Abstract
A new long terminal repeat (LTR) retrotransposon, named REM1, has been identified in the green alga Chlamydomonas reinhardtii. It was found in low copy number, highly methylated, and with an inducible transpositional activity. This retrotransposon is phylogenetically related to Ty3-gypsy LTR retrotransposons and possesses new and unusual structural features. A regulatory module, ORF3p, is present in an inverse transcriptional orientation to that of the polyprotein and contains PHD-finger and chromodomains, which might confer specificity of the target site and are highly conserved in proteins involved in transcriptional regulation by chromatin remodeling. By using different wild-type and mutant strains, we show that CrREM1 was active with a strong transcriptional activity and amplified its copy number in strains that underwent foreign DNA integration and/or genetic crosses. However, integration of CrREM1 was restricted to these events even though the expression of its full-length transcripts remained highly activated. A regulatory mechanism of CrREM1 retrotransposition which would help to minimize its deleterious effects in the host genome is proposed.
Transposable elements are a powerful source of change in eukaryotic and prokaryotic genomes since they provide an important flexibility and evolutionary capacity on the host genome. Their capacity for duplication and mobilization from one site to another, causing mutations and reorganizations, affects the regulation of genes where they integrate, so that they have been positively selected and maintained during evolution (4, 16).
Class I of mobile elements includes long terminal repeat (LTR) retrotransposons whose genome has structural features very similar to those of retroviruses. They are bounded by two long terminal repeats and possess a coding region that is normally divided in a single gag-pol or two separate gag and pol open reading frames (ORFs) that determine several mature proteins. The former, named gag (specific antigen group), encodes an RNA binding protein (Gag) and other proteins involved in maturation and packaging of retrotransposons, and it usually presents zinc finger CCHC motifs similar to those described in eukaryotic transcription factors. The latter, named pol (polymerase), encodes several proteins, such as protease, integrase, reverse transcriptase, and RNase H. The retrotranscriptase and RNase H activities are required for cDNA synthesis and the integrase activity is required for the integration of the cDNA into a new chromosomal location (3). The pol-encoded protease is necessary for cleavage of the polyproteins into functional peptides.
Two families of LTR elements (Ty1-copia and Ty3-gypsy) have been differentiated in terms of structure and known protein sequences (mainly reverse transcriptase). Both of them are ancient and ubiquitous constituents of eukaryotic genomes. The main difference among them concerns the relative position of the integrase domain with respect to the ORF homologous to the pol gene (28). Some members of the Ty3-gypsy group from invertebrates have recently been suggested to be endogenous retroviruses, since they show a third additional ORF related to the retroviral env gene (5, 27) and at least one of those elements, mdg-4 of Drosophila melanogaster, is infectious (17).
Retrotransposons are particularly abundant in plants and account for about 50 to 80% of repetitive DNA in maize and about 90% in wheat (2, 20). Genome size variations correlate with both the total mass of retrotransposons and the number of different retrotransposons families, like the 13,000 copies of the del family in Lilium speciosum (22) and the 50,000 copies of the Bare-1 element in barley (26). Both LTR and non-LTR retrotransposons account for 70 to 85% of the nuclear genome in maize (42). In contrast to these plants, the Arabidopsis nuclear genome contains few repeated sequences, and therefore it is likely that the content of retrotransposons is about 4 to 10% of total DNA, although many types of both LTR and non-LTR retrotransposons have been reported and appear concentrated on centromeric regions (20).
It is estimated that 30% of the nuclear genome of Chlamydomonas spp. contains repetitive DNA sequences and examples of each of the two main classes of mobile elements (11). In this eukaryotic alga, a TOC1 element classified as an LTR retrotransposon contains an unusual rearrangement of the long terminal repeats (7, 8), and a DNA fragment named CRRE1 encodes a protein (21) with homology to the rvt sequence.
One of the exciting topics in retrotransposons biology concerns the control of activation of its transcription and retrotransposition. Most plant retrotransposons are inactive during normal growth and development but are transcriptionally activated by biotic and abiotic stress factors (10). In addition, the activation of retrotransposons by introduction of foreign DNA is a phenomenon observed in hybrids of marsupials (32), rice (24), and Arabidopsis thaliana (13). Moreover, the retrotransposon activation by DNA introduction is followed by rapid DNA methylation and repression (24). In Drosophila, where DNA methylation does not exist, retrotransposons are located mainly in regions of the heterochromatin (36) and gene silencing, including that of mobile endogenous elements, occurs by a group of polycomb proteins (45).
In this work, we have identified a new LTR retrotransposon in Chlamydomonas reinhardtii named REM1, phylogenetically related to the gypsy family elements, that possesses unique structural and expression features not still described in mobile elements of this group. Foreign DNA integration and/or genetic crosses activated retrotransposition and transcription of REM1. However, integration of CrREM1 was restricted to those events even though the expression of its primary transcript remained highly activated. A regulation of retrotransposition mostly dependent on the DNA integration step is proposed.
MATERIALS AND METHODS
Chlamydomonas reinhardtii strains and growth conditions.
The Chlamydomonas reinhardtii strains used in this work are listed in Table 1. Cells were grown at 25°C under continuous light in liquid minimal medium (11) containing 7.5 mM ammonium chloride and bubbled with 4% (vol/vol) CO2-enriched air. Cells were collected at mid-exponential phase of growth (4,000 × g, 5 min) and transferred to medium containing the specified nitrogen source. After the indicated times, cells were collected and processed immediately for DNA or RNA extraction, or other analyses. Solid media contained 1.5% agar and 8 mM of NH4Cl or 4 mM KNO3. When chlorate sensitivity in the presence of ammonium (referred as CSA phenotype) was tested, 100 mM KClO3 and 1 mM NH4Cl were included in the solid medium containing 4 mM KNO3.
TABLE 1.
Chlamydomonas strains useda
| Strain | Relevant genotype | Phenotype | Reference |
|---|---|---|---|
| 6145c | mt− | Nit+ CRA | 11 |
| 21gr | Nit5 mt+ | Nit+ CRA | 11 |
| 305CW15 | Nia1 cw15 mt+ | Nit− CW− CRA | 11 |
| N1 | 305cw15::pMN24 (Nia1) Nrg1 mt+ | Nit+ CSA | 37 |
| N10 | 305cw15::pMN24 (Nia1) Nrg3 mt+ | Nit+ CSA | This work |
| N24 | 305cw15::pMN24 (Nia1) Nrg4 mt+ | Nit+ CSA | This work |
| 305 | Nia1 mft | Nit− | 11 |
| Tx-7 | Nia1::pMN24 (Nia1) mt− | Nit+ CRA | 18 |
| PAD305-2 | 305cw15::pAD35 (Nia1) mt+ | Nit+ CW− | 40 |
| A54 | ac17 sr-1 cw mt+ | Nit+ | 38 |
| E18 | ac17 sr-1 cw (ΔNia1) mt+ | Nit− | 38 |
| P3 | ΔNia1 | Nit− | 30 |
| P3-NR-2 | P3::pMN24 (Nia1) | Nit+ CRA | 30 |
| P3-NR-4 | P3::pMN24 (Nia1) | Nit+ CRA | 30 |
Abbreviations: Nit+/Nit−: ability to grow with nitrate; mt: sexual competence; cw15, absence of cell wall (phenotype cw); Nrg, CSA phenotype (sensitivity to chlorate in the presence of ammonium); CRA, resistance to chlorate in the presence of ammonium; ac-17, acetate requirement.
Inverse PCR on genomic DNA from N1.
Religated DNA (50 ng) was used as a template for inverse PCR experiments using internal primers deduced from the Nia1 gene promoter sequence (NUP5, 5′-TCCTGCTTGGTGCTCCACTTCAC-3′; NDO5, 5′-CAGCGTCCTATGGAAGCGAATGC-3′). Genomic DNA of mutant N1 was previously digested with XhoI and religated. The internal primers NDO5 and NUP5 were used to amplify the genomic DNA at the insertion site of pMN24 that had originated the phenotype of chlorate sensitivity in the presence of ammonium (CSA) (37). The PCR mixture (50 μl) contained 2 μl of religated DNA, 0.5 μl primer (50 μM), 1.25 μl deoxynucleoside triphosphates (20 mM), 5 μl buffer (10×), and 0.75 μl Taq (Expand Long Template PCR System, Boehringer Mannheim). The program profile used was as follows: 94°C for 2 min; 10 cycles of 94°C for 15 s, 69°C for 30 s (−0.5°C/cycle), and 68°C for 2 min; 25 cycles of 94°C for 15 s, 64°C for 30 s (+10 s/cycle), and 68°C 2 min; and 14°C overnight.
Screening procedures, DNA and RNA isolation, and hybridization analysis.
The λEMBL4 genomic DNA library (Carolyn F. Silflow, University of Minnesota) was screened according to previously reported methods (38). For Southern blot experiments, chromosomal DNA was extracted after lysis of the cells with 2% sodium dodecyl sulfate (SDS) in 100 mM Tris-HCl (pH 8.0), 400 mM NaCl, and 50 mM EDTA. Following phenol extraction, DNA was precipitated with 2.5 volumes of 95% ethanol, washed in 70% ethanol, and resuspended in 10 mM Tris-HCl, pH 8.0. Upon digestion, 2 μg of DNA per lane was loaded on 0.8% agarose gels. Total RNA was isolated as described by Schloss et al. (43).
Northern blots were performed by loading and fractionation of 15 to 20 μg RNA per sample on 1.2% agarose gels containing 17.5% formaldehyde and transferred onto nylon membranes (Hybond-N+; Amersham) (41). After blotting onto the above-mentioned nylon membranes the fixation was carried out at 80°C for 2 h. Probes used were radioactively labeled with [α-32P]dCTP by the random primer method. The membranes were prehybridized and hybridized in 50% formamide solutions at 42°C (41, 43) and washes were performed at 65°C, with 0.2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.2% (wt/vol) SDS solution. Probes used were Gag-1, a 650-bp PvuII-EcoRV fragment of the gag gene; Gag-2, a 719-bp SmaI/SmaI fragment corresponding to ORF1 of CrREM1; GP-CrREM1, a 1,753-bp fragment isolated by PvuII/EcoRV digestion from pN1EV plasmid corresponding to the 3′ end of ORF1 and 5′ of ORF2; reverse transcriptase, a 1,433-bp SalI/SalI fragment from pB3 in the reverse transcriptase coding region of CrREM1; and Nii1, a 642-bp SalI/SalI fragment of the Nii1 gene (39). The intensity of hybridization signals was analyzed with Data Processing Program Quantity-One.
DNA sequencing.
Plasmid isolations were obtained in prepacked purification columns (QIAGEN Tip-20; QIAGEN). DNA sequencing was performed with fluorescence-labeled terminators for an automated sequencer (model ABI Prism 373XL; Applied Biosystems) in the Central Service for Research Support of the University of Córdoba. Sequences were normalized using the DNAStar for Windows and MAC software (Lasergene Navigator) and Chromas program and subsequently analyzed as indicated below in “Bioinformatic tools for analysis.”
Bioinformatics tools for analysis.
Sequence search homologies were conducted using the BLASTN (nucleotide versus nucleotide) and TBLASTX (nucleotide versus trans nucleotide) program available at DDBJ (http://www.ddbj.nig.ac.jp) and ChlamyDB (http://genome.jgi-psf.org/chlre1/chlre1.home.html; http://bahama.jgi-psf.org/prod/bin/chlamy/home.chlamy.cgi). Structural analysis and protein domains described in this article were identified using RPsBLAST or CDD from NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and other programs whose links are in the database http://molbiol-tools.ca. Multiple alignments of protein sequences were created by BioEdit Sequence Alignment Editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Phylogenetic analysis was conducted using the MegAlign program within the DNAStar software.
Nucleotide sequence accession numbers.
Sequence data in this article have been deposited in the EMBL/GenBank data libraries under accession numbers as follows: for nucleotide sequences N1, AY227353, and CrREM1, AY227352; and for translation products of pB3 and ORF1p, AA073550; ORF2p, AA073551; and ORF3p, AA073552.
RESULTS
Isolation of CrREM1 from a genomic library of C. reinhardtii.
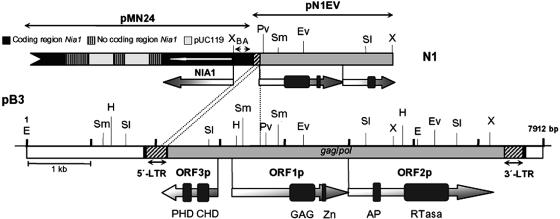
C. reinhardtii strain N1 was isolated by insertional mutagenesis and is affected in a locus, named Nrg1, which corresponds to a regulatory gene mediating the negative effect of ammonium on the expression of nitrate assimilation genes. This mutant has a phenotype of chlorate sensitivity in the presence of ammonium (37). To isolate the DNA region adjacent to the inserted plasmid pMN24 used in the insertional mutagenesis, inverse PCR was performed based on the restriction map of the integration site. An amplification band of 2,475 bp was obtained. This product, pN1EV (Fig. 1), was cloned in pBKSII−. An XhoI-ApaI fragment of 1.2 kb was subcloned from pN1EV and used as a probe in Southern blot to check for the expected restriction fragment length polymorphisms between the parental and the N1 mutant strains at the insertion site, as it was found (results not shown). This indicates that the isolated fragment belongs to the region affected in mutant N1.
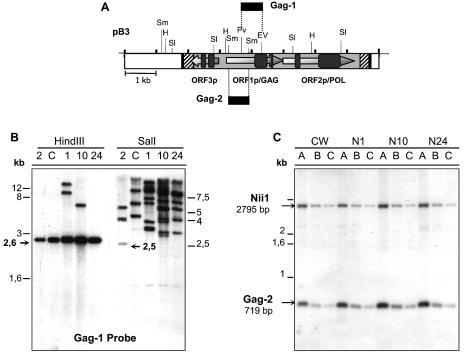
FIG. 1.
Structure of pN1EV and pB3 clones. CrREM1 is located between positions 1819 and 7627 of pB3 (shadowed box). Black boxes indicate direct repeats of target site integration, striped boxes are 5′ and 3′ LTRs and the gray box is the central coding region of CrREM1. This figure shows the three ORFs identified with ORF-Finder of NCBI and the domains that contain each protein. GAG, Retro-gag (pfam03732); Zn, zinc finger type C2H2 (smart00355); AP, aspartic protease (pfam00077); RTase, reverse transcriptase (pfam00078); PHD, PHD-finger domain (pfam00628, smart00249); CHD: chromatin organization modifier domain (smart00298). Ap, ApaI; E, EcoRI; EV, EcoRV; H, HindIII; Pv, PvuII; Sl, SalI; Sm, SmaI; X, XhoI. Plasmid pMN24 is not drawn to scale. Primers deduced from the NR promoter and used in inverse PCR: NDO5 (A) and NUP5 (B).
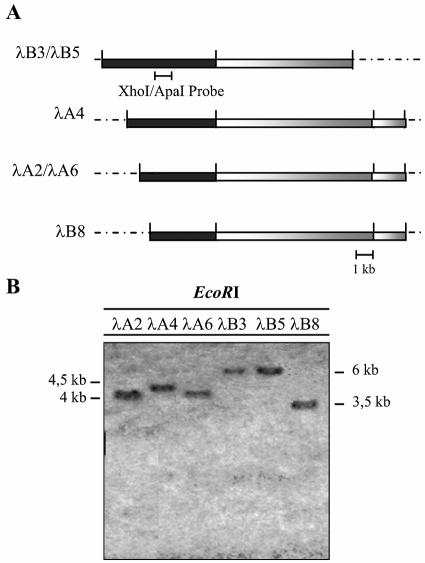
A λEMBL4 genomic DNA library of the wild-type strain 21gr was screened with the XhoI-ApaI probe. Among 50,000 phages screened, six overlapping clones were recovered and named λA2, λA4, λA6, λB3, λB5, and λB8. Subsequently, the clones were analyzed by EcoRI digestions that released their genomic DNA inserts from the arms of phage λEMBL4 (Fig. 2A), and hybridized with the probe used in the screening of the genomic DNA library to study the fragments which contained the XhoI-ApaI sequence (Fig. 2B). These fragments were cloned in pBKSII-, previously linearized and dephosphorylated. The physical maps of each clone were deduced from band patterns of double and simple digestions with restriction enzymes and the interesting fragments were subcloned.
FIG. 2.
Analysis of six phage-positive clones from strain 21gr of C. reinhardtii. (A) Restriction map from phage clones with endonuclease EcoRI. The black bars correspond to fragments hybridizing to the probe used in the screening of the library. The restriction sites of EcoRI at the ends come from the lambda cloning vector (dashed lines). (B) The 1.2-kb XhoI-ApaI fragment from pN1EV was labeled with digoxigenin-dUTP and used as a probe in the Southern blot analysis of phages. Six hybridization signals corresponding to a fragment of 6 kb in clones λB3 and λB5 and between 4.5 and 3.5 kb in λA2, λA4, λA6, and λB8 are shown.
At first, we subcloned and sequenced the largest one of 6054 bp (Fig. 2, black bar in λB3). A BLAST database search did not allow us to find significant identity with any known gene or protein. Because of this fact, an adjacent fragment from phage λB3 was also subcloned and sequenced to expand the region affected by the insertion to 7912 bp whose restriction map is shown in the Fig. 1. A new BLAST search revealed the presence of two long direct terminal repeats that would be flanking a long retrotransposon-like sequence. The translation product of pB3 in the databases of GenBank/EMBL/DDBJ/PDB at the NCBI produced a significant degree of similarity with gene products of some LTR retrotransposons of the gypsy type, especially at region 6090 to 6763 of clone pB3 that contains a reverse transcriptase domain. A comparison with the Chlamydomonas genome database (http://genome.jgi-psf.org/chlre1/chlre1.home.html) revealed that this sequence had not yet been reported and corresponds to a new gypsy LTR retrotransposon from C. reinhardtii that we have named REM1.
When the sequence of pN1EV was compared to that of pB3 (Fig. 1) it was found that plasmid pMN24 had inserted at a central region of CrREM1 causing a deletion of 1,590 bp at the 5′ end and still maintaining only 52 bp of the 5′ LTR (Fig. 1). We also sequenced genomic regions adjacent at both the 5′ and 3′ LTRs and internal regions of the retrotransposon from λA2, λA4, λA6, and λB8. It was found that the sequences obtained were identical to those in pB3. Thus, these DNAs had the same structural organization and correspond to the same genomic localization, suggesting that strain 21gr bears a single copy of REM1.
Structural characteristics of CrREM1.
In general, CrREM1 showed most of the structural characteristics of LTR retrotransposons with some important differences. The analysis of the sequence showed that CrREM1 is a mobile element of 5,812 bp delimited by two LTRs of 286 bp. Sequences at the 5′ LTR and 3′ LTR of CrREM1 are 100% identical and the LTR tips have the retroviral consensus inverted repeat terminal sequences 5′TG… CA3′ that determine the beginning and end of the retrotransposon. CrREM1 is also flanked by the 5-bp direct repeat sequence ATGCT that seems to correspond to the target integration site that duplicated during retrotransposition. The signals controlling the start and end of the transcription of the retrotransposons are found in its LTRs excepting the position of the primer binding site that could not be identified unambiguously in CrREM1.
Three ORFs were also identified (Fig. 1). Coding regions of CrREM1 were confirmed by reverse transcription-PCR with primers designed to anneal to predicted exons by computer analysis (results not shown). The hypothetical protein products in ORF1 and ORF2 of 556 and 776 amino acids, respectively, exhibit significant identity with gene products of known retroelements, especially with plant, fungal, and Drosophila gypsy retrotransposons and would correspond with the genes gag and pol, respectively. ORF1p contains a domain of 96 amino acids (326 to 421) named Retro-gag or the WCCH domain. There is a central motif, QGXXEXXXXXFXXLXXH, common to Gag proteins and a region of 20 amino acids (507 to 526) with the characteristic zinc finger ZnF-C2H2 present in transcription activator/repressor proteins (34). The N-terminal portion of ORF2p includes a folding of 24 amino acids (residues 163 to 186) which show homology with the single domain retroviral aspartic protease (RVP) from retroviruses, retrotransposons, and hepadnaviruses (plant dsDNA viruses). The RVP of CrREM1 shows the highly conserved domain DTGA characteristic of this type of protease that usually form part of a larger polyprotein Pol.
An RVT sequence of 231 amino acids (Fig. 1) was subsequently identified in a final portion of ORF2p (residues 414 to 644), which contains the seven characteristic domains of the retrotranscriptase (46) as well as the conserved sequence YXDDIL that seems to be located at the active site of the reverse transcriptase (44). In the alignment of the Ty3-gypsy type retrotranscriptases from eukaryotic organisms and that from CrREM1 (results not shown), we observed that they share 42% to 49% (using nr, not redundant database) or 34 to 36% (Swissprot database) amino acid identity to reverse transcriptases from photosynthetic eukaryotes.
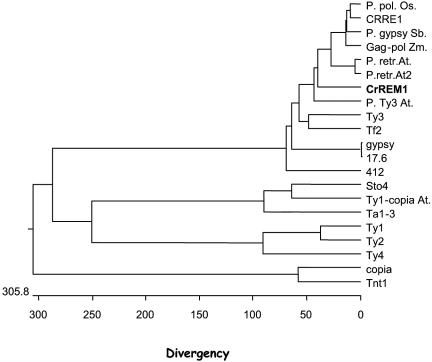
Analysis of the 3′ end of ORF2p failed to identify any RNase H and integrase motifs in CrREM1. Therefore, the domain order did not allow us to confirm that CrREM1 is related to a copia or gypsy retrotransposon. However, an evolutionary study of CrREM1 comparing the amino acid sequence of reverse transcriptases from both groups showed a higher evolutionary distance with retroelements of the group copia than with those of the group gypsy (Fig. 3). Moreover, the translation products of the identified ORFs in CrREM1 present an important similarity with the gene products of gypsy-like LTR retrotransposons, especially with putative polyproteins from C. reinhardtii, Zea mays, and Oryza sativa (Fig. 3 and data not shown).
FIG. 3.
Phylogenetic tree of LTR retrotransposons based on the reverse transcriptases sequence of the following retroelements: retrotransposons in the gypsy group: Oryza sativa P. polyprotein (BAB68106); C. reinhardtii CRRE1 (AB033239); Streptococcus bicolor gypsy (AAD22153); Zea mays Gag-Pol (AAL59229); Arabidopsis thaliana P. Gypsy/Ty3 (1) (NP_671762); Arabidopsis thaliana P. Gypsy/Ty3 (2) (NP_178767); ORF2p of C. reinhardtii REM1 (AA073551); Arabidopsis thaliana Ty3/gypsy (AAG51046); Saccharomyces cerevisiae Ty3B (CAA97115); Schizosaccharomyces pombe Tf2 (Q05654); Drosophila virilis gypsy (P10401); Drosophila melanogaster 17.6 (P04323); and Arabidopsis thaliana 412 (NP_671762). Retrotransposons in the copia group: Zea mays Sto-4 (T17429); Arabidopsis thaliana Ty1/copia (AG51258); Arabidopsis thaliana Ta1-3 (X13291); Saccharomyces cerevisiae Ty1-H3 (B28097); Saccharomyces cerevisiae Ty2B (P25384); Saccharomyces cerevisiae Ty4B (P47024); copia from Drosophila melanogaster (P04146); and Tnt1 from Neurospora crassa (CAA32025).
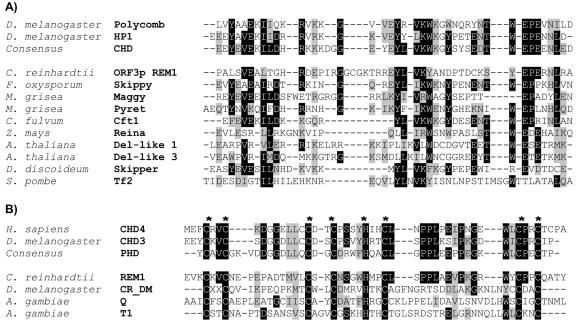
Surprisingly, another open reading frame (ORF3) was identified between the 5′ LTR and the start of CrREM1 ORF1 that shows an opposite transcriptional orientation to that of the polyprotein Gag-Pol (Fig. 1). This ORF3p (270 amino acids) contains two particular motifs: a chromodomain (chromatin organization modifier domain) and a zinc finger named PHD-finger (1, 6) of 61 and 45 amino acids, respectively. Figures 4A and B show the alignment with chromodomains and PHD from a variety of chromatin-related activity proteins and putative chromodomains and PHD from various LTR retrotransposons and non-LTR retrotransposons, respectively. Neither the presence of PHD finger domain nor the cluster of both domains PHD and chromodomain has been reported in proteins encoded by LTR retrotransposons.
FIG. 4.
Alignment of chromodomains and PHD module from chromatin modifiers proteins and various gypsy-type LTR retrotransposons. (A) The upper group includes authentic chromodomains of chromatin-modifying proteins polycomb (P26017) and HP1 (P05205) of Drosophila melanogaster and the consensus sequence of chromodomain available at the NCBI conserved domain database. The lower group represents chromodomains of CrREM1 and 3′-end integrase of Ty3-gypsy class of LTR retrotransposons. The accession numbers of retrotransposons which have been translated from their DNA sequence are as follows: ORF3p REM1, AA073552; Reina, U69258 and Pyret, AB062507. The accession numbers of retrotransposons available in the protein databases are as follows: Del-like 1 and 3, AC002534 and Z97342; Cft1, AF051915; Maggy, AAA33420; Skippy, AAA88791; Tf2, T38401; and Skipper, AAC48336. (B) The upper block includes authentic PHD-finger domain from chromatin-modifying proteins: chromo domain helicase-DNA-binding protein 4 (CHD4) of Homo sapiens (Q14839), CHD3 of Drosophila melanogaster (AAF49162), and consensus sequence of PHD available in the NCBI conserved domain database. The lower block contains PHD conserved in ORF3p encoded by REM1 of C. reinhardtii (AA073552), ORF1p encoded by non-LTR retrotransposons (CR_DM of Drosophila melanogaster, [15]), and the retrotransposons Q (T43019) and T1 (M93689) of Anopheles gambiae. The asterisks show conserved cysteine and histidine residues matching the PHD domain. Black boxes, identical amino acids; gray boxes, similar amino acids.
Analysis of transposition/integration of CrREM1 in wild-type and mutant C. reinhardtii.
CrREM1 was isolated from the adjacent region to the insertion of plasmid PMN24 that generated the CSA mutant N1 (37). Transposition of CrREM1 was studied by Southern blot in strain N1, and in two other newly isolated insertional mutants that were affected at the negative signal of ammonium (N24 and N10), which have the same phenotype as N1 and are defective each at unlinked loci as deduced from the genetic analysis (results not shown). The parental strain 305CW15 and the wild-type 21gr were used as controls so we could study whether mobilization of CrREM1 had been affected in these three independent mutants that had undergone the same phenomenon of DNA integration.
Filters containing DNA of each strain digested with endonucleases HindIII and SalI were hybridized with the Gag-1 probe (Fig. 5A). The expected 2.6-kb HindIII band was obtained in all the strains. Nevertheless, two additional bands of 8 and 12 kb were identified in N1 and another of 5.8 kb in N10. In the lanes of the SalI digestion, the expected 2.5-kb band was only observed in the wild-type strain 21gr together with additional bands. Larger fragments were found in all the other strains.
FIG. 5.
Study of the transposition and copy number of CrREM1 in strains of C. reinhardtii. (A) The locations of the probes used and the restriction map of CrREM1 are indicated. (B) DNA (2 μg) from mutants N1 (1), N10 (10), and N24 (24), wild-type 21gr (2), and strain 305CW15 (C) was digested with endonuclease HindIII or SalI and analyzed by Southern blot using the Gag-1 probe. (C) gDNA of the indicated strains (2 μg) was digested with SmaI. The different lanes in each strain correspond to different dilutions of SmaI digestion: A, 1; B, 1/5; C, 1/10. The probes used were Gag-2 and Nii1. These probes were subsequently purified, quantified and added together (each at 106 cpm/ml) to the same hybridization solution. The expected band sizes are indicated with arrows.
Moreover, it can be deduced from the intensity and the number of hybridizing bands that the copy number of CrREM1 has increased significantly in the CSA mutants (Fig. 5B). The expected bands corresponded to the restriction map shown in Fig. 1 in strain 21gr. The extra bands can be explained on the basis of different insertion sites in the mutants in comparison to the 21gr strain or methylation modifications that generate SalI sites resistant to digestion in copies of CrREM1 in those strains.
The CrREM1 copy number in these CSA mutants was compared to that in the parental strain 305CW15. A Southern blot of SmaI digestions of DNA from these strains was hybridized with two probes of similar size: one of the Nii1 gene (a single-copy gene control) and another from ORF1 of CrREM1 (Gag-2) (Fig. 5C). Quantification of radioactive signals indicated that strain 305CW15 possesses one to two copies of CrREM1in the genome and the mutants CSA about 1.5 to 2 times more. Thus, after the insertion of the pMN24 plasmid, these three CSA mutants had increased CrREM1 copy numbers.
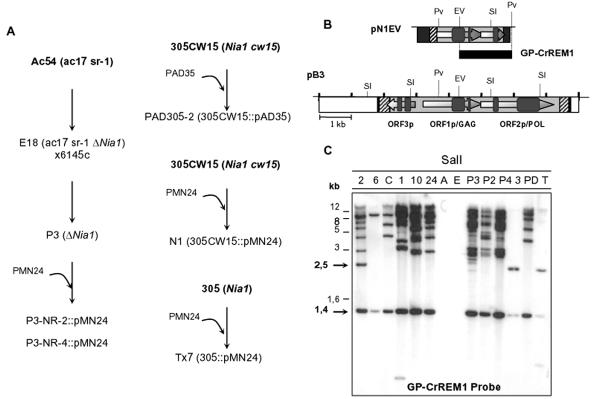
Thereby, to know whether DNA introduction caused transposition of CrREM1, we studied its mobilization in different strains that, like N1, N24 and N10, had been transformed with plasmids (PAD35 and pMN24) containing the Nia1 gene (Fig. 6A). A Southern blot of total DNA digestions with SalI was hybridized with the GP-CrREM1 probe (Fig. 6B). As shown in Fig. 6C, an increase in CrREM1 copy number and hybridizing bands was again observed in the three CSA mutants, but also in strains that had undergone a greater number of changes in its genome (genetic crosses and transformation), as deduced from comparing strain P3 with its parental strains E18 and 6145c (Fig. 6C). A54 and E18 do not possess copies of CrREM1 since they have a genetic background different from the other strains analyzed. However, this increase in CrREM1 copy number does not occur in all transformants like Tx-7 that showed identical hybridization signals than its parental strains 305 (Fig. 6C). Moreover, the expected 1,429-bp SalI band is observed in all strains while the 2,500-bp band is only observed in strains 21gr, 305, and Tx-7. These results suggest that in those strains with a higher copy number of CrREM1, the region analyzed with the GP-CrREM1 probe would have undergone methylations, to which SalI is sensitive (GTm5CGAC, GTCGm6AC, and Ghm5UCGAC). Some differences in bands intensity, like those between P3 and P2 (Fig. 6C), might also be due to differences in DNA amounts loaded on the gels.
FIG. 6.
Study of CrREM1 mobilization in strains of C. reinhardtii. (A) Relationships of parental-transformant-segregant strains used in the Southern blot analysis. Plasmids pMN24 and pAD35 contain the structural gene of the NR. (B) Probe GP-CrREM1 (a 1,753-bp fragment) was isolated by PvuII/EcoRV digestion from plasmid pN1EV. (C) DNA (1 μg) from the following strains was digested with restriction endonuclease SalI: 2, 21gr; 6, 6145c; C, 305CW15; 1, N1; 10, N10; 24, N24; A, A54; E, E18; P3, P3; P2, P3-NR-2; P4, P3-NR-4; 3, 305; PD, PAD305-2; and T: Tx-7. DNA hybridization was carried out with the GP-CrREM1 probe. The expected band sizes are indicated with arrows.
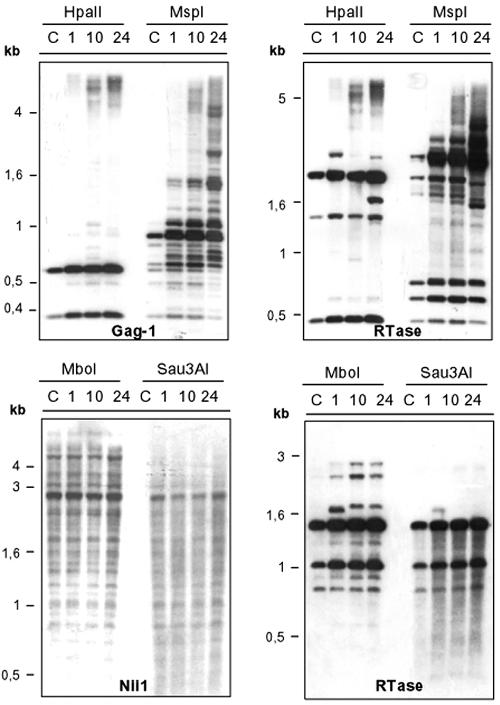
Therefore, this retroelement seems to have a high transpositional activity (Fig. 5B and 6C). Its integration into different genomic regions could explain the complex hybridization patterns observed in Southern blots. In addition, these patterns can be caused by de novo methylation modifications in new copies of CrREM1 or by varying the methylation background after transposition of this retroelement (Fig. 6C). Hence, we decided to study methylation patterns in strains N1, N10, and N24 and their parental strain 305CW15 using methylation-sensitive restriction endonucleases. HpaII and MspI recognize the target site 5′-CCGG-3′. HpaII does not cleave DNA when both the inner and outer C residues are methylated (m4CCGG, m5CCGG, Cm4CCGG, and Cm5CCGG) although it cleaves the hydroxymethylated sequence hm5Chm5CGG. In contrast, MspI is inhibited by 5-methylcytosine in the outer C (m5CCGG) and 5-hydroxymethylcytosine in both C residues (hm5Chm5CGG) but is not sensitive to methylations at m4CCGG, Cm4CGG, Cm5CGG.
Figure 7 shows hybridization with the probe Gag-1, where we should have observed two bands of 600 and 329 bp if CrREM1 is not methylated. Both hybridization signals were identified in all the lines although in the MspI-digested samples the intensity of these bands was very weak. Moreover, in MspI lines we observed bands with larger sizes than expected (Fig. 7). These results suggest that there exist copies of CrREM1 whose first C residue at the CCGG target site, within the studied region with the Gag-1 probe, would present the methylation type conferring inhibition of MspI digestion. A different methylation pattern was also observed when this hybridization was performed with the reverse transcriptase probe (Fig. 7). We expected three bands, one of 1371 bp only detected in HpaII digestion, another of 481 bp detected in both digestions but less intense in MspI lines, and a band of 403 bp which was not identified probably because it was not transferred to the nylon membrane. In both digestions again we can observe larger bands, indicating that reverse transcriptase of CrREM1 copies, especially in mutants N1, N10, and N24, are highly methylated and that DNA methylation occurred in different regions of this retroelement.
FIG. 7.
Analysis of methylation at regions of CrREM1. Total DNA (2 μg) from strains 305CW15 (C), N1 (1), N10 (10), and N24 (24) were digested with the endonucleases HpaII, MspI, Sau3AI, and MboI, which vary in their sensitivity to methylation, as detailed in the text. The probes used were Gag-1, RTase, and Nii1.
We also digested total DNA with the isoschizomer couple of endonucleases MboI/Sau3AI. They recognize the sequence 5′-GATC-3′, but MboI is inactive on methylation of type GmATC. As shown in Fig. 7, the hybridization patterns of MboI and Sau3AI were similar when this filter was hybridized with the NiR (endogenous gene) probe being identified the expected 2,687-bp band. This suggests that is region is not methylated and thus it is a valid control to assess the completeness of the restriction digestions. When the same filter was hybridized with reverse transcriptase probe (Fig. 7), the expected hybridization pattern (1,497 and 1,053 bp) was observed with both endonucleases together with some extra bands in the CSA mutants. These results indicate that the analyzed sequences of CrREM1 show a very partial methylation of type GmATC and very extensive cytosine methylation.
Analysis of CrREM1 expression.
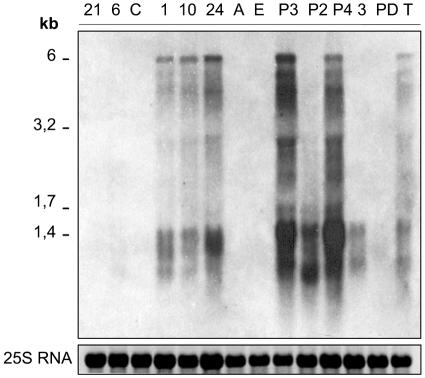
The first step to control retrotransposition activity of a retrotransposon is by regulation of its transcriptional initiation (12, 20). Thus, we studied the expression of CrREM1 transcripts from 14 parallel subcultures of the wild-type types and transformant strains analyzed above. Figure 8 shows RNA hybridization using the Gag-CrREM1 probe. We detected the overexpression of a transcript in the CSA mutants N1, N10, and N24 and the strains P3, P3-NR-4 and Tx-7 that would correspond with the expected size of a full primary transcript encoding CrREM1 polyproteins (6 kb). We have observed that this overexpression pattern is stably maintained in many of the above strains during several years of growth. In the lane of mutant P3-NR-2 bands corresponding to smaller transcripts were observed that were less intense in strain 305. No transcripts of the expected size were detected in any of the other analyzed strains. However, after a long overexposure of the filters the band of 6 kb could be observed in most of them (results not shown). The transcripts observed in Northern blot analysis showed discrete bands of a hybridizing RNA smear, presumably because of mRNA degradation. We obtained very similar results by using the reverse transcriptase probe (results not shown).
FIG. 8.
Expression of CrREM1 transcripts in wild-type and mutants strains of C. reinhardtii. Cells were grown in TAP medium containing 8 mM ammonium. Total RNA of the indicated strains (abbreviations are shown in the legend to Fig. 6) was extracted and subsequently analyzed in RNA transfer hybridizations using the specific GP-CrREM1 probe (Fig. 6B).
The Tx-7 strain possesses only one copy of CrREM1, which seems to be transcriptionally very active (Fig. 7). Therefore, no clear correlation exists between the increased copy number and the overexpression of transcripts detected with different specific regions of CrREM1 used as a probe. These results suggest that regulatory mechanisms of expression and integration of CrREM1 are independent.
The expression of the putative ORF3 was also studied by using an antisense single strand DNA probe; however, no transcript signal was detected. This result suggests that ORF3 would have a very low expression not easily detectable in Northern blots.
DISCUSSION
Structure and organization of CrREM1.
The attempt to identify the C. reinhardtii locus affected in mutant N1 isolated by insertional mutagenesis led us to detect an LTR retrotransposon of 5,812 bp that we named REM1, flanked by two identical 286-bp LTRs. The internal region of CrREM1 contained two ORFs encoded by gag and pol genes indicating that REM1 would synthesize a Gag-Pol polyprotein. The copy number of CrREM1 seems to be low like the families of endogenous retrotransposons in Arabidopsis thaliana, an organism with a genome size similar to that of C. reinhardtii, in agreement with the proposal that the abundance and distribution or retrotransposons would correlate with genome size differences (20).
The hypothetical protein products of the ORFs of CrREM1 include the domains Retro-gag, zinc finger, protease and reverse transcriptase conserved among autonomously active retrotransposons and retroviruses (3, 29), though typical RNase H and integrase domains could not be identified (Fig. 1). In spite of this fact, CrREM1 is functional since its primary transcript is overexpressed and multiple copies can be integrated. On the basis of these results, sequence comparison and phylogenetic study, we conclude that a new and unusual LTR retrotransposon related to the Ty3-gypsy family elements has been identified in C. reinhardtii.
The distinctive feature of CrREM1 is a particular coding region, named ORF3, which possesses a structural order and translational orientation different to that reported in LTR retrotransposons. This ORF3 was identified between the 5′ LTR and the gag gene and its transcriptional orientation is reverse to that of the REM1 polyprotein (Fig. 1). In addition ORF3p presents two specific domains, named PHD-finger and chromodomain; each of these functional domains have been described separately in some mobile elements. The characterization of DNA sequences adjacent to the 5′ and 3′ LTRs in all the isolated phages showed that we had isolated the same copy of CrREM1, probably the only one, present in the 21gr strain of C. reinhardtii. However, if another copy of REM1 with another gene structure is present in this strain, we would have been unable to detect it and PCR experiments designed to that purpose (not shown) would have failed.
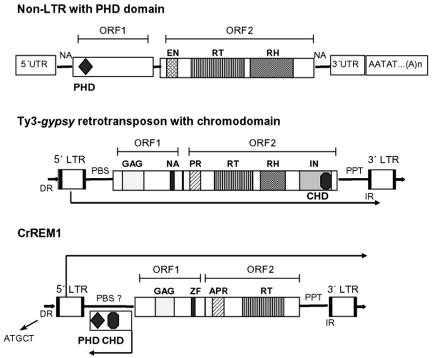
The PHD finger has been identified in the ORF1 proteins encoded by CR1 from fruit flies and T1 and Q non-LTR retrotransposons from the African malaria mosquito (15), and the chromodomain is encoded by the Ty3-gypsy group of retrotransposons at the integrase within the C-terminal end of pol (25) (Fig. 9). In the isolated copy of CrREM1, we could not identify the integrase in the order defined for that family of retroelements. Nevertheless, we could identify a chromodomain associated with a PHD-finger domain at the C terminus of ORF3p (Fig. 9) that could correspond to the integrase from CrREM1. A structural organization of ORFs or enzymatic modules similar to those in CrREM1 has not been described in other retrotransposons. This would imply that the presence of both domains in CrREM1 is highly divergent. Alternatively, CrREM1 could borrow integrase and RNase H proteins from other endogenous retrotransposons for its functionality.
FIG. 9.
Schematic structure of CrREM1, non-LTR and LTR retrotransposon with PHD and chromodomain. The boxes represent the ORFs in which the positions of the different domains are shown. PBS, primer binding sites; PPT, polypurine tract; NA, nucleic acid binding motif; ZF, zinc finger motif type C2H2; EN, endonuclease; CP, capsid-like protein; GAG, retro-Gag domain; PR, protease; RT, retrotranscriptase; RH, RNase H; IN, integrase; 5′-UTR, 5′ untranslated region; 3′-UTR, 3′ untranslated region; IR, inverted direct repeats; DR, flanking target direct repeat; CHD, chromatin organization modifier domain; and PHD, PHD-finger domain. The transcription senses of the LTR retrotransposon, CrREM1 primary transcript, and ORF3 transcript are shown by arrows.
The chromodomain is a conserved motif of about 60 amino acids that was originally identified with the cloning of the modifiers of position effect variegation Polycomb (33) and HP1 (14) in Drosophila melanogaster. The PHD-finger is a C4HC3-type zinc-finger motif reminiscent of, but distinct from, the C3HC4-type RING finger (6). The chromodomain is found in association with a striking variety of other functional domains (9). Specifically, in the subfamily of CHD3 proteins it is found clustered with PHD-type zinc fingers (31). The PHD and/or chromodomain proteins are involved in chromatin-mediated transcription control (1, 19), altering transcriptional activation or repression indirectly via protein-protein interactions (9, 23, 47). Therefore, its specificity on particular DNA regions depends on the combination of motifs present in each protein. Based on sequence similarity and functional analogy, PHD and chromodomain of CrREM1 might be implicated in the retrotransposition/integration of this retroelement by interaction with other proteins in processes of chromatin restructuring. The genetic inactivity has been associated with a heterochromatin state in many higher eukaryotes, including plants. Therefore, it is possible that PHD and chromodomain can minimize the deleterious effects of CrREM1 by channeling its integration to heterochromatin regions so that its transcription is inhibited.
We failed to find a sequence similar to CrREM1 in a search against the C. reinhardtii genome database. We have to consider that about 5% of the C. reinhardtii genome sequence not yet released could include regions of heterochromatin, telomeres, repetitive sequences, etc. CrREM1 copies might be at these chromosomal locations. In addition, the sequencing of the Chlamydomonas reinhardtii genome was carried out from the cw92 strain and we obtained the copies of CrREM1 from a genomic DNA library of the wild-type strain 21gr.
Transposition and transcription of CrREM1.
REM1 is overexpressed in some C. reinhardtii strains while in others its transcripts could hardly be detected (Fig. 8), only after overexposing the films. These results seem to indicate that the regulation/silencing mechanism of this retroelement is not active in all strains. On the other hand, it could be observed that REM1 is a distinctively low copy number retrotransposon in the C. reinhardtii strains studied and in some mutant strains, mainly in those with major changes in their genome (transformation and genetic crosses), REM1 has increased its copy number. This low copy number might reflect a highly efficient mechanism for inhibiting its transposition and integration in spite of very active transcription.
These results open several questions: Why and how does CrREM1 copy number increase? A54 and E18 do not have any CrREM1 copies in their genome, whereas wild-type strains 21gr and 6145c probably possess a single copy since they come from different genetic backgrounds. Strain P3, a segregant from the genetic cross between 6145c and E18, presents six to eight copies in its genome. Mutants N1, N10, and N24 also possess a CrREM1 copy number higher than that of their parental strain 305CW15, which was transformed with the pMN24 plasmid to generate these CSA mutants (Fig. 6C). In plants, biotic and abiotic stress factors activate transcriptionally retrotransposons that are inactive during normal growth (10). However, the C. reinhardtii strains showing amplification of REM1 had not been exposed to any of these factors. Therefore, the only common feature among those strains is that all underwent either transformation and/or genetic crossover events.
Activation of retrotransposons by introduction of foreign DNA has also been observed in marsupials (32) and plants (13, 24). In fact, when Arabidopsis thaliana is transformed with tobacco retrotransposon Tto1, transpositional activation of Tto1 and the endogenous retrotransposon Tar17 is observed (13). However, the process of transformation or generation of hybrid lines is not always associated to the mobilization of retrotransposons (35). In agreement with this, we also found that the C. reinhardtii transformant strain Tx-7 did not increase the single REM1 copy present in its parental strain 305. Our results lead us to propose that factors participating in the insertion of foreign DNA during transformation of C. reinhardtii and/or meiotic recombination promote transposition of REM1 in a process that might or not be productive in this copy number increase. However, mitotic events do not appear to favor REM1 transposition since cells of the studied strains have grown vegetatively during a very long time (years) with a stable copy number.
Another question is why CrREM1 is overexpressed in some strains. Since retrotransposons transpose by an RNA intermediate, the inhibition of their transcription would be an effective way to minimize and regulate the possible deleterious effects of retrotransposition in the host. DNA methylation might prevent retrotransposition of transposable elements (13, 32, 48). In fact, retrotransposon activation by DNA introduction in rice is followed by rapid repression and DNA methylation (24). The methylation analysis of CrREM1 regions using Gag-1 and reverse transcriptase probes in strains overexpressing CrREM1 transcript indicate that they contain hypermethylated and demethylated copies of CrREM1 and copies which might have internal deletions. Hirochika et al. (13) have shown that Tto1 of Arabidopsis, initially silenced by hypermethylation, became transcriptional and transpositionally reactivated when introduced in a homozygous ddm1 (“decrease DNA methylation 1”) background. Therefore, they suggest that this mechanism is necessary for silencing of repeated genes. Liu and Wendel (24) have also proposed this way of silencing by DNA methylation to regulate the reactivation of the element Tos17.
Methylation of CrREM1 might also serve as a control mechanism of its transposition. However, not all CrREM1 copies were silenced, since it was transcriptionally very active. This implies that methylation is not the only mean of silencing retrotransposon in C. reinhardtii. In Drosophila melanogaster, retrotransposons are located mainly in regions of the heterochromatin (36) and are silenced by a group of polycomb proteins (45).
The increase in CrREM1 copy number could be associated with cytosine changes as described in rice (24) but this phenomenon should be more extensively studied. In hybrid lines of rice (Oryza sativa × Zea latifolia), it has been observed that occasional internal deletion of a gypsy retrotransposon is accompanied by the activation of the element (24). This could explain CrREM1 overexpression in mutant N1, where the CrREM1 copy isolated by inverse PCR had a deletion of 1,590 bp which affected partially to the 5′ LTR and the gag gene and was probably caused by the pMN24 insertion. This phenomenon could have also occurred in other mutant strains that overexpressed CrREM1 and have been transformed with selectable markers.
In conclusion, we have isolated a new LTR retrotransposon in C. reinhardtii (REM1) which possesses structural features not described to date, and is phylogenetically related to those of the Ty3-gypsy type. An ORF containing a PHD-finger and a chromodomain is present in CrREM1 with a transcriptional orientation reverse to those of the primary retrotransposon and gag-pol transcripts. CrREM1 can amplify its copy number in strains that undergo phenomena of foreign DNA integration and/or genetic crosses. Though extensive methylation might occur in some copies of the retrotransposon another copies are highly active transcriptionally in a stable fashion. However, this fact does not imply an increase in the retrotransposon copy number that is also stably maintained during very long periods of cell growth. It is proposed that elements involved in the integration of foreign DNA and/or meiotic recombination are needed for integration and transposition of this retrotransposon, probably directed to heterochromatin locations.
Acknowledgments
We thank M. I. Macías for technical support, C. Santos and I. Molina for secretarial assistance, and A. Galván for critical reading of the manuscript.
This work was supported by Ministerio de Ciencia y Tecnología, Spain (MCYT grant no. BMC2002-03325), Junta de Andalucía, Spain (Plan Andaluz de Investigación, CVI-0128), and the Vth Framework Program of the EU (HPRN-CT-2002-00247).
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Bennetzen, J. L. 2000. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42:251-269. [PubMed] [Google Scholar]
- 3.Boeke, J. D., and V. G. Corces. 1989. Transcription and Reverse Transcription of Retrotransposons. Annu. Rev. Microbiol. 43:403-433. [DOI] [PubMed] [Google Scholar]
- 4.Bowen, N. J., and I. K. Jordan. 2002. Transposable elements and the evolution of eukaryotic complexity. Curr. Issues Mol. Biol. 4:65-76. [PubMed] [Google Scholar]
- 5.Britten, R. J. 1995. Active gypsy/Ty3 retrotransposons or retroviruses in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 92:599-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capilli, A. D., D. C. Schultz, F. J. Rauscher, and K. L. B. Borden. 2001. Solution structure of the PHD domain from KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day, A., M. Schimer-Rahire, M. R. Kuchka, S. P. Mayfield, and J. D. Rochaix. 1988. A transposon with an unusual arrangement of long terminal repeats in the green alga Chlamydomonas reinhardtii. EMBO J. 7:1917-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day, A., and J. D. Rochaix. 1991. Structure and inheritance of sense and anti-sense transcripts from a transposon in the green alga Chlamydomonas reinhardtii. J. Mol. Biol. 218:273-291. [DOI] [PubMed] [Google Scholar]
- 9.Eissenberg, J. C. 2001. Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene 275:19-29. [DOI] [PubMed] [Google Scholar]
- 10.Grandbastein, M. A. 1998. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3:181-187. [Google Scholar]
- 11.Harris, E. H. 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Academic Press, New York, N.Y. [DOI] [PubMed]
- 12.Hirochika, H. 1997. Retrotransposons of rice: their regulation and use for genome analysis. Plant Mol. Biol. 35:231-240. [PubMed] [Google Scholar]
- 13.Hirochika, H., H. Okamoto, and T. Kakutani. 2000. Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12:357-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James, T. C., and S. C. Elgin. 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6:3862-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapitonov, V. V., and J. Jurka. 2003. The Esterase and PHD Domains in CR1-Like Non-LTR Retrotransposons. Mol. Biol. Evol. 20:38-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidwell, M. G., and D. R. Lisch. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55:1-24. [DOI] [PubMed] [Google Scholar]
- 17.Kim, A. I., C. Terzian, P. Santamaria, A. Pélisson,. N. Prud'homme, and A. Bucheton. 1994. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91:1285-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kindle, K. L., R. A. Schnell, E. Fernández, and P. A. Lefebvre. 1989. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 109:2589-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin, E. V., S. Zhou, and J. C. Lucchesi. 1995. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 23:4229-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar, A., and J. L. Bennetzen. 1999. Plant retrotransposons. Annu. Rev. Genet. 33:479-532. [DOI] [PubMed] [Google Scholar]
- 21.Kumekawa, N., H. Ohtsubo, T. Horiuchi, and E. Ohtsubo. 1999. Identification and characterization of novel retrotransposons of the gypsy type in rice. Mol. Gen. Genet. 260:593-602. [DOI] [PubMed] [Google Scholar]
- 22.Leeton, P. R., and D. R. Smyth. 1993. An abundant LINE-like element amplifies in the genome of Lilium speciosum. Mol. Gen. Genet. 237:97-104. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., D. A. Kirschmann, and L. L. Wallrath. 2002. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. USA 99:16462-16469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, B., and J. F. Wendel. 2000. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43:874-880. [PubMed] [Google Scholar]
- 25.Malik, H. S., and T. H. Eickbush. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 73:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manninen, I., and A. H. Schulman. 1993. BARE-1, a copia-like retroelement in barley (Hordeum vulgare L.). Plant Mol. Biol. 22:829-846. [DOI] [PubMed] [Google Scholar]
- 27.Marsano, R. M., R. Moschetti, C. Caggese, C. Lanave, P. Barsanti, and R. Caizzi. 2000. The complete Tirant transposable element in Drosophila melanogaster shows a structural relationship with retrovirus-like retrotransposons. Gene 247:87-95. [DOI] [PubMed] [Google Scholar]
- 28.Mount, S. M., and G. M. Rubin. 1995. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol. Cell. Biol. 5:1630-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayashiki, H., H. Matsuo, I. Chuma, K. Ikeda, S. Betsuyaku, M. Kusaba, Y. Tosa, and S. Mayama. 2001. Pyret, a Ty3/Gypsy retrotransposon in Magnaporthe grisea contains an extra domain between the nucleocapsid and protease domains. Nucleic Acids Res. 29:4106-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navarro, M. T., E. Guerra, E. Fernández, and A. Galván. 2000. Nitrite reductase mutants as an approach to understanding nitrite assimilation in Chlamydomonas reinhardtii. Plant Physiol. 122:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogas, J., S. Kaufmann, J. Henderson, and C. Somerville. 1999. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96:13839-13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O′Neill, R. J. M., M. J. O′Neill, and J. A. M. Graves. 1998. Under-methylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 393:68-72. [DOI] [PubMed] [Google Scholar]
- 33.Paro, R., and D. S. Hogness. 1991. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. USA 88:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parraga, G., S. J. Horvath, A. Eisen, W. E. Taylor, L. Hood, E. T. Young, and R. E. Klevit. 1988. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science 241:1489-1492. [DOI] [PubMed] [Google Scholar]
- 35.Pearce, S. R., G. Harrison, J. S. Heslop-Harrison, A. J. Flavell, and A. Kumar. 1997. Characterization and genomic organization of the Ty1-copia group retrotransposons in rye (Secale cereale). Genome 40:617-625. [DOI] [PubMed] [Google Scholar]
- 36.Pimpinelli, S., M. Berloco, L. Fanti, P. Dimitri, S. Bonaccorsi, E. Marchetti, R. Caizzi, C. Caggese, and M. Gatti. 1995. Transposable elements are stable structural components of the Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci. USA 92:3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prieto, R., A. Dubus, A. Galván, and E. Fernández. 1996. Isolation and characterization of two regulatory mutants for nitrate assimilation in Chlamydomonas reinhardtii. Mol. Gen. Genet. 251:461-471. [DOI] [PubMed] [Google Scholar]
- 38.Quesada, A., A. Galván, R. Schnell, P. A. Lefebvre, and E. Fernández. 1993. Five nitrate assimilation related loci are clustered in Chlamydomonas reinhardtii. Mol. Gen. Genet. 240:387-394. [DOI] [PubMed] [Google Scholar]
- 39.Quesada, A., I. Gomez, and E. Fernández. 1998. Clustering of the nitrite reductase gene and a light-regulated gene with nitrate assimilation loci in Chlamydomonas reinhardtii. Planta 206:259-265. [DOI] [PubMed] [Google Scholar]
- 40.Rexach, J., E. Fernández, and A. Galván. 2000. The Chlamydomonas reinhardtii Nar1 gene encodes a cloroplast membrane protein involved in nitrite transport. Plant Cell 12:1441-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sanmiguel, P., and J. L. Bennetzen. 1998. Evidence that a recent increase maize genome size was caused by the massive amplification of integrase retrotransposons. Ann. Bot. 81:37-44. [Google Scholar]
- 43.Schloss, J. A., C. D. Silflow, and J. L. Rosenbaum. 1984. mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol. Cell. Biol. 4:424-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varmus, H., and P. Brown. 1989. Mobile DNA, p. 53-108. In D. E. Berg and M. M. Howe (ed.), Retroviruses. American Society for Microbiology, Washington, D.C.
- 45.Wolffe, A. P., and M. A. Matzke. 1999. Epigenetics: Regulation through repression. Science. 286:481-486. [DOI] [PubMed] [Google Scholar]
- 46.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yochum, G. S., and D. E. Ayer. 2001. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol. 21:4110-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, Y., E. B. Cambareri, and J. A. Kinsey. 2001. DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Mol. Genet. Genomics 265:748-754. [DOI] [PubMed] [Google Scholar]