Abstract
Enzymatic activities that cleave Holliday junctions are required for the resolution of recombination intermediates and for the restart of stalled replication forks. Here we show that human cell-free extracts possess two distinct endonucleases that can cleave Holliday junctions. The first cleaves Holliday junctions in a structure- and sequence-specific manner, and associates with an ATP-dependent branch migration activity. Together, these activities promote branch migration/resolution reactions similar to those catalysed by the Escherichia coli RuvABC resolvasome. Like RuvC-mediated resolution, the products can be religated. The second, containing Mus81 protein, cuts Holliday junctions but the products are mostly non-ligatable. Each nuclease has a defined substrate specificity: the branch migration-associated resolvase is highly specific for Holliday junctions, whereas the Mus81-associated endonuclease is one order of magnitude more active upon replication fork and 3′-flap structures. Thus, both nucleases are capable of cutting Holliday junctions formed during recombination or through the regression of stalled replication forks. However, the Mus81-associated endonuclease may play a more direct role in replication fork collapse by catalysing the cleavage of stalled fork structures.
Keywords: DNA repair/genome instability/Mus81/recombination/replication restart
Introduction
Holliday junctions are formed when homologous duplexes exchange strands or when stalled replication forks regress (Holliday, 1964; Szostak et al., 1983; Cox, 2001; Flores et al., 2001; Kuzminov, 2001). The enzymatic activities that promote Holliday junction formation and resolution have been extensively characterized in bacteria. DNA strand exchange between homologues is catalysed by RecA and the resulting Holliday junctions are acted upon by RuvABC (West, 1992, 1997; Sharples et al., 1999). The three Ruv proteins interact to form a ‘resolvasome’ that catalyses the extension of heteroduplex DNA by branch migration and then resolves the junction by nucleolytic cleavage. In this complex, RuvA acts as the specificity factor that targets two hexameric rings of RuvB to the junction, the RuvB rings provide the motor for ATP-driven branch migration and RuvC promotes endonucleolytic resolution. RuvC is a highly specific Holliday junction resolvase that cleaves junctions by the introduction of symmetrically related nicks in two opposing strands of like polarity. Selectivity for the Holliday junction is enhanced by sequence specificity, such that RuvC scans for cleavable sequences as the DNA passes through the ternary RuvABC–Holliday junction complex (van Gool et al., 1998).
Much less is known about Holliday junction resolution in eukaryotic cells. Resolvases have been identified in Saccharomyces cerevisiae (Cce1) and Schizosaccharo myces pombe (Ydc1), but are thought to be catalytically active only in mitochondria (Symington and Kolodner, 1985; Kleff et al., 1992; White and Lilley, 1996; Oram et al., 1998; Whitby and Dixon, 1998). Resolvase activities have also been detected in mammalian cell-free extracts (Elborough and West, 1990; Hyde et al., 1994; Chen et al., 2001). The first (Elborough and West, 1990), which remains unidentified, was shown to fit the bacterial RuvABC resolvasome paradigm by its functional association with an ATP-dependent branch migration activity (Constantinou et al., 2001). Unlike the yeast resolvases, this activity was not compartmentalized to the mitochondria.
Recently, three groups reported a new endonuclease called Mus81 from fission yeast (Boddy et al., 2001), budding yeast (Kaliraman et al., 2001) and humans (Chen et al., 2001) that is involved in the maturation of recombination intermediates. Two-hybrid screens have shown that Mus81 interacts with the recombination protein Rad54 in S.cerevisiae (Interthal and Heyer, 2000) and with the FHA domain of the checkpoint kinase Cds1 in S.pombe (Boddy et al., 2000). Mutants in MUS81 are sensitive to the DNA-damaging agents methyl methane sulfonate (MMS) and UV, but are resistant to X-ray damage. Saccharomyces cerevisiae mus81 mutants were also identified as synthetic lethals in combination with sgs1, which encodes a protein belonging to the RecQ helicase family that is thought to act at stalled replication forks (Mullen et al., 2001). A similar synthetic lethal phenotype was observed with the S.pombe mus81 rqh1 double mutant (Boddy et al., 2000; Doe et al., 2002). The phenotypic properties of mus81 mutants are therefore suggestive of a connection between Mus81 and replication restart.
Mus81 protein exhibits sequence homology with the Rad1 subunit of the S.cerevisiae structure-specific endonuclease complex Rad1–Rad10 (in humans XPF– ERCC1), which is involved in nucleotide excision repair (Boddy et al., 2000; Interthal and Heyer, 2000). Schizosaccharomyces pombe Mus81, isolated as part of a complex with Eme1, was shown to cleave Holliday junctions and fork structures in vitro (Boddy et al., 2001). However, recombinant Mus81–Eme1 prepared from Escherichia coli exhibited a much greater cleavage activity with fork and flap structures compared with Holliday junctions (Doe et al., 2002). Similar results have been observed with recombinant S.cerevisiae Mus81, which associates with Mms4 (a protein that shares limited sequence homology with Eme1) (Kaliraman et al., 2001). At the present time, it is not clear whether the apparent differences between the recombinant and native proteins are due to the presence of peptide tags or simply relate to the purity of the enzyme preparations.
Schizosaccharomyces pombe mus81 and eme1 mutants exhibit meiotic lethality, indicating that Mus81 plays an important role in meiosis (Boddy et al., 2001). The meiotic phenotype can be reduced by suppression of meiotic recombination or by overexpression of the bacterial RusA protein (Boddy et al., 2001), which specifically promotes Holliday junction cleavage in vitro (Bolt and Lloyd, 2002). These observations led to the suggestion that Mus81 is required to resolve Holliday junctions in vivo in fission yeast (Boddy et al., 2001). However, the possibility that Mus81 acts on structures that arise prior to Holliday junction formation was not excluded. In S.cerevisiae, meiotic recombination defects in mus81 and mms4 mutants are less obvious (de los Santos et al., 2001). Indeed, the role that Mus81 plays in replication fork maintenance, and/or in the processing of recombination intermediates, is controversial, as indicated by the recent ‘Fuss about Mus81’ review (Haber and Heyer, 2001).
Like its yeast homologue, human Mus81 protein interacts with Cds1 (Chen et al., 2001). Mus81 increases in abundance following exposure to agents that block replication fork progression, and immunoprecipitated Mus81-containing complexes have been shown to cleave Holliday junctions in vitro.
In this work, we have determined whether Mus81 is related to the previously reported resolvase that co-purifies with a branch migration activity. To enable this, HeLa cell-free extracts were fractionated extensively, ultimately revealing the presence of two distinct Holliday junction cleavage activities in human cells. We found that the branch migration-associated resolvase was independent of Mus81 and specific for Holliday junctions. In contrast, the second junction-cutting activity, which was Mus81 associated, preferentially cleaved substrates that mimic replication forks and 3′-flap structures. The human Mus81-associated endonuclease may play a more generalized role in replication repair by acting directly upon stalled replication forks and other replication intermediates.
Results
Holliday junction resolvase activities in HeLa extracts
To determine the relationship, if any, between the mammalian branch migration/resolution activities (Constantinou et al., 2001) and Mus81 (Chen et al., 2001), HeLa extracts were extensively fractionated to reveal two junction-cutting activities. To assay for branch migration and Holliday junction resolution, we used small synthetic Holliday junctions made by annealing four oligonucleotides (Constantinou et al., 2001). The junction contained a homologous core of 26 bp, flanked by terminal regions of heterology, and was 5′-32P-labelled in one strand only. With this substrate, ATP-dependent branch migration leads to the formation of linear duplexes with splayed arms, whereas resolution leads to the formation of nicked duplex DNA products. During purification, we also followed the elution profile of Mus81 protein using affinity-purified anti-Mus81 polyclonal antibodies. Moreover, we carried out functional assays for Mus81-associated Holliday junction resolution activity using pull-downs in which immunopurified Mus81 was immobilized on beads (Chen et al., 2001).
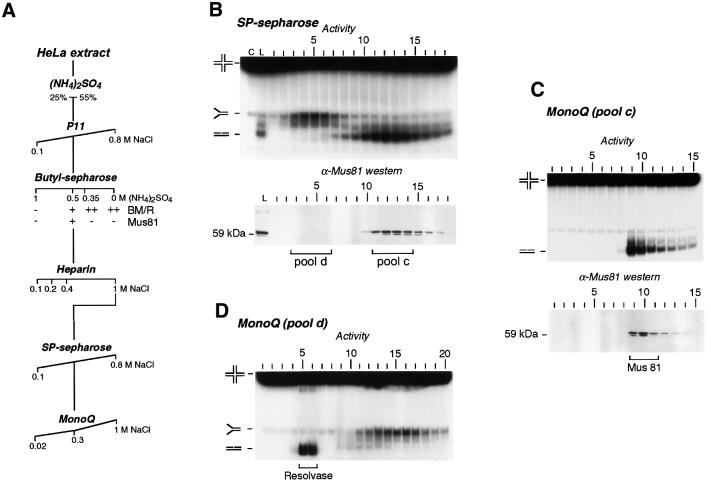
The purification scheme is shown in Figure 1A, and described in detail in Materials and methods. Following ammonium sulfate precipitation and phosphocellulose chromatography, fractions that catalysed branch migration/resolution were purified further on a butyl–Sepharose column that was eluted with three ammonium sulfate step washes. Mus81 protein was found in the 0.5 M (NH4)2SO4 eluate, whereas branch migration activity was detected in all washes. This 0.5 M step was therefore applied to a heparin column. Proteins that eluted in a 0.4–1.0 M NaCl wash were then further purified using an SP-Sepharose column, which was eluted with a 0.1–0.8 M salt gradient. For the first time, two distinct peaks of branch migration and resolvase activities were observed (Figure 1B, upper panel). The peak of resolution activity coincided with Mus81, as determined by western blotting for the 59 kDa protein (Figure 1B, lower panel) and confirmed by the Mus81 functional pull-down assay (data not shown). Fractions containing the Mus81-associated endonuclease (pool c) were then further purified by MonoQ chromatography. Holliday junction cleavage activity (Figure 1C, upper panel) again co-purified with the peak of Mus81 protein (Figure 1C, lower panel).
Fig. 1. Fractionation of HeLa cell-free extract. (A) Purification scheme (see Materials and methods for details). The presence or absence of Mus81 protein, branch migration (BM) and resolution (R) activities in the butyl–Sepharose fractions are indicated with (+) and (–). (B) Characterization of the SP-Sepharose fractions. Fractions were analysed for the presence of branch migration and resolution activities (upper panel), and for Mus81 protein by western blotting (lower panel). Fractions with relevant activities (pools d and c, respectively) were purified further as indicated. The products of branch migration and resolution (splayed arm and nicked duplexes, respectively) are indicated. Lane C, control reaction from which protein was omitted. Lane L, reaction containing the elutate from heparin that was loaded onto SP-Sepharose. (C) Fractionation of SP-Sepharose pool c on MonoQ. Fractions were analysed for the presence of Holliday junction resolvase activity (upper panel) and for Mus81 by western blotting (lower panel). (D) Fractionation of SP-Sepharose pool d on MonoQ.
The peak of branch migration activity from SP-Sepharose (Figure 1B, pool d) was also fractionated through MonoQ. Holliday junction resolvase activity became apparent at this stage (Figure 1D, lanes 5 and 6), and was followed by fractions containing branch migration activity (lanes 13–17). Mus81 could not be detected in any of these fractions by western blotting (data not shown). The abrupt appearance of the branch migration-associated resolvase activity at the MonoQ stage was not unexpected, since the nature of the coupled branch migration/resolution complex often masks resolution (or branch migration). Indeed, it is only when the two activities separate after extensive purification that they are observed as distinct entities.
In summary, after six purification steps, two activities capable of Holliday junction resolution have been isolated from human cells. Subsequent characterization studies were carried out with the most purified MonoQ fractions (fractions 5 and 6 of Figure 1D for the Mus81-independent resolvase, and fractions 9 and 10 of Figure 1C for the Mus81-associated endonuclease). For simplicity, we refer to the branch migration-associated (Mus81-independent) resolvase as resolvase A.
Two distinct activities that can resolve Holliday junctions in vitro
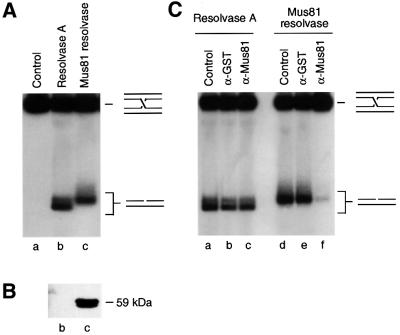
Resolvase A promotes Holliday junction resolution, leading to the formation of nicked duplex products (Figure 2A, lane b). As seen previously with E.coli RuvC (van Gool et al., 1998), two distinct product bands were observed by native PAGE. As detailed later, these products correspond to cleavage at selected major sites within the 26 bp homologous core, leading to the formation of products with different mobilities defined by the positions of the nicks. Mus81 could not be detected in this fraction by western blotting (Figure 2B, lane b). In contrast to resolvase A, the Mus81-associated endonuclease (Figure 2B, lane c) produced a single band of resolution products (Figure 2A, lane c).
Fig. 2. Two distinct Holliday junction endonucleases. (A) Holliday junction resolution reactions contained either the branch migration-associated resolvase A or the Mus81-associated endonuclease. Nicked duplex products were analysed by neutral agarose gel electrophoresis. Reactions were carried out in the absence of ATP, and with the most highly purified fractions (0.5 µl) from MonoQ. (B) Detection of Mus81 by Western blotting using aliquots (20 µl) of the fractions used in (A). (C) Resolvase fractions from MonoQ were immunodepleted using anti-Mus81 or anti-GST antibodies coupled to protein A–Sepharose beads, as described in Materials and methods. The beads were then precipitated and the supernatants incubated with 32P-labelled Holliday junction DNA. Lanes a and d, controls in which the resolvases were incubated on ice without Sepharose beads; lanes b and e, after pre-incubation with anti-GST–Sepharose beads; lanes c and f, after pre-incubation with anti-Mus81–Sepharose beads.
Having separated resolvase A from the Mus81-associated activity, we next confirmed their distinct identities by immunodepletion using Mus81 antibodies coupled to protein A–Sepharose beads. As expected, immunodepletion of Mus81 from the fraction containing the Mus81-associated endonuclease cleared it of almost all Holliday junction resolvase activity (Figure 2C, compare lanes d and f), whereas a GST control pull-down had no effect (lane e). In contrast, Mus81 antibodies failed to remove the equivalent activity from fractions containing resolvase A (compare lanes a and c). These results confirm the distinct identities of the two activities capable of Holliday junction resolution.
Characterization of the Holliday junction resolution reactions
All Holliday junction resolvases characterized to date introduce nicks at sites that are symmetrically related in two strands of like polarity. However, in contrast to the branch migration-associated resolvase (Elborough and West, 1990), initial studies with Mus81 pull-downs indicated that cleavage by Mus81 may not be restricted to symmetrically related sites (Boddy et al., 2001; Chen et al., 2001). To analyse the mechanism of Holliday junction cleavage, and to compare the incision activities of resolvase A and the Mus81-associated activity directly, the products of resolution reactions were analysed by denaturing PAGE. In these experiments, the Holliday junction containing the 26 bp homologous core was 5′-32P-labelled in strand 1, 2, 3 or 4 to permit observations of symmetrical cleavage.
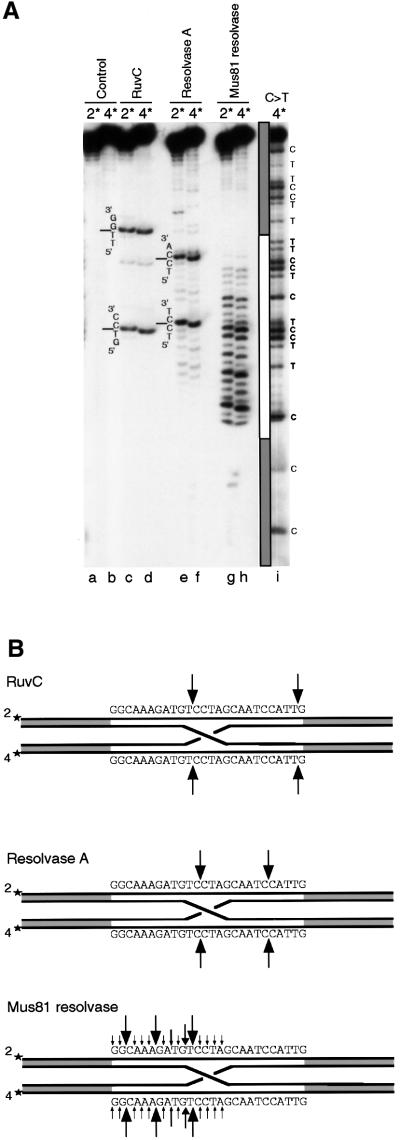
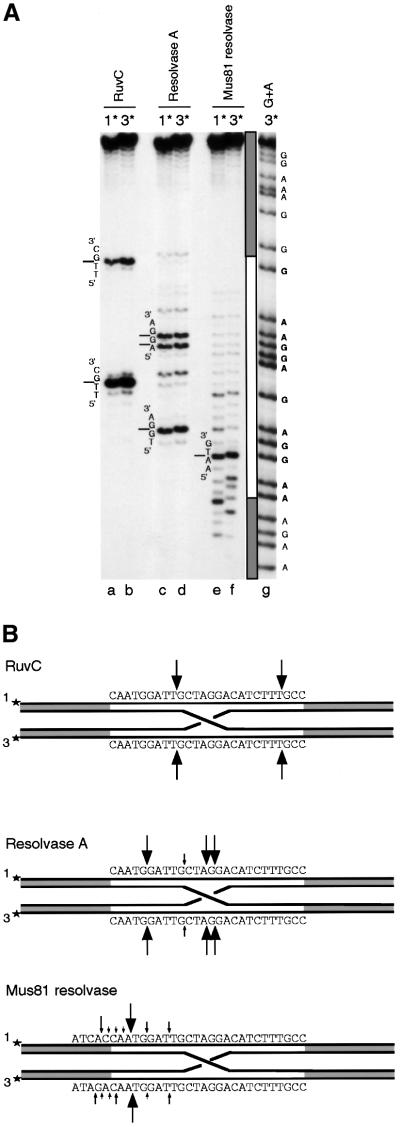
In control reactions, shown in Figure 3A (lanes c and d), Figure 4A (lanes a and b) and summarized in Figures 3B and 4B, E.coli RuvC protein cleaved the junction at two major sites in strands 2/4 and 1/3 within the homologous core. As expected, the incisions were introduced with perfect symmetry.
Fig. 3. Holliday junction resolution by RuvC, resolvase A and Mus81-associated endonuclease: incisions in strands 2 and strand 4. (A) The synthetic Holliday junction containing a 26 bp homologous core, 5′-32P-end-labelled either in strand 2 (2*) or strand 4 (4*), was incubated without proteins (lanes a and b), with 100 nM RuvC (lanes c and d), with 0.5 µl of MonoQ-purified resolvase A (lanes e and f) or with 0.5 µl of MonoQ-purified Mus81-associated endonuclease (lanes g and h). Cleavage products were analysed by denaturing PAGE and run alongside a C>T sequence ladder produced from 5′-32P-end-labelled strand 4. The homologous core and the terminal regions of heterology are indicated schematically by white and grey boxes, respectively. (B) Schematic representation of the incision sites. The nucleotide sequence of the homologous core region is shown, as are the sites of strong and weak cleavage indicated by the data presented in (A).
Fig. 4. Holliday junction resolution by RuvC, resolvase A and Mus81-associated endonuclease: incisions in strands 1 and 3. (A) Reactions were conducted as in Figure 3A using synthetic Holliday junctions labelled either in strand 1 (1*) or strand 3 (3*). Resolution by RuvC (lanes a and b), resolvase A (lanes c and d) and Mus81-associated endonuclease (lanes e and f). Lane g is a G+A sequence ladder produced from strand 3. (B) Schematic representation of the major and minor cleavage sites.
The patterns observed with resolvase A were remarkably similar: nicks were introduced with perfect symmetry at two distinct cleavage sites in strands 2/4 (Figure 3A, lanes e and f, and summarized in B) and at three major sites in strands 1/3 (Figure 4A, lanes c and d, and summarized in B). Cleavage at a limited number of sites is characteristic of a sequence/structure preference and it may be noteworthy that incision occurred between two cytosine residues (5′-TC↓CT-3′ and 5′-TC↓CA-3′) in strands 2/4. Interestingly, the corresponding sites in the complementary strands 1/3 were also preferentially cleaved (5′-TG↓GA-3′ and 5′-A↓G↓GA-3′).
In contrast to RuvC and resolvase A, the Mus81-associated endonuclease did not show any pronounced sequence preferences in strands 2/4 (Figure 3A, lanes g and h, and summarized in B); instead, the incisions were distributed throughout the homologous core of the junction. However, a distinct preference for cleavage on the 5′-side of the homologous core was apparent. The incision profiles observed in strands 2 and 4 were similar. The major cleavage sites in strands 1 and 3 also appeared to be symmetric, with one site (5′-AA↓TG-3′) being particularly favoured (Figure 4A, lanes e and f, and B). As observed with strands 2/4, a strong bias for cleavage towards the 5′-side of the homologous core was observed in strands 1/3 (Figure 4B). This bias was sufficiently pronounced to produce a few asymmetric nicks at the border between homologous and heterologous sequences of the junction. The observed cleavage patterns obtained with resolvase A and the Mus81-associated resolvase further confirm the presence of two distinct activities that can promote Holliday junction resolution in human cells.
Nick repair of the resolution products
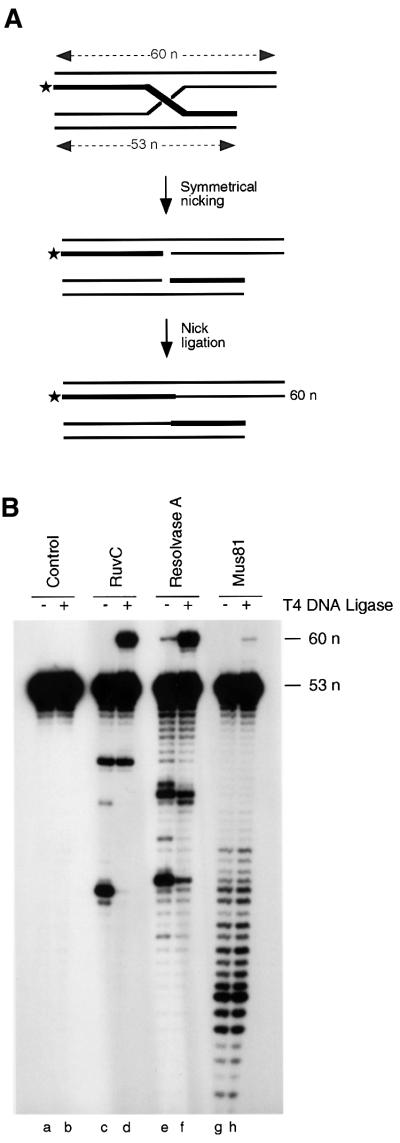
We next determined whether the two human endonucleases were bona fide Holliday junction resolvases, i.e. that resolution occurs by the introduction of symmetrical nicks in strands of like polarity to form nicked duplex products. To achieve this, we constructed a synthetic Holliday junction with arms of different length: nicking of a 53-nucleotide-long 5′-32P-labelled strand, followed by religation, would result in its conversion to 60 nucleotides, as indicated schematically in Figure 5A.
Fig. 5. Nick repair by DNA ligase. (A) Schematic representation indicating the synthetic Holliday junction with one short arm. Resolution of the junction by symmetrical cleavage followed by nick ligation converts the 53-nucleotide-long 5′-32P-labelled strand into one that is 60 nucleotides in length. The 32P-label is indicated by the asterisk. (B) The synthetic Holliday junction shown in (A) was incubated with RuvC, resolvase A or Mus81-associated endonuclease, and the reactions were supplemented with T4 DNA ligase, where indicated. Control, reactions without endonuclease. 32P-labelled DNA products were analysed by denaturing PAGE followed by autoradiography.
The junction was treated with RuvC, resolvase A or the Mus81-associated endonuclease and an aliquot of each resolution reaction was supplemented with T4 DNA ligase. The products were then analysed by denaturing gel electrophoresis. As shown in Figure 5B, addition of DNA ligase to the products of the RuvC cleavage reaction resulted in an additional band 60 nucleotides in length (lane d). Similar products were observed with resolvase A (lane f). In contrast, we observed very poor nick ligation following cleavage by the Mus81-associated endonuclease (lane h). These results show that a significant proportion of the resolution products produced by RuvC or resolvase A have readily ligatable nicks as a consequence of symmetrical cleavage. In contrast, the products of the Mus81 cleavage reactions are mostly non-ligatable.
Substrate specificities
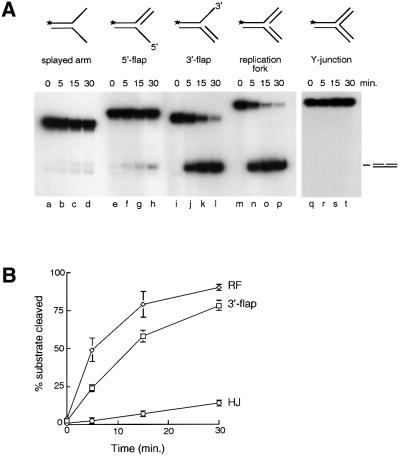
To determine the substrate specificities of the two endonucleases, a series of branched DNA substrates was generated. These included splayed arm duplexes, 5′-flap structures, 3′-flap structures, replication fork structures and Y-junctions (Figure 6A). We found that the 3′-flap structure served as a very good substrate for the Mus81-associated endonuclease, which removed the single-stranded DNA arm to give rise to nicked duplex products (Figure 6A, lanes i–l). Analysis by denaturing PAGE using 5′-32P-end labels revealed that cleavage occurred at the junction between single- and double-stranded DNA (data not shown). Moreover, the Mus81-associated endonuclease cut the replication fork structure particularly efficiently (lanes m–p). That this fork cleavage activity was related to the Holliday junction cleavage activity of Mus81 was confirmed by immunodepletion using Mus81 antibodies (data not shown). In contrast, at the same protein concentration, little cleavage was observed with the splayed arm structure (lanes a–d) or the 5′-flap substrate (lanes e–h). Cleavage of the three-armed Y-structure was not observed (lanes q–t).
Fig. 6. Substrate specificity of Mus81-associated endonuclease. (A) Activity of Mus81-associated endonuclease on the branched substrates indicated. Reactions were carried out as described in Materials and methods, and DNA products were analysed by neutral PAGE. All substrates were 5′-32P-end-labelled on a common oligonucleotide. (B) Quantification of the efficiency of cleavage by Mus81-associated resolvase with replication fork (RF), 3′-flap and Holliday junction (HJ) substrates. At the times indicated, aliquots were withdrawn and the products analysed by neutral PAGE. Cleavage products were quantified by phosphoimaging and expressed as a percentage of total radiolabel. The values presented are an average of three independent time courses.
We next determined the efficiency of cleavage of the flap and fork substrates relative to Holliday junctions. To achieve this, Mus81-associated endonuclease was incubated with the 3′-flap, the replication fork and the Holliday junction substrate under identical reaction conditions. We observed over a 30 min time course that replication forks and 3′-flaps were the preferred substrates for this nuclease (Figure 6B). Indeed, the Mus81-associated activity cut replication forks and 3′-flap structures at initial rates that were 24- and 12-fold greater than that observed with the Holliday junction substrate.
In contrast to the Mus81-associated nuclease, resolvase A showed little or no ability to cleave the 3′-flap or replication fork structures (data not shown). A 5′-flap endonuclease activity was observed in the resolvase A fractions after chromatography on MonoQ. This was subsequently found to be due to a minor contamination with the flap endonuclease FEN-1. 5′-flap cleavage activity, but not Holliday junction resolvase activity, could be removed from these fractions by immunodepletion using antibodies raised against FEN-1. The fractions also contain some ligase activity, as can be seen in the data presented in Figure 5B. These results show that the two human activities that cleave Holliday junctions exhibit distinct specificities on branched DNA substrates. Particularly noteworthy is the specificity of resolvase A for Holliday junctions, as opposed to the versatility of the Mus81-associated endonuclease, which preferentially cleaves replication forks and 3′-flaps.
Discussion
The primary conclusion of this work is that human cells contain two distinct activities that can promote Holliday junction cleavage. The two endonucleases are, however, different in a number of ways. First, the branch migration-associated resolvase A is highly specific for Holliday junctions and exhibits sequence selectivity at the incision step. This cleavage reaction is, therefore, very similar to that promoted by E.coli RuvC, in which structure and sequence requirements limit the actions of the resolvase to a freely branch migratable Holliday junction intermediate. Moreover, the products of resolution can be readily religated, indicating that cleavage occurs by the introduction of symmetrically related nicks in strands of opposite polarity. In contrast, the Mus81-associated resolvase exhibits less selectivity for sequence, and instead cleaves at several sites located towards the 5′-side of the region of homology. The products could not be religated, indicating that the majority of Holliday junctions were not cleaved by a mechanism involving symmetrical nicking. The lack of sequence specificity and symmetrical cleavage leads to the expectation that this nuclease would be more promiscuous, as observed when the substrate specificities of the two nucleases were analysed. For example, we found that the initial rate of cleavage of replication forks and 3′-flap structures by the Mus81-associated endonuclease was 24- and 12-fold greater, respectively, than that observed with the Holliday junction substrate. In this regard, the specificity of the human Mus81 complex was similar to the recombinant S.cerevisiae Mus81–Mms4 and S.pombe Mus81–Eme1 complexes (Kaliraman et al., 2001; Doe et al., 2002). In contrast, resolvase A failed to cut 3′-flap or replication fork structures, indicative of a high specificity for the Holliday junction.
Resolution activities and associated proteins
At the present time, the precise protein constitution of the two Holliday junction-cutting endonucleases is unknown, and even after six purification steps neither activity is homogeneous (data not shown). As indicated by studies of yeast Mus81 and its associated partner(s) Eme1 or Mms4 (Boddy et al., 2001; Kaliraman et al., 2001), it is anticipated that the human Mus81-associated endonuclease will include the equivalent of Eme1 or Mms4 (Chen et al., 2001). However, the limited ability of recombinant Mus81–Mms4 and Mus81–Eme1 complexes to cleave Holliday junctions may indicate that the heterodimer is insufficient for full Holliday junction cleavage activity (Kaliraman et al., 2001; Doe et al., 2002), and that other factors remain to be identified in this partially purified fraction from HeLa cells.
The fuss about Mus81? Structure-specific endonucleases with specialized functions
As discussed elsewhere (Haber and Heyer, 2001), several observations in S.cerevisiae do not appear consistent with Mus81 being the yeast Holliday junction resolvase. First, in mms4 mutants, meiotic crossovers are reduced only 2-fold compared with wild type (de los Santos et al., 2001), indicating that budding yeast may contain an alternative resolvase to the Mus81-associated activity. In contrast, spore viability is severely reduced in S.pombe mus81 mutants, an effect that can be partially overcome by overexpression of RusA (Boddy et al., 2001), suggestive of a greater meiotic dependence upon Mus81 cleavage activity. Such dependence may be due to the lack of an alternative resolvase, or may indicate differences in the mechanisms of recombination that are utilized in budding and fission yeast.
Secondly, mus81 deletion mutants are sensitive to MMS and UV, but not to γ-radiation or double-strand breaks (DSBs) induced by HO-endonuclease (Boddy et al., 2000; Interthal and Heyer, 2000). In S.pombe, hypersensitivity to agents that cause replication blocks can be suppressed by expression of the RusA Holliday junction resolvase (Doe et al., 2002). Resistance to DSB-inducing agents may arise by functional redundancy of two resolvases or, alternatively, it is possible that the Mus81-associated endonuclease does not participate in the repair of DSBs in mitotic cells. It is possible that the repair of DSBs by mitotic recombination may occur by pathways that may not involve Holliday junction resolution. In this regard, synthesis-dependent strand annealing or break-induced replication are now thought to provide mechanisms for DSB repair that more closely reflect the intimate links between replication and recombination (Holmes and Haber, 1999; Paques and Haber, 1999).
So what is the mitotic function of Mus81? Like its S.cerevisiae and S.pombe counterparts (Kaliraman et al., 2001; Doe et al., 2002), we found that the human Mus81-associated endonuclease cleaves substrates that resemble replication forks. A role in the maintenance or restart of replication forks stalled by persistent DNA lesions would therefore appear likely, especially given that Mus81 interacts with the replication checkpoint kinase Cds1/Chk2/Rad53 (Boddy et al., 2000; Chen et al., 2001; Ho et al., 2002). Using its structure-specific endonuclease activity, Mus81 may cleave the leading strand template to produce a DSB that can be used to reinitiate replication by a recombination-dependent process. Alternatively, the 3′-flap endonuclease activity may act to remove structures that are formed during synthesis-dependent strand annealing (Allers and Lichten, 2001; Kaliraman et al., 2001). Thus, in contrast to the branch migration-associated resolvase, it is likely that the actions of Mus81 are not restricted to Holliday junctions.
Multiple pathways for replication fork restart
Mus81-mediated DSB formation at a replication fork may be regarded as a rather drastic solution to the problems caused by fork stalling. A more conservative mechanism involves RecQ family DNA helicases, such as yeast Sgs1 and human BLM and WRN, which are able to migrate Holliday junctions (Bennett et al., 1998; Karow et al., 2000; Constantinou et al., 2001). When progression of a replication fork is blocked, both the leading and lagging strands may anneal such that a ‘chicken foot’ structure containing a Holliday junction is formed at the fork. The benefit of fork regression is that it permits repair of the blocking lesion or lesion bypass. Once repair has been completed, however, it is thought that RecQ-family helicases migrate the Holliday junction back to a replication fork to permit the reinitiation of DNA replication. The synthetic lethality of mus81 with sgs1/rqh1 in yeast (Boddy et al., 2000; Mullen et al., 2001) implies that Mus81 is essential for processing replication forks in cells that are defective in RecQ helicase function. These helicases are therefore important to limit recombination, as indicated by the hyper-recombination phenotype of sgs1 mutants (Gangloff et al., 1994; Watt et al., 1996), and by the high levels of sister chromatid exchanges observed in cell lines derived from individuals with Bloom’s syndrome (Chaganti et al., 1974).
In summary, there appear to be at least three alternative mechanisms for replication fork restart: (i) fork regression and repair; (ii) regression and Holliday junction cleavage; and (iii) cleavage of the stalled fork itself. The enzymatic properties of the human Mus81-associated endonuclease, which show that it can cleave both replication fork structures and Holliday junctions, indicate that it may function in two of these pathways and thereby play a key role in the restoration of stalled fork structures in vivo. Defects in either of these pathways would be expected to result in synthetic lethality in combination with Sgs1/BLM mutations.
Materials and methods
DNA substrates
The synthetic Holliday junction (X26) was made by annealing four oligonucleotides, as described previously (Constantinou et al., 2001). The splayed arm substrate was generated by annealing oligo 1 (5′-GACG CTGCCGAATTCTACCAGTGCCTTGCTAGGACATCTTTGCCCAC CTGCAGGTTCACCC-3′) with oligo 4 (5′-ATCGATAGTCGGATC CTCTAGACAGCTCCATGTAGCAAGGCACTGGTAGAATTCGGC AGCGT-3′). The 5′-flap structure contained oligo 1, oligo 2 (5′-TG GGTGAACCTGCAGGTGGGCAAAGATGTCC-3′) and oligo 4. The 3′-flap structure contained oligo 1, oligo 3 (5′-CATGGAGCTGTC TAGAGGATCCGACTATCGA-3′) and oligo 4. The replication fork substrate contained oligo 1, oligo 2, oligo 3 and oligo 4. The Y-junction contained oligo 1, oligo 5 (5′-TGGGTGAACCTGCAGGTGGGCAAA GATGTCCCATGGAGCTGTCTAGAGGATCCGACTATCGA-3′) and oligo 4. Unless stated otherwise, oligo 1 was 5′-32P-end-labelled and annealing was carried out as described previously (Elborough and West, 1990). All substrates were purified by gel electrophoresis.
Extract fractionation
HeLa S3 cells were grown in RPMI 1640 supplemented with 5% fetal calf serum, penicillin (100 000 U/l) and streptomycin (100 mg/l). Exponentially growing cells (50 l at 8 × 105 cells/ml) were harvested and washed three times in ice-cold PBS and once in hypotonic lysis buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 2 mM DTT) (Baumann and West, 1998). The cell pellet was then resuspended in 2 vols of lysis buffer, left on ice for 20 min and lysed using an ‘A’ pestle (25 strokes) in the presence of protease inhibitors (1 mM PMSF, 1.5 µg/ml aprotinin, 1 µg/ml each leupeptin, pepstatin A, chymostatin and Nα-p-tosyl-l-lysine chloromethyl-ketone). After 20 min on ice, 0.5 vol. of high salt buffer (10 mM Tris–HCl pH 8.0, 1 M KCl, 1 mM EDTA, 2 mM DTT) was added and the extract centrifuged for 3 h at 42 000 r.p.m. in a Beckman 45Ti rotor. Solid ammonium sulfate (25%; 134 g/l) was added to the supernatant and dissolved by gentle stirring on ice for 30 min. Insoluble materials were removed by low speed centrifugation (30 min at 8000 r.p.m.). The ammonium sulfate concentration was then raised to 55% (an additional 179 g/l), and the proteins were precipitated during 30 min stirring on ice. The precipitate (∼1 g) was recovered by centrifugation, resuspended in 100 ml of buffer A (50 mM Na2HPO4/NaH2PO4 pH 6.8, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.01% NP-40) containing 100 mM NaCl, and dialysed against the same buffer before loading onto a 100 cm3 phosphocellulose column (Whatman P11). Proteins were eluted with a 1 l gradient in buffer A supplemented with 0.8 M NaCl. Fractions (40 ml) with resolvase activity were pooled and supplemented with solid ammonium sulfate to a final concentration of 1 M before loading onto a 20 cm3 butyl–Sepharose 4 fast flow column equilibrated in buffer A containing 1 M (NH4)2SO4. The column was washed, and proteins eluted stepwise with buffer A containing 0.5, 0.35 and 0 M (NH4)2SO4. Mus81 eluted in 0.5 M wash, whereas branch migration/resolution activity was found in all three fractions. Each fraction (30 ml) was therefore processed independently during subsequent steps.
Fractions were desalted on a HiPrep™ 26/10 desalting column with buffer A containing 100 mM NaCl before loading onto a 5 cm3 heparin high trap column. The column was washed stepwise with buffer A containing 0.1, 0.2, 0.4 and finally 1.0 M NaCl. Mus81 and the resolvasome activities were recovered in the final step. Pooled fractions (10 ml) were desalted and then fractionated on a 1 cm3 SP-Sepharose high trap column. This was washed with 5 column vols of buffer A supplemented with 0.1 M NaCl, and activities were eluted with a 20 ml gradient to 0.8 M NaCl. Fractions (0.5 ml) with branch migration activity or Mus81 protein were pooled and dialysed against buffer B (20 mM Tris–HCl pH 8.0, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.01% NP-40) containing 20 mM NaCl. The pooled fractions were further purified on MonoQ PC 1.6/5. Proteins were separated with a two-step gradient of 1.5 ml of buffer B to 0.3 M NaCl, followed by 1 ml to 0.6 M NaCl. Active fractions were stored at –80°C.
Immunoblotting and immunopurification
Polyclonal antibodies were raised against GST–Mus81 as described previously (Chen et al., 2001). Mus81 was detected by western blotting following 10% SDS–PAGE. Monoclonal antibodies against FEN-1 (ab462) were purchased from Genentex (Cambridge).
Mus81 was immunopurified by mixing 100 µl fractions with 10 µl of protein A–Sepharose beads and 3 µl of anti-Mus81 polyclonal antibody. After 3 h at 4°C, the beads were washed twice in 1 ml of RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP-40, 2 mM EDTA, 50 mM NaF) and twice in phosphate buffer (60 mM Na2HPO4/NaH2PO4 pH 7.4, 5 mM MgCl2, 5% glycerol). The MusA antibody–protein A–Sepharose beads were then resuspended in 20 µl for use in resolution assays.
For Mus81 immunodepletion experiments, 20 µl reaction mixtures (lacking DNA) were incubated with Mus81 antibody–protein A– Sepharose beads (5 µl) for 1 h at 4°C. The beads were then removed by centrifugation and the supernatants analysed for their ability to cleave 32P-labelled DNA substrates.
Cleavage assays
Holliday junction branch migration and resolution assays were carried out as described previously (Constantinou et al., 2001). Substrate specificity experiments were carried out under similar conditions. Reactions (20 µl) contained 32P-labelled substrates (∼1 nM) and 0.5 µl aliquots of the indicated fractions in phosphate buffer (60 mM Na2HPO4/NaH2PO4 pH 7.4, 5 mM MgCl2, 1 mM DTT, 100 µg/ml BSA) supplemented with 2 mM ATP where indicated. Reactions with RuvC (100 nM), purified as described previously (Eggleston et al., 1997), were carried out in 20 mM Tris–acetate pH 7.5, 15 mM Mg(OAc)2, 1 mM DTT and 100 µg/ml BSA (van Gool et al., 1999).
Reactions were incubated for 30 min at 37°C and the DNA products deproteinized for 15 min at 37°C using 2 mg/ml proteinase K and 0.4% SDS. Labelled DNAs were analysed by 10% neutral PAGE. To map the resolution sites, DNA products were phenol extracted, ethanol precipitated and analysed on 8% denaturing gels containing 7 M urea. Sites of cleavage were determined by comparison with sequencing ladders (Constantinou et al., 2001).
Nick repair assay
For this experiment, a synthetic Holliday junction was constructed by annealing oligonucleotide 1 (5′-CCGCTACCAGTGATCACCAATGG ATTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCC-3′), oligo nucleotide 2 (5′-TGGGTGAACCTGCAGGTGGGCAAAGATGTCCT AGCAATCCATTGTCTATGACG-3′), oligonucleotide 3 (5′-CGTCAT AGACAATGGATTGCTAGGACATCTTTGCCGTCTTGTCAATATC GGC-3′), and oligonucleotide 4 (5′-TGCCGATATTGACAAGACGGC AAAGATGTCCTAGCAATCCATTGGTGATCACTGGTAGCGG-3′). The junction was 5′-32P-labelled in strand 2.
Holliday junction cleavage assays were carried out essentially as described above, except that the buffer was 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 2 mM ATP, 1 mM DTT and 100 µg/ml BSA. After 30 min incubation at 37°C, T4 DNA ligase (400 U; NEB) was added and the reactions were left at room temperature for 3 h. The products were phenol extracted, ethanol precipitated and analysed by denaturing PAGE.
Acknowledgments
Acknowledgements
We thank the members of the West laboratory for suggestions, Ruth Peat and the ICRF cell production unit for cell culture. This work was supported by Cancer Research UK. A.C. was supported by a European Science Exchange Fellowship and a fellowship from the Swiss Foundation for Grants in Biology and Medicine.
References
- Allers T. and Lichten,M. (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell, 106, 47–57. [DOI] [PubMed] [Google Scholar]
- Baumann P. and West,S.C. (1998) DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.J., Sharp,J.A. and Wang,J.C. (1998) Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem., 273, 9644–9650. [DOI] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard,P.H.L., McDonald,W.H., Shanahan,P., Yates,J.R. and Russell,P. (2001) Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell, 107, 537–548. [DOI] [PubMed] [Google Scholar]
- Boddy M.N., Lopez-Girona,A., Shanahan,P., Interthal,H., Heyer,W.D. and Russell,P. (2000) Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol., 20, 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt E.L. and Lloyd,R.G. (2002) Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell, 10, 187–198. [DOI] [PubMed] [Google Scholar]
- Chaganti R.S., Schonberg,S. and German,J. (1974) A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl Acad. Sci. USA, 71, 4508–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.B. et al. (2001) Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell, 8, 1117–1127. [DOI] [PubMed] [Google Scholar]
- Constantinou A., Davies,A.A. and West,S.C. (2001) Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell, 104, 259–268. [DOI] [PubMed] [Google Scholar]
- Cox M.M. (2001) Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet., 35, 53–82. [DOI] [PubMed] [Google Scholar]
- de los Santos T., Loidl,J., Larkin,B. and Hollingsworth,N.M. (2001) A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics, 159, 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Ahn,J.S., Dixon,J. and Whitby,M.C. (2002) Mus81–Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem., 277, 32753–32759. [DOI] [PubMed] [Google Scholar]
- Eggleston A.K., Mitchell,A.H. and West,S.C. (1997) In vitro reconstitution of the late steps of genetic recombination in E.coli. Cell, 89, 607–617. [DOI] [PubMed] [Google Scholar]
- Elborough K.M. and West,S.C. (1990) Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J., 9, 2931–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M.J., Bierne,H., Ehrlich,S.D. and Michel,B. (2001) Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J., 20, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type-I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog—a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. and Heyer,W.D. (2001) The fuss about Mus81. Cell, 107, 551–554. [DOI] [PubMed] [Google Scholar]
- Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Holliday R. (1964) A mechanism for gene conversion in fungi. Genet. Res. Camb., 5, 282–304. [DOI] [PubMed] [Google Scholar]
- Holmes A.M. and Haber,J.E. (1999) Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell, 96, 415–424. [DOI] [PubMed] [Google Scholar]
- Hyde H., Davies,A.A., Benson,F.E. and West,S.C. (1994) Resolution of recombination intermediates by a mammalian endonuclease activity functionally analogous to Escherichia coli RuvC resolvase. J. Biol. Chem., 269, 5202–5209. [PubMed] [Google Scholar]
- Interthal H. and Heyer,W.D. (2000) MUS81 encodes a novel helix–hairpin–helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet., 263, 812–827. [DOI] [PubMed] [Google Scholar]
- Kaliraman V., Mullen,J.R., Fricke,W.M., Bastin-Shanower,S.A. and Brill,S.J. (2001) Functional overlap between Sgs1–Top3 and the Mms4–Mus81 endonuclease. Genes Dev., 15, 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow J.K., Constantinou,A., Li,J.-L., West,S.C. and Hickson,I.D. (2000) The Bloom’s Syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S., Kemper,B. and Sternglanz,R. (1992) Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J., 11, 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. (2001) DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl Acad. Sci. USA, 98, 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman,V., Ibrahim,S.S. and Brill,S.J. (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics, 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram M., Keeley,A. and Tsaneva,I. (1998) Holliday junction resolvase in Schizosaccharomyces pombe has identical endonuclease activity to the Cce1 homolog Ydc2. Nucleic Acids Res., 26, 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples G.J., Ingleston,S.M. and Lloyd,R.G. (1999) Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG and RusA. J. Bacteriol., 181, 5543–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L.S. and Kolodner,R. (1985) Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc. Natl Acad. Sci. USA, 82, 7247–7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver,T.L., Rothstein,R.J. and Stahl,F.W. (1983) The double-strand break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- van Gool A.J., Shah,R., Mézard,C. and West,S.C. (1998) Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J., 17, 1838–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool A.J., Hajibagheri,N.M.A., Stasiak,A. and West,S.C. (1999) Assembly of the Escherichia coli RuvABC resolvasome directs the orientation of Holliday junction resolution. Genes Dev., 13, 1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt P.M., Hickson,I.D., Borts,R.H. and Louis,E.J. (1996) SGS1, a homolog of the Blooms and Werners syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics, 144, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C. (1992) Enzymes and molecular mechanisms of homologous recombination. Annu. Rev. Biochem., 61, 603–640. [DOI] [PubMed] [Google Scholar]
- West S.C. (1997) Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet., 31, 213–244. [DOI] [PubMed] [Google Scholar]
- Whitby M.C. and Dixon,J. (1998) Substrate specificity of the SpCCE1 Holliday junction resolvase of Schizosaccharomyces pombe. J. Biol. Chem., 273, 35063–35073. [DOI] [PubMed] [Google Scholar]
- White M.F. and Lilley,D.M.J. (1996) The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J. Mol. Biol., 257, 330–341. [DOI] [PubMed] [Google Scholar]