRSK peptide, whereas phosphorylation of the activation loop by pre-incubation with PDK1 resulted in stimulation of kinase activity. However, the combination of pHMRSK and activation loop phosphorylation resulted in synergistic stimulation of RSK21–373 catalytic activity. Half-maximal activation was observed at 20–30 µM pHMRSK (data not shown). In contrast, the non-phosphorylated hydrophobic motif peptide, HMRSK, showed no ability to stimulate RSK21–373 up to 350 µM peptide, the highest concentration possible here. To assess the role of the predicted phosphoserine-binding Arg119 in the C-helix, similar experiments were performed with RSK21–373 in which Arg119 had been mutated to alanine. RSK21–373R119A showed normal stimulation upon activation loop phosphorylation. However, pHMRSK failed to stimulate RSK21–373R119A catalytic activity in synergy with activation loop phosphorylation. This suggests that activation of RSK21–373 by pHM requires binding of pHM to Arg119 and, moreover, that the R119A mutation does not compromise tertiary structure, since RSK21–373R119A was activated normally by PDK1.
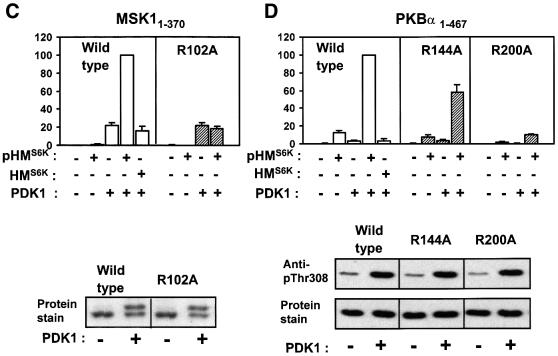
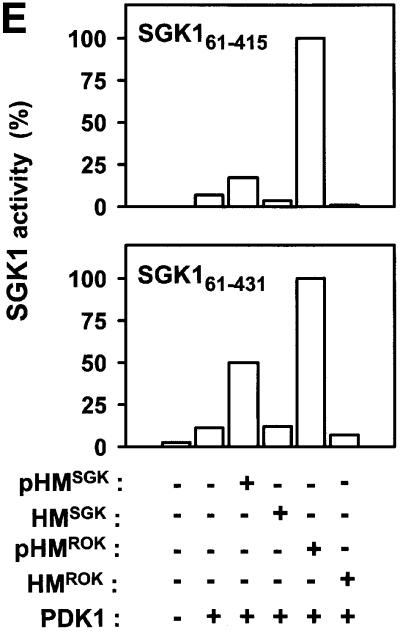
Fig. 6. Evidence for intramolecular activation of AGC kinases by the phosphorylated hydrophobic motif. (A–E) Upper panels: wild-type or point-mutated kinase domains of RSK2, S6K1, MSK1, PKBα and SGK1, all lacking their hydrophobic motif (except SGK161–431), were pre-phosphorylated or not by PDK1. Thereafter, PDK1 was removed (except in the RSK2 assay). The activity of each kinase was then determined in the absence or presence of phosphorylated (pHM) or non-phosphorylated (HM) hydrophobic motif peptide at 170 µM (A, B and E) or 360 µM (C and D) and expressed as a percentage of maximal activity. Data are means ± SD of (A) 4–10 observations from 2–4 independent experiments, (B–D) three independent experiments or (E) means of duplicates with <5% difference between duplicate samples from one experiment performed twice with similar results (A–D, lower panels). To control for an equal amount of protein and PDK1-induced phosphorylation of wild-type and mutant kinase, aliquots of the reactions were subjected to SDS–PAGE and protein staining/anti-HA blotting or to blotting with phospho-specific antibody to the PDK1 site. However, in (A), phosphorylation of HA-RSK21–373 was assessed by including [γ-32P]ATP during pre-incubation with PDK1, followed by precipitation with anti-HA antibody, SDS–PAGE and autoradiography.
Remarkably similar results were obtained in experiments performed with the isolated kinase domains of S6K1, MSK1 and PKBα (S6K11–364, MSK11–370 and PKBα1–467 in Figure 6B–D, respectively). The experiments shown with MSK1 and PKB were performed with pHM peptides derived from S6K1, since pHMMSK peptide was not available and pHMPKB peptide induced only a 2- to 3-fold activation of PKBα1–467 (data not shown). In contrast to the other kinases, pHM activated PKBα1–467, although slightly, in the absence of PDK1 co-expression, most probably because PKBα1–467 was partially phosphorylated by endogenous PDK1 (Figure 6D, lower panel). In S6K11–364 and PKBα1–467, we also mutated the N-terminal phosphate-binding residue, Lys62 and Arg144, respectively. This resulted in 50% reduced activation of S6K11–364 by pHMS6K (SD = 12%, n = 3; data not shown) and 40% reduced activation of PKBα1–467 by pHMS6K (Figure 6D). EC50 experiments suggested that these mutants had a 3- to 4-fold lower affinity towards pHM compared with the wild-type kinase (data not shown). Control experiments showed that all mutants of the phosphate-binding basic residues were expressed normally and were phosphorylated in the activation loop by PDK1 similarly to the wild-type enzymes (Figure 6A–D, lower panels). Phosphorylation of MSK11–370 was evaluated by mobility shift in prolonged electrophoresis, since no phosphospecific antibody to the activation loop site has been reported. The maximal specific activities of RSK21–373, S6K11–364, MSK11–370 and PKBα1–467 obtained by combined activation loop phosphorylation and pHM were similar or several fold higher than that of the full-length, recombinant proteins activated by EGF or insulin in vivo (data not shown). This may reflect that the in vitro experiments were performed under conditions with a higher degree of phosphorylation of the hydrophobic motif and the activation loop than obtained in vivo.
Also, SGK1 was activated by its phosphorylated, but not unphosphorylated, hydrophobic motif (Figure 6E). This was observed both with SGK161–415, in which the hydrophobic motif was deleted, and with SGK161–431, which retained its hydrophobic motif. Both SGK1 constructs lack residues 1–60, as the full-length protein is not expressed at levels sufficient for protein purification (Kobayashi and Cohen, 1999). SGK1 could be activated even further by pHM peptide from Rho kinase-α (also an AGC kinase), but not by HMROK. In contrast, pHM from RSK2, S6K1, PKBα or myotonic dystrophy kinase (DMK), another AGC kinase, or HM from PRK2 or PKCζ showed little ability to activate SGK1 (data not shown). Thus, in some instances, an AGC kinase can be activated by pHM from another AGC kinase. However, since the EC50 values for heterologous activation were always >10 µM, such activation may not be physiological.
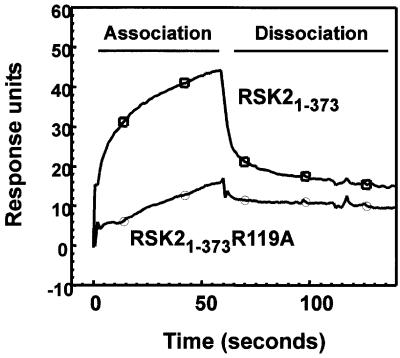
Surface plasmon resonance analysis showed that pHMRSK is able to bind RSK21–373, but not RSK21–373 R119A (Figure 7). The affinity, however, was too low for quantitative estimates of the dissociation constant in a BiaCore instrument, in agreement with the EC50 value of 20–30 µM pHMRSK in catalytic activation of RSK21–373. HMRSK showed no binding to RSK21–373 or RSK21–373 R119A (data not shown).
Fig. 7. Interaction of RSK2 with its phosphorylated hydrophobic motif. The interaction of RSK2 with its own hydrophobic motif was analysed by surface plasmon resonance measurements. Biotinylated pHMRSK peptide was used to coat Sensor Chips SA (500 response units) and tested for binding to GST–HA-RSK21–373 or GST–HA-RSK21–373 R119A. The experiments were performed several times with similar results.
In conclusion, these experiments demonstrate that the phosphorylated hydrophobic motif stimulates the catalytic activity of the growth factor-activated AGC kinases in synergy with the phosphorylation event in the activation loop. Moreover, the data suggest that binding and activation by pHM requires the conserved arginine in the C-helix and, to a lesser degree, the conserved basic residue N-terminal to the kinase domain.
