Abstract
We have initiated a biochemical analysis of splicing complexes in extracts from the fission yeast Schizosaccharomyces pombe. Extracts of S.pombe contain high levels of the spliceosome-like U2/5/6 tri-snRNP, which dissociates into mono-snRNPs in the presence of ATP, and supports binding of U2 snRNP to the 3′ end of introns, yielding a weak ATP-independent E complex and the stable ATP-dependent complex A. The requirements for S.pombe complex A formation (pre-mRNA sequence elements, protein splicing factors, SF1/BBP and both subunits of U2AF) are analogous to those of mammalian complex A. The S.pombe SF1/BBP, U2AF59 and U2AF23 are tightly associated in a novel complex that is required for complex A formation. This pre-formed SF1– U2AF59–U2AF23 complex may represent a streamlined mechanism for recognition of the branch site, pyrimidine tract and 3′ splice site at the 3′ end of introns.
Keywords: S.pombe/SF1/BBP/U2AF/U2 snRNP/U2/5/6 tri-snRNP
Introduction
The study of pre-mRNA splicing and of spliceosome assembly has benefited enormously from biochemistry in mammalian systems, the combination of facile genetics and biochemistry available in the yeast Saccharomyces cerevisiae, and comparisons between these systems. Yet, important aspects of splice site recognition seen in mammals are not paralleled in S.cerevisiae. These include some of the earliest interactions required for intron–exon recognition, such as the involvement of SR proteins, enhancer elements and U2AF.
The fission yeast Schizosaccharomyces pombe conserves many of the mammalian interactions and provides a genetically tractable system that has already been used to identify components of the splicing machinery (e.g. Potashkin et al., 1989, 1993). Notably, the S.pombe genome contains many introns (>4700), and individual genes typically contain multiple introns (up to 15), some of which may be subject to alternative splicing (Okazaki and Niwa, 2000; Wood et al., 2002). Unlike in S.cerevisiae, most splice site sequences do not conform strictly to a consensus. Splicing factors in S.pombe typically are more similar to their mammalian counterparts, e.g. the snRNAs and many proteins, than are their S.cerevisiae homologs (e.g. Brennwald et al., 1988). Furthermore, S.pombe contains SR proteins and highly conserved homologs of both subunits of U2AF, which are critical for early interactions in the mammalian system but absent or non-essential in S.cerevisiae (reviewed in Burge et al., 1999; Käufer and Potashkin, 2000).
In metazoan and S.cerevisiae in vitro systems, splicing components recognize sequence elements in the pre-mRNA through an ordered series of interactions. The first of these form the early or commitment complex (E or CC) and are ATP independent. In this complex, the 5′ splice site (SS) is recognized by U1 snRNP. Mammalian proteins SF1 and U2AF65 bind the branch region and pyrimidine tract, respectively; in S.cerevisiae, the ortholog of SF1, BBP, also binds the branch region, whereas the ortholog of U2AF, Mud2p, fulfills a role similar to that of U2AF65, although how it interacts with pre-mRNA is not clear (Abovich and Rosbash, 1997; Berglund et al., 1997). In mammals, U2AF65 is tightly complexed with U2AF35, whose binding to the 3′SS AG is critical for recognition of ‘AG-dependent’ introns, so called because the AG is required for early complex formation and the first step of splicing (Merendino et al., 1999; Wu et al., 1999; Zorio and Blumenthal, 1999); in S.cerevisiae, no homolog of U2AF35 has been found, and consistently all introns seem to be AG independent (reviewed in Umen and Guthrie, 1995). In the mammalian system, U2 snRNP is already loosely associated in the E complex, but not stably engaged (Das et al., 2000), whereas such an association has not been detected in S.cerevisiae CC (Liao et al., 1992). In both mammals and S.cerevisiae, the first ATP-dependent step is the stable binding of U2 snRNP to the intron branch site, in part through U2 snRNA–branch region base pairing (reviewed in Moore et al., 1993). This forms complex A in the mammalian system, or pre-spliceosomes in S.cerevisiae. Subsequently, a larger complex, B, is formed by association of U4/5/6 tri-snRNP. Complex C follows B after significant rearrangements and is the active spliceosome, containing U2/5/6 snRNPs and splicing intermediates (reviewed in Moore et al., 1993).
To facilitate studies in which genetics and biochemistry may be combined easily to elucidate early intron–exon recognition events, we initiated biochemical analysis of splicing complex formation in extracts from the fission yeast S.pombe. Unlike S.cerevisiae or metazoan systems, S.pombe extracts contain endogenous spliceosome-like U2/5/6 particles. Upon incubation with ATP, these particles dissociate, releasing U2 snRNP. Subsequently, these extracts efficiently form a U2 snRNP-containing complex on a variety of pre-mRNAs; this complex is analogous to mammalian complex A. Surprisingly, SF1, U2AF59 and U2AF23 are tightly associated in S.pombe. Thus, a pre-formed SF1–U2AF59–U2AF23 complex may represent a streamlined mechanism for simultaneous recognition of multiple sequence elements at the 3′ end of introns.
Results
Extracts of S.pombe efficiently form a complex on pre-mRNAs
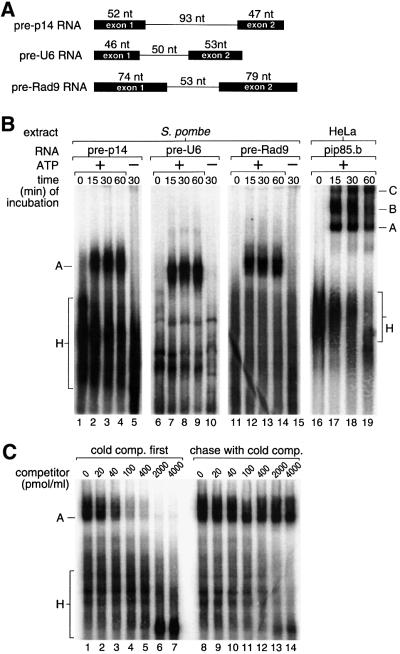
To test whether extracts prepared from S.pombe cells could form splicing complexes, we prepared a variety of in vitro transcribed S.pombe pre-mRNAs (Figure 1A). All formed one discrete complex detected by native gel electrophoresis after incubation in S.pombe extract, shown in Figure 1B on three pre-mRNAs (pre-p14, pre-U6 and pre-Rad9). This complex, once formed, was stable to chase with excess unlabeled pre-p14 competitor RNA, at saturating concentrations of the competitor (Figure 1C). We also assayed for in vitro splicing of pre-mRNA by direct gel analysis or by primer extension; however, no spliced products were detected (data not shown). For comparison, complex formation is shown in HeLa nuclear extract on an adenovirus-based substrate (Figure 1B).
Fig. 1. Extracts of S.pombe efficiently form a complex on S.pombe pre-mRNAs. (A) Schematic of S.pombe pre-mRNA constructs used in this study. Pre-p14 is derived from the gene for the U2 snRNP p14 component (Will et al., 2001; SPBC29A3.07c), pre-U6 from the gene for U6 snRNA (X14196, M55650) and pre-Rad9 from the gene for DNA repair protein Rad9 (SPAC664.07c). Solid boxes represent exons, and lines represent introns. (B) Formation of complexes on pre-p14 RNA (lanes 1–5), pre-U6 RNA (lanes 6–10) and pre-Rad9 RNA (lanes 11–15) in S.pombe extract, or for comparison on adenovirus-derived pre-mRNA in a HeLa cell nuclear extract (lanes 16–19). RNAs were incubated in extract at 30°C for the times indicated, adjusted to 0.5 mg of heparin/ml, separated on a native 4% polyacrylamide gel and visualized by phosphoimaging. A, U2 snRNP complexes containing pre-mRNA; B, spliceosomal complex B containing U2/4/5/6 snRNPs and pre-mRNA; C, spliceosomal complex C containing U2/5/6 snRNPs and splicing intermediates; H, non-specific complexes. (C) Stability of S.pombe complex A. Unlabeled competitor pre-p14 RNA was added either before (lanes 1–7) or after (lanes 8–14) formation of complex A on labeled pre-p14 RNA and reincubated at 30°C for 30 min. Complexes were analyzed as in (B).
The S.pombe complex requires and contains U2 snRNP
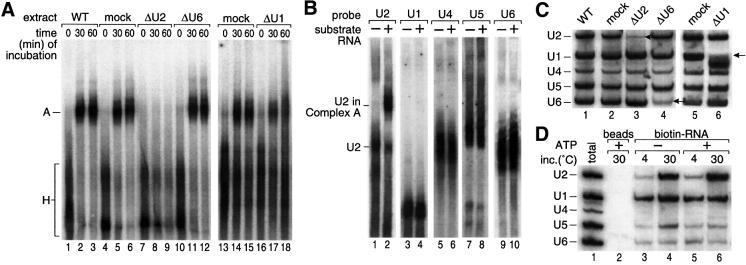
The defining characteristic of both mammalian complex A and S.cerevisiae pre-spliceosomes is ATP-dependent binding of U2 snRNP to the branch region. The S.pombe complex required both ATP and incubation at 30°C (Figure 1B), suggesting that it is related to complex A or later complexes and not to E/CC, which is ATP independent. To test the requirement for snRNPs, individual snRNAs were targeted for degradation by antisense DNA oligonucleotides and RNase H, and their degradation was monitored by northern blot analysis (Figure 2C). Targeting of U2 completely abrogated complex formation, whereas reactions degrading U6 or mock reactions (targeting U3) did not decrease the amount of complex (Figure 2A). Degrading the 5′ end of U1 resulted in a modest decrease in complex formation; although U1 was difficult to degrade completely, reactions in which ∼95% of U1 was degraded yielded an ∼50% decrease in complex formation (Figure 2A, lanes 16–18, and C, lane 6). This is consistent with results presented below showing a stimulatory, but not requisite, role of the 5′SS.
Fig. 2. The complex formed in extracts contains U2 snRNP, analogously to complex A. (A) snRNA requirement. Formation of complexes on pre-p14 RNA using extracts in which individual snRNAs were targeted for degradation by incubation with an antisense DNA oligonucleotide and RNase H. Untreated extract (lanes 1–3); mock-treated extract (lanes 4–6 and 13–15); extracts treated with oligonucleotides complementary to the branch site pairing region of U2 snRNA (nucleotides 28–42; lanes 7–9), U6 snRNA (nucleotides 21–69; lanes 10–12) or the 5′ end of U1 snRNA (nucleotides 1–14; lanes 16–18). Complexes were analyzed as in Figure 1B. (B) Native northern blot analysis of the complex. Extract was incubated with or without 400 pmol/ml unlabeled pre-p14 RNA at 30°C for 30 min (with ATP), separated on a native gel and transferred to Hybond N membrane, which was probed sequentially using riboprobes complementary to U2, U1, U4, U5 and U6 snRNAs. (C) Northern blot analysis of targeted snRNAs. Aliquots of extracts used in (A) were deproteinized, separated by 10% PAGE, blotted to Nytran and hybridized with antisense oligonucleotides to assess the extent of targeted degradation. (D) snRNA composition. Biotinylated pre-p14 RNA was incubated in extracts as indicated above each lane, bound to streptavidin–agarose beads and washed. Bound complexes were eluted and the RNAs separated and probed as in (C).
To test whether the complex visualized on native gels not only required but also contained U2 snRNP, we incubated unlabeled substrate RNA in extract, separated complexes on a native gel as before, and probed for snRNAs by northern blot analysis. U2 snRNA was shifted efficiently to a slower form that co-migrated with com plex A formed on labeled substrates (Figure 2B, lanes 1–2; data not shown). In contrast, U1 snRNA was not found in this complex, nor were U4, U5 or U6 snRNAs (lanes 3–10). Consistent with the ATP dependence of complexes formed on labeled substrates, this shift of U2 was observed only in the presence of ATP (data not shown; Figure 3A). We conclude that the S.pombe complex requires and contains U2 snRNP and is dependent on ATP, consistent with it being an analog of complex A or pre-spliceosomes. We henceforth refer to it as S.pombe complex A.
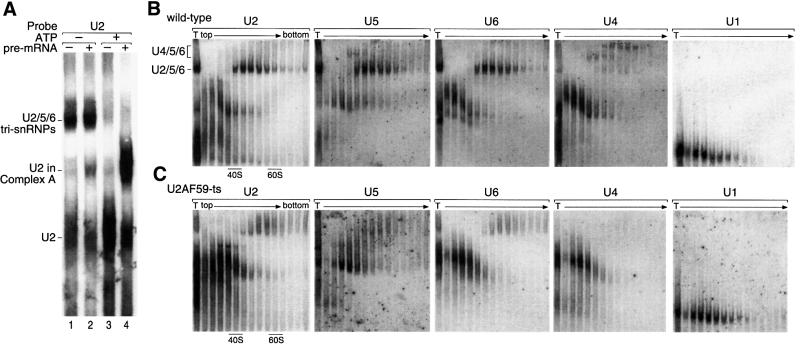
Fig. 3. Analysis of S.pombe snRNP distribution: S.pombe extracts contain U2/5/6 tri-snRNPs and few U4/5/6 tri-snRNPs. (A) Dissociation of U2/5/6 upon incubation with ATP, and accumulation of U2 in complex A. Native northern blot analysis of the complex. Extract was incubated with or without ATP and with or without 400 pmol/ml unlabeled pre-U6 RNA at 30°C for 30 min as indicated, separated on a native gel and transferred to Hybond N membrane. The membrane was probed using a riboprobe complementary to U2 snRNA. (B) Glycerol gradient fractionation and subsequent native gel separation of extracts from wild-type cells. Extract was fractionated through a 10–30% glycerol gradient; fractions were loaded directly onto a native gel, which was blotted and probed consecutively for each snRNA as indicated. The positions of 40S and 60S ribosomal subunits from parallel gradients are indicated. (C) Analysis as in (B) of U2AF59ts mutant extract, after growth at the non-permissive temperature for 2 h.
Analysis of S.pombe snRNP distribution
Northern blot analysis of U2 snRNP indicated a striking redistribution upon incubation with ATP (Figure 3A, lanes 1 and 3). Surprisingly, without ATP treatment, a large portion of U2 was present in a slow migrating complex, which subsequent probing for other snRNAs revealed to contain additionally U5 and U6, but not U4 or U1 (data not shown; see below). To characterize the distribution of U2 and other snRNPs in the S.pombe extract further, we separated the snRNPs by glycerol gradient followed by native gel electrophoresis, and then analyzed them by northern blotting (Figure 3B). A large portion of snRNAs was found in the top gradient fractions, representing either individual mono-snRNPs (U1, U2 or U5) or possibly di-snRNPs (U4/6). As suggested from the native gel northern blot (Figure 3A, lane 1), another large portion of the U2, U5 and U6 snRNAs sedimented together in an ∼45S complex. In contrast, only a small amount of co-sedimenting U4, U5 and U6 snRNAs was detected. Thus, the extract contained mostly U2/5/6 tri-snRNP and mono- or di-snRNPs, but little U4/5/6 tri-snRNP. This low level of U4/5/6 tri-snRNP may explain why no spliceosome formation (complex B and C) or splicing has been observed in S.pombe extracts.
To test whether the U2/5/6 complex was a product of the endogenous splicing machinery or fortuitously formed upon extract preparation, we used a temperature-sensitive (ts) mutant (prp2.1; Potashkin et al., 1993) of the large subunit of essential splicing factor U2AF (U2AF59), whose mammalian counterpart is required early in spliceosome assembly. Extracts prepared from these cells after shift to the non-permissive temperature did not contain U2/5/6 tri-snRNP (Figure 3C), whereas extracts from the U2AF59ts cells grown at permissive temperature and from wild-type cells that were temperature shifted did contain U2/5/6 tri-snRNP (data not shown). The same pattern was also observed with a second U2AF59ts mutant (prp2.2; Beales et al., 2000; data not shown). We conclude that active U2AF59 is required for the formation (or stability) of the U2/5/6 tri-snRNP, and thus it is likely to be a product of the endogenous splicing pathway.
Upon incubation with ATP, the U2/5/6 tri-snRNP decreased in abundance and faster migrating forms of U2, U5 and U6 increased (Figure 3A, lanes 1 and 3; data not shown). In particular, the faster migrating U2 was shifted into complex A by addition of pre-mRNA (lane 4). Thus, U2 snRNP that participated in forming complex A was probably first released from U2/5/6 in the extract. Although we could not test this directly, the change in migration of the bulk of U2 is consistent with this scenario; it is also possible that some free U2 contributed to for mation of complex A. Because release of U2 from U2/5/6 required incubation with ATP, it was possible that the subsequent formation of complex A did not require ATP. However, this was not the case, as pre-incubation with ATP, which dissociated U2/5/6, followed by ATP depletion and subsequent addition of pre-mRNA, resulted in no discernible complex A (data not shown). We conclude that ATP mediates at least two distinct events: release of U2 from U2/5/6 tri-snRNP, and complex A formation per se.
U2 snRNP binds pre-mRNA independently of ATP, but engages more stably with ATP
As an independent test of the snRNP composition of complex A and to detect weaker binding, biotinylated pre-mRNA was incubated with extract, and the bound complex selected using streptavidin–agarose. Selected snRNAs were then detected by northern blot analysis. To detect ATP-independent interactions, extract was first incubated with ATP to release U2 from U2/5/6 complexes, then depleted of ATP. Incubation on ice, either with or without ATP, enriched for U1, whereas incubation at 30°C with ATP enriched for U1 and U2 (Figure 2D), consistent with the result of similar selection from mammalian and S.cerevisiae extracts. Intriguingly, incubation at 30°C without ATP also enriched for U1 and U2 (lane 4) (a low level of U5 was also observed, perhaps related to previous observations of early weak U5 associations in metazoan systems; Chabot et al., 1985; Das et al., 2000). Thus, incubation without ATP resulted in weak U2 binding (stable to biotin–avidin pull-down, but not to gel electrophoresis), whereas incubation with ATP resulted in strong U2 binding (stable to both pull-down and gel electrophoresis). This is similar to observations of weak U2 binding in the ATP-independent mammalian E complex (Das et al., 2000), which becomes engaged more stably in the transition to the ATP-dependent complex A. Under all conditions, a weak binding of U1 was also observed.
Schizosaccharomyces pombe complex A requires the branch site but not 5′SS
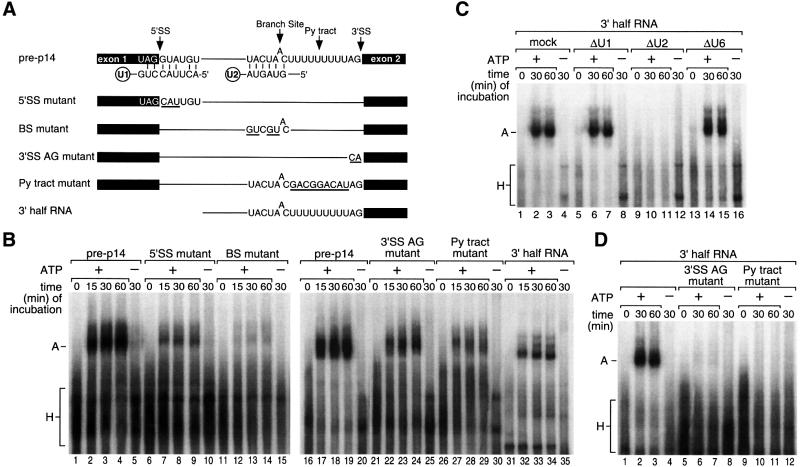
Mammalian complex A is dependent on base pairing interactions between U2 snRNA and the branch region, and is stimulated by 5′SS; the analogous S.cerevisiae complex strictly requires both these sequence elements. To test the contribution of these and other elements to formation of the S.pombe complex, we individually mutated the 5′SS, branch region, pyrimidine tract and 3′SS of the pre-p14 RNA construct (Figure 4A); similar results were obtained for branch region and 3′SS mutants of pre-Rad9 (data not shown).
Fig. 4. Sequence elements required for S.pombe complex A formation. (A) Schematic of pre-p14 constructs. SS, splice site; BS, branch site; Py tract, pyrimidine tract. (B) Formation of complexes on mutant pre-p14 RNAs. RNAs represented in (A) were incubated in S.pombe extract at 30°C for the times indicated, and separated on a native gel, as in Figure 1B. (C) Formation of complexes on the 3′ half pre-p14 RNA using extracts in which individual snRNAs were targeted for degradation by RNase H as in Figure 2A. (D) Formation of complexes on 3′ half pre-p14 RNAs containing 3′SS or Py tract mutations.
Branch region. Most importantly, mutation from 5′-UACUAAC-3′ to 5′-GUCGUAC-3′, decreasing the pairing potential with U2 snRNA from 6 to 2 bp, resulted in <8% complex A, in comparison with the control (Figure 4B, compare lanes 11–15 with lanes 1–5). This is as expected for complex A and consistent with the U2 requirement shown above.
5′SS. Three nucleotides at the 5′ end of the intron were mutated from 5′-GUA-3′ to 5′-CAU-3′. This substrate formed complex A to ∼30% of the control level (Figure 4B, compare lanes 6–10 with lanes 1–5), an effect similar to that of U1 5′ end degradation (Figure 2). As a different test of the contribution of 5′SS, exon 1, the 5′SS and first 13 nucleotides of the intron were deleted (‘3′ half RNA’; Figure 4A); this RNA also formed complex A in a yield similar to the 5′SS mutant (40% relative to the control; Figure 4B, lanes 31–35). Because potential binding sites for U1 might remain, extracts in which the 5′ end of U1 had been degraded by RNase H were also tested with this 3′ half RNA; U1 degradation had no detectable effect relative to mock or U6 degradation, whereas U2 degradation abrogated complex formation (Figure 4C). Thus, the 5′SS stimulates S.pombe complex A formation, but it is not strictly required.
3′SS AG. In mammalian introns, the 3′SS AG is required for efficient formation of E complex and subsequently complex A on AG-dependent introns. In contrast, AG-independent introns do not require the AG for either formation of splicing complexes or catalytic step 1, and S.cerevisiae introns are AG independent. To test the importance of the 3′SS AG in the S.pombe system, the p14 intron AG was changed to CA. This formed complex A to <50% relative to the control (Figure 4B, compare lanes 21–25 with lanes 16–20). As a more sensitive test, we also assayed this 3′SS mutation in the context of the 3′ half substrate; the 3′ half substrate was more AG dependent than the full-length pre-mRNA and formed no detectable complex A (Figure 4D, lanes 5–8). Thus, the 3′SS AG can contribute significantly to complex A formation in S.pombe, and thus at least some S.pombe introns are AG dependent.
Pyrimidine tract. This sequence element is required for splicing of mammalian introns and functions early as a binding site for U2AF65. The U9 sequence of the pre-p14 RNA was replaced with 5′-GACGGACAU-3′ or 5′-UGGUAAG-3′, which we expected to disrupt interactions with pyrimidine tract-binding proteins. These substrates formed complex A to <20% relative to the control (Figure 4B, compare lanes 26–30 with lanes 16–20; data not shown). In the context of the 3′ half substrate, these two pyrimidine tract mutants produced no detectable complex A (Figure 4D, lanes 9–12; data not shown). Thus, the pyrimidine tract is an important element for U2 binding in S.pombe.
In summary, mutational analysis of 5′SS, branch site, pyrimidine tract and 3′SS demonstrates that all four sites contribute to recognition steps in formation of S.pombe complex A. In particular, the stimulatory effect of 5′SS–U1 interaction and the 3′SS AG dependence are parallel to the contributions of these elements to mammalian complex A.
SF1/BBP and U2AF are required for S.pombe complex A and are tightly associated
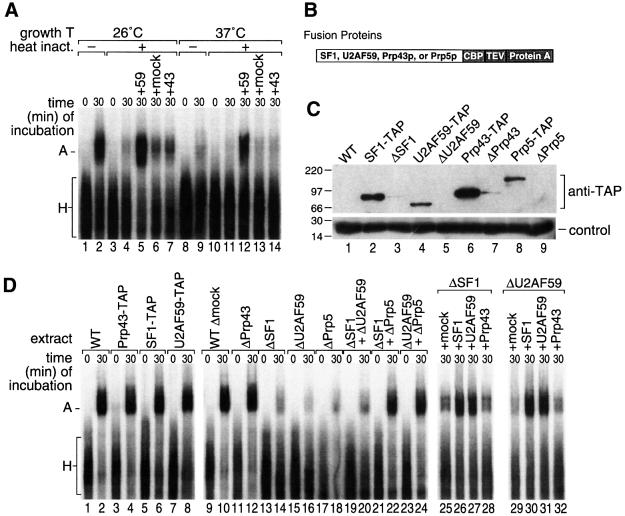
Mammalian SF1/mBBP and U2AF bind pre-mRNA at and adjacent to the branch site, respectively, and are required for U2 recruitment. To test their roles in S.pombe, we used several ts strains of U2AF59. Extracts from the prp2.2 strain formed complex A when grown at the permissive temperature (26°C), but little complex A after shift to the non-permissive temperature (37°C; Figure 5A, lanes 1 and 2, and 8 and 9). In addition, pre-treatment of the active prp2.2 extract in vitro at elevated temperature (37°C) resulted in loss of complex formation (lane 4). In contrast, the wild-type strain grown at either temperature efficiently formed complex A, and in vitro pre-treatment at 37°C resulted in a <20% decrease in activity (data not shown). Similar results were obtained with two other U2AF59ts strains (prp2.1 and prp2.3; data not shown). Furthermore, the inactivated extracts could be complemented by a partially purified preparation of U2AF (see below), in comparison with either mock-purified protein or similarly purified Prp43p (lanes 5–7 and 12–14). This is consistent with a requirement for active U2AF.
Fig. 5. The S.pombe complex A requires U2AF. (A) Formation of complex A on pre-p14 RNA in extracts from a U2AF ts mutant (prp2.2; Beales et al., 2000), grown at permissive temperature (lanes 1–7) or shifted for 2 h to the non-permissive temperature (lanes 8–14). Extracts were pre-incubated for 30 min either on ice (lanes 1, 2, 8 and 9) or at 37°C to heat inactivate (lanes 3–7 and 10–14). Lanes 5 and 12, addition of U2AF-TAP purified complexes; lanes 6 and 13, addition of mock-purified material; lanes 7 and 14, addition of Prp43-TAP purified material. Complexes were analyzed as in Figure 1B. (B) Schematic of TAP fusion proteins. CBP, calmodulin-binding peptide; TEV, TEV protease cleavage site; protein A, two copies of the IgG-binding domain derived from protein A (Puig et al., 2001). (C) Extent of tagged protein depletion. Extracts containing TAP-tagged SF1 (SPCC962.06c), U2AF59 (SPBC146.07), Prp43p (SPBC16H5.10c) or Prp5p (SPCC10H11.01) (lanes 2, 4, 6 and 8) were depleted of tagged proteins using IgG–Sepharose (lanes 3, 5, 7 and 9), separated on a 10% polyacrylamide gel, western blotted and probed for the TAP tag. WT, extract from untagged wild-type cells. Control, probed with antiserum to spindle checkpoint protein Slp1 as a loading control. (D) Depletion of U2AF and SF1/BBP and reconstitution of complex A. Formation of complexes on pre-p14 RNA using extracts in which various proteins had been TAP tagged, depleted and supplemented with partially purified protein complexes. Wild-type untagged extract (lanes 1 and 2), extracts in which Prp43p, SF1 or U2AF59 were TAP tagged (lanes 3 and 4, 5 and 6, and 7 and 8, respectively); extract that was mock-treated under depletion conditions (lanes 9 and 10), extracts depleted of Prp43p-TAP, SF1-TAP, U2AF59-TAP and Prp5p-TAP (lanes 11 and 12, 13 and 14, 15 and 16, and 17 and 18, respectively); mixture of SF1-depleted and U2AF59-depleted extracts (lanes 19 and 20); mixture of SF1-depleted and Prp5p-depleted extracts (lanes 21 and 22); mixture of U2AF59-depleted and Prp5p-depleted extracts (lanes 23 and 24). Lanes 25–32: SF1-depleted extract or U2AF59-depleted extract was supplemented with mock-purified, SF1-purified, U2AF59-purified or Prp43p-purified fractions.
To develop a system for more facile depletion and reconstitution of proteins involved in S.pombe complex A formation, we prepared strains containing the tandem affinity purification (TAP) tag at the C-terminus of the endogenous SF1 and U2AF59 proteins by homologous recombination (Figure 5B; Puig et al., 2001) or on Prp43p as a control (a DExH-box protein implicated in spliceosome disassembly; Arenas and Abelson, 1997; Martin et al., 2002). The presence of the TAP tags did not affect complex A formation (Figure 5D, compare lanes 4, 6 and 8 with lane 2). The TAP tag was then used to deplete extracts of each protein. Extracts depleted of SF1 or of U2AF59 made significantly less complex A than did wild-type (i.e. untagged) cell extract (compare lanes 14 and 16 with lane 2). In contrast, mock-depleted extract (extract from untagged cells that underwent the same procedures) and Prp43p-depleted extract were unaffected in comparison with wild-type (compare lanes 10 and 12 with lane 2). The small amount of complex A formed in the depleted extracts may be due to some residual SF1 or U2AF59 remaining in the extract or a low level of SF1- or U2AF-independent complex formation.
The depleted extracts could be complemented with partially purified SF1 or U2AF59. Proteins were partially purified in 1 M KCl through an IgG affinity column (see Materials and methods). This SF1 fraction complemented SF1-depleted extract, whereas mock-purified protein (similarly purified material from untagged cells) or Prp43p purified in this manner did not (Figure 5D, lanes 25, 26 and 28). Similarly purified U2AF59 complemented U2AF59-depleted extract, whereas mock-purified protein or Prp43p did not (lanes 29, 31 and 32). Surprisingly, the U2AF59 preparation also complemented SF1-depleted extract, and the SF1 preparation complemented U2AF59-depleted extract (lanes 27 and 30), suggesting that SF1 and U2AF59 were co-purified from S.pombe extract as a stable complex.
Consistent with this possibility, U2AF59-depleted and SF1-depleted extracts did not complement each other when mixed (Figure 5D, lane 20), although each was complemented when mixed with extract depleted of a different factor required for complex A formation, the DEAD-box protein SpPrp5p (lanes 18, 22 and 24; Y.-Z.Xu and C.C.Query, submitted). Together, these results argue that each depleted extract was ‘active’, but that a common component was co-depleted by SF1-TAP and by U2AF59-TAP.
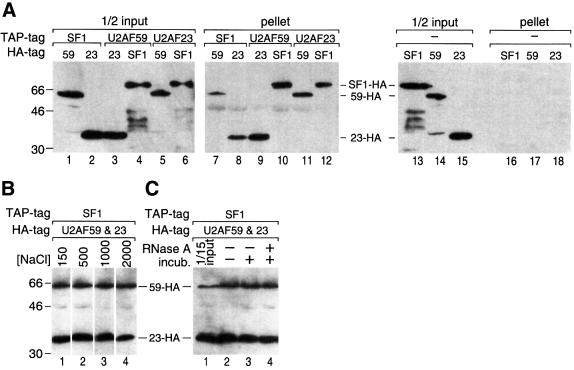
To test the possibility of a stable complex of SF1 and U2AF, we prepared S.pombe strains containing combinations of the TAP and hemagglutinin (HA) tags on SF1, U2AF59 and U2AF23, or on Prp43p as a control. Based on the stable U2AF65/35 dimer in mammalian cells, we expected a stable association of S.pombe U2AF59/23. In each case, using the TAP tag, we affinity selected complexes from extracts and then probed the purified material using anti-HA antibody. Affinity-selected SF1-TAP co-purified both U2AF59-HA and U2AF23-HA (Figure 6A, lanes 7 and 8); U2AF59-TAP co-purified U2AF23-HA and SF1-HA (lanes 9 and 10); and U2AF23-TAP co-purified U2AF59-HA and SF1-HA (lanes 11 and 12). In the absence of the TAP-tagged component, no HA-tagged proteins were selected (lanes 16–18); nor was Prp43-HA co-purified with U2AF59-TAP or with SF1-TAP from cells containing these tagged pairs (data not shown). Thus, SF1, U2AF59 and U2AF23 exist together in a complex.
Fig. 6. U2AF and SF1/BBP are tightly complexed. (A) Affinity selection of the U2AF–SF1 complex. Lysates from double-tagged cells, containing TAP- and HA-tagged proteins as indicated, were incubated with IgG–Sepharose beads to bind the TAP moiety. Co-purifying proteins were analyzed by western blot using anti-HA antibodies. Lanes 1–6, half of the input lysate; lanes 7–12, HA-tagged proteins present in the purified material; lanes 13–18, half of the input and IgG-selected material from cells lacking any TAP-tagged protein as a control; 59, U2AF59; 23, U2AF23. (B) The U2AF–SF1 complex is stable to high salt. Lysate from triple-tagged cells (SF1-TAP, U2AF59-HA, U2AF23-HA) was incubated with IgG beads and washed at salt concentrations as indicated (mM). Affinity-selected proteins were analyzed as in (A). (C) The U2AF–SF1 complex is resistant to RNase. Lysate from triple-tagged cells as in (B) was incubated with 0.5 mg/ml RNase A for 30 min at 30°C prior to incubation with IgG beads. Affinity-selected proteins were analyzed as in (A). Lane 1, 1/15th of input lysate; lane 2, untreated lysate; lane 3, mock treatment; lane 4, lysates incubated with RNase.
The association of these three factors was stable at high salt. Essentially identical results of TAP affinity selection were obtained using either 150 mM or 1 M NaCl conditions from the strains used in Figure 6A (data not shown). Using a strain containing three tags, SF1-TAP, U2AF59-HA and U2AF23-HA, co-purification of U2AF59-HA and U2AF23-HA with SF1-TAP was essentially the same from 150 mM to 2 M NaCl (Figure 6B). Because such an association might be due to a network of interactions dependent on RNA, we treated the extracts with RNase prior to affinity selection. Both U2AF59-HA and U2AF23-HA were co-purified with SF1-TAP at similar efficiency with and without incubation with RNase (Figure 6C, compare lanes 2 and 4). The extent of RNase digestion was monitored by northern blotting samples of these reactions and probing for snRNAs, which were no longer detectable after digestion (data not shown). Thus, the association of SF1 with U2AF59 and U2AF23 was not dependent on either mRNA or the snRNAs.
To test whether the size of the SF1–U2AF complex is consistent with a 1:1:1 stoichiometry, we fractionated extract from the triply tagged strain by 10–30% glycerol gradient centrifugation. SF1, U2AF59 and U2AF23 co-sedimented between 150 and 200 kDa, consistent with a 1:1:1 complex, whose predicted mass is 172 kDa. In addition, silver staining of the SF1–U2AF complex purified from a U2AF59-TAP-tagged strain was also consistent with a 1:1:1 ratio of these proteins (data not shown). We conclude that S.pombe SF1/BBP, U2AF59 and U2AF23 are tightly associated and that this complex is required for complex A formation.
Discussion
To understand better the molecular mechanisms underlying spliceosome assembly, we have focused on early recognition steps at the 3′ end of introns that result in engagement of U2 snRNP around the branch site region. Here, we describe the first in vitro analysis of intron–snRNP interactions in the fission yeast S.pombe. This organism provides a valuable system for this study, because of its manageability and shared similarities with metazoan splicing. In particular, the close correspondence between S.pombe and metazoan homologs of SF1 and both large and small subunits of U2AF may provide further insight into mechanisms underlying recognition of diverse sequences found at the 3′ end of most metazoan and S.pombe introns.
Schizosaccharomyces pombe complex A
A U2 snRNP-containing complex efficiently assembled on a variety of pre-mRNAs using S.pombe extracts. This complex is analogous to complex A because it requires U2 snRNP, ATP and incubation at 30°C, protein factors U2AF and SF1, and expected pre-mRNA sequence elements, most importantly, a consensus branch site. Due to current limitations in the extract, possibly limiting U4/5/6, we cannot yet chase this complex into active spliceosomes. Nonetheless, and above all, we designate this complex as S.pombe complex A because it represents a stable binding of U2 snRNP on the 3′ end of introns. These characteristics parallel those of both mammalian complex A and S.cerevisiae pre-spliceosomes.
Two other important characteristics more closely resemble the mammalian than the S.cerevisiae system. First, while formation of S.pombe complex A is stimulated by U1 snRNP and a 5′SS, the U1–5′SS interaction is not essential for the stable binding of U2 snRNP. Deletion of the 5′ end of U1 snRNA resulted in only a modest decrease of complex A formation, as did mutation or deletion of the 5′SS. On 3′ half RNAs that lack the ‘defined’ 5′SS, deletion of the 5′ end of U1 had no effect, indicating that no cryptic 5′SS was recruiting U1 (Figure 4C). Thus, U1–5′SS interaction stimulates, but is not required for S.pombe complex A. This is similar to the mammalian system, whereas U1 and the 5′SS are essential for pre-spliceosome assembly in S.cerevisiae (reviewed in Moore et al., 1993).
Secondly, in S.pombe extracts, the 3′SS AG contributes to the formation of complex A. This is consistent with the presence of S.pombe U2AF23, a U2AF35 homolog (Wentz-Hunter and Potashkin, 1996). In mammals, U2AF35 recognizes the 3′SS AG and is critical for pre-spliceosome assembly on AG-dependent introns. Consistent with our data, there is evidence for AG dependence of some S.pombe introns in vivo (Romfo and Wise, 1997). In contrast, S.cerevisiae introns are AG independent. Collectively, these data suggest that S.pombe provides a system in which early factor–intron interactions more closely parallel the early recognition events of mammalian AG-dependent introns and that S.pombe U2AF23 plays a key role in 3′SS recognition and recruitment of U2 snRNP. As discussed below, this 3′SS AG recognition is most probably facilitated by the binding of the pre-formed SF1–U2AF59–U2AF23 complex.
SF1/BBP and U2AF are tightly complexed
One of the advantages of using an S.pombe system for the study of U2AF is the availability of ts strains and the ability to create tagged genes easily. Neither heat-inactivated extracts from strains ts in U2AF59 nor extracts depleted of U2AF59 using the TAP tag were competent for formation of complex A. Both could be complemented by addition of partially purified U2AF. Surprisingly, extracts depleted of U2AF and extracts depleted of SF1 could not complement each other, because each was depleted of a common stable complex of SF1–U2AF59–U2AF23. This complex was stable to high salt and was insensitive to RNase. Thus, S.pombe SF1, U2AF59 and U2AF23 are tightly associated, and this complex is required for complex A formation.
Interactions between mammalian SF1/mBBP and U2AF65, and between S.cerevisiae ySF1/BBP and Mud2p were observed previously using recombinant proteins and yeast two-hybrid assays (Abovich and Rosbash, 1997; Rain et al., 1998), consistent with cooperative interactions between U2AF65 and SF1/mBBP in binding to mammalian introns (Berglund et al., 1998). However, such interactions were not detected by co-immunoprecipitation in extracts, and when these interactions occurred remained unclear. Because the SF1–U2AF59–U2AF23 complex represented most of these proteins in S.pombe extract, and because SF1-depleted and U2AF59-depleted extracts did not complement one another, we conclude that in S.pombe the interaction between SF1 and U2AF occurs before E/CC complex formation. This pre-formed SF1–U2AF59– U2AF23 complex may facilitate recognition of the 3′ end of introns, which in S.pombe contain degenerate branch regions and often lack pyrimidine-rich tracts upstream of the 3′SS. Cooperative binding among these three proteins would increase the affinity of the complex over that of each protein individually for its respective site. Thus, S.pombe has developed a streamlined mechanism simultaneously to recognize multiple points of the 3′ end of introns (Figure 7).
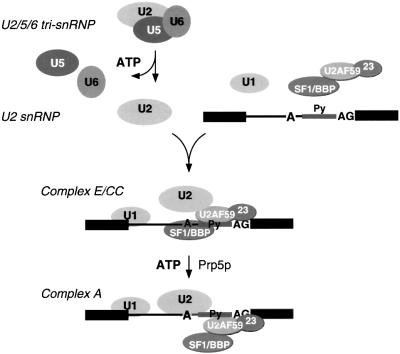
Fig. 7. Schematic model of snRNP dynamics observed in S.pombe extract. U2/5/6 tri-snRNP undergoes an ATP-dependent dissociation, resulting in an increase in the apparent free pool of U2, U5 and U6. This free pool of U2 participates in weak binding to pre-mRNA in an ATP-independent E complex, which is dependent on SF1–U2AF59– U2AF23 complex. A second ATP-dependent step results in stable engagement of U2 on the pre-mRNA; the extent of concomitant displacement of SF1–U2AF59–U2AF23 is uncertain.
The domain of mammalian SF1 that interacts with U2AF65 lies within the N-terminal 137 amino acids, and the N-terminal 28 amino acids are required for binding (Rain et al., 1998). Phosphorylation of Ser20 has been demonstrated to interfere with SF1–U2AF65 interaction (Wang et al., 1999), possibly inhibiting pre-spliceosome assembly. Alternatively, this phosphorylation may trigger displacement of SF1 from the branch site, which is a step thought to be required for subsequent U2 binding. Unlike SF1 homologs in mammals, Drosophila melanogaster and Caenorhabditis elegans (Mazroui et al., 1999), S.pombe SF1 does not contain a serine at the corresponding position. Perhaps, then, SF1–U2AF interactions in S.pombe are not regulated or remain associated throughout the splicing cycle.
Depletion of the SF1–U2AF59–U2AF23 complex, by either tagged SF1 or tagged U2AF59, resulted in little complex A formation. In S.cerevisiae, no significant effect on pre-spliceosome formation was observed in either BBP- or Mud2p-depleted extracts (Rutz and Séraphin, 1999). Depletion of SF1/mBBP from HeLa nuclear extract has variable results: although SF1/mBBP originally was identified and purified by complementation for complex A formation (Krämer and Utans, 1991; Krämer, 1992), its depletion had little effect on complex A assembly (Guth and Valcárcel, 2000). Because the interaction between SF1 and U2AF was stable in S.pombe, we were unable to deplete these factors individually; this co-depletion of SF1, U2AF59 and U2AF23 may explain the dramatic decrease of complex A formation upon depletion of SF1. In either case, we conclude that the SF1–U2AF59–U2AF23 complex is required for recruiting U2 snRNP in S.pombe.
Schizosaccharomyces pombe extracts contain U2/5/6 tri-snRNPs—spliceosomes
The S.pombe extracts are enriched in an abundant new particle, U2/5/6 tri-snRNP, resembling the composition of the mature spliceosome. Because this particle is not observed in extracts from strains ts in U2AF, it is likely to be a product of the endogenous splicing machinery. A similarly composed particle has been reported associated with the myb-related Cdc5p (Ohi et al., 2002). In our extracts, this complex represents a major fraction of the snRNAs. Thus, unlike S.cerevisiae or metazoan systems, S.pombe extracts contain endogenous spliceosome-like particles. Upon incubation with ATP, U2/5/6 disassembled into mono-snRNPs, representing a step in spliceosome disassembly and snRNP–snRNP rearrangement not previously observable or understood.
It has been suggested that spliceosome disassembly is required to recycle snRNPs for the next round of splicing. Although the recent description of penta-snRNPs in S.cerevisiae challenges the dogma of stepwise assembly and the requirement for disassembly (Stevens et al., 2002), extensive rearrangements would still occur in spliceosomes at the end of splicing to release products and reset the snRNAs. After the release of mRNA, Prp43p is proposed to release intron-lariat and subsequently disassemble spliceosomes (Arenas and Abelson, 1997). Recently, S.cerevisiae Prp43p has been demonstrated to mediate intron release in vitro (Martin et al., 2002). Interestingly, depletion of Prp43p from S.pombe extracts did not result in a defect in U2/5/6 disassembly (data not shown). This suggests that an additional ATP-requiring step exists after the function of Prp43p. A tempting model would be that Prp43p releases intron-lariat from spliceosomes, while additional ATPases are needed to unwind helices between U2 and U6 snRNAs and reset U2, U5 and U6 snRNAs for the next round of splicing. Consistent with this model, proteomic analyses of purified spliceosomes have revealed additional DExD/H ATPases (Jurica et al., 2002; Stevens et al., 2002). Alternatively, any ATPase that acts early in spliceosome assembly might have a second function in disassembly, which would not have been detected in previous experiments, but would be testable in the S.pombe in vitro system.
Dynamics of U2 snRNP in S.pombe extract
U2/5/6 tri-snRNP disassembly resulted in a large increase in a free form of U2, which subsequently was depleted upon formation of complex A (Figure 3A). This suggests that much of the U2 that participated in complex A formation derived from pre-existing U2/5/6. It is also possible that release of U2 from U2/5/6 was not a prerequisite for complex A assembly, or that some complex A formed from pre-existing free U2 snRNP. However, because the bulk of U2 was initially in U2/5/6, because extracts deficient in disassembly of U2/5/6 did not form complex A (data not shown) and because the bulk of U2 could be chased into complex A, we favor the former scenario and conclude that U2 in the extract cycled from U2/5/6 into complex A.
We detected two stages of U2 association in the formation of complex A: first, an ATP-independent association of U2 with the pre-mRNA, consistent with the weak binding of U2 observed in mammalian E complex; and, secondly, an ATP-dependent stable binding of U2. In separate work, we have shown that this ATP-dependent step requires the DEAD-box ATPase Prp5p (Y.-Z.Xu and C.C.Query, submitted). Taken together, the major pathway of U2 cycling we observed in S.pombe extracts is the ATP-dependent dissociation of U2/5/6, which releases U2, followed by an ATP-independent weak association with the pre-mRNA (E complex), followed by the ATP-dependent formation of complex A in which U2 is stably bound (Figure 7).
Study of evolutionarily distant systems can provide new insights and perspectives into the mechanisms of complex systems. Because of differences between S.pombe and the well-characterized S.cerevisiae system, and at the same time similarities between S.pombe and metazoans in factors and flexibility of intron recognition, the S.pombe system provides a new perspective on intron recognition. In addition to extending the analysis of U2 engagement and of the ATPases required for this process, the system can be used to analyze mechanistic aspects of U2/5/6 dissociation. Further, as we understand these partial assembly reactions and snRNP–snRNP interactions better, our analysis will probably lead to development of a system capable of splicing catalysis in vitro.
Materials and methods
RNA substrates
Pre-mRNAs were derived from genes p14 (SPBC29A3.07c), U6 (X14196, M55650) and RAD9 (SPAC664.07c). Sequences are: pre-p14, 5′-GGAGCUUGCGAAAAGCUUUCUGGCUACAACUUCAUGGAU AGAUACUUGGUAG↓GUAUGUUAUCUUAGGGAAUGUAUUAG CUACUAUUAAUUUUAAUUUCUCCUUUCUAACGGCUUUUUA AUAUCUUCUUACUAACUUUUUUUUUAG↓UGCAUUAUUAUA ACCCUGAAAGAGCUAAGGUUGAUGGUCAAGAUUUG-3′; pre-U6, 5′-GAUCUUCGGAUCACUUUGGUCAAAUUGAAACGAUAC AGAGAAGAUU↓GUAAGUAACAAUAUUUACCAAGGUUCGAG UCAUACUAACUCGUUGUUUAG↓AGCAUGGCCCCUGCACAA GGAUGACACUGCGACAUUGAGAGAAAACCCAUUUU-3′; and pre- Rad9, 5′-GGGACCUCGCAAGGAUCUUUACAAAUCUUUCUAGAA UCGAUGAUGCUGUCAACUGGGAAAUUAACAAAAAUCAG↓GU GUGUUGGAACUUUUUUCAAACCUUACUAAACAUUGAUACUA ACUGGUAAAG↓AUAGAGAUUACAUGUUUAAAUUCUUCUAGG UCAGGAUUUAGCAUGGUGACUUUAAAAAAGGCAUUUUUUGA CAAGUACA-3′. The 5′ and 3′ splice sites (SS) are indicated by arrows; branch sites are in bold. Pre-p14 5′SS was optimized to UAG↓GUAUGU. Pre-p14 and pre-Rad9 branch sites were optimized to UACUAAC. 3′-half pre-p14 RNA started at intron position 14. Other pre-p14 derivatives are described in Figure 4. Transcripts were synthesized in vitro using T7 RNA polymerase (Promega) and PCR-derived templates.
Strains and media
Strains used were: 972 (h–) and 401 (h+ leu1-32), gifts from Dr T.Matsumoto; prp2.1 (h– leu1.32 prp2.1), prp2.2 (h– ade6.M216 prp2.2) and mis11-453 (h– leu1.32 ade6.704 prp2.3), gifts from Dr J.Potashkin (Beales et al., 2000); and tagged derivatives of 972 (h–) described below. prp2.1, prp2.2 and mis11-453 were grown in YEA medium at 26°C or shifted to 37°C for 2 h. All other strains were grown in YE medium at 30°C.
Proteins of interest were tagged at their C-termini by PCR-based gene targeting (Bahler et al., 1998). pFA6a-3HA-kanMX6 was used as template for 3HA tagging. For TAP tagging (Puig et al., 2001), pFA6a-TAP-kanMX6 was used as template, in which GST sequences in pFA6a-GST-kanMX6 were substituted with TAP sequences (a gift of M.Lisbin). pFA6a-3HA-kanMX6 and pFA6a-GST-kanMX6 were gifts from Dr J.Bahler. Genomic tagging was confirmed by PCR and western blot analysis. Multiply tagged strains were made by crosses and subsequent random spore analysis.
Preparation of extracts from S.pombe
Extracts were prepared as described in Ansari and Schwer (1995) for S.cerevisiae with the following modifications. Two liter cultures of S.pombe cells were grown at 30°C to 2 × 107 cells/ml and harvested by centrifugation in a JA-10 rotor at 5000 r.p.m. for 5 min at 4°C. Cell pellets were washed in cold water, then in cold AGK400 buffer [10 mM HEPES-KOH pH 7.9, 400 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF) and 10% glycerol], and were resuspended in AGK400 to 1 g cells/ml. Cell suspensions were frozen in liquid N2 and stored at –80°C. Frozen cell pellets were ground to a fine powder using a mortar and pestle, thawed on ice and spun in a JA-20 rotor at 15 000 r.p.m. for 30 min. The supernatant was spun in a TLA-100.3 rotor at 56 000 r.p.m. for 20 min and dialyzed twice for 1.5 h against buffer D (20 mM HEPES-KOH pH 7.9, 0.2 mM EDTA, 100 mM KCl, 0.5 mM DTT, 1 mM PMSF and 20% glycerol).
Depletion of SF1/BBP and U2AF, and affinity selections
Depletions were based on TAP purification (Puig et al., 2001). Prior to dialysis, extracts as described above were adjusted to 1 M KCl, incubated twice for 1.5 h at 4°C with tumbling with IgG–Sepharose beads (Pharmacia) pre-washed with IPP150, and then dialyzed against buffer D.
For complementation experiments, beads from the first incubation were washed three times with 10 ml of IPP150 and once with 10 ml of TEV cleavage buffer (Puig et al., 2001). TAP tags were cleaved using 80 U of TEV protease (Gibco) in 500 µl of TEV cleavage buffer for 2 h at 16°C. A 1 µl aliquot of released material was used in complementations.
TAP affinity selections were performed in cell lysates produced as follows: cell pellets from 50 ml cultures were resuspended in 200 µl of lysis buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 0.1% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 100 µM Na3VO4, 2 µg of leupeptin per ml) and disrupted using glass beads. Lysates were incubated with IgG–Sepharose beads for 2 h at 4°C with tumbling. Beads were washed five times in IPP150 (or salt concentrations described in the text) for 5 min each. Samples were separated on a 10% SDS–polyacrylamide gel for western blot analysis. Western blots were probed using anti-HA monoclonal antibody 12AC5 (Boehringer) or anti-Slp1 (gift of T.Matsumoto) and horseradish peroxidase (HRP)-conjugated sheep anti-mouse (Amersham); or rabbit anti-chicken–HRP (Pierce) to bind protein A of TAP.
Native gel analysis
Complex A assembly reactions contained 20 mM KOAc, 2 mM MgOAc2, 0.05 mg/ml Escherichia coli tRNA, 0.1 mM DTT, 0.5 U/µl RNasin, 1 mM ATP, 5 mM creatine phosphate and 40% S.pombe extracts similar to standard conditions used with HeLa nuclear extracts (Grabowski et al., 1984). Hexokinase (Roche) and 2 mM glucose were added to deplete ATP. Complexes were analyzed by electrophoresis in native 4% acrylamide gels (80:1) run in 50 mM Tris–glycine (Konarska and Sharp, 1987). For RNase H digestions, 1 µg of oligonucleotide was added to 4 µl of extracts, with 0.5 U/µl RNase H (USB) for 30 min at 30°C, targeting nucleotides 1–14 of U1, 28–42 of U2, 21–69 of U6 or 106–130 of U3.
Glycerol gradients
Schizosaccharomyces pombe extracts, depleted of ATP from both wild-type and prp2.1 extracts, were loaded onto 10–30% glycerol gradients in 50 mM Tris–glycine, 1 mM EDTA, and spun at 49 000 r.p.m. for 3 h at 4°C in an SW50.1 rotor (Konarska and Sharp, 1987).
Acknowledgments
Acknowledgements
We thank J.Potashkin and T.Matsumoto for generous sharing of strains and reagents, J.R.Warner for generous sharing of resources, and M.M.Konarska, M.J.Moore, B.Schwer and J.R.Warner for discussions and critical review of the manuscript. J.V. was supported by NIH grant GM25532 to J.R.Warner. This work was supported in part by NIH grant GM57829 and by an HHMI New Faculty Award to C.C.Q., and by a Cancer Center Support (core) grant from the NCI.
References
- Abovich N. and Rosbash,M. (1997) Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell, 89, 403–412. [DOI] [PubMed] [Google Scholar]
- Ansari A. and Schwer,B. (1995) SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J., 14, 4001–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas J.E. and Abelson,J.N. (1997) Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl Acad. Sci. USA, 94, 11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J., Wu,J.Q., Longtine,M.S., Shah,N.G., McKenzie,A.,III, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Beales M., Flay,N., McKinney,R., Habara,Y., Ohshima,Y., Tani,T. and Potashkin,J. (2000) Mutations in the large subunit of U2AF disrupt pre-mRNA splicing, cell cycle progression and nuclear structure. Yeast, 16, 1001–1013. [DOI] [PubMed] [Google Scholar]
- Berglund J.A., Chua,K., Abovich,N., Reed,R. and Rosbash,M. (1997) The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell, 89, 781–787. [DOI] [PubMed] [Google Scholar]
- Berglund J.A., Abovich,N. and Rosbash,M. (1998) A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev., 12, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P., Porter,G. and Wise,J.A. (1988) U2 small nuclear RNA is remarkably conserved between Schizosaccharomyces pombe and mammals. Mol. Cell. Biol., 8, 5575–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T.H. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Chabot B., Black,D.L., LeMaster,D.M. and Steitz,J.A. (1985) The 3′ splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science, 230, 1344–1349. [DOI] [PubMed] [Google Scholar]
- Das R., Zhou,Z. and Reed,R. (2000) Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell, 5, 779–787. [DOI] [PubMed] [Google Scholar]
- Grabowski P.J., Padgett,R.A. and Sharp,P.A. (1984) Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell, 37, 415–427. [DOI] [PubMed] [Google Scholar]
- Guth S. and Valcárcel,J. (2000) Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem., 275, 38059–38066. [DOI] [PubMed] [Google Scholar]
- Jurica M.S., Licklider,L.J., Gygi,S.R., Grigorieff,N. and Moore,M.J. (2002) Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA, 8, 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käufer N.F. and Potashkin,J. (2000) Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res., 28, 3003–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M.M. and Sharp,P.A. (1987) Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell, 49, 763–774. [DOI] [PubMed] [Google Scholar]
- Krämer A. (1992) Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol. Cell. Biol., 12, 4545–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. and Utans,U. (1991) Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J., 10, 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.C., Colot,H.V., Wang,Y. and Rosbash,M. (1992) Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res., 20, 4237–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Schneider,S. and Schwer,B. (2002) Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem., 277, 17743–17750. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Puoti,A. and Krämer,A. (1999) Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA, 5, 1615–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L., Guth,S., Bilbao,D., Martinez,C. and Valcárcel,J. (1999) Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature, 402, 838–841. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Query,C.C. and Sharp,P.A. (1993) Splicing of precursors to mRNA by the spliceosome. In Gesteland,R. and Atkins,J. (eds.), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, p. 303–357.
- Ohi M.D., Link,A.J., Ren,L., Jennings,J.L., McDonald,W.H. and Gould,K.L. (2002) Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors and snRNAs. Mol. Cell. Biol., 22, 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K. and Niwa,O. (2000) mRNAs encoding zinc finger protein isoforms are expressed by alternative splicing of an in-frame intron in fission yeast. DNA Res., 7, 27–30. [DOI] [PubMed] [Google Scholar]
- Potashkin J., Li,R. and Frendewey,D. (1989) Pre-mRNA splicing mutants of Schizosaccharomyces pombe. EMBO J., 8, 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin J., Naik,K. and Wentz-Hunter,K. (1993) U2AF homolog required for splicing in vivo. Science, 262, 573–575. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary,F., Rigaut,G., Rutz,B., Bouveret,E., Bragado-Nilsson,E., Wilm,M. and Séraphin,B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods, 24, 218–229. [DOI] [PubMed] [Google Scholar]
- Rain J.C., Rafi,Z., Rhani,Z., Legrain,P. and Krämer,A. (1998) Conservation of functional domains involved in RNA binding and protein–protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA, 4, 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romfo C.M. and Wise,J.A. (1997) Both the polypyrimidine tract and the 3′ splice site function prior to the first step of splicing in fission yeast. Nucleic Acids Res., 25, 4658–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B. and Séraphin,B. (1999) Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA, 5, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S.W., Ryan,D.E., Ge,H.Y., Moore,R.E., Young,M.K., Lee,T.D. and Abelson,J. (2002) Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell, 9, 31–44. [DOI] [PubMed] [Google Scholar]
- Umen J.G. and Guthrie,C. (1995) The second catalytic step of pre-mRNA splicing. RNA, 1, 869–885. [PMC free article] [PubMed] [Google Scholar]
- Wang X., Bruderer,S., Rafi,Z., Xue,J., Milburn,P.J., Krämer,A. and Robinson,P.J. (1999) Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J., 18, 4549–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz-Hunter K. and Potashkin,J. (1996) The small subunit of the splicing factor U2AF is conserved in fission yeast. Nucleic Acids Res., 24, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Schneider,C., MacMillan,A.M., Katopodis,N.F., Neubauer,G., Wilm,M., Lührmann,R. and Query,C.C. (2001) A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J., 20, 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- Wu S., Romfo,C.M., Nilsen,T.W. and Green,M.R. (1999) Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature, 402, 832–835. [DOI] [PubMed] [Google Scholar]
- Zorio D.A. and Blumenthal,T. (1999) Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature, 402, 835–838. [DOI] [PubMed] [Google Scholar]