Abstract
Aerobic metabolism produces reactive oxygen species, including superoxide anions, which cause DNA damage unless removed by scavengers such as superoxide dismutases. We show that loss of the Cu,Zn-dependent superoxide dismutase, SOD1, or its copper chaperone, LYS7, confers oxygen-dependent sensitivity to replication arrest and DNA damage in Saccharomyces cerevisiae. We also find that sod1Δ strains, and to a lesser extent lys7Δ strains, when arrested with hydroxyurea (HU) show reduced induction of the MEC1 pathway effector Rnr3p and of Hug1p. The HU sensitivity of sod1Δ and lys7Δ strains is suppressed by overexpression of TKL1, a transketolase that generates NADPH, which balances redox in the cell and is required for ribonucleotide reductase activity. Our results suggest that the MEC1 pathway in sod1Δ mutant strains is sensitive to the altered cellular redox state due to increased superoxide anions and establish a new relationship between SOD1, LYS7, and the MEC1-mediated checkpoint response to replication arrest and DNA damage in S. cerevisiae.
Reactive oxygen species (ROS), which include superoxide anions (O2−) and hydroxyl free radicals (OH−), are by-products of aerobic life (36). Both nonenzymatic and enzymatic mechanisms can remove ROS and maintain a reducing environment. Nonenzymatic defenses include production of glutathione, glutaredoxins, thioredoxins, and NADPH (14). Enzymatic defenses include superoxide dismutases (SOD), which are responsible for O2− scavenging and dismutation to hydrogen peroxide (H2O2) and O2, and peroxidases, including thiol-dependent peroxiredoxins and thiol-independent catalases, which dissociate H2O2 to water and O2 (4). Failure to detoxify ROS leads directly or indirectly to DNA damage, oxidation of protein and lipids, increased mutation rates, aneuploidy, and, ultimately, cell death (47, 60). In mammalian cells, accumulation of ROS induced by ionizing radiation or exogenous H2O2 activates the ATM/ATR pathway, resulting in increased expression of the tumor suppressor p53 followed by cell cycle arrest and/or apoptosis (4, 35, 62). Patients with defective DNA damage signaling pathway syndromes, such as ataxia telangiectasia, or DNA repair deficiency syndromes, such as Fanconi's anemia or Bloom's syndrome, are susceptible to DNA damage and increased oxidative stress (55).
As befits their role as the first line of defense against ROS and oxidative stress in the cell, SODs are conserved evolutionarily, although the enzymes vary in their subcellular localization and requirements for metal ion cofactors (14). In humans, mutations in cytosolic SOD1 have been implicated in 20% of cases of the familial form of the motor neuron disease amyotrophic lateral sclerosis (Lou Gehrig's disease) (43, 44). Decreased affinities for zinc and copper ions have been implicated in the protein dimer destabilization and toxic gain-of-function properties of SOD1 protein (3, 13, 24, 41). In Saccharomyces cerevisiae, SOD is found in two forms: Sod1p, which depends on copper and zinc for its activity and localizes to the cytosol, the nuclei, and the inner membrane spaces of mitochondria (51), and Sod2p, which relies on manganese for its activity and localizes exclusively to the mitochondria (40). Sod1p activity is exquisitely responsive to intracellular copper levels and depends on the copper chaperone, Lys7p, for copper loading (16). Strains lacking SOD1 and LYS7 exhibit a variety of oxygen-dependent phenotypes, including slow growth, sensitivity to hyperoxia and superoxide-generating agents such as menadione and paraquat (50), increased spontaneous mutation rates (23), and methionine and lysine auxotrophy (14). The auxotrophies arise due to disruption of biosynthetic pathways of these amino acids by an altered redox environment. Decreased NADPH levels and oxidation of a putative iron-sulfur (4Fe-4S) reactive center of homoaconitase (Lys4p) in sod1Δ strains lead to blockage of methionine and lysine biosynthetic pathways, respectively (14, 49, 58).
As little is known about the molecular relationships between accumulation of excess superoxides and DNA damage checkpoint pathways, we used budding yeast as a model system to dissect these relationships. In S. cerevisiae, replication blocks and DNA damage generate signals that activate the protein kinase Mec1p, the ortholog of human ATM/ATR, which transmits these signals through two downstream pathways. In the first pathway, MEC1-dependent phosphorylation and activation of Rad53p, the ortholog of human Chk2, leads to phosphorylation and activation of Dun1p, which mediates the transcriptional induction of gene products involved in DNA synthesis and repair, including the subunits of ribonucleotide reductase (RNR), and Hug1p (6, 18). In the second pathway, Mec1p activation results in the Rad53p-independent activation of Chk1p and Pds1p (11, 21). Both pathways mediate a cell cycle arrest allowing sufficient time for the cell to repair the replication block or DNA damage (21, 45).
In this paper, we investigate the roles of SOD1 and LYS7 in the MEC1-dependent DNA damage checkpoint response and the oxygen-dependent phenotypes of sod1Δ and lys7Δ strains. We demonstrate that, in the absence of SOD1 and LYS7, induction of Rnr3p and Hug1 in response to replication arrest or DNA damage is dramatically reduced in the presence of oxygen. These null strains also exhibit oxygen-dependent sensitivity to hydroxyurea (HU) and DNA-damaging agents, suggesting that increased oxidative stress due to elevated levels of superoxide anions compromises the MEC1-dependent response to replication arrest and DNA damage. The oxygen-dependent HU sensitivity of these strains was suppressed by addition of N-acetyl-l-cysteine (NAC), an antioxidant that scavenges ROS (2). These data led us to screen for high-copy-number suppressors of the oxygen-dependent HU sensitivity of sod1Δ strains. In this screen we identified TKL1, a transketolase of the nonoxidative branch of the pentose phosphate pathway, which restores NADPH levels and a cellular reducing environment. We have also demonstrated that TKL1 partially restores Rnr3p and Hug1p induction in sod1Δ strains treated with HU under normoxic conditions. These results suggest that certain aspects of the MEC1 pathway are sensitive to the altered cellular redox state created by excess superoxide anions and point to a functional relationship between SOD1 and LYS7 and the MEC1-mediated checkpoint response to replication arrest and DNA damage in S. cerevisiae.
MATERIALS AND METHODS
Media, strains, plasmids, and general methods.
Media and methodology used for yeast growth were as described previously (1, 28, 48), except where indicated. All yeast strains are listed in Table 1. For growth under hypoxic conditions, strains were incubated at 30°C in BBL GasPak anaerobic culture jars (Becton-Dickinson Labware, Lincoln Park, NJ), and oxygen was depleted using a CO2-generating system with palladium catalyst (50). Normoxic conditions were equivalent to standard growth conditions at 30°C. Plasmids pLJ175 (pSOD1/CEN/URA3), pLS113 (pLYS7/CEN/HIS3) and pKS10 (pTKL1/2μm/LEU2) were generous gifts of V. Culotta, and pBAD54 (GAP vector) and pBAD790 (GAP-RNR1) were generous gifts of S. Elledge. KS103 was engineered with the sod1Δ::TRP1 plasmid pKS3 in strain background FY250 (15). The sod1Δ sml1Δ strains were constructed by mating strains U952-3C and BY4741/sod1Δ, and the meiotic progeny of two independent diploids were analyzed.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BY4741 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics (Huntsville, AL) |
| BY4741 (sod1Δ) | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sod1Δ::KANMX4 | Research Genetics (Huntsville, AL) |
| BY4741 (lys7Δ) | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lys7Δ::KANMX4 | Research Genetics (Huntsville, AL) |
| BY4741 (skn7Δ) | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 skn7Δ::KANMX4 | Research Genetics (Huntsville, AL) |
| BY4741 (tsa1Δ) | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tsa1Δ::KANMX4 | Research Genetics (Huntsville, AL) |
| BY4741 (yap1Δ) | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yap1Δ:KANMX4 | Research Genetics (Huntsville, AL) |
| W1588-4A | mataleu2-3,112 ade2-1 can1-100 his3-11,15 ura3-1 trp1-1 RAD5 | 64 |
| U952-3C | mataleu2-3,112 ade2-1 can1-100 his3-11,15 ura3-1 trp1-1 RAD5 sml1Δ::HIS3 | 64 |
| U953-61D | mataleu2-3,112 ade2-1 can1-100 his3-11,15 ura3-1 trp1-1 RAD5 mec1Δ::TRP1 sml1Δ::HIS3 | 64 |
| U960-5C | matα leu2-3,112 ade2-1 can1-100 his3-11,15 ura3-1 trp1-1 RAD5 rad53Δ::HIS3 sml1-1 | R. Rothstein |
| EG103 | matα leu2-3,112 his3Δ1 trp1-289a ura3-52 | 24 |
| KS101 | matα leu2-3,112 his3Δ1 trp1-289a ura3-52 sod1Δ::LEU2 | 17 |
| KS103 | matα leu2-3,112 his3Δ1 trp1-289a ura3-52 lys7Δ::LEU2 | V. Culotta |
| 1783 | mataleu2-3,112 his4 trp1-1 ura3-52 can1r | 51 |
| KS105 | mataleu2-3,112 his4 trp1-1 ura3-52 can1r sod1Δ::TRP1 | 51 |
| KS113 | mataleu2-3,112 his4 trp1-1 ura3-52 can1r zwf1Δ::URA3 | 51 |
| KS117 | mataleu2-3,112 his4 trp1-1 ura3-52 can1r sod1Δ::TRP1 zwf1Δ::URA3 | 51 |
| YMB3233 | mataleu2 ura3 his3 | This study |
| YMB3234 | mataleu2 ura3 his3 trp1 sml1Δ::HIS3 | This study |
| YMB3235 | mataleu2 ura3 his3 sod1Δ::KANMX4 | This study |
| YMB3236 | mataleu2 ura3 his3 trp1 smll1Δ::HIS3 sod1Δ::KANMX4 | This study |
Sensitivity to replication arrest and DNA damage.