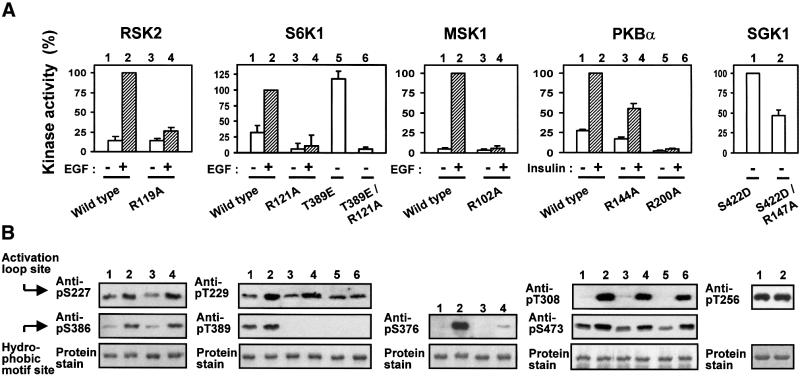
We next analysed the role of the phosphate-binding residues for in vivo activation (Figure 8A) and phosphorylation state (Figure 8B) of full-length AGC kinases in response to physiological stimuli. In RSK2, EGF-induced activation was abolished by mutation of Arg119. RSK2-R119A showed normal expression and phosphorylation of the activation loop and the hydrophobic motif, as demonstrated by immunoblotting. Mutation of the N-terminal basic residue, Lys62, reduced kinase activity by 21% (SD = 7.0%, n = 3; data not shown). In S6K1, basal and EGF-induced activity was largely abolished by mutation of Arg121 in the C-helix. S6K1-R121A showed normal expression and phosphorylation of the activation loop but, interestingly, phosphorylation of the hydrophobic motif was largely absent. The loss of phosphorylation occurred in vivo and not during immunoprecipitation, since S6K1-R121A from cells lysed with Laemmli buffer after EGF treatment showed a similar lack of phosphorylation in the hydrophobic motif (data not shown). This suggests that in S6K1, the hydrophobic motif phosphorylation site is subject to rapid dephosphorylation when not bound to the hydrophobic pocket and thereby protected from phosphatase action. To evaluate the role of Arg121 for S6K1 activity in vivo, we therefore generated a S6K1-T389E mutant, in which the phosphorylation site in the hydrophobic motif was mutated to glutamic acid. The residue Thr389 is the same hydrophobic motif residue as Thr412 in Figure 4, but the numbering differs according to the long and short S6K1 splice variants used in Figures 4 and 8, respectively. S6K1-T389E had high basal activity that was not increased much by EGF under the conditions used (data not shown). Introduction of the R121A mutation in S6K1-T389E abolished kinase activity without affecting the expression level or phosphorylation of the activation loop. In MSK1, mutation of Arg102 in the C-helix resulted in complete loss of kinase activity as well as phosphorylation of the hydrophobic motif, whereas the expression level was unaffected. Mutation of the N-terminal basic residue, K43, had no effect on kinase activity (data not shown). In PKBα, mutation of the N-terminal Arg144 and C-helix Arg200 resulted in partial and complete loss of kinase activity, respectively. Both mutations resulted in somewhat reduced phosphorylation of the activation loop and the hydrophobic motif, without affecting the expression level. Finally, since SGK1 was not activated consistently by insulin or EGF under the conditions used, we generated a mutant which contains aspartic acid at the phosphorylation site of the hydrophobic motif. SGK1-S422D showed basal activity 3–4 times that of stimulated wild-type SGK1 (data not shown). Mutation of the C-helix Arg147 in SGK1-S422D significantly reduced kinase activity without affecting the expression level or phosphorylation of the activation loop.
Fig. 8. Role of the phosphate-binding arginines in activation and phosphorylation of AGC kinases in vivo. COS7 cells were transfected with plasmids expressing HA- or GST-tagged wild-type or mutant kinase. After 48 h and a final 4 h serum starvation period, cells were exposed to 20 nM EGF for 20 min or 1 µM insulin for 8 min as indicated, and then lysed. Thereafter, the kinases were precipitated from the cell lysates with antibody to the HA tag or with glutathione beads. (A) Kinase activity was determined and expressed as a percentage of the wild-type stimulated value. Data are means ± SD of three (RSK2, S6K1, MSK1 and SGK1) or four (PKBα) independent experiments performed in duplicate. (B) Precipitated kinases from (A) were subjected to SDS–PAGE. The gel was subjected to immunoblotting with the indicated anti-phosphopeptide antibody or stained for protein.
These findings show that the phosphate-binding basic residues are important for the in vivo activity of the growth factor-activated AGC kinases. Moreover, the results suggest that the phosphate-binding basic residues may have a dual role by promoting catalytic activation as well as protecting the hydrophobic motif from dephosphorylation by intracellular phosphatases.
Discussion
The mechanism by which phosphorylation of the hydrophobic motif stimulates the activity of AGC kinases has been elusive. In the present work, we identify a binding site for the phosphoserine/phosphothreonine of the hydrophobic motif that functions to promote interaction of the hydrophobic motif with the hydrophobic pocket in these kinases. Moreover, we propose a mechanism by which phosphorylation of the hydrophobic motif and the activation loop can synergize to stimulate catalytic activity.
Although we identified the phosphate-binding site in PDK1, this kinase is an exception to the rule. PDK1 has no hydrophobic motif and uses the phosphate-binding pocket for docking to the hydrophobic motif of certain of its substrates in a phosphorylation-dependent manner. We have previously described how phosphorylation- dependent docking enables PDK1 to activate RSK2 (Frödin et al., 2000), S6K1 and SGK1 (Biondi et al., 2001), and here we describe the structural basis of this phenomenon. Taken together, our findings suggest a central role for the phosphate-binding pocket in regulation of the growth factor-activated AGC kinases by mediating intermolecular as well as intramolecular activation steps (Figure 9).
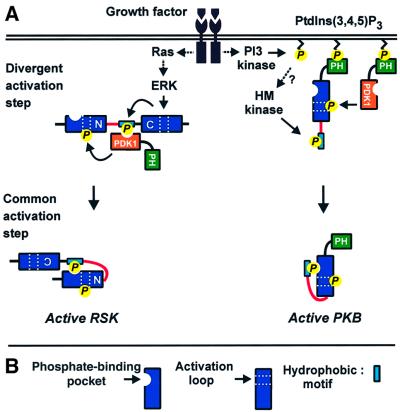
Fig. 9. Roles of the phosphate-binding pocket in regulation of the growth factor-activated AGC kinases. (A) The activation mechanism of AGC kinases may be divided into a divergent and a common activation step. In both steps, phosphoserine/phosphothreonine recognition by the phosphate-binding pocket may play a key role, as exemplified here with RSK and PKB. (1) In the divergent step, the kinases are subject to pathway-specific regulation via signalling modules flanking the kinase domain and which leads to phosphorylation of the hydrophobic motif (in RSK by the C-terminal kinase domain and in PKB by an as yet unknown kinase). In RSK (and S6K or SGK), the phosphate of the hydrophobic motif is then recognized by the phosphate-binding pocket of PDK1, resulting in recruitment of PDK1 and subsequent phosphorylation of the activation loop. Thus, the regulatory phosphorylations occur in a mandatory order. In PKB, PDK1 is recruited via PH domain-mediated co-localization at the cell membrane, and thus the order of the regulatory phosphorylations does not appear mandatory. (2) The second activation step may be common to all AGC kinases that contain a phosphorylatable hydrophobic motif, thus also PKC, Rho kinase and Ndr not studied here. In this step, the phosphate-binding site promotes intramolecular binding of the hydrophobic motif to the hydrophobic pocket within the AGC kinase domain. In concert with activation loop phosphorylation, this brings about a conformational change in the kinase domain, leading to catalytic activation. (B) Key to major symbols used in (A).
Structural modelling and biochemical analysis performed here suggest that the recognition of phosphoserine/threonine by the phosphate-binding site is similar in the kinases studied. When the hydrophobic motif is docked in the pocket, the phosphogroup is grabbed by two oppositely positioned arginine or lysine residues located N-terminal to the kinase domain and in the C-helix, respectively. The two phosphate-binding residues do not appear equally important, since mutation of the N-terminal arginine/lysine and the C-helix arginine typically resulted in partial and complete loss of kinase activation, respectively. The phosphate-binding pocket of PDK1 may be different from that of the other kinases, since PDK1 contains four N-terminal basic residues, of which three may be involved in docking to the hydrophobic motif as judged from the mutational analysis. The cluster of positive charges near to the pocket may function to attract the phosphate in the motif of the target kinases followed by docking of the motif in the pocket. In agreement with this possibility, PDK1 has much higher affinity for the phosphorylated hydrophobic motif of its target kinases than the kinases have for their own motif, as evidenced by BiaCore and EC50 measurements. Presumably, the affinity in the intramolecular interaction can be low because the effective concentration of pHM is high. Moreover, it may be important to have a low affinity in the intramolecular binding reaction in order to allow exposure of the phosphorylated motif for dephosphorylation and inactivation of the kinase.
Our characterization of the phosphate-binding pocket of PDK1 was supported later by the recently solved PDK1 crystal structure (Biondi et al., 2002). In the PDK1 structure, a sulfate ion from the crystalization buffer (probably mimicking a phosphogroup) binds two of the basic residues (Arg131 and Lys76) identified here, as well as Gln150 and Thr148. In our model, the backbone of the hydrophobic motif is located between Thr148 and the phosphogroup, which should preclude their interaction. In contrast, our model would allow binding of Gln150 to the phosphogroup. Thus, Gln150 may constitute a new phosphate-binding residue, in addition to those described here.
The present study provides direct evidence that the phosphorylated hydrophobic motif can stimulate catalytic activity of the growth factor-activated AGC kinases by an intramolecular mechanism. This was demonstrated using an in vitro reconstitution assay with isolated RSK2, S6K1, MSK1, PKBα and SGK1 kinase domains and addition of hydrophobic motif peptides. The results demonstrate that the phosphorylated hydrophobic motif cannot induce activation alone, whereas phosphorylation of the activation loop turns on the activity many fold. However, the combined effect of the phosphorylated hydrophobic motif and phosphorylated activation loop results in synergistic stimulation of catalytic activity. These findings are consistent with previous, but more indirect, studies on the effect of these phosphorylation sites on the activity of PKB (Alessi et al., 1996), S6K (Alessi et al., 1998; Dennis et al., 1998), RSK (Jensen et al., 1999) and SGK1 (Kobayashi and Cohen, 1999; Park et al., 1999).
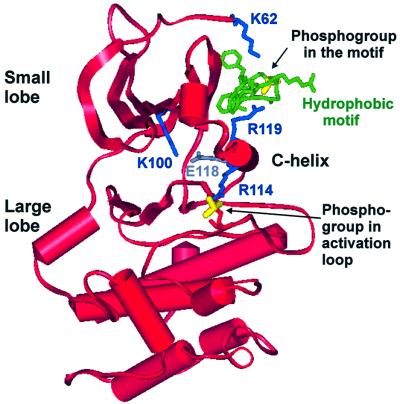
The question is how the two phosphorylation sites can act in a concerted manner to stimulate catalysis. Here, we show that the phosphate in the hydrophobic motif interacts with an arginine residue in the C-helix, which may provide an answer to this question. Preceding the arginine residue, the C-helix contains a conserved glutamic acid residue required for the catalytic activity of protein kinases. The many kinase structures now solved have revealed that the catalytically active conformation of kinases is similar, whereas the inactive conformations differ (reviewed in Huse and Kuriyan, 2002). In the active conformation, the glutamic acid residue in the C-helix (Glu 91 in PKA) forms an ion pair with a conserved lysine residue (Lys72 in PKA), that functions to position the phosphate of ATP for phosphotransfer in the kinase reaction. In the inactive conformation of many kinases, the C-helix is displaced, which disrupts the glutamic acid–lysine ion pair. A key step in activation of such kinases involves repositioning of the C-helix, e.g. by regulatory subunits, in order to establish the glutamic acid–lysine ion pair. We therefore suggest that the phosphorylated hydrophobic motif interacts with the C-helix in order to stabilize the conserved glutamic acid residue for optimal ion pair formation with the ATP-binding lysine residue (Figure 10). Furthermore, in crystals of PKA in the closed, active conformation, the phosphate in the activation loop binds His87 in the C-helix. This interaction is thought to stabilize the closed and active conformation, in which the small and large lobes of the kinase domain approach one another and in which the residues of the active site are fixed in the proper position for catalysis (Johnson and Lewis, 2001; Johnson et al., 2001). The growth factor-activated AGC kinases also contain histidine or arginine at the position equivalent to His87 in the C-helix of PKA (Figure 5A), that probably binds the phosphate in the activation loop. Referring to the RSK2 model in Figure 10, we therefore suggest that the activation loop phosphate and the phosphorylated hydrophobic motif act via the C-helix to induce their synergistic stimulation of kinase activity: first, the activation loop and the hydrophobic motif may cooperate to stabilize C-helix Glu118 in a position optimal for ion pairing with the ATP-binding Lys100. Moreover, occupancy of the pocket by the motif will restrict the mobility of the C-helix and thereby favour binding of C-helix Arg114 to the activation loop phosphate, which will stabilize the kinase in the closed, active conformation.
Fig. 10. Model of how the phosphates of the hydrophobic motif and the activation loop cooperate to activate AGC kinases. Ribbon representation of the model in Figure 5B of RSK2 interacting with its hydrophobic motif, but viewed from another angle and showing the entire RSK2 kinase domain. The figure illustrates how the phosphates of the hydrophobic motif and the activation loop can cooperate to position the C-helix for optimal ion pairing of Glu118 and the ATP-binding Lys100. Moreover, the C-helix, fixed by the hydrophobic motif, can function as a bridge between the small and large lobes to stabilize the kinase in the closed (active) conformation, which is shown here.
After submission of our manuscript, Yang et al. (2002) published a PKB structure with a non-phosphorylated hydrophobic motif consistent with the present findings. In the crystal, both the hydrophobic motif and the C-helix are not visible, suggesting that they are highly mobile, because the motif is not docked in the pocket when not phosphorylated. In vitro, pHMPKB and HMPRK2 activated PKB 4- and 15-fold, respectively, but not PKB-R200D, pre-phosphorylated by PDK1. Finally, calorimetric measurements suggested that HMPRK2 induced order in PKB, interpreted as stabilization of the C-helix and the active kinase conformation in general.
We also obtained evidence for a role for the phosphate-binding residues in promoting the phosphorylation state of the hydrophobic motif of the AGC kinases. The importance of this function varied, being high in S6K1 and MSK1, minor in PKBα and not observed in RSK2. Most probably, the basic residues promote the phosphorylation state of the motif by protecting the phosphoserine/phosphothreonine from dephosphorylation, in agreement with the buried position of the phosphate between the basic residues suggested by the structural models.
The present study was performed with kinases that are members of five different AGC kinase subfamilies. It is therefore reasonable to assume that the mechanism described here is a general activation mechanism among AGC kinases that contain a phosphorylatable hydrophobic motif. Sequence alignment shows that the hydrophobic motif phosphorylation site and the arginine/lysine residue in the C-helix are strictly co-conserved in AGC kinase subfamilies not studied here, such as PKCs, Rho kinases and Ndr-related kinases. Only in rare cases does the negative charge in the hydrophobic motif appear not to be required for kinase activity. An example is the glutamic acid residue in the PKCζ hydrophobic motif, but the residue increases thermal stability, suggesting that binding to the pocket does occur and has a functional role (Balendran et al., 2000; Parekh et al., 2000). The N-terminal basic residue is conserved in ∼80% of AGC kinases with a phosphorylatable hydrophobic motif. In other kinase families, the two phosphate-binding basic residues are absent, suggesting that the basic residues do not have a structural function in the kinase fold, but only serve to bind phosphate.
In conclusion, the intramolecular activation mechanism based on phosphorylation-dependent interaction between the hydrophobic motif and the hydrophobic pocket may be general within AGC kinases. If so, the mechanism would represent the most general activation mechanism described so far in protein kinases, next to activation loop phosphorylation.
Materials and methods
Antibodies, cDNAs, immunoprecipitation/blotting and protein staining are described in the Supplementary data available at The EMBO Journal Online.
Materials
Peptide sequences were: S6 peptide (RRLSSLRA), CREBtide (EILSRRPSYRK), crosstide (GRPRTSSFAEG), pHMRSK (KKPPSANAHQLFRGFSFVAITSDDE of mRSK2), pHMS6K (TLSESANQVFLGFTYVAPSC of rS6K1), pHMSGK (VKEAAEAFLGFSYAPPTP of hSGK1) and pHMROK (VGNQLPFIGFTYFRENL of hROKα), with underlined residues phosphorylated.
Transfection and immunoprecipitation
Kinases were expressed and purified from COS7 cells as described in Frödin et al. (2000) and in the Supplementary data.
In vivo AGC kinase assays
Beads with precipitated kinase were resuspended in 20 µl of 1.5× buffer A (30 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol). The kinase reaction was initiated by the addition of 10 µl (final concentrations) of ATP (200 µM, 0.2 µCi of [γ-32P]ATP) and peptide substrate: 800 µM S6 peptide (RSK, S6K), 166 µM crosstide (PKB, SGK) or 200 µM CREBtide (MSK). After 10 min at 28°C with vigorous shaking (the reaction was linear with time), 20 µl of the supernatant was removed (leaving behind precipitated kinase) and spotted onto phosphocellulose paper (Whatman p81). After washing with 150 mM orthophosphoric acid, [32P]phosphate incorporated into protein substrate was quantified on a PhosphorImager (Molecular Dynamics Inc.).
In vitro AGC kinase assays
In the RSK2 assay, 200 ng of GST–haemagglutinin (HA)-RSK21–373 (wild-type or R119A) were incubated in 20 µl of buffer A containing 60 µM MgATP with or without 100 ng of His-PDK1 for 1 h at 28°C in vitro. At this time, phosphorylation of RSK by PDK1 had reached saturation, as determined by time course experiments. In the S6K1, MSK1 and PKBα assays, COS7 cells were co-transfected with plasmids expressing HA-S6K1–364, HA-MSK11–370, HA-PKBα1–467 or point mutants thereof together with mycPDK1 or empty vector. S6K1, MSK1 or PKBα were then purified from the lysed cells using anti-HA antibody as described above and resuspended in 20 µl of buffer A. In the SGK1 assay, GST–SGK161–431 or GST–SGK161–415 was incubated with or without His-PDK1 and 100 µM MgATP for 1 h in vitro, whereafter His-PDK1 was removed by precipitation with Ni–agarose beads. GST–SGK161–431 (40 ng) or GST–SGK161–415 (400 ng) were then resuspended in 20 µl of buffer A. RSK, S6K, MSK, PKB or SGK activity was then determined in the absence or presence of the indicated hydrophobic motif peptide using kinase assay conditions as above. For RSK, a reaction blank (PDK1 alone) was subtracted from all values. Phosphorylation of His-S6K11–421 by PDK1 was performed as described (Biondi et al., 2001).
Surface plasmon resonance measurements
BiaCore analysis was performed as described in the Supplementary data or in Biondi et al. (2000, 2001).
Modelling procedures
Homology modelling and energy minimization of human PDK175–351, mouse RSK262–351 and human PKBα144–417 sequences were performed using the Homology and Discover modules as implemented in INSIGHT II (97.0) (Biosym/MSI, San Diego). The template for the modelling experiments was PKA (1YDR) (Engh et al., 1996), which has 39–45% sequence identity and 60–64% similarity to the targets. Energy minimization was performed using a steepest descents algorithm as implemented in Discover. The calculations were performed ignoring solvent interactions. The force field used was AMBER as implemented in INSIGHT II. Hydrophobic motif peptides were docked into the hydrophobic pockets using the program AUTODOCK (Ver. 3.0.3) from the Scripps Research Institute and Molecular Graphics Laboratory with default parameters, and subsequently were refined manually. The hydrophobic motif peptides were considered rigid with no flexibility of the torsion angles. The program GRID (Ver. 20) (Goodford, 1985) was used to compute the hydrophobic surface contours, which were used to predict the extent of the hydrophobic pocket. The program PROCHECK (Laskowski et al., 1993) was used to validate the geometry of the energy-minimized structures.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Birte Kofoed for expert technical assistance, Drs Sine Larsen, Anne Mølgaard and Lydia Tabernero for advice on structures, and Dr George Thomas for pRK5-GST–S6K1. This work was supported by grants from the Danish Cancer Society (DP01004), the Novo Nordisk Foundation, Denmark and the Danish Research Center for Growth and Regeneration.
References
- Alessi D.R., Andjelkovic,M., Caudwell,B., Cron,P., Morrice,N., Cohen,P. and Hemmings,B.A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J., 15, 6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Alessi D.R., James,S.R., Downes,C.P., Holmes,A.B., Gaffney,P.R., Reese,C.B. and Cohen,P. (1997) Characterization of a 3-phospho inositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol., 7, 261–269. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Kozlowski,M.T., Weng,Q.P., Morrice,N. and Avruch,J. (1998) 3-phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol., 8, 69–81. [DOI] [PubMed] [Google Scholar]
- Balendran A., Biondi,R.M., Cheung,P.C., Casamayor,A., Deak,M. and Alessi,D.R. (2000) A 3-phosphoinositide-dependent protein kinase-1 (PDK1) docking site is required for the phosphorylation of protein kinase Cζ (PKCζ) and PKC-related kinase 2 by PDK1. J. Biol. Chem., 275, 20806–20813. [DOI] [PubMed] [Google Scholar]
- Biondi R.M., Cheung,P.C., Casamayor,A., Deak,M., Currie,R.A. and Alessi,D.R. (2000) Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J., 19, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi R.M., Kieloch,A., Currie,R.A., Deak,M. and Alessi,D.R. (2001) The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J., 20, 4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi R.M., Komander,D., Thomas,C.C., Lizcano,J.M., Deak,M., Alessi,D.R. and van Aalten,D.M.F. (2002) High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J., 21, 4219–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak M., Clifton,A.D., Lucocq,L.M. and Alessi,D.R. (1998) Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38 and may mediate activation of CREB. EMBO J., 17, 4426–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P.B., Pullen,N., Pearson,R.B., Kozma,S.C. and Thomas,G. (1998) Phosphorylation sites in the autoinhibitory domain participate in p70s6k activation loop phosphorylation. J. Biol. Chem., 273, 14845–14852. [DOI] [PubMed] [Google Scholar]
- Engh R.A., Girod,A., Kinzel,V., Huber,R. and Bossemeyer,D. (1996) Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8 and H89. Structural implications for selectivity. J. Biol. Chem., 271, 26157–26164. [DOI] [PubMed] [Google Scholar]
- Etchebehere L.C., van Bemmelen,M.X.P., Anjard,C., Traincard,F., Assemat,K., Reymond,C. and Veron,M. (1997) The catalytic subunit of Dictyostelium cAMP-dependent protein kinase. Role of the N-terminal domain and of the C-terminal residues in catalytic activity and stability. Eur. J. Biochem., 248, 820–826. [DOI] [PubMed] [Google Scholar]
- Flynn P., Mellor,H., Casamassima,A. and Parker,P.J. (2000) Rho GTPase control of protein kinase C-related protein kinase activation by 3-phosphoinositide-dependent protein kinase. J. Biol. Chem., 275, 11064–11070. [DOI] [PubMed] [Google Scholar]
- Franke T.F., Yang,S.I., Chan,T.O., Datta,K., Kazlauskas,A., Morrison,D.K., Kaplan,D.R. and Tsichlis,P.N. (1995) The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell, 81, 727–736. [DOI] [PubMed] [Google Scholar]
- Frödin M, Jensen,C.J., Merienne,K. and Gammeltoft,S. (2000) A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J., 19, 2924–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodford P.J. (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem., 28, 849–857. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Huse M. and Kuriyan,J. (2002) The conformational plasticity of protein kinases. Cell, 109, 275–282. [DOI] [PubMed] [Google Scholar]
- Jensen C.J., Buch,M.B., Krag,T.O., Hemmings,B.A., Gammeltoft,S. and Frödin,M. (1999) 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J. Biol. Chem., 274, 27168–27176. [DOI] [PubMed] [Google Scholar]
- Johnson D.A., Akamine,P., Radzio-Andzelm,E., Madhusudan,M. and Taylor,S.S. (2001) Dynamics of cAMP-dependent protein kinase. Chem. Rev., 101, 2243–2270. [DOI] [PubMed] [Google Scholar]
- Johnson L.N. and Lewis,R.J. (2001) Structural basis for control by phosphorylation. Chem. Rev., 101, 2209–2242. [DOI] [PubMed] [Google Scholar]
- Knighton D.R., Zheng,J.H., Teneyck,L.F., Ashford,V.A., Xuong,N.H., Taylor,S.S. and Sowadski,J.M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science, 253, 407–414. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Cohen,P. (1999) Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J., 339, 319–328. [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Ming X.F., Burgering,B.M., Wennstrom,S., Claesson-Welsh,L., Heldin,C.H., Bos,J.L., Kozma,S.C. and Thomas,G. (1994) Activation of p70/p85 S6 kinase by a pathway independent of p21ras. Nature, 371, 426–429. [DOI] [PubMed] [Google Scholar]
- Parekh D.B., Ziegler,W. and Parker,P.J. (2000) Multiple pathways control protein kinase C phosphorylation. EMBO J., 19, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Leong,M.L., Buse,P., Maiyar,A.C., Firestone,G.L. and Hemmings,B.A. (1999) Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J., 18, 3024–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen N., Dennis,P.B., Andjelkovic,M., Dufner,A., Kozma,S.C., Hemmings,B.A. and Thomas,G. (1998) Phosphorylation and activation of p70s6k by PDK1. Science, 279, 707–710. [DOI] [PubMed] [Google Scholar]
- Richards S.A., Fu,J., Romanelli,A., Shimamura,A. and Blenis,J. (1999) Ribosomal S6 kinase 1 (RSK1) activation requires signals dependent on and independent of the MAP kinase ERK. Curr. Biol., 9, 810–820. [DOI] [PubMed] [Google Scholar]
- Stokoe D., Stephens,L.R., Copeland,T., Gaffney,P.R., Reese,C.B., Painter,G.F., Holmes,A.B., McCormick,G. and Hawkins,P.T. (1997) Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science, 277, 567–570. [DOI] [PubMed] [Google Scholar]
- Sturgill T.W., Ray,B.L., Erikson,E. and Maller,J.L. (1988) Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinase II. Nature, 334, 715–718. [DOI] [PubMed] [Google Scholar]
- Vik T.A. and Ryder,J.W. (1997) Identification of serine 380 as the major site of autophosphorylation of Xenopus pp90rsk. Biochem. Biophys. Res. Commun., 235, 398–402. [DOI] [PubMed] [Google Scholar]
- Williams M.R., Arthur,J.S., Balendran,A., van der Kaay,J., Poli,V., Cohen,P. and Alessi,D.R. (2000) The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol., 10, 439–448. [DOI] [PubMed] [Google Scholar]
- Yang J., Cron,P., Thompson,V., Good,V.M., Hess,D., Hemmings,B.A. and Barford,D. (2002) Molecular mechanism for the regulation of protein kinase B/AKT by hydrophobic motif phosphorylation. Mol. Cell, 9, 1227–1240. [DOI] [PubMed] [Google Scholar]