Abstract
The downstream effectors of the Drosophila sex determination cascade are mostly unknown and thought to mediate all aspects of sexual differentiation, physiology and behavior. Here, we employed serial analysis of gene expression (SAGE) to identify male and female effectors expressed in the head, and report 46 sex-biased genes (>4-fold/P < 0.01). We characterized four novel, male- or female-specific genes and found that all are expressed mainly in the fat cells in the head. Tsx (turn on sex-specificity), sxe1 and sxe2 (sex-specific enzyme 1/2) are expressed in males, but not females, and are dependent on the known sex determination pathway, specifically transformer (tra) and its downstream target doublesex (dsx). Female-specific expression of the fourth gene, fit (female-specific independent of transformer), is not controlled by tra and dsx, suggesting an alternative pathway for the regulation of some effector genes. Our results indicate that fat cells in the head express sex-specific effectors, thereby generating distinct physiological conditions in the male and female head. We suggest that these differences have consequences on the male and female brain by modulating sex-specific neuronal processes.
Keywords: Drosophila/fat cells/mating behavior/SAGE/sex-specific genes
Introduction
In Drosophila, sex determination is mediated by the ratio of X chromosomes to sets of autosomes (X:A ratio). A ratio of 1 (2X:2A) initiates a regulatory cascade involving the splicing regulators Sex-lethal (Sxl), transformer (tra) and transformer2 (tra2), and ultimately leads to the generation of sex-specific transcription factors encoded by doublesex (dsx) and fruitless (fru; reviewed by McKeown, 1994; Cline and Meyer, 1996). Sxl is transcriptionally active in both sexes, but only the female embryos produce functional SXL protein by a mechanism of alternative splicing which is maintained by an autoregulatory feedback mechanism (Bell et al., 1991). SXL also regulates female-specific splicing of tra pre-mRNA to allow production of functional TRA protein (Sosnowski et al., 1989; Inoue et al., 1990). TRA and the constitutively expressed TRA2 (Amrein et al., 1988; Goralski et al., 1989) positively regulate dsx pre-mRNA splicing to generate a female-specific transcript encoding DSXf protein (Burtis and Baker, 1989). In males, which have an X:A ratio of 0.5 (1X:2A), the absence of SXL, and consequently of TRA, results in constitutive splicing of dsx pre-mRNA to produce a transcript that encodes a male-specific protein, DSXm. The absence of TRA in males also results in the alternative splicing of fru pre-mRNA, leading to expression of a male-specific FRUm protein, which is essential for female-directed male courtship behavior (Ryner et al., 1996). The function of TRA and TRA2 for female differentiation is dramatically revealed in XX flies homozygous mutant for either gene; these flies are sexually transformed and develop as so-called ψ (pseudo)-males, which are indistinguishable from normal XY males in morphology and behavior, but are sterile (Baker and Ridge, 1980; Belote and Baker, 1987). Three other regulatory genes, intersex (ix), hermaphrodite (her) and dissatisfaction (dsf), are also involved in the regulation of sex determination. However, they are likely to mediate their male and female functions through other sex-specific factors, since at least HER and DSF appear to be expressed equally in both sexes (Chase and Baker, 1995; Finley et al., 1998; Li and Baker, 1998).
Whereas the regulatory splicing pathway of Sxl– tra–dsx/fru is well understood, much less is known about the downstream targets (effectors). The only sex-specific effectors identified to date are the female-specifically expressed yolk protein genes yp1, yp2 and yp3, at least two of which (yp1 and yp2) are directly regulated by the DSX proteins (Burtis et al., 1991; Coschigano and Wensink, 1993; An and Wensink, 1995a,b). DSXf and DSXm share identical DNA-binding domains, but differ in their C-termini, which function as protein–protein interaction domains (Coschigano and Wensink, 1993; Erdman and Burtis, 1993). This observation, together with in vivo analysis of yp1 promoter–lacZ reporter constructs (Abrahamsen et al., 1993), has led to a model in which DSXf acts as a transcriptional activator and DSXm as a repressor of the yp1 and yp2 genes. No other downstream target genes of DSX or FRU are known.
To understand the molecular, sex-specific processes that operate during development and control adult behavior, genome-wide expression analysis will be necessary. Here we report a first step towards this goal. To identify effector genes with sex-specific or sex-biased expression in the adult, we carried out serial analysis of gene expression (SAGE) and generated a transcript inventory of the Drosophila male and female head. Analysis of >7000 unique tags (i.e. transcripts) revealed the presence of 46 sex-biased candidate genes. Northern blot analysis of eight novel candidates confirmed sex specificity/bias for all of them. Interestingly, four of the genes were expressed in either males or females only, most notably in the fat cells of the head. Three of these genes are virtually male specific and are dependent on the sex determination cascade, particularly on dsx. Surprisingly, expression of the fourth gene, which is highly female biased, is dependent on Sxl, but independent of tra, tra2 and dsx, suggesting the existence of an alternative pathway important for sex-specific gene expression. We propose that the fat cells in the head, and perhaps other non-neuronal cells, play an important role in establishing sex-specific differences in physiology, which might modulate brain function and ultimately influence male and female behaviors, including courtship and mating.
Results
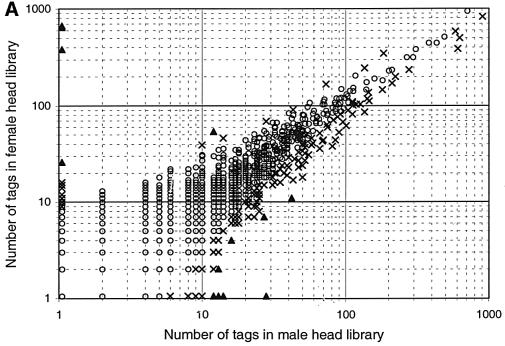
We sought to identify genes that are expressed at significantly different levels in adult males and females. We reasoned that SAGE (Velculescu et al., 1995) could be used efficiently to investigate differences in gene expression in the male and female head and, thus, should allow us to identify sex-specific genes that might be involved in behavior. We decided to use RNA from heads, as opposed to whole flies or dissected brains, for two main reasons. First, it eliminates a large number of genes expressed in the testes and ovaries. Secondly, even though the brain plays the central role in behavior, it may not necessarily be the major source of male- and female-specific genes, but rather be modulated by ‘instructive signals’ such as peptides and/or hormones synthesized in other tissues. Therefore, we generated SAGE libraries from RNA of wild-type male and female heads and compiled a data set of >110 000 individual transcripts (67 000 female tags and 49 000 male tags) representing ∼7000 genes (for details, see Materials and methods). The vast majority of tags are present at similar levels in both sexes (Figure 1A).
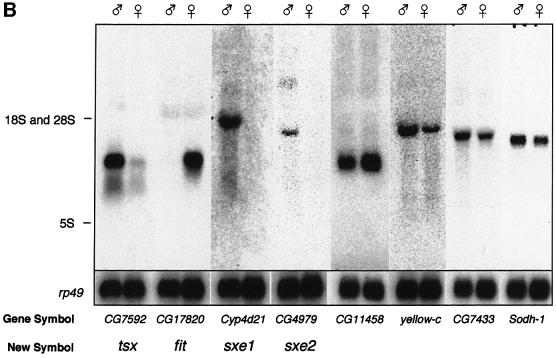
Fig. 1. Identification of sex-biased SAGE tags and expression of candidate genes. (A) Distribution of SAGE tags from male and female head libraries. The number of times each unique SAGE tag was observed was plotted on a logarithmic scale. Tags not found in a library were converted to a value of 1. Male tag numbers are normalized to female tag numbers (coefficient is 1.4). Triangles indicate tags with P < 0.001; crosses indicate tags with 0.001 ≤ P < 0.01; circles indicate tags with P ≥ 0.01. (B) Northern blot analysis of eight novel, sex-biased genes in male and female heads. Total RNA was isolated from dissected male and female heads. Blots were hybridized with probes to all eight novel genes, represented by SAGE tags that are at least four times more abundant in one library versus the other (≥4-fold; P < 0.001). 18S/28S and 5S RNA markers are indicated on the left and were derived from ethidium bromide-stained gels. All blots contained 5 µg of male and female RNA per lane, respectively, and were re-hybridized with a probe against the rp49 gene to provide an RNA loading control.
A small set of sex-biased transcripts identified by SAGE
We focused our studies on genes that were expressed at least 4-fold more abundantly in one sex versus the other. Applying moderate (P < 0.01) or stringent (P < 0.001) probability criteria, we identified 46 or 13 genes, respectively (Figure 1A; Table I; see Materials and methods), including all five previously known sex-specific genes, the three yolk protein (yp) and both roX genes. roX1 and roX2 were found exclusively in males and were represented by 13 and 12 tags, respectively. (Note that throughout this paper, the terms ‘sex-specific’, ‘male-specific’ and ‘female-specific’ refer to expression differences of at least 10-fold between the sexes as analyzed by northern blot analysis and phosphoimaging.) Tags for the yp genes were found several hundred times in the female library, but only once or never in the male library (Table I). Sex-biased expression of the eight novel candidates was confirmed by northern blot analysis on male and female head RNA (Figure 1B). We focused our investigations on the four novel, sex-specific genes CG7592, CG17820, cyp4d21 and CG4979. The entire data set from our SAGE can be accessed at http://rush.genetics.duke.edu/amrein/sage/
Table I. List of sex-biased tags in the adult fly head.
| Tag sequence | Male | Female | P chance | Gene symbol | Function/note | CG number | Location |
|---|---|---|---|---|---|---|---|
| GCCACGCCCC | 0 | 667 | < 0.0001 | Yp3 | Yolk protein | 11129 | 12B8 |
| CGAGCGAACC | 0 | 644 | < 0.0001 | Yp2 | Yolk protein | 2979 | 9B1 |
| GGCATCGATA | 1 | 381 | < 0.0001 | Yp1 | Yolk protein | 2985 | 9B1 |
| GATCCAGCCA | 28 | 1 | < 0.0001 | CG7592 (tsx)a | Ligand binding or carrier | 7592 | 99B8 |
| TCATTCATTC | 0 | 26 | < 0.0001 | CG17820 (fit)a | Unknown | 17820 | 93F8–9 |
| CCAGGAGCAA | 14 | 0 | < 0.0001 | Cyp4d21 (sxe1)a | Cytochrome P450 | 6730 | 28B4 |
| TTTTGACAGA | 13 | 0 | < 0.0001 | roX1 | Non-coding RNA, dosage compensation | – | 3F |
| GTTGACGCGC | 12 | 0 | < 0.0001 | roX2 | Non-coding RNA, dosage compensation | – | 10B17 |
| GGCGGTGAAC | 13 | 2 | 0.0005 | CG4979 (sxe2)a | Phosphatidylserine-specific phospholipase A1 | 4979 | 89B6 |
| GAGGCGGCGG | 12 | 54 | < 0.0001 | CG11458a | Unknown | 11458 | 77F4 |
| CTGTGTTTAT | 16 | 4 | 0.0008 | yellow-ca | yellow family | 4182 | 35B8 |
| CCAACATCGT | 27 | 7 | < 0.0001 | CG7433a | 4-aminobutyrate aminotransferase | 7433 | 76D8–E1 |
| ATCCACGTCC | 42 | 11 | < 0.0001 | Sodh-1a | l-iditol 2-dehydrogenase | 1982 | 84B2 |
| CACTTCGTAG | 8 | 0 | 0.001 | CG15219b | Testis-specific transcript | 15219 | 40B1 |
| GCCTATTCGT | 8 | 0 | 0.001 | AI946554b | Testis-specific transcript | – | 60E12 |
| TAATAACAAC | 10 | 1 | 0.001 | CG10135 | Calcium binding, calcium sensing | 10135 | 87D1 |
| TGCGATTCCA | 12 | 2 | 0.001 | CG5288 | Galactose kinase | 5288 | 66E6 |
| TTAAAATTAA | 8 | 0 | 0.001 | CG15425 | Unknown | 15425 | 24E1 |
| TTTAGTTCTT | 10 | 1 | 0.001 | RH08983.3′ | – | – | 33E9 |
| TACAGCGTAA | 13 | 3 | 0.002 | Pebb | Testis-specific transcript | 2665 | 60F5 |
| TACTTATGTC | 9 | 1 | 0.002 | CG3488 | Esterase/lipase/thioesterase family | 3488 | 23D4 |
| ACGTCAGAAC | 12 | 3 | 0.003 | BcDNA:LD06023 | Unknown | – | 102F4 |
| GTTCTTTTTC | 10 | 2 | 0.005 | CG2736 | Scavenger receptor, defense response | 2736 | 60F4 |
| TGAGGATGAA | 10 | 2 | 0.005 | Actn | Calcium binding, cytoskeletal anchor protein | 4376 | 2C4–7 |
| TTTGAACATT | 10 | 2 | 0.005 | AE003804/AE003542 | – | – | 53E10–54B8/68F1–69A3 |
| AAAACAACAG | 8 | 1 | 0.006 | AE003803 | – | – | 54B8-C11 |
| ACGATGTTGA | 6 | 0 | 0.006 | CG1764 | Dimethylargininase | 1764 | 11E9 |
| ATGATTACCA | 6 | 0 | 0.006 | CG5344 | RAB GTPase activator | 5344 | 86E15 |
| ATGGTATGCC | 8 | 1 | 0.006 | CG17661 | Ligand binding or carrier, calcium binding | 17661 | Unknown |
| CGGATTGCTG | 6 | 0 | 0.006 | AE003677/AE003736 | – | – | 84E4-F2/93F3–F12 |
| CTGAAACATC | 8 | 1 | 0.006 | GM06073.5′ | – | – | 67E1–3 |
| GACCCTAACC | 6 | 0 | 0.006 | Mio | Transcription factor | 18362 | 39C1–3 |
| GCGGCAGTGG | 8 | 1 | 0.006 | CG11236 | d-aspartate oxidase | 11236 | 27B1 |
| TATTGAATAT | 8 | 1 | 0.006 | DIP1 | dsRNA binding, chromatin binding | 17686 | 20A |
| TTAGAATGGT | 8 | 1 | 0.006 | HL04936.5′ | – | – | 82C3 |
| TTTGGAATAA | 6 | 0 | 0.006 | CG2131 | Procollagen N-endopeptidase, | 2131 | 39F1–3 |
| TTTTCCAATT | 8 | 1 | 0.006 | CG12012 | Unknown | 12012 | 63D2 |
| ACCAAGGCAC | 0 | 9 | 0.007 | CG7461 | Very long-chain acyl-CoA dehydrogenase | 7461 | 56D3 |
| CGAGCAGAAA | 0 | 9 | 0.007 | AE003649 | – | – | 35D7–F1 |
| CTGTCAACCT | 0 | 9 | 0.007 | fau | Anoxia up-regulated protein | 6544 | 86C4 |
| ACTTTTATGT | 1 | 12 | 0.008 | RH19272.5′ | – | – | 24A5 |
| ACAATATACG | 9 | 2 | 0.009 | CG30069 | Unknown | 30069 | 50E8 |
| GATCGCTACC | 9 | 2 | 0.009 | msta | Unknown | 18033 | 2E2 |
| TATATCTATA | 9 | 2 | 0.009 | cmp44E | Plasma membrane protein | 8739 | 44E4 |
| TATTACTGCC | 9 | 2 | 0.009 | CG9645 | Enteropeptidase | 9645 | 88A12 |
| TTCTTGAATG | 9 | 2 | 0.009 | CG11572 | Unknown | 11572 | 102A7–8 |
Male tag numbers are normalized to female tag numbers (coefficient is 1.4). Tags are listed as P < 0.01, and fold difference ≥4. P was calculated using Monte Carlo Simulation in the SAGE 2000 Software v4.12. Gene symbol, predicted function, CG number and location are retrieved from FlyBase at http://flybase.bio.indiana.edu. If a tag matches multiple ESTs from the same gene, only one EST is listed. Due to biological (alternative splicing, multiple sites of polyadenylation) and technical (incomplete digestion of cDNAs with NlaIII) reasons, several tags may be annotated to a single gene. In these cases, only the most abundant tag is listed in this table. Genes known previously to be expressed in a sex-biased manner are in bold.
aNorthern blot analyses for these tags are shown in Figure 2B.
bBy Northern blot analysis, these tags were found to be expressed only in the male abdomen, but not the head (data not shown).
The tags representing the male-specific genes encode members of three distinct protein families (Table I): an odorant-/pheromone-binding protein (opbp; CG7592), a cytochrome P450 enzyme (CYPs; cyp4d21) and a phospholipase (CG4979). The female-specific tag identified a gene of unknown biochemical function (CG17820). We shall refer to these genes as turn-on sex-specificity (tsx), sex-specific enzyme 1 and 2 (sxe1 and sxe2) and female-specific independent of transformer (fit), respectively.
Tissue and developmental expression of the sex-specific genes
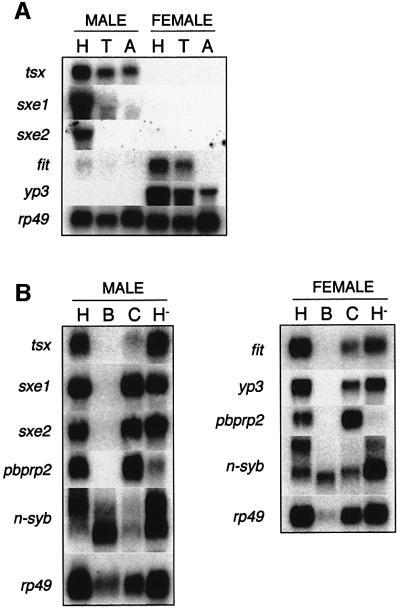
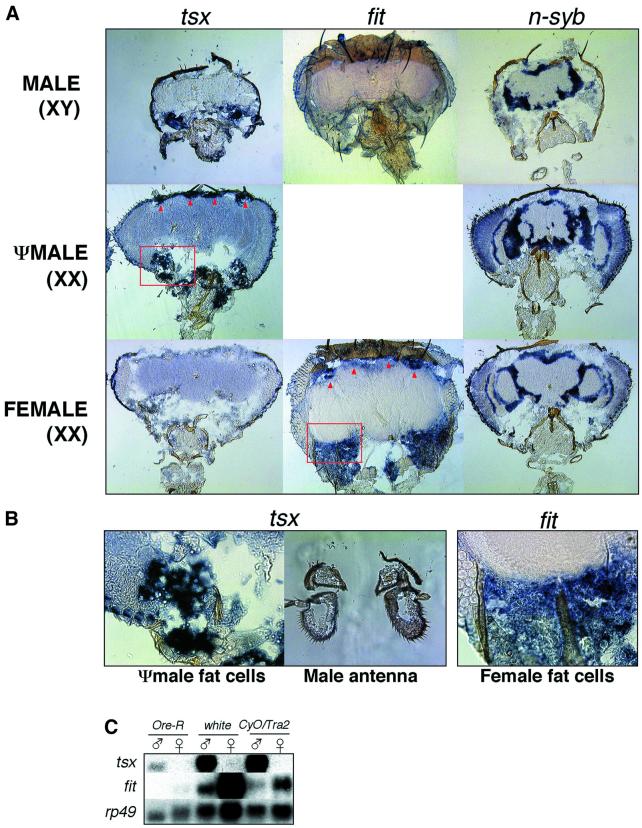
We investigated the tissue-specific expression of tsx, sxe1, sxe2 and fit using northern blot hybridization with RNA isolated from the head, thorax and abdomen of male and female adult flies (Figure 2A). Northern blot analysis confirmed our results from the SAGE screen; all four genes were expressed in the head in a sex-specific manner. In addition, tsx was expressed in all major body segments of the adult, whereas sxe1 and fit are expressed at much lower levels in the thorax than the head and not at all in the abdomen. Finally, sxe2 is expressed exclusively in the male head. Initially, in situ hybridization experiments for all four genes did not show defined and localized signals within the head, whereas control experiments with the male-specific roX2 gene (Amrein and Axel, 1997), the neuron-specific synaptobrevin gene (n-syb; Sudhof et al., 1989) and the fat body-specific yp3 gene (Garabedian et al., 1987) revealed the expected sex and tissue specificity (Figure 3, right panel; data not shown). This observation suggested that the expression levels of the novel genes are too low for detection by this technique. However, during the course of this study, we identified two strains (w1118 and Bs;CyO/cn tra2 bw) in which tsx and fit are expressed at significantly higher levels than in wild-type Ore-R flies (Figure 3C). We therefore performed in situ hybridization experiments on tissue sections of fly heads from these strains for both tsx and fit. Hybridization signals of tsx transcripts were localized to the (deep) fat cells in the ventral and dorsal regions of the head but not the brain, the eyes, the antenna, the maxillary palps or the labellum of CyO males or tra2 ψ-males (Figure 3A, left panel, and B). As expected, no hybridization signals were observed in any parts of female heads (Figure 3A, bottom of left panel). For fit, we used heads from males and females of the w1118 strain (Figure 3A, middle panel). Hybridization signals were only found in females, but not males, and were also restricted to deep fat cells in both the dorsal and ventral part of the head. Again, we noticed no fit-positive cells in the brain, the visual or the chemosensory systems. Thus, these data showed that tsx and fit are expressed mainly in the fat cells of the male or female head, respectively.
Fig. 2. Expression of tsx, sxe1, sxe2 and fit is restricted mainly to the head. (A) Northern blot analysis was carried out with RNA isolated from head (H), thorax (T) and abdomen (A) to determine sex- and tissue-specific expression. Approximately 5 µg of total RNA were loaded in each lane. Identical northern blots were hybridized with probes for tsx, sxe1, sxe2 and fit. An rp49 probe was used as an RNA loading control. (B) Northern blot analysis of various head tissues from males and females: brains without the optic lobes (B) were dissected from the remaining parts of the head carcass (C), which consisted mainly of the cuticle (containing no RNA) and the fat cells, as well as the visual, olfactory (antenna and maxillary palps) and taste sensory systems (labellum). Note that deep fat cells in the head were partly lost in the dissection procedure and are therefore reduced in either B or C. The H– lane contained RNA isolated from heads in which all chemosensory organs (antenna, maxillary palps and labellum) were removed. Identical northern blots were hybridized with probes for tsx, sxe1, sxe2 and control genes (pbprp2, n-syb and rp49) or for fit and control genes (yp3, pbprp2, n-syb and rp49), respectively.
Fig. 3. tsx and fit are expressed in the fat cells of the head. (A) In situ hybridization of digoxigenin-labeled antisense RNA derived from a tsx and fit cDNA clone reveals specific signals in large fat cells in the ventral and dorsal region (red arrows) of the head capsule of CyO males, tra2 ψ-males (tsx) and w1118 females (fit), respectively. Note the absence of signal in the brain and the visual system. The female (tsx) and the male (fit) head show no hybridization signals. Sections of males, ψ-males and females were also hybridized with antisense RNA probes derived from n-syb (right panel) to indicate the location of neuron cell bodies in the brain. Strains used were BsY; CyO/tra2 (tsx; n-syb) and w1118 (fit). (B) Enlargement of the ventral region of the head containing a large number of deep fat cells located below the brain (first and third frame; see red rectangles in A). Also note the absence of tsx hybridization signals in the antenna (middle frame). (C) Northern blot of males and females of the three strains used for the in situ hybridization experiment. Note the several fold higher levels of fit and tsx RNA in both w1118 and CyO/tra2 strains when compared with Ore-R. Also note that low fit expression can be observed in males of these strains.
To examine tissue specificity further, we performed northern blot analysis of RNA from dissected heads (Figure 2B). The ‘purity’ of the dissected tissue was examined by northern hybridization experiments with probes for tissue-specific genes, which demonstrated that the desired tissue was either successfully enriched for or removed. For example, brain-specific n-syb transcripts (Sudhof et al., 1989) were highly enriched in dissected brain (B) when compared with carcass (C), whereas fat body-specific yp3 expression (Garabedian et al., 1987) was found abundantly in the carcasses and heads lacking chemosensory organs (H–), but not the brain. Similarly, pbprp2 expression, which is restricted to the chemosensory system (Pikielny et al., 1994), was almost entirely lost in the H– fraction (Figure 2B, lower panels). When these northern blots were hybridized with probes for tsx, sxe1, sxe2 and fit, we found that none was expressed in the brain (Figure 2B, top panels, lane B). All genes were expressed in the C and H– preparations, suggesting that the head carcass, but not the chemosensory system, is the main source of transcripts for all four genes (Figure 2B, top panels, lanes C and H–). These findings are consistent with the in situ hybridization experiments that detected tsx and fit transcripts in the deep fat cells in the head (Figure 3A).
To investigate whether tsx, sxe1, sxe2 and fit are expressed exclusively in adults, we hybridized probes for each of them to RNA from various embryonic, larval and pupal stages. The only gene expressed prior to the adult stage was tsx, which was first observed in the third instar larvae, but then gradually decreases during pupal stages (data not shown). Interestingly, expression in larvae is non-sex specific and higher than in adult males.
The male-specifically expressed genes are dependent on tra, tra2 and dsx function
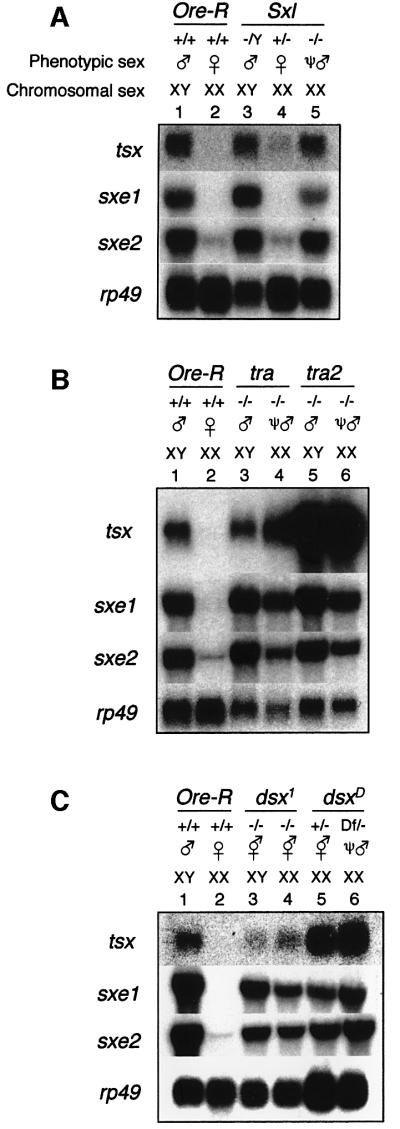
To explore the regulation of the four sex-specific genes, we carried out a series of northern blot analyses using flies with mutations in various sex determination genes. For example, XX animals homozygous mutant for tra or tra2 are sexually transformed into ψ-males, which are indistinguishable from wild-type, tra or tra2 XY males in their sexual morphology and behavior. Thus, male-specific genes that are downstream of Sxl and tra/tra2 in the sex determination cascade should be expressed in ψ-males and, conversely, female-specific genes should be repressed. We first analyzed expression of tsx, sxe1 and sxe2 in heterozygous males and females of these various strains, which confirmed that these genes were only expressed in males but not females (Figure 3C; data not shown). We then investigated the tsx, sxe1 and sxe2 expression in XX flies homozygous mutant for Sxl, tra and tra2, and found that all were expressed in these ψ-males (Figure 4A and B). Expression levels for tsx were much higher in tra2 ψ-males and X/B Y;CyO/tra2 males than tra ψ-males or wild-type males, suggesting that increased expression is caused by the genomic background in the CyO/tra2 strain, but not the tra2 mutation itself. In any case, there was no significant difference in expression levels of all three genes between XY males and XX ψ-males within a given strain (Figure 4A, compare lanes 3 and 5; B, compare lanes 3 and 4, and 5 and 6), indicating that tsx, sxe1 and sxe2 are regulated by Sxl, tra and tra2. These results are also consistent with the RNA in situ hybridization experiments, which showed tsx expression in tra2 ψ-males (Figure 3A).
Fig. 4. Expression of tsx, sxe1 and sxe2 is dependent on Sxl, tra and dsx. Blots were hybridized with RNA from manually dissected heads. Approximately 5 µg of RNA were loaded per lane. All blots were re-hybridized with an rp49 probe to monitor RNA loading. Head RNA from Ore-R males and females was used for comparison in each experiment (lanes 1 and 2). (A) Expression of tsx, sxe1 and sxe2 in Sxl mutant flies: Sxl ψ-males express all three male-specific genes, as indicated by strong hybridization signals in lane 5. No expression was observed in heterozygous Sxl females (lane 4). Genotypes were: y cm Sxlf7M1ct v/Y males (lane 3); w SxlM1f3sn/Binsinscy females (lane 4); and y cm Sxlf7M1ct v/w SxlM1,f3sn ψ-males (lane 5). (B) Expression of tsx, sxe1 and sxe2 in tra and tra2 mutant flies: both tra and tra2 ψ-males strongly express all three male-specific genes, as indicated by the strong hybridization signals in lanes 4 and 6, respectively. Genotypes were: X/BsY; th st tra cp ri pp/tra e ca males (lane 3); X/X; th st tra cp ri pp/tra e ca ψ-males (lane 4); BsY; cn tra2 bw/cn tra2 bw males (lane 5); and X/X; cn tra2 bw/ cn tra2 bw ψ-males (lane 6). (C) Expression of tsx, sxe1 and sxe2 in various dsx mutant flies: XX flies with various dsx mutations show a low to intermediate level of expression of tsx, sxe1 and sxe2. Note the dramatic increase of tsx expression in flies expressing DSXm (lanes 5 and 6) when compared with flies with no functional DSX protein, suggesting an activating role for dsx on tsx. Genotypes were: X/BsY; dsx1/dsx1 intersexes (lane 3); X/X; dsx1/dsx1 intersexes (lane 4); X/X; TM6/dsxD intersexes; and X/X; Df[dsx]/dsxD ψ-males; X/X; TM6/dsxD intersexes (lane 5); and X/X; Df[dsx]/dsxD ψ-males (lane 6).
The dominant Y-linked eye mutation Barstone (Bs) is present in the tra and tra2 strains to distinguish sexually transformed XX ψ-males from XY males. Bs reduces the size of the eye and the visual brain centers by ∼90% (Childress, 1973). As shown in Figure 4B, males with wild-type and Bs eyes have similar transcript levels of all three male-specific genes (compare lanes 1 and 3), indicating that the visual system cannot be the major site of expression for tsx, sxe1 and sxe2. This observation is consistent with previous northern blots and in situ hybridization experiments (Figures 2B, 3A and B).
To test whether tsx, sxe1 and sxe2 were indeed dependent on dsx, we investigated their expression in flies with different dsx mutations. dsx1 is a null mutation, generating no functional DSX protein (Nöthiger et al., 1987; Burtis, 1993). Thus, dsx1 homozygous mutant flies are intersexual in phenotype, regardless of their karyotype. dsxD is an insertion of a transposable DNA element in the fourth, female-specific exon, leading to TRA-independent, male-specific dsx splicing (Nagoshi and Baker, 1990). Therefore, in hemizygous X/X; dsxD/Df[dsx] flies, only DSXm but no DSXf is produced, whereas in heterozygous X/X; dsxD/dsx+ flies, both forms of DSX are generated (DSXf from the dsx+ allele and DSXm from the dsxD allele). Northern blot analysis of RNA isolated from heads of these various dsx mutant flies indicated that all three genes are controlled by dsx (Figure 4C). First, tsx, sxe1 and sxe2 were expressed, albeit at reduced levels, in dsx1 homozygous XX and XY intersexes, suggesting that the absence of functional DSX leads to basal expression of these genes (Figure 4C, lanes 3 and 4). Secondly, X/X; dsxD/Df ψ-males (producing only DSXm) and X/X; dsxD/dsx+ intersexes (producing both DSXm and DSXf) also express all three genes (Figure 4C, lanes 5 and 6). Thus, our data demonstrate that tsx, sxe1 and sxe2 are downstream effectors of the sex determination cascade and may be direct targets of dsx.
fit is dependent on Sxl, but not on tra and tra2
As for the male-specific genes, we investigated the regulation of the female-specific gene fit using flies mutant for the sex determination genes. Genes normally expressed only in females are expected to be repressed in any ψ-male, if regulated by the sex determination cascade. We found that fit expression was indeed abolished in Sxl ψ-males (Figure 5A, lane 5). Sxl/Y males showed no fit expression, just like wild-type males, because males do not produce any SXL protein (Figure 5A, lane 3). This result suggested that fit expression is dependent on Sxl function, just as observed for the three male-specific genes.
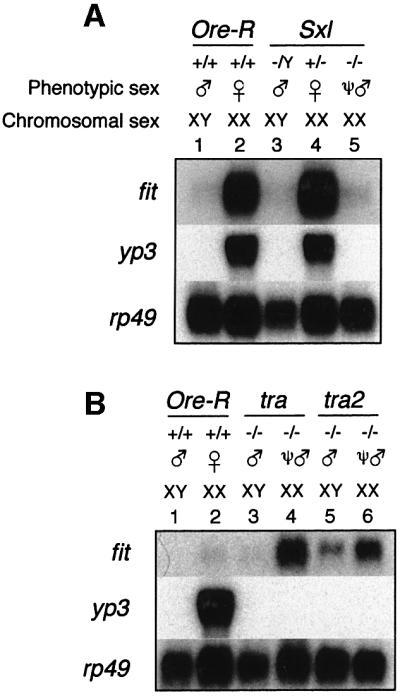
Fig. 5. fit is not regulated by tra and tra2. Blots were hybridized with RNA from manually dissected heads. Approximately 5 µg of RNA were loaded per lane. All blots were re-hybridized with an rp49 probe to monitor RNA loading. RNA from Ore-R males and females was used for comparison in each experiment (lanes 1 and 2). (A) Expression of fit in Sxl mutant flies. Sxl ψ-males do not express the fit gene, as indicated by complete lack of signal in lane 5. Note that yp3 is also dependent on SLX. Genotypes were: y cm Sxlf7,M1ct v/Y males (lane 3); w SxlM1,f3sn /Binsinscy females (lane 4); and y cm Sxlf7,M1ct v/w SxlM1,f3sn ψ-males (lane 5). (B) Expression of fit in tra and tra2 mutant flies. Both tra and tra2 ψ-males express significant levels of fit RNA, similar to that observed in heterozygous females of these strains (see Figure 3C), as indicated by clear hybridization signals in lanes 4 and 6, respectively. A very low level of fit expression was observed in both homozygous XY males (lanes 3 and 5) and heterozygous males (Figure 3C, lane 5). Genotypes were X/BsY; th st tra cp ri pp/ tra e ca males (lane 3); X/X; th st tra cp ri pp/ tra e ca ψ-males (lane 4); BsY; cn tra2 bw/cn tra2 bw males (lane 5); and X/X; cn tra2 bw/ cn tra2 bw ψ-males (lane 6).
We then asked whether fit is also dependent on regulators downstream of Sxl. Northern blot analysis of homozygous mutant tra and tra2 flies revealed that ψ-males showed high levels of fit expression that typically are observed in heterozygous females of these strains (Figures 5B, lanes 4 and 6, and 3C, lane 6; data not shown). Similarly, homozygous males showed a very low level of fit expression, similar to that observed for heterozygous XY males of these strains (Figures 5B, lanes 3 and 5, and 3C, lane 5; data not shown). Taken together, these data indicate that fit expression is independent of tra, tra2 and, hence, the downstream regulators dsx and fru. In addition, analysis of RNA from different strains revealed rather large variability in expression levels of fit in females. In strains with elevated levels of female fit expression, low levels were observed in males (Figures 3C and 5B).
Ectopic expression of TSX in females reduces receptivity
To investigate a potential role for TSX in behavior, we constructed GAL4-dependent UAS reporters with epitope-tagged (V5; see Materials and methods) cDNAs for these genes and generated transgenic flies. The TSX protein was stably expressed under the control of a ubiquitously expressed GAL4 driver (actin:GAL4; data not shown). We tested whether ectopic expression of TSX caused any effects on mating behavior and measured mating performance of males and females by determining latency time in single mating set-ups (for details, see Materials and methods). No obvious effects on mating performance were observed in males with ubiquitous TSX expression, but we observed that TSX-expressing females were negatively affected (Table II). In both lines tested, significantly fewer TSX-expressing females mated during the observation period (15 min) when compared with control females. The increase in non-maters is probably caused by an elevated escape response, as TSX-expressing females often run away from an approaching male. Taken together, our experiments suggest that ectopic TSX expression interferes with normal female mating response.
Table II. Reduced mating performance of females ectopically expressing TSX protein.
| Genotype | n | Fraction of females that fail to copulate (%) | Latencies (min) |
|---|---|---|---|
| Act:GAL4/UAS:tsx_V5.4 | 60 | 41.7 | 5.9 ± 0.7 |
| UAS:tsx_V5.4/+ | 64 | 19.4 | 5.6 ± 0.6 |
| Act:GAL4/UAS:tsx_V5.5 | 60 | 26.7 | 5.7 ± 0.6 |
| UAS:tsx_V5.5 /+ | 55 | 10.9 | 5.8 ± 0.5 |
| Act:GAL4/+ | 63 | 15.9 | 5.8 ± 0.6 |
| White | 63 | 12.7 | 5.5 ± 0.5 |
| Wild type | 63 | 15.9 | 3.8 ± 0.6 |
Individual virgin females (3–6 days old) of the genotype indicated were placed with a single Ore-R male (3–8 days old) in a mating chamber (see Materials and methods). Each pair was observed for 15 min. The time to copulation (latency) was recorded and averaged. The mean latencies and SEMs were calculated only for pairs that mated within the 15 min observation period. Note that the fraction of females that fail to copulate within the 15 min observation period is significantly larger in the experimental females (genotypes in bold) than in the various controls.
Discussion
Genetic investigations have led to an understanding of the molecular splicing cascade that culminates in the expression of sex-specific transcription factors encoded by dsx and fru. We hypothesized that a detailed analysis of gene expression differences between adult males and females could lead to an understanding of the critical sex-specific physiological events downstream of these regulators. The characterization of the first few candidates identified by SAGE has established the value of this approach.
fit is regulated by a tra-independent pathway
An unexpected result was the identification of a Sxl-dependent, but tra- and tra2-independent pathway that mediates female-specific fit expression. It is generally believed that most aspects of sexual differentiation and behavior are mediated by tra and tra2 (Cline and Meyer, 1996; Baker et al., 2001). However, this assumption has not been tested rigorously for female mating behaviors.
How is fit expression controlled in females? First, fit could be regulated directly by SXL, which seems unlikely, because SXL functions post-transcriptionally (Bell et al., 1988, 1991; Salz et al., 1989; Valcarcel et al., 1993; Bashaw and Baker, 1995). The known RNA targets of SXL, Sxl, tra and msl2 pre-mRNAs, are stably expressed in the absence of SXL in males (McKeown et al., 1987; Bell et al., 1988; Bashaw and Baker, 1995; Kelley et al., 1995; Zhou et al., 1995). Therefore, if fit RNA were a target of SXL, we would expect to find stable fit RNA precursor in males as well as conserved SXL-binding sites within the fit pre-mRNA. Neither is the case for fit. A second possibility is that fit is controlled through a Sxl-dependent, unknown regulator. We identified several strains in which females expressed higher levels of fit RNA than wild-type Ore-R females (Figure 3C). Variations in fit expression are probably caused by differences in genetic backgrounds, which could include both cis-regulatory elements in the fit promoter and trans-acting factors. Identifying these elements might provide an opportunity to elucidate the nature of the tra-independent pathway that regulates fit expression. Naturally, exogenous factors, such as temperature, diet, population density, etc., may also influence fit expression.
Sex-specific functions may be mediated by the fat cells in the head
Another intriguing result from our investigations is the observation that none of the four genes is expressed at significant levels in the brain. Instead, our studies show that tsx, sxe1, sxe2 and fit are expressed mainly in the fat cells of the head (Figures 2 and 3), which suggests that these genes create different physiological conditions in the adult male and female head important for sex-specific functions.
We propose that fit, sxe1 and sxe2, which are expressed mainly in the head, may exert their functions on the brain. Similar to the pituitary in the head of mammals, the fat cells could play the role of an endocrine organ. Such a function has been shown for the fat cells of females, where the YPs are synthesized and released into the hemolymph (Butterworth et al., 1991, 1999).
Based on the predicted protein sequences, we can envisage sex-specific roles for sxe1 and tsx. tsx encodes a member of the opbp gene family. Oderant/pheromone binding proteins (OPBPs) are expressed generally in support cells of chemosensory sensilla and secreted into the extracellular lymph space, where they interact with odor and taste ligands to increase their solubility, protect them from degradation or remove them from the lymph space (Pikielny et al., 1994; Galindo and Smith, 2001). We suggest that TSX has been co-opted for a role to interact with and transport small molecules in the head. Upon release from the fat cells, TSX bound to a ligand may reach a target organ, for example the brain, to exert its physiological effects. The reduced mating activity of females ectopically expressing TSX is consistent with such a role (Table II). In addition, a precedent for a protein related to Drosophila OPBPs with a putative function unrelated to chemosensation has been reported in rats (Schoentgen and Jolles, 1995).
Expression of all Drosophila opbp genes, including tsx, has been analyzed previously using the GAL4/UAS system (Galindo and Smith, 2001). These experiments revealed tsx expression in chemosensory organs of both adult sexes. In contrast, our northern blot analysis and RNA in situ hybridization experiments showed that tsx transcripts in the adult are male specific and found mainly in the fat cells of the head, but not the chemosensory organs (Figures 2B and 3). Moreover, we observed abundant non-sex-specific expression in the larvae and pupae. The most likely explanation for these discordant results is the lack of multiple, essential regulatory promoter elements in the GAL4 driver used in the previous study (Galindo and Smith, 2001), which would therefore inaccurately represent tsx expression. However, our studies do not exclude the possibility of low tsx expression in the chemosensory system, which would be consistent with a role in pheromone detection. Such a role is appealing, because Drosophila mating behavior is mediated by taste cues recognized by sensory bristles located in the labellum and the forelegs (Nayak and Singh, 1983). In addition, strain-specific differences might also account for the non-sex-specific expression observed in the tsx:Gal4 driver.
The second gene for which we wish to propose a function, sxe1, encodes a cytochrome P450 protein (CYP), members of which have been studied extensively in mammals and insects. One major role of these enzymes is liver detoxification, whereby toxic, water-insoluble metabolites are rendered sufficiently water soluble to be excreted in the urine. A second, important function for CYPs is their role in steroid hormone metabolism, in both mammals and insects. Of particular interest in this regard is cytochrome P450arom (CYP19), which has been widely implicated in sex-specific functions in vertebrates (Roselli and Resko, 1997; Fisher et al., 1998; Conley and Hinshelwood, 2001).
In insects, CYPs are involved in ecdysone metabolism, specifically in hydroxylation of cholesterol precursors (Mayer et al., 1978; Smith et al., 1979; Grieneisen et al., 1993; Adams et al., 2000). Disembodied (dib), the only studied Cyp gene in Drosophila, is involved in ecdysone metabolism during embryogenesis (Chavez et al., 2000). However, ecdysone triggers most transition phases during development, including larval molts and various differentiation processes in the pupae during metamorphosis (Mitsui and Riddiford, 1978; Riddiford, 1993). A reported function of ecdysone in the adult is its involvement in yp gene regulation in the fat body of females (Shirk et al., 1983).
The expression of sxe1 in the fat cells of the male head suggests the intriguing possibility that small molecules (e.g. steroid hormones) might be synthesized in a sex-specific fashion. Released into the circulatory system, they could reach any organ in the adult male fly, including the brain, and hence mediate sex-specific physiological states that could affect behaviors. One target of such a male-specific hormone might be the neurons in the brain expressing DSF, an orphan nuclear hormone receptor, which controls different male- and female-specific behaviors in adult flies (Finley et al., 1997, 1998).
A role for sex-specific genes in behavior
Expression and regulation of the genes reported here provide a means to generate distinct physiological states in the head of the two sexes. What are the likely consequences of these differences? What aspects of maleness and femaleness are affected by the presence/absence of these proteins and enzymes? It is tempting to speculate that these genes play specific roles in sexual behaviors, simply because of their implicated biochemical function (i.e. tsx and sxe1). However, our knowledge about the extent of behavioral differences between sexes is limited by the way we study fruit flies, namely in confined spaces with abundant food sources and mating choices. For example, very little is known about foraging, energy consumption, food intake, grooming, locomotion, etc. in wild fruit flies in general, and with regard to potential differences between the sexes in particular. Thus, some of the sex-specific genes described here may be involved in aspects of physiology and behavior not related to courtship and mating. In this regard, it is interesting to note that two of the eight genes, sxe1 and sodh-1 (Figure 1; Table I), were also isolated in a microarray analysis for genes under circadian control, and tsx was found in the same study to be under the control of the circadian regulator CLK (McDonald and Rosbash, 2001). Moreover, recent behavioral studies reported sex-specific differences in locomotor activity between males and females (Helfrich-Forster et al., 2001). Thus, these observations suggest that sex-specific physiological differences in adult Drosophila may influence a variety of behaviors not linked to courtship.
Materials and methods
Fly strains and genetics
The following fly strains were used: Ore-R, w1118, w SxlM1,f3sn/Y∧X y f, y cm Sxlf7M1, ct v/Binsinscy, BsY; th st tra cp ri pp/TM3, tra e ca/TM6, BsY; cn tra2 bw/CyO, Df [dsx]/TM3, BsY; dsx1/TM3, BsY; hs→traf Df[3L] st tra pp dsx/dsxD Sb e/TM6. Crosses were performed at 25°C. For the generation of SAGE libraries, we used manually dissected male or female heads of 2- to 4-day-old Ore-R flies.
SAGE
SAGE libraries were generated as described in ‘Serial Analysis of Gene Expression’, version 1.0d with minor modifications. The protocol and analysis software, SAGE 2000 Software Version 4.12, were obtained from The Johns Hopkins University (see also http://www.sagenet.org). Poly(A)+ RNA was isolated from collected heads with RNAzol B (TEL-TEST, Inc.) and oligo(dT) columns (Stratagene). Dynabeads oligo(dT)25 (Dynal) were used as primer to synthesize cDNA from 1.0 µg of poly(A)+ RNA. Double-stranded cDNA was digested with NlaIII (NEB) and collected by a magnet. After linker ligation, DNA fragments were digested with the tagging enzyme, BsmFI (NEB), and ligated. PCR amplification (26 cycles) was carried out with biotinylated primers and 1% of the ligation products as template. After NlaIII (NEB) digestion of the 102 bp PCR products, the released 26 bp fragments were purified by the streptavidin-linked magnetic beads and PAGE. After 1.5 h concatenation, concatemers were cloned into the SphI site of pZero-1 (Invitrogen). Purified plasmids from cultured transformants were screened for long inserts (>800 bp) by XbaI and XhoI digestion. We sequenced 2236 (screened 8640) and 2496 (screened 10 848) plasmids in the male and female head libraries, respectively. Sequencing was performed on the ABI 3700 automated DNA sequencer (Applied Biosystems).
Analysis and annotation of SAGE tags
Sequence data were analyzed using SAGE 2000 Software Version 4.12. To build the database for annotation, we downloaded data sets from The Berkeley Drosophila Genome Project (BDGP; http://www.fruitfly.org) on November 23, 2001 as the following: na_geno.dros.RELEASE2 (The Celera/BDGP whole-genome shotgun sequence from Release 2), na_gadfly.dros.RELEASE2 (nucleic acid sequence for every predicted transcript cDNA from Release 2), na_EST.dros [Drosophila expressed sequence tag (EST) sequences from BDGP], na_cDNA.dros (Drosophila full-length cDNA sequences from BDGP) and na_gb.dros [Drosophila sequences from GenBank (minus BDGP and EDGP sequences)]. Using the ‘Genomic mode’ of SAGE 2000 Software, we extracted all the 10 bp sequences downstream of the CATG site. We annotated tags to genes using the following criteria: (i) tags matched to the 3′-most CATG site of the full-length cDNA or 3′ EST; (ii) tags matched to the 3′-most CATG site of the predicted gene; and (iii) tags matched to only genomic DNA sequence and the matched CATG site is located within 2 kb downstream of the predicted gene.
Northern blot analysis
Total RNA was used for all northern blot analysis in this report. RNA was loaded on formamide–agarose gels (1%) and blotted to Hybond-N+ membranes (Amersham/Pharmacia). In Figure 1B, oligo DNA and PCR probes were used. The StarFire oligo DNAs (50 bases) were labeled with [32P]dATP using the StarFire DNA Labeling System (Integrated DNA Technologies). The names of probes and their sequences are the following:
StF-Cyp4d21 (5′-TTAAGTTTGCTCCTGGCATGTAACATAAAAT AAATACAAAAAAATGTAAA-3′);
StF-yellow-c (5′-TAATGTCATAATGTAAATACAAAAATGCACC GCGTCATAAACACAGCATG-3′);
StF-CG4979 (5′-CACCGCCCATGAACTCGTTGCCGGTGCATGT ATTGACTGCATATTTTTCG-3′);
StF-CG17820 (5′-TGAATGAATGACATGGTGTATCCAATCGAA CGCCTAACGCAGTGCACGTC-3′). Other probes (coding region of each genes) were labeled with [32P]dCTP by random priming. The cDNA clones for Sodh-1 (LP12301, gb;AI297864), CG7433 (GM13560, gb;AA803585), CG7592 (LP05187, gb;AI261107) and CG11458 (GH15115, gb;AI238960) were kindly provided by Dr Todd Laverty.
RNA in situ hybridization
RNA in situ hybridization was carried out essentially as described (Schaeren-Wiemers and Gerfin-Moser, 1993). This protocol was modified to include detergents in most steps, thereby increasing sensitivity and reducing background. The hybridization buffer contained 50% formamide, 5× SSC, 5× Denhardt’s, 250 µg/ml yeast tRNA, 500 µg/ml herring sperm DNA, 50 µg/ml heparin, 2.5 mM EDTA, 0.1% Tween-20 and 0.25% CHAPS. Anti-DIG antibody (Boehringer Mannheim) steps were in the presence of 0.1% Triton X-100, and the reaction was developed for ∼18 h in buffer containing 0.1% Tween-20 using the NBT/BCIP kit from Promega. Slides were mounted in Glycergel (Dako) and viewed with Nomarski optics.
Tissue dissections
Flies were anesthetized with CO2, decapitated and placed in a depression well dish in 1× phosphate-buffered saline (PBS) at room temperature. Brains were dissected out using fine forceps (no. 4 or finer) and placed directly into a microcentrifuge tube with 0.5 ml of ice-cold 1× PBS. The remaining carcasses were gathered into a separate tube and also placed on ice. When finished with all flies, tubes were quickly (∼10 s) spun down on a tabletop picofuge and PBS removed. Parts were immediately frozen on dry ice and then stored at –80°C until RNA was prepared. For the H– fraction, labella and antennae were carefully removed from heads using forceps and then processed in the same manner as above.
Cloning of expression constructs and generation of transgenic flies
A cDNA of the tsx gene was obtained by RT–PCR from male head RNA and cloned into the vector pET-DEST42 (Invitrogen). The V5-tagged TSX fusion gene was then cloned into the pUAST vector (Brand and Perrimon, 1993) and transgenic flies were generated according to Amrein and Axel (1997).
Mating assay
Males and females in mating assays were collected within 8 h after hatching and reared on standard food in isolation. They were kept on a 12 h light:12 h dark cycle for 3–8 days. Matings were performed in mating chambers (small plastic cuvettes) and were observed until copulation occurred, but not longer than 15 min. The genotype of the experimental males was generated from the following crosses: Act:GAL4/UAS-tsx_V5.4 (or V5.5): yw/w; act:GAL4/TM3SerGFP × w; UAS:tsx_V5.4 (or V5.5); UAS:tsx_V5.4 (or V5.5)/+: OreR × w; UAS:tsx_V5.4 (or V5.5); Act:GAL4/+: OreR × yw/w; act:GAL4/TM3SerGFP.
Acknowledgments
Acknowledgements
We are indebted to Caroline Chromey for excellent technical assistance and editing, Nathalie Velarde for help with template preparation and sequencing, and Corum McNealy and Whitney Jones for preparing RNA samples. We would like to thank Gregg Riggins for helpful advice on SAGE, and Fred Dietrich for providing support with sequence analysis. We appreciate the generosity of Tom Cline, Monica Steinmann, the Stock Center at Indiana University in Bloomington (fly strains) and Todd Laverty (EST clones) for providing essential materials used in this study. Raquel Sitcheran, Joe Nevins, Robin Wharton and members of the Amrein laboratory made insightful comments on the manuscript. This work was supported by an NIH grant awarded to H.A. (GM-60234-02).
References
- Abrahamsen N., Martinez,A., Kjaer,T., Sondergaard,L. and Bownes,M. (1993) Cis-regulatory sequences leading to female-specific expression of yolk protein genes 1 and 2 in the fat body of Drosophila melanogaster. Mol. Gen. Genet., 237, 41–48. [DOI] [PubMed] [Google Scholar]
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Amrein H. and Axel,R. (1997) Genes expressed in neurons of adult male Drosophila. Cell, 88, 459–469. [DOI] [PubMed] [Google Scholar]
- Amrein H., Gorman,M. and Nöthiger,R. (1988) The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell, 55, 1025–1035. [DOI] [PubMed] [Google Scholar]
- An W. and Wensink,P.C. (1995a) Integrating sex- and tissue-specific regulation within a single Drosophila enhancer. Genes Dev., 9, 256–266. [DOI] [PubMed] [Google Scholar]
- An W. and Wensink,P.C. (1995b) Three protein binding sites form an enhancer that regulates sex- and fat body-specific transcription of Drosophila yolk protein genes. EMBO J., 14, 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.S. and Ridge,K.A. (1980) Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics, 94, 383–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.S., Taylor,B.J. and Hall,J.C. (2001) Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell, 105, 13–24. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J. and Baker,B.S. (1995) The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development, 121, 3245–3258. [DOI] [PubMed] [Google Scholar]
- Bell L.R., Maine,E.M., Schedl,P. and Cline,T.W. (1988) Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell, 55, 1037–1046. [DOI] [PubMed] [Google Scholar]
- Bell L.R., Horabin,J.I., Schedl,P. and Cline,T.W. (1991) Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell, 65, 229–239. [DOI] [PubMed] [Google Scholar]
- Belote J.M. and Baker,B.S. (1987) Sexual behavior: its genetic control during development and adulthood in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 84, 8026–8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Burtis K.C. (1993) The regulation of sex determination and sexually dimorphic differentiation in Drosophila. Curr. Opin. Cell Biol., 5, 1006–1014. [DOI] [PubMed] [Google Scholar]
- Burtis K.C. and Baker,B.S. (1989) Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell, 56, 997–1010. [DOI] [PubMed] [Google Scholar]
- Burtis K.C., Coschigano,K.T., Baker,B.S. and Wensink,P.C. (1991) The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J., 10, 2577–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth F.M., Bownes,M. and Burde,V.S. (1991) Genetically modified yolk proteins precipitate in the adult Drosophila fat body. J. Cell Biol., 112, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth F.M., Burde,V.S., Mauchline,D. and Bownes,M. (1999) A yolk protein mutant leads to defects in the secretion machinery of Drosophila melanogaster. Tissue Cell, 31, 212–222. [DOI] [PubMed] [Google Scholar]
- Chase B.A. and Baker,B.S. (1995) A genetic analysis of intersex, a gene regulating sexual differentiation in Drosophila melanogaster females. Genetics, 139, 1649–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez V.M., Marques,G., Delbecque,J.P., Kobayashi,K., Hollingsworth, M., Burr,J., Natzle,J.E. and O’Connor,M.B. (2000) The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development, 127, 4115–4126. [DOI] [PubMed] [Google Scholar]
- Childress D. (1973) The inter- and intrachromosomal effects of the bar–stone translocation in Drosophila melanogaster. Mol. Gen. Genet., 121, 133–138. [DOI] [PubMed] [Google Scholar]
- Cline T.W. and Meyer,B.J. (1996) Vive la difference: males vs females in flies vs worms. Annu. Rev. Genet., 30, 637–702. [DOI] [PubMed] [Google Scholar]
- Conley A. and Hinshelwood,M. (2001) Mammalian aromatases. Reproduction, 121, 685–695. [DOI] [PubMed] [Google Scholar]
- Coschigano K.T. and Wensink,P.C. (1993) Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev., 7, 42–54. [DOI] [PubMed] [Google Scholar]
- Erdman S.E. and Burtis,K.C. (1993) The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J., 12, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley K.D., Taylor,B.J., Milstein,M. and McKeown,M. (1997) dissatisfaction, a gene involved in sex-specific behavior and neural development of Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 94, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley K.D., Edeen,P.T., Foss,M., Gross,E., Ghbeish,N., Palmer,R.H., Taylor,B.J. and McKeown,M. (1998) Dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron, 21, 1363–1374. [DOI] [PubMed] [Google Scholar]
- Fisher C.R., Graves,K.H., Parlow,A.F. and Simpson,E.R. (1998) Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cyp19 gene. Proc. Natl Acad. Sci. USA, 95, 6965–6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo K. and Smith,D.P. (2001) A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics, 159, 1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian M.J., Shirras,A.D., Bownes,M. and Wensink,P.C. (1987) The nucleotide sequence of the gene coding for Drosophila melanogaster yolk protein 3. Gene, 55, 1–8. [DOI] [PubMed] [Google Scholar]
- Goralski T.J., Edstrom,J.E. and Baker,B.S. (1989) The sex determination locus transformer-2 of Drosophila encodes a polypeptide with similarity to RNA binding proteins. Cell, 56, 1011–1018. [DOI] [PubMed] [Google Scholar]
- Grieneisen M.L., Warren,J.T. and Gilbert,L.I. (1993) Early steps in ecdysteroid biosynthesis: evidence for the involvement of cytochrome P-450 enzymes. Insect Biochem. Mol. Biol., 23, 13–23. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C., Winter,C., Hofbauer,A., Hall,J.C. and Stanewsky,R. (2001) The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron, 30, 249–261. [DOI] [PubMed] [Google Scholar]
- Inoue K., Hoshijima,K., Sakamoto,H. and Shimura,Y. (1990) Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature, 344, 461–463. [DOI] [PubMed] [Google Scholar]
- Kelley R.L., Solovyeva,I., Lyman,L.M., Richman,R., Solovyev,V. and Kuroda,M.I. (1995) Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell, 81, 867–877. [DOI] [PubMed] [Google Scholar]
- Li H. and Baker,B.S. (1998) Her, a gene required for sexual differentiation in Drosophila, encodes a zinc finger protein with characteristics of ZFY-like proteins and is expressed independently of the sex determination hierarchy. Development, 125, 225–235. [DOI] [PubMed] [Google Scholar]
- Mayer R.T., Svoboda,J.A. and Weirich,G.F. (1978) Ecdysone 20-hydroxylase in midgut mitochondria of Manduca sexta (L.). Hoppe Seyler’s Z. Physiol. Chem., 359, 1247–1257. [DOI] [PubMed] [Google Scholar]
- McDonald M.J. and Rosbash,M. (2001) Microarray analysis and organization of circadian gene expression in Drosophila. Cell, 107, 567–578. [DOI] [PubMed] [Google Scholar]
- McKeown M. (1994) Sex determination and differentiation. Dev. Genet., 15, 201–204. [DOI] [PubMed] [Google Scholar]
- McKeown M., Belote,J.M. and Baker,B.S. (1987) A molecular analysis of transformer, a gene in Drosophila melanogaster that controls female sexual differentiation. Cell, 48, 489–499. [DOI] [PubMed] [Google Scholar]
- Mitsui T. and Riddiford,L.M. (1978) Hormonal requirements for the larval–pupal transformation of the epidermis of Manduca sexta in vitro. Dev. Biol., 62, 193–205. [DOI] [PubMed] [Google Scholar]
- Nagoshi R.N. and Baker,B.S. (1990) Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev., 4, 89–97. [DOI] [PubMed] [Google Scholar]
- Nayak S.V. and Singh,R.N. (1983) Sensilla on the tarsal segments and the mouthparts of adult Drosophila melanogaster. Int. J. Insect Morphol. Embryol., 12, 273–291. [Google Scholar]
- Nöthiger R., Leuthold,M., Anderesen,N., Gerschwiler,P., Grueter,A., Keller,W., Leist,C., Roost,M. and Schmid,H. (1987) Genetic and developmental analysis of the sex-determining gene doublesex (dsx) of Drosophila melanogaster. Genet. Res., 50, 113–123. [Google Scholar]
- Pikielny C.W., Hasan,G., Rouyer,F. and Rosbash,M. (1994) Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron, 12, 35–49. [DOI] [PubMed] [Google Scholar]
- Riddiford L.M. (1993) Hormone receptors and the regulation of insect metamorphosis. Receptor, 3, 203–209. [PubMed] [Google Scholar]
- Roselli C.E. and Resko,J.A. (1997) Sex differences in androgen-regulated expression of cytochrome P450 aromatase in the rat brain. J. Steroid Biochem. Mol. Biol., 61, 365–374. [PubMed] [Google Scholar]
- Ryner L.C., Goodwin,S.F., Castrillon,D.H., Anand,A., Villella,A., Baker, B.S., Hall,J.C., Taylor,B.J. and Wasserman,S.A. (1996) Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell, 87, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Salz H.K., Maine,E.M., Keyes,L.N., Samuels,M.E., Cline,T.W. and Schedl,P. (1989) The Drosophila female-specific sex-determination gene, Sex-lethal, has stage-, tissue- and sex-specific RNAs suggesting multiple modes of regulation. Genes Dev., 3, 708–719. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N. and Gerfin-Moser,A. (1993) A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry, 100, 431–440. [DOI] [PubMed] [Google Scholar]
- Schoentgen F. and Jolles,P. (1995) From structure to function: possible biological roles of a new widespread protein family binding hydrophobic ligands and displaying a nucleotide binding site. FEBS Lett., 369, 22–26. [DOI] [PubMed] [Google Scholar]
- Shirk P.D., Minoo,P. and Postlethwait,J.H. (1983) 20-hydroxyecdysone stimulates the accumulation of translatable yolk polypeptide gene transcript in adult male Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 80, 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.L., Bollenbacher,W.E., Cooper,D.Y., Schleyer,H., Wielgus,J.J. and Gilbert,L.I. (1979) Ecdysone 20-monooxygenase: characterization of an insect cytochrome p-450 dependent steroid hydroxylase. Mol. Cell. Endocrinol., 15, 111–133. [DOI] [PubMed] [Google Scholar]
- Sosnowski B.A., Belote,J.M. and McKeown,M. (1989) Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell, 58, 449–459. [DOI] [PubMed] [Google Scholar]
- Sudhof T.C., Baumert,M., Perin,M.S. and Jahn,R. (1989) A synaptic vesicle membrane protein is conserved from mammals to Drosophila. Neuron, 2, 1475–1481. [DOI] [PubMed] [Google Scholar]
- Valcarcel J., Singh,R., Zamore,P.D. and Green,M.R. (1993) The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature, 362, 171–175. [DOI] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- Zhou S. et al. (1995) Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J., 14, 2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]