All strains were grown overnight at 30°C under hypoxic conditions and then diluted in fresh medium to obtain a logarithmic phase culture under hypoxic conditions. For growth spotting assays, three microliters of serial dilutions (1:5) of logarithmic phase cultures was spotted on yeast-peptone-dextrose agar (YPD) or YPD containing HU (H8627; Sigma, St. Louis, MO), methane methylsulfonate (MMS) (64294; Fluka Chemika, Switzerland), 4-nitroquinoline-1-oxide (N8141; Sigma, St. Louis, MO) or bleomycin (BLM) (3154-01; Bristol-Myers Squibb Co., Princeton, NJ) at indicated concentrations. For NAC suppression assays, cells were spotted on YPD or YPD containing HU with or without NAC (A7250; Sigma, St. Louis, MO) at indicated concentrations. For viability measurements, logarithmic phase cultures grown under hypoxic conditions were plated in triplicate on YPD or YPD containing 100 mM HU or 0.02% MMS and then incubated under normoxic and hypoxic conditions. For recovery experiments, strains were treated with 100 mM HU for 3, 6 or 9 hours, cells were washed, and appropriate dilutions were plated on YPD. Results (see Fig. 2) are averages from three experiments. Wild-type strain viabilities were taken as 100%.
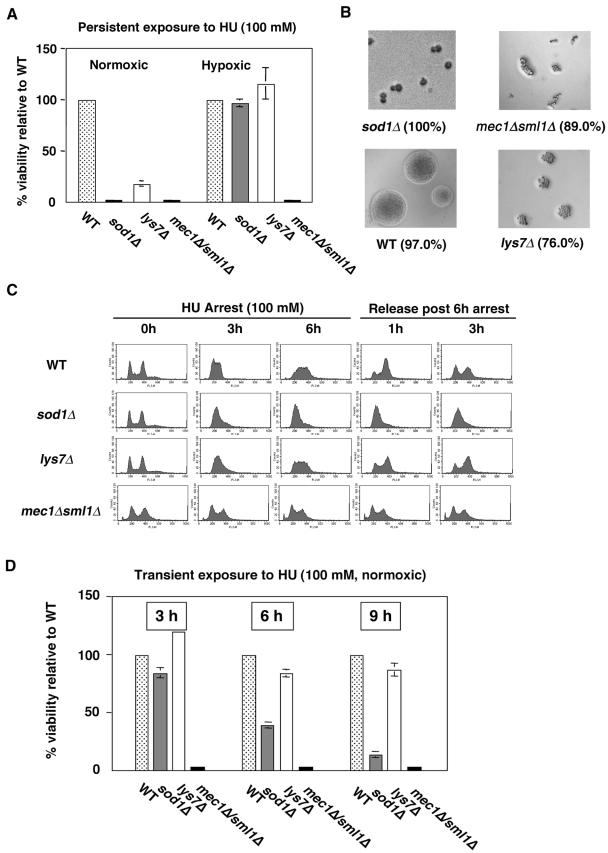
FIG. 2.
Strains lacking SOD1 and LYS7 arrest, but sod1Δ strains fail to produce viable colonies on HU medium. (A) Wild-type (WT; BY4741), sod1Δ, lys7Δ, and mec1Δ sml1Δ (U953-61D) strains were plated on YPD containing 100 mM HU, and plates were incubated under hypoxic or normoxic conditions for 3 to 6 days. lys7Δ strains showed small colonies only upon prolonged incubation for 8 days at 30°C. (B) Terminal phenotypes of strains from panel A grown on HU-containing medium under normoxic conditions were recorded using a light microscope. The percentage of cells with the depicted morphology is indicated in parentheses for each strain. (C) DNA content analysis by flow cytometry. Wild-type (BY4741), sod1Δ, lys7Δ, and mec1Δ sml1Δ (U953-61D) strains were grown under normoxic conditions to logarithmic phase (column 1) and treated with 100 mM HU for 3 hours (column 2) or 6 hours (column 3). Strains arrested with 100 mM HU for 6 hours were released into drug-free medium, and an aliquot of cells was removed every 30 min for up to 4 hours. The DNA contents of cells from 1 (column 4) and 3 hours (column 5) post-HU release are shown. Cells from these samples were analyzed by flow cytometry, and DNA fluorescence units per cell were measured. (D) Wild-type (BY4741), sod1Δ, lys7Δ, and mec1Δ sml1Δ (U953-61D) strains were grown under normoxic conditions to logarithmic phase and treated with 100 mM HU for 6 hours. Cells were washed, and appropriate dilutions were plated on YPD, allowed to recover under normoxic conditions, and counted after 2 to 3 days of incubation at 30°C. For panels A and D, at least 2,500 cells per strain were plated per experiment. For each experiment, the percent viability of wild-type strains was considered to be 100%. Plating efficiency for wild-type cells was 36% on YPD containing HU versus YPD alone. Numbers are averages from three experiments, and error bars represent standard deviation.
Cell cycle arrest.
Logarithmic phase cultures were treated with 100 mM HU for 3 or 6 hours and then analyzed by flow cytometry as described previously (5), using a Becton-Dickinson FACSort flow cytometer and CellQuest software (BD Biosciences, Boston, MA). For arrest and release experiments, logarithmic phase cultures were treated with 100 mM HU for 6 hours and then washed and resuspended in YPD lacking drug. Aliquots were taken at indicated times and analyzed by flow cytometry as described above. At least 10,000 cells were counted for each sample. Microtubule morphology was examined by tubulin staining, which was performed as described previously (27), using cells arrested by treatment with 100 mM HU for 3 hours.
Expression analysis by Western blotting.
For Western blot analysis of Rad53p, strains were grown to logarithmic phase in hypoxic conditions and then treated as indicated, and whole-cell extracts (WCE) were isolated using a trichloroacetic acid extraction method as follows. Cell pellets were resuspended in 2 N NaOH-1.2 M 2-mercaptoethanol and incubated on ice for 10 min. An equal volume of 50% TCA was added, and samples were further incubated on ice. Samples were then centrifuged (4°C, 1 min, 13,000 rpm), supernatant was discarded, and pellets were washed with ice-cold acetone and recentrifuged. Cell pellets were resuspended in 5% sodium dodecyl sulfate, boiled at 100°C for 3 minutes, and centrifuged, and supernatant was collected as WCE. The Bio-Rad RC/DC protein assay kit (500-0113; Bio-Rad Laboratories, Hercules, CA) was used to determine protein concentration according to the manufacturer's protocol. Forty micrograms of WCE was separated on NuPAGE 4 to 12% Bis-Tris polyacrylamide gels (Invitrogen, Carlsbad, CA) with MOPS (morpholinepropanesulfonic acid) buffer and transferred to Protran nitrocellulose (pore size, 0.2 μm) (BA83; Schleicher and Schuell, Keene, NH). Membranes were blocked with 5% nonfat milk in 1× Tris-buffered saline containing 0.3% Triton X-100 and then probed using a goat polyclonal antibody (yC-19) raised against the C terminus of Rad53p (1:500) (sc-6749; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) followed by horseradish peroxidase (HRP)-conjugated bovine anti-goat immunoglobulin G secondary antibody (1:5,000) (sc-2350; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). For RNR subunit and Hug1p Western blot analyses, indicated strains were grown as described above, and WCE were made by the boil-freeze method as previously described (34). Extracts were separated using 10% Tris-Tricine sodium dodecyl sulfate-polyacrylamide gels and transferred to Immobilon polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 5% nonfat milk in 1× phosphate-buffered saline (pH 7.0) and then probed using antibodies to Rnr1p (1:10,000), Rnr2p (1:20,000), Rnr3p (1:10,000), Hug1p (1:500), or Tub2p (1:50,000) followed by HRP-conjugated donkey anti-rabbit immunoglobulin G secondary antibody (1:7,500) (NA934V; Amersham Biosciences, Piscataway, NJ). All antibodies were diluted in blocking buffer. Extracts were also tested separately with a rabbit polyclonal antibody raised to full-length Rad53p kindly provided by David Stern (53; data not shown). RNR antibodies were a kind gift of J. Stubbe. The rabbit polyclonal antibody to Tub2p was generously provided by D. Koshland. HRP activity was detected using the Super Signal West Pico chemiluminescent substrate kit (Pierce Biotechnology Inc., Rockford, IL) followed by exposure to BioMax Light film (Eastman Kodak Co., Rochester, NY).
High-copy-number suppression screen.
BY4741/sod1Δ was transformed with a pRS202-based library (2μm/URA3) (12) and incubated at 30°C under normoxic conditions on medium lacking uracil. Approximately 2,000 transformants were replica plated to YPD containing 100 mM HU, and two colonies were verified as HU resistant. The library plasmids rescued from these strains complemented the HU sensitivity of the BY4741/sod1Δ strain. DNA sequencing of the two plasmids indicated that the complementing clones contained a 7.7-kb insert from chromosome XVI (nucleotides 687952 to 695669) containing TKL1.
RESULTS
Loss of SOD1 and LYS7 sensitizes cells to replication arrest and DNA damage in an oxygen-dependent manner.
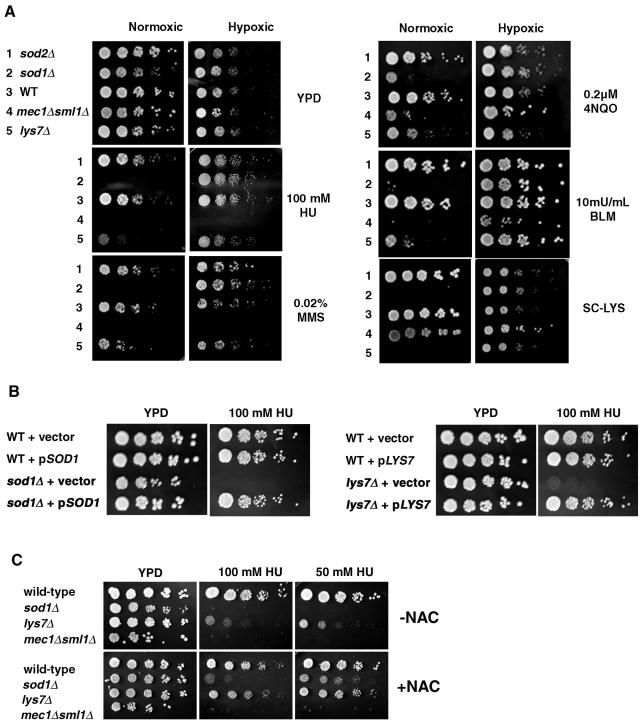
A genome-wide screen for suppressors of genomic instability by Kolodner and coworkers identified SOD1 and LYS7 and three other oxidative stress response genes (TSA1, YAP1, and SKN7) as candidates (25, 26). SOD1 and LYS7 were also identified in two separate genome-wide screens of S. cerevisiae haploid deletion strains for sensitivity to MMS and HU (10, 38). As both sod1Δ and lys7Δ strains exhibit several oxygen-dependent phenotypes, including lysine and methionine auxotrophies (50), we tested whether the sensitivity of these strains to replication arrest and DNA damage was also oxygen dependent. We also tested a null strain of the mitochondrial superoxide dismutase, a SOD2 strain, and a MEC1 pathway mutant, a mec1Δ sml1Δ strain. Under normoxic conditions, where oxygen levels are normal, sod2Δ, mec1Δ sml1Δ, and lys7Δ strains grew as well as the isogenic wild-type strain on YPD (Fig. 1A), while the sod1Δ strain exhibited slightly reduced growth as previously reported (23). On YPD containing the replication arrest agent, HU, or DNA-damaging agent 4-nitroquinoline-N-oxide, BLM, or MMS, or on minimal medium lacking lysine, sod2Δ strains grew as well as wild-type strains, but sod1Δ and lys7Δ strains showed growth inhibition (Fig. 1A). These phenotypes of sod1Δ and lys7Δ are not observed under hypoxic conditions, where levels of oxygen are reduced (Fig. 1A). The lys7Δ strain was less sensitive to these agents than the sod1Δ strain. We also confirmed that the phenotypes observed for sod1Δ and lys7Δ were due to loss of SOD1 and LYS7, since plasmid-borne copies of SOD1 or LYS7 complement HU sensitivity of sod1Δ and lys7Δ strains, respectively (Fig. 1B). As the growth inhibition phenotypes were oxygen dependent and thus may depend on increased levels of ROS that occur in the absence of superoxide anion scavenging, we sought to determine whether the antioxidant NAC suppressed HU sensitivity (Fig. 1C). Addition of NAC to the medium containing HU allowed some growth of sod1Δ and lys7Δ strains in 100 mM HU and significant growth of both strains in 50 mM HU. Addition of NAC, however, did not suppress the HU sensitivity of mec1Δ sml1Δ strains.
FIG. 1.
Sensitivity to replication arrest and DNA damage is oxygen dependent in the absence of SOD1 or LYS7. (A) Three microliters of fivefold serial dilutions of sod2Δ, sod1Δ, wild-type (BY4741), mec1Δ sml1Δ (U953-61D), or lys7Δ strains was spotted onto YPD or YPD containing 100 mM HU, 0.02% MMS, 0.2 μM 4NQO, or 10 mU/ml BLM or minimal medium lacking lysine (SC-LYS) and incubated under normoxic or hypoxic conditions at 30°C for 3 to 5 days. (B) Wild-type (BY4741), sod1Δ, and lys7Δ strains were transformed with vector alone, pLJ175 (pSOD1/CEN/URA3), or pLS113 (pLYS7/CEN/URA3). Three microliters of fivefold serial dilutions of each transformant was spotted onto YPD with or without 100 mM HU and incubated under normoxic conditions at 30°C for 2 to 3 days. (C) Wild-type (BY4741), sod1Δ, lys7Δ, and mec1Δ sml1Δ (U953-61D) strains were spotted onto YPD or YPD containing 100 mM or 50 mM HU with or without 100 mM NAC and incubated under normoxic conditions at 30°C for 3 to 5 days. WT indicates wild-type strains.
We reasoned that the HU and DNA damage sensitivity phenotypes of sod1Δ and lys7Δ strains might be due to presence of excess superoxide anions caused by Sod1p and Lys7p deficiency and not due to an excess of other forms of ROS. We tested strains lacking genes that mediate the oxidative stress response to H2O2 (SKN7, TSA1, and YAP1) (26) and determined that these strains were able to grow on medium containing HU or MMS (see Fig. S1 in the supplemental material). Taken together, these results suggest that SOD1 and LYS7 are required for oxygen-dependent resistance to replication arrest and DNA damage and that the sensitivity of these strains to HU and MMS may be due to excess superoxide anions that arise in the absence of these superoxide anion scavengers.
Strains lacking SOD1 arrest but fail to produce viable colonies on HU-containing medium.
Our results have shown that sod1Δ and to a lesser extent lys7Δ strains exhibit sensitivity to growth on HU-containing medium similar to the checkpoint mutant (mec1Δ sml1Δ). To determine whether these strains were similar to or distinct from mec1Δ sml1Δ in terms of growth phenotype, we analyzed these strains for (i) viability and terminal phenotype in the persistent presence of HU, (ii) ability to arrest the cell cycle in response to HU treatment, and (iii) cell cycle progression and viability upon release from transient HU treatment. Quantitative analyses showed that on HU-containing medium sod1Δ strains formed no visible colonies under normoxic conditions and a small percentage (12.5%) of lys7Δ cells formed very small colonies only after prolonged incubation (8 days) under normoxic conditions (Fig. 2A). This effect was oxygen dependent as the viability of sod1Δ strains was only slightly reduced and that of lys7Δ strains was unaffected on HU-containing medium under hypoxic conditions. As expected, the control strain mec1Δ sml1Δ produced no viable colonies under both normoxic and hypoxic conditions. To determine whether the terminal phenotypes of the sod1Δ and lys7Δ strains on HU-containing medium under normoxic conditions were similar to mec1Δ sml1Δ strains, we assessed the cell morphology of the sod1Δ and lys7Δ strains in the persistent presence of HU. On HU-containing medium, wild-type strains arrest in S phase of the cell cycle and then adapt and resume growth to form colonies. A mec1Δ sml1Δ strain, however, does not arrest and instead continues to divide for two to three divisions, giving rise to abnormally shaped microcolonies of cells (59).Our results showed that on HU-containing medium, 100% of the cells plated from the sod1Δ strains failed to grow beyond the large-budded stage, and a large portion (76%) of the lys7Δ cells that failed to produce a visible colony gave rise to microcolonies of about 25 to 30 normally shaped cells (Fig. 2B). As expected, almost all (97%) of the cells from the wild-type strain produced colonies of normal size and shape whereas a majority (89%) of cells from the mec1Δ sml1Δ strain gave rise to clumps of less than 15 irregularly shaped cells. These results showed that while sod1Δ, lys7Δ, and mec1Δ sml1Δ strains are all sensitive to growth on HU-containing medium, the terminal growth phenotypes of sod1Δ and lys7Δ strains are distinct from those of both the wild-type and mec1Δ sml1Δ strains.
We next investigated the cell cycle arrest phenotypes by examining the nuclear DNA content of the strains treated with HU (100 mM for 3 and 6 hours) and upon release from a 6-hour HU treatment. Unlike wild-type cells that arrest in G1/S phase with a short spindle, mec1Δ sml1Δ strains fail to arrest, and cells show an elongated spindle after 3 hours of HU treatment (19, 30, 59). Flow cytometry analysis showed that similar to wild-type strains, sod1Δ and lys7Δ strains arrested in response to HU treatment (3 hours) under normoxic conditions (Fig. 2C, column 2). In addition, DAPI (4′,6′-diamidino-2-phenylindole) and tubulin staining showed that sod1Δ and lys7Δ strains with large-bud arrest morphology maintained short spindles similar to wild-type strains when treated with HU (100 mM for 3 hours) (data not shown). Consistent with the terminal phenotypes that we observed on HU-containing medium, the sod1Δ strains maintained G1/S-phase arrest after 6 hours (Fig. 2C, column 3) and 9 hours (data not shown) of treatment with HU. The wild-type and lys7Δ strains exhibited broader S-phase peaks and some cells with 2N DNA content (Fig. 2C, column 3) after 6 hours in HU, consistent with progression into the cell cycle. The checkpoint mutant mec1Δ sml1Δ failed to arrest in response to HU treatment as evidenced by both S-phase and 2N peaks after 3, 6 (Fig. 2C, column 2), and 9 hours (data not shown) in HU. To determine whether sod1Δ and lys7Δ strains arrested with HU progress into the cell cycle when released from that arrest, we performed an arrest release experiment by resuspending HU-treated cells (100 mM, 6 hours) in drug-free medium and analyzing aliquots at 30-minute intervals for up to 4 hours. Wild-type and lys7Δ strains progressed through the cell cycle starting at 30 minutes (data not shown) and as depicted in Fig. 2C for representative time points of 1 (column 4) and 3 (column 5) hours post-HU release. In contrast, sod1Δ strains progressed only slightly through the cell cycle following release from HU treatment. Examination of the flow cytometry samples of the sod1Δ strain under the microscope from 1 and 3 hours post-HU release showed that all the cells displayed the morphology seen in Fig. 2B. The mec1Δ sml1Δ strain failed to arrest in response to HU treatment, resulting in catastrophic divisions for a few generations with multibudded cells. These results suggest that the loss of viability of sod1Δ strains on HU-containing medium is not due simply to a failure to arrest and that the checkpoint-mediated cell cycle arrest seems to be intact in the sod1Δ strains.
Our results showed that sod1Δ strains are inviable in the presence of HU and maintained G1/S-phase arrest even upon prolonged incubation (Fig. 2B). Hence, we wanted to determine whether sod1Δ strains were viable after release from transient HU treatment. When treated with HU (100 mM) under normoxic conditions, the viability of sod1Δ strains was 84.6%, 40.0%, and 13.5% that of wild-type strains following 3, 6 and 9 hours of treatment, respectively (Fig. 2D). The inviable sod1Δ cells did not proceed beyond the large-budded stage (data not shown), similar to the cell morphology shown in Fig. 2B. The increased loss of viability of sod1Δ strains in response to increasing times of HU treatment may be due to an inability of the cells to reenter the cell cycle as shown in Fig. 2C. This effect was oxygen dependent, because the viabilities of the sod1Δ strains were near those of wild-type strains under hypoxic conditions (data not shown). The viability of lys7Δ strains was similar to that of the wild type under both normoxic (Fig. 2D) and hypoxic (data not shown) conditions. In comparison, the mec1Δ sml1Δ strain failed to produce colonies under both normoxic (Fig. 2D) and hypoxic (data not shown) conditions when released from 3, 6, or 9 hours of HU treatment. From these results we conclude that the growth phenotypes of sod1Δ and lys7Δ strains treated with HU are not similar to those observed for checkpoint mutants such as mec1Δ sml1Δ and that the cell cycle arrest aspect of the DNA damage checkpoint seems intact in the absence of SOD1 and LYS7.
Rad53p is expressed and phosphorylated in sod1Δ and lys7Δ strains treated with HU under normoxic conditions.
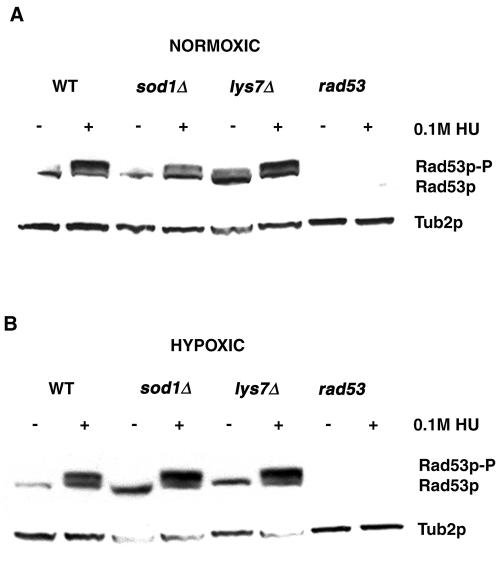
An indicator of activation of the MEC1-dependent DNA damage checkpoint pathway is the phosphorylation of Rad53p, which mediates cell cycle arrest and transcriptional induction of downstream effectors in response to DNA damage or replication arrest (46, 52). Our results for the cell cycle arrest phenotype of the sod1Δ and lys7Δ strains in response to HU treatment led us to investigate the expression of Rad53p in these strains. We analyzed WCE of wild-type, sod1Δ, and lys7Δ strains grown under normoxic or hypoxic conditions in the presence or absence of HU. Extracts were analyzed by Western blotting using an antibody to the C terminus of Rad53p, and extracts from a rad53Δ sml1-1 strain served as a negative control. Our results showed that like the wild-type strain, sod1Δ and lys7Δ strains treated with HU under normoxic (Fig. 3A) or hypoxic (Fig. 3B) conditions express both unphosphorylated and phosphorylated forms of Rad53p, as indicated by multiple bands. We did observe, however, that the mobility shift of Rad53p that occurs upon HU treatment of sod1Δ strains is not identical to that of wild-type and lys7Δ strains.
FIG. 3.
Rad53p is expressed and phosphorylated following replication arrest and DNA damage. Wild-type (WT; BY4741), sod1Δ, lys7Δ, and rad53Δ sml1-1 (U960-5C) strains were grown in YPD in the presence or absence of HU (100 mM, 3 hours) under normoxic (A) or hypoxic (B) conditions as indicated. WCE were subjected to Western blotting using a Rad53p C-terminal polyclonal antibody, and a Tub2p polyclonal antibody was used as a loading control.
Induction of MEC1 pathway effector protein Rnr3p and of Hug1p in the presence of HU under normoxic conditions is dependent on SOD1 and LYS7.
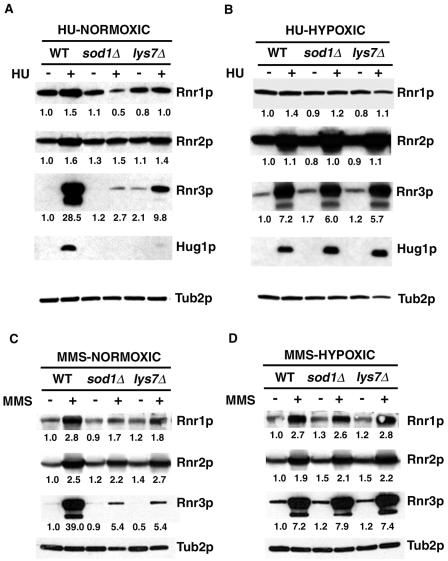
Another indicator of signaling through the MEC1 pathway is transcriptional induction of effectors such as RNR1, RNR2, RNR3, and RNR4 (32) and of HUG1 (6) in response to replication arrest or DNA damage. The DNA damage and HU sensitivity phenotypes of sod1Δ and lys7Δ strains prompted us to investigate, by Western blot analysis, whether expression of these effectors was altered in the sod1Δ and lys7Δ strains. Following HU exposure under normoxic conditions (Fig. 4A), Rnr3p and Hug1p, and to a lesser extent, Rnr1p and Rnr2p, were induced in wild-type cells. In sod1Δ strains, however, we observed a striking lack of induction of Rnr3p or Hug1p following HU treatment under normoxic conditions compared to wild-type strains (for Rnr3p, 2.7-fold versus 28.5-fold). In addition, these proteins were less induced in lys7Δ strains compared to the wild-type strains (for Rnr3p, 9.8-fold versus 28.5-fold). Rnr2p was induced to approximately the same levels in sod1Δ, lys7Δ, and wild-type strains while Rnr1p was slightly less induced in sod1Δ strains than in lys7Δ and wild-type strains. The reliance on SOD1 and, to a lesser extent, LYS7 for Rnr3p and Hug1p induction is entirely dependent on the presence of oxygen, as Rnr3p and Hug1p were each expressed similarly in wild-type, sod1Δ, and lys7Δ strains under hypoxic conditions (Fig. 4B). Exposure of sod1Δ and lys7Δ strains to MMS (0.02%) under normoxic conditions also resulted in a sharp decrease in Rnr3p induction compared to that in wild-type strains (5.4-fold versus 39.0-fold) (Fig. 4C). No changes in induction of Rnr3p compared to wild-type strains were found following MMS exposure under hypoxic conditions (Fig. 4D), however, and Hug1p was not detected under either condition (data not shown). We conclude that in sod1Δ and lys7Δ strains exposed to replication arrest under normoxic but not hypoxic conditions, induction of the MEC1 pathway effector Rnr3p and of Hug1p is significantly defective, suggesting that this aspect of the MEC1-dependent DNA damage checkpoint is not fully functional under normoxic conditions in the absence of SOD1 and LYS7.
FIG. 4.
sod1Δ and lys7Δ strains show altered levels of MEC1 checkpoint pathway effector protein Rnr3p and of Hug1p after replication arrest or DNA damage. Wild-type (BY4741), sod1Δ, or lys7Δ strains were grown for 3 hours in YPD with or without 100 mM HU (A and B) or 0.02% MMS (C and D) under normoxic (A and C) or hypoxic (B and D) conditions. WCE were subjected to Western blotting using Rnr1p, Rnr2p, Rnr3p, Hug1p, or Tub2p polyclonal antibodies. The value below each band indicates the ratio of the signal intensity of each protein band to the signal intensity of the Tub2p band compared to the ratio of these bands in the control lane (untreated WT) for each blot. Signal intensities were compared using ImageQuant TL software (Amersham Biosciences, Piscataway, NJ). Quantitation of proteins from a second set of blots gave similar normalized values. No values are given for Hug1p, since the HUG1 transcript is not present in the absence of replication arrest or DNA damage (6).
Overexpression of RNR1 or deletion of SML1 does not suppress the HU sensitivity of sod1Δ strains.
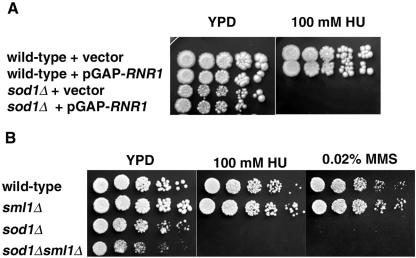
We reasoned that the growth inhibition phenotypes of sod1Δ strains in response to replication arrest and DNA damage may be due to lower levels of dinucleoside triphosphates (dNTPs) being available for DNA synthesis and repair, which occurs when levels and/or activity of RNR is altered (9, 33). The lack of induction of Rnr3p upon HU and MMS treatment prompted us to test whether increasing the abundance of Rnr3p would suppress these phenotypes in the sod1Δ and lys7Δ strains. However, we found that overexpression of RNR3 rendered wild-type cells sensitive to HU and resulted in extremely slow growth of the sod1Δ strains even in the absence of HU (data not shown). The slight reduction that we see in the induction of Rnr1p in the sod1Δ and lys7Δ strains under these conditions might be sufficient to affect the replication arrest and DNA damage sensitivity phenotypes of these strains, since a single copy of RNR1 can rescue the lethality of mec1 and rad53 mutants (17). Since Rnr1p is the rate-limiting subunit for RNR activity (9), we reasoned that increasing RNR1 levels might be sufficient to overcome the HU sensitivity of the sod1Δ strains. However, neither sod1Δ (Fig. 5A) nor lys7Δ strains (data not shown) transformed with RNR1 under the control of a constitutive glyceraldehyde-3-phosphate promoter (GAP-RNR1) were able to grow on HU-containing medium, even though Rnr1p was constitutively expressed in these strains as analyzed by Western blotting (data not shown). An alternate method to increase RNR activity is the removal of Sml1p, which binds to Rnr1p and renders it unavailable for incorporation into active RNR (8, 61). We constructed a strain lacking both SML1 and SOD1. While wild-type and sml1Δ strains grew on HU- and MMS-containing media, sod1Δ and sod1Δ sml1Δ strains did not (Fig. 5B). Combined, these data lead us to suggest that increasing the amount of Rnr1p, either by overexpression of RNR1 or removal of SML1, is not sufficient to suppress the replication arrest and DNA damage sensitivity of sod1Δ strains.
FIG. 5.
Overexpression of RNR1 and deletion of SML1 do not suppress sod1Δ phenotypes. (A) Three microliters of fivefold serial dilutions of wild-type (EG103) or sod1Δ (KS101) strains transformed with either vector alone (pBAD54) or RNR1 expressed from a constitutive promoter (pBAD790) was spotted on YPD and YPD containing 100 mM HU. At least two independent transformants were tested for each strain, and expression of RNR1 was verified by Western blot analysis (data not shown). (B) Three microliters of fivefold serial dilutions of wild-type (YMB3233), sml1Δ (YMB3234), sod1Δ (YMB3235), and sod1Δ sml1Δ strains (YMB3236) was spotted onto YPD, YPD containing 100 mM HU, or 0.02% MMS and then incubated under normoxic conditions at 30°C for 2 to 3 days. At least two independent strains were tested for each genotype.
Screen for suppressors of sod1Δ and lys7Δ replication arrest and DNA damage sensitivity identifies TKL1.
We established that addition of an antioxidant to the medium suppressed the oxygen-dependent HU sensitivity of the sod1Δ and lys7Δ strains (Fig. 1D). This prompted us to screen for cellular factors that might mimic this antioxidant effect by suppressing the HU sensitivity phenotype of sod1Δ and lys7Δ strains when expressed at high copy number. We transformed sod1Δ with a 2μm-URA3 library and screened the primary transformants for ability to grow on YPD containing HU. Of 2000 colonies screened, we identified two Ura+/HU-resistant transformants that each carried a plasmid containing TKL1. Interestingly, Tkl1p is known to suppress the methionine auxotrophy and oxygen sensitivity, but not the lysine auxotrophy, of sod1Δ strains through its ability to increase intracellular levels of NADPH (49).
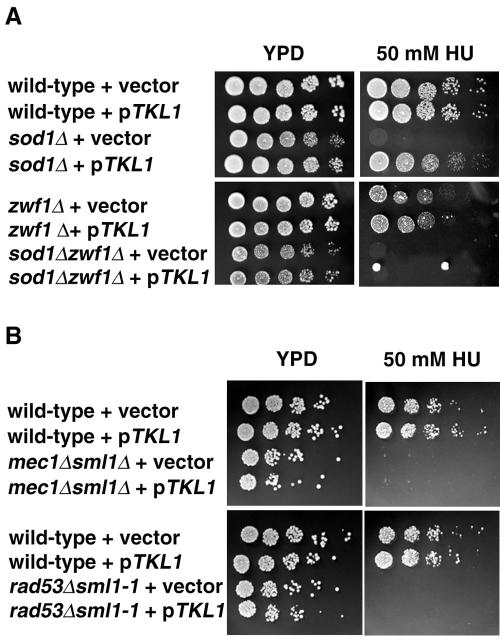
We verified that TKL1 was responsible for the suppression of the HU sensitivity since sod1Δ transformed with a 2μm TKL1 plasmid grew on HU-containing medium (Fig. 6A). TKL1 similarly suppressed the MMS sensitivity of sod1Δ strains and the HU and MMS sensitivities of lys7Δ strains (data not shown). One explanation for the suppression of sod1Δ phenotypes by TKL1 may be its ability to increase NADPH levels in the cell through its role in the nonoxidative branch of the pentose phosphate pathway (54). If suppression of sod1Δ phenotypes by TKL1 is due to increased NADPH, then the suppression should be dependent on Zwf1p, which catalyzes the rate-limiting step of the pentose phosphate pathway (49). Therefore, we tested the phenotypes of zwf1Δ and sod1Δ zwf1Δ strains carrying plasmids bearing TKL1. While zwf1Δ strains were slightly more sensitive to HU than wild-type strains, TKL1 in these strains did not affect their growth on medium containing HU (Fig. 6A). The sod1Δ zwf1Δ strains were as sensitive to HU as the sod1Δ strains, but TKL1 did not confer HU resistance to the double mutant, verifying the strains' dependence on ZWF1 for suppression of HU sensitivity (Fig. 6A). The ability of TKL1 to restore HU resistance to sod1Δ strains was not a general effect on a compromised MEC1 pathway, since TKL1 did not suppress the HU sensitivity of mec1Δ sml1Δ or rad53Δ sml1-1 strains (Fig. 6B).
FIG. 6.
Overexpression of TKL1 in sod1Δ strains, but not in mec1 or rad53 strains, restores HU resistance. (A) Three microliters of fivefold serial dilutions of wild-type (1783), sod1Δ (KS105), zwf1Δ (KS113), or sod1Δ zwf1Δ (KS117) strains transformed with vector alone or pKS10 (pTKL1/2μm/LEU2) was spotted onto YPD with or without 50 mM HU and incubated under normoxic conditions at 30°C for 2 days. At least two independent transformants were tested for each strain. Expression of pKS10 (pTKL1/2μm/LEU2) in lys7Δ strains also complemented the HU sensitivity of these strains (data not shown). (B) Three microliters of fivefold serial dilutions of wild-type (W1588-4A), mec1Δ sml1Δ (U953-61D), or rad53Δ sml1-1 (U960-5C) strains transformed with vector alone or pKS10 (pTKL1/2μm/LEU2) was spotted onto YPD with or without 50 mM HU and incubated under normoxic conditions at 30°C for 2 days.
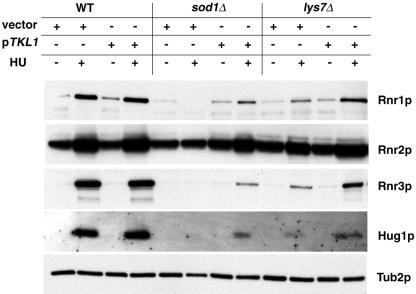
We next asked whether suppression of sod1Δ phenotypes by TKL1 restored expression of MEC1 pathway effectors. WCE made from wild-type, sod1Δ and lys7Δ strains transformed with vector alone or TKL1 and treated with HU were subjected to Western blotting and probed with polyclonal antibodies to Rnr1p, Rnr2p, Rnr3p, and Hug1p. In HU-treated sod1Δ and lys7Δ strains carrying TKL1, Rnr3p and Hug1p expression was increased compared to that in strains carrying vector alone, although not to wild-type levels (Fig. 7). Taken together with results shown in Fig. 6, TKL1 partially restored the expression of effectors of the MEC1-dependent signaling pathway in sod1Δ and lys7Δ strains and made these strains resistant to replication arrest and DNA damaging agents, suggesting that alteration of NADPH levels in the cell may counteract the elevated levels of superoxide anions present in strains lacking SOD1 and LYS7.
FIG. 7.
Overexpression of TKL1 in sod1Δ and lys7Δ strains partially restores expression of the MEC1 pathway effector protein Rnr3p and of Hug1p. Wild-type (WT; BY4741), sod1Δ, or lys7Δ strains transformed with vector alone or with pTKL1 (pKS10) were grown under hypoxic conditions overnight in minimal medium lacking leucine. Cultures were diluted, grown to logarithmic phase, and then grown for 3 hours in YPD with or without 100 mM HU under normoxic conditions. WCE were subjected to Western blotting. Blots were probed with polyclonal antibodies to Rnr1p, Rnr2p, Rnr3p, Hug1p, and Tub2p.
DISCUSSION
In this study, we show that loss of the primary cytosolic superoxide anion scavenger, Sod1p, or its copper chaperone Lys7p, leads to a defect in signaling of the replication arrest and DNA damage mediated by the MEC1 pathway and that this defect correlates with an inability to cope with genotoxic stress. We have determined that under normoxic, but not hypoxic, conditions, the absence of S. cerevisiae SOD1, and to a lesser extent the absence of LYS7, leads to (i) sensitivity to replication stress and DNA damaging agents and (ii) defective induction of an effector of the MEC1 pathway, Rnr3p, and of Hug1p, following HU treatment. These results suggest that HU and MMS sensitivities of sod1Δ and lys7Δ strains coincide with an impaired response of the MEC1-dependent checkpoint pathway. A high-copy-number suppressor screen for genes that suppress the oxygen-dependent HU sensitivity of sod1Δ strains identified TKL1, which was capable of restoring HU and MMS resistance and partial induction of Rnr3p and Hug1p in sod1Δ and lys7Δ strains. Based on these results we propose that these sod1Δ phenotypes may arise in part due to defective induction through the MEC1 pathway and low NADPH levels resulting from excess superoxide anions and that TKL1 complements these phenotypes by increasing NADPH and restoring a normal redox balance. Our results establish that the MEC1 pathway is sensitive to an altered cellular redox state due to an overabundance of superoxide anions and suggests a functional relationship between SOD1 and LYS7 and the MEC1-mediated checkpoint response to replication arrest and DNA damage in S. cerevisiae.
Analysis of the cell cycle arrest phenotype in response to HU treatment shows that the sod1Δ strains arrest in G1/S phase unlike checkpoint mutants that do not arrest. In addition, the viability of sod1Δ released from a HU treatment decreases as the time of treatment is increased, and the terminal phenotype of HU-treated sod1Δ strains is distinct from that of mec1Δ sml1Δ strains. From these data we infer that the checkpoint mechanism that leads to cell cycle arrest in response to HU seems to be intact in the sod1Δ strains. Furthermore, we have also observed that the cell cycle arrest response is not HU-specific, since sod1Δ strains treated with MMS (0.02%) for up to 5 hours show S-phase morphology as evidenced by flow cytometry (data not shown) unlike mec1Δ sml1Δ strains, which progress through the cell cycle (39). The cell cycle arrest in the sod1Δ strains is most likely mediated by the Rad53p branch of the MEC1 pathway as Rad53p is expressed and phosphorylated in these strains. Additional support for this conclusion is based on the observation that the arrest phenotype of sod1Δ is not due to the CHK1-dependent, RAD53-independent branch of the MEC1 pathway, since sod1Δ pds1Δ strains also arrest in response to HU treatment (data not shown).
Our data showed that the mobility shift of Rad53p that occurs upon HU treatment of sod1Δ strains is not identical to that observed for wild-type and lys7Δ strains. To our knowledge, previous reports have not established what levels of phosphorylated Rad53p are necessary for cell cycle arrest and induction of downstream effectors of the MEC1 pathway. The levels of Rad53p in the sod1Δ strains seem to be sufficient for cell cycle arrest in response to HU but may affect the optimal induction of Rnr3p and Hug1p.
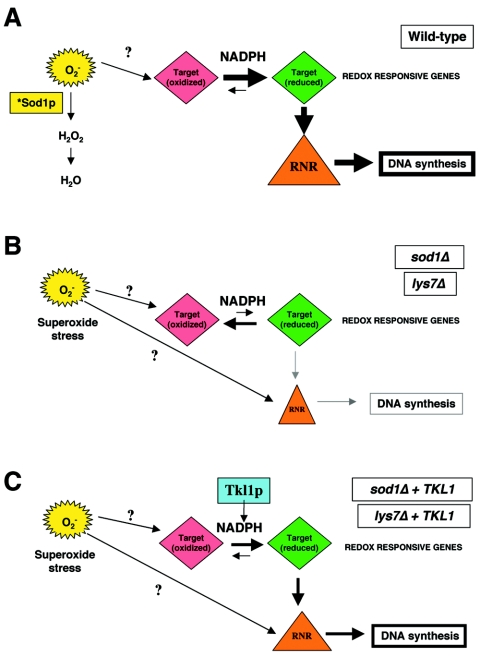
While the cell cycle arrest response of sod1Δ strains in the presence of HU under normoxic conditions is seemingly wild type, sod1Δ strains show increased sensitivity on HU-containing medium, reduced viability following transient HU exposure, and defective induction of Rnr3p and Hug1p. These phenotypes may be due to altered activity of the MEC1 pathway enzymes or other cellular targets in response to excess superoxide anions. Alternatively, sod1Δ strains may fail to reenter the cell cycle and thus fail to survive after a prolonged arrest, thereby resulting in increased HU sensitivity. We do not favor the latter possibility, since sod1Δ strains treated with MMS (0.02% for 3 hours), which causes a slowed progression in S phase, showed only a slight (<10%) reduction in viability when plated on medium lacking MMS (data not shown). The phenotypes of sod1Δ strains may also stem from a decrease in dNTP levels due to a compromised signaling through the MEC1 pathway. Our attempts to increase dNTP levels by overexpression of RNR1 or removal of the negative regulator of RNR1 activity, SML1, did not complement the mutant phenotypes of sod1Δ strains. Since mere alteration of Rnr1p levels available for incorporation into active RNR does not seem to suppress the HU-sensitive sod1Δ strains, we hypothesize that sod1Δ strains may have altered RNR activity, no matter the level of Rnr1p. Several factors regulate the activity of RNRs, including the cellular redox state and the availability of NADPH (29, 42). Class I RNR enzymes from eukaryotes and Escherichia coli are extremely sensitive to ROS (20, 22). Oxidative stress also leads to decreased levels of reduced forms of thioredoxins and glutathione, cofactors for RNR that utilize NADPH (7). In E. coli, superoxide anions can also directly inactivate the small subunit of RNR (R2) by irreversible oxidation of a stable tyrosyl radical essential for its activity (20). In addition, the large subunit (R1) contains three redox-active cysteine residues that participate in ribonucleotide reduction; these redox-sensitive moieties exist in eukaryotic RNR as well (20). The ZWF1-dependent suppression of sod1Δ phenotypes by TKL1 is consistent with our hypothesis that RNR activity is affected in sod1Δ strains, since NAPDH is required for RNR activity. We hypothesize that in wild-type cells (Fig. 8A) Sod1p scavenges superoxide anions while a reducing environment is maintained in the cytosol by a certain level of NADPH (31, 37, 47). In the absence of SOD1 (Fig. 8B), however, the increased levels of superoxide anions lead either directly or indirectly (through conversion to hydroxyl radicals) to oxidation of redox-sensitive cellular targets, such as thioredoxins and glutaredoxins and perhaps MEC1 pathway components, thereby requiring more NADPH to maintain these targets in a reduced and active state. The decreased general availability of NADPH in sod1Δ strains would lead to decreased RNR activity, resulting in lowered dNTP levels and replication arrest and DNA damage sensitivity. We propose that TKL1 (Fig. 8C) suppresses the sod1Δ phenotypes via an alteration of the cellular redox environment, perhaps through a concomitant increase of NADPH levels that restores signaling through the MEC1 pathway. This model is further supported by our data that the antioxidant NAC suppresses the HU sensitivity phenotype of sod1Δ and lys7Δ strains, presumably by removing hydroxyl radicals which are created from superoxide anions (2) and that lead to the more oxidized state of the cell.
FIG. 8.
Model for relationship between superoxide anions, NADPH levels, and RNR activity. (A) In wild-type strains, superoxide anions are scavenged by active Sod1p (*Sod1p), and sufficient levels of NADPH are provided through the pentose phosphate shunt to maintain redox-sensitive proteins in a reduced state. Redox-sensitive cellular targets, such as thioredoxins and glutaredoxins, transfer reductive capacity to RNR, and sufficient levels of active RNR are therefore available for DNA synthesis and repair. (B) In sod1Δ and lys7Δ strains, increases in superoxide anions and lowered NADPH levels shift the equilibrium of redox-sensitive cellular targets to more oxidized forms, resulting in lower RNR activity, decreased DNA synthesis and repair, and sensitivity to replication arrest and DNA damaging agents. Activity of RNR and induction of the MEC1 pathway effector Rnr3p and of Hug1p may also be decreased by the redox sensitivity of MEC1 pathway components. (C) Overexpression of TKL1 in sod1Δ or lys7Δ strains increases the available NADPH in a ZWF1-dependent manner and shifts the equilibrium of redox-sensitive cellular targets towards their reduced forms, thereby increasing RNR activity and suppressing the replication arrest and DNA damage phenotypes of sod1Δ and lys7Δ strains. In addition, induction of Rnr3p and Hug1p is partially restored in the sod1Δ and lys7Δ strains.
Our observation that the MEC1-dependent response to replication arrest and DNA damage requires SOD1 but shows less dependence on the presence of LYS7 suggests that the sensitivity of sod1Δ mutants does not rely entirely on the Lys7p-dependent activity of Sod1p. Induction of Rnr3p and Hug1p, and to a lesser extent Rnr1p, upon HU exposure and viability after release from replication arrest also seem to differ between the sod1Δ and lys7Δ strains. Taken together, these data support either a nonenzymatic role for Sod1p or the presence of a low level of active Sod1p in the absence of Lys7p. A low level of active Sod1p might be present in sod1Δ strains due to unassisted copper loading or copper loading dependent on a cellular protein other than Lys7p. In addition, a role for Sod1p that is independent of its Lys7-dependent SOD activity in response to replication arrest and DNA damage may exist. While both SOD1 and LYS7 have been reported to exhibit genetic interactions with some genes involved in human disease and DNA synthesis and repair, including MRE11, RAD27, RAD50, and RAD52, SOD1 alone showed genetic interactions with POL32, RTT107/ESC4, and SGS1 (56, 57), further supporting the stronger phenotypes we have observed for the sod1Δ strains.
We have identified an important relationship between alteration of cellular redox state due to loss of superoxide anion scavengers and the MEC1-dependent checkpoint response in S. cerevisiae. TKL1, which may alter the cellular redox state by increasing intracellular NADPH levels, suppresses HU sensitivity caused by the absence of SOD1 and restores expression of the MEC1 pathway effector, Rnr3p, and of Hug1p, in sod1Δ strains. Future experiments addressing the involvement of S. cerevisiae Sod1p and Lys7p with the MEC1 pathway may shed light on the role of a compromised redox state due to excess superoxide anions in signaling through checkpoint pathways responding to replication arrest and DNA damage in other systems.
Supplementary Material
Acknowledgments
We thank the following: M. Lichten, K. J. Myung, N. Dhillon, and members of the Basrai lab for comments on the manuscript; V. Culotta, S. Elledge, L. Jensen, D. Koshland, R. Rothstein, K. Slekar, D. Stern, and J. Stubbe for strains, plasmids, or antibodies; V. Culotta and C. Mann for advice and comments on the manuscript; and O. Kerscher and B. Todd for initiating this project in our laboratory.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 2.Aruoma, O. I., B. Halliwell, B. M. Hoey, and J. Butler. 1989. The antioxidant action of N-acetylcysteine: its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Radic. Biol. Med. 6:593-597. [DOI] [PubMed] [Google Scholar]
- 3.Assfalg, M., L. Banci, I. Bertini, P. Turano, and P. R. Vasos. 2003. Superoxide dismutase folding/unfolding pathway: role of the metal ions in modulating structural and dynamical features. J. Mol. Biol. 330:145-158. [DOI] [PubMed] [Google Scholar]
- 4.Barzilai, A., G. Rotman, and Y. Shiloh. 2002. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair 1:3-25. [DOI] [PubMed] [Google Scholar]
- 5.Basrai, M. A., J. Kingsbury, D. Koshland, F. Spencer, and P. Hieter. 1996. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2838-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basrai, M. A., V. E. Velculescu, K. W. Kinzler, and P. Hieter. 1999. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7041-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 8.Chabes, A., V. Domkin, and L. Thelander. 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274:36679-36683. [DOI] [PubMed] [Google Scholar]
- 9.Chabes, A., B. Georgieva, V. Domkin, X. Zhao, R. Rothstein, and L. Thelander. 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112:391-401. [DOI] [PubMed] [Google Scholar]
- 10.Chang, M., M. Bellaoui, C. Boone, and G. W. Brown. 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA 99:16934-16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen-Fix, O., and D. Koshland. 1997. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc. Natl. Acad. Sci. USA 94:14361-14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crow, J. P., J. B. Sampson, Y. Zhuang, J. A. Thompson, and J. S. Beckman. 1997. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J. Neurochem. 69:1936-1944. [DOI] [PubMed] [Google Scholar]
- 14.Culotta, V. C. 2000. Superoxide dismutase, oxidative stress, and cell metabolism. Curr. Top. Cell. Regul. 36:117-132. [DOI] [PubMed] [Google Scholar]
- 15.Culotta, V. C., H. D. Joh, S. J. Lin, K. H. Slekar, and J. Strain. 1995. A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J. Biol. Chem. 270:29991-29997. [DOI] [PubMed] [Google Scholar]
- 16.Culotta, V. C., L. W. Klomp, J. Strain, R. L. Casareno, B. Krems, and J. D. Gitlin. 1997. The copper chaperone for superoxide dismutase. J. Biol. Chem. 272:23469-23472. [DOI] [PubMed] [Google Scholar]
- 17.Desany, B. A., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elledge, S. J., Z. Zhou, J. B. Allen, and T. A. Navas. 1993. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 15:333-339. [DOI] [PubMed] [Google Scholar]
- 19.Fay, D. S., Z. Sun, and D. F. Stern. 1997. Mutations in SPK1/RAD53 that specifically abolish checkpoint but not growth-related functions. Curr. Genet. 31:97-105. [DOI] [PubMed] [Google Scholar]
- 20.Fontecave, M. 1998. Ribonucleotide reductases and radical reactions. Cell. Mol. Life Sci. 54:684-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner, R., C. W. Putnam, and T. Weinert. 1999. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 18:3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudu, P., V. Niviere, Y. Petillot, B. Kauppi, and M. Fontecave. 1996. The irreversible inactivation of ribonucleotide reductase from Escherichia coli by superoxide radicals. FEBS Lett. 387:137-140. [DOI] [PubMed] [Google Scholar]
- 23.Gralla, E. B., and J. S. Valentine. 1991. Null mutants of Saccharomyces cerevisiae Cu,Zn superoxide dismutase: characterization and spontaneous mutation rates. J. Bacteriol. 173:5918-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hough, M. A., J. G. Grossmann, S. V. Antonyuk, R. W. Strange, P. A. Doucette, J. A. Rodriguez, L. J. Whitson, P. J. Hart, L. J. Hayward, J. S. Valentine, and S. S. Hasnain. 2004. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc. Natl. Acad. Sci. USA 101:5976-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, M. E., and R. D. Kolodner. 2005. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol. Cell 17:709-720. [DOI] [PubMed] [Google Scholar]
- 26.Huang, M. E., A. G. Rio, A. Nicolas, and R. D. Kolodner. 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100:11529-11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iouk, T., O. Kerscher, R. J. Scott, M. A. Basrai, and R. W. Wozniak. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159:807-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S., and T. A. Weinert. 1997. Characterization of the checkpoint gene RAD53/MEC2 in Saccharomyces cerevisiae. Yeast 13:735-745. [DOI] [PubMed] [Google Scholar]
- 31.Kirsch, M., and H. De Groot. 2001. NAD(P)H, a directly operating antioxidant? FASEB J. 15:1569-1574. [DOI] [PubMed] [Google Scholar]
- 32.Kiser, G. L., and T. A. Weinert. 1996. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol. Biol. Cell 7:703-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koc, A., L. J. Wheeler, C. K. Mathews, and G. F. Merrill. 2004. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 279:223-230. [DOI] [PubMed] [Google Scholar]
- 34.Lamb, J. R., W. A. Michaud, R. S. Sikorski, and P. A. Hieter. 1994. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 13:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macip, S., M. Igarashi, P. Berggren, J. Yu, S. W. Lee, and S. A. Aaronson. 2003. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol. Cell. Biol. 23:8576-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marnett, L. J. 2000. Oxyradicals and DNA damage. Carcinogenesis 21:361-370. [DOI] [PubMed] [Google Scholar]
- 37.Moreira dos Santos, M., G. Thygesen, P. Kotter, L. Olsson, and J. Nielsen. 2003. Aerobic physiology of redox-engineered Saccharomyces cerevisiae strains modified in the ammonium assimilation for increased NADPH availability. FEMS Yeast Res. 4:59-68. [DOI] [PubMed] [Google Scholar]
- 38.Parsons, A. B., R. Geyer, T. R. Hughes, and C. Boone. 2003. Yeast genomics and proteomics in drug discovery and target validation. Prog. Cell Cycle Res. 5:159-166. [PubMed] [Google Scholar]
- 39.Paulovich, A. G., R. U. Margulies, B. M. Garvik, and L. H. Hartwell. 1997. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 145:45-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravindranath, S. D., and I. Fridovich. 1975. Isolation and characterization of a manganese-containing superoxide dismutase from yeast. J. Biol. Chem. 250:6107-6112. [PubMed] [Google Scholar]
- 41.Ray, S. S., and P. T. Lansbury, Jr. 2004. A possible therapeutic target for Lou Gehrig's disease. Proc. Natl. Acad. Sci. USA 101:5701-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichard, P. 2002. Ribonucleotide reductases: the evolution of allosteric regulation. Arch. Biochem. Biophys. 397:149-155. [DOI] [PubMed] [Google Scholar]
- 43.Rosen, D. R. 1993. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 364:362. [DOI] [PubMed] [Google Scholar]
- 44.Rowland, L. P., and N. A. Shneider. 2001. Amyotrophic lateral sclerosis. N. Engl. J. Med. 344:1688-1700. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 47.Shackelford, R. E., W. K. Kaufmann, and R. S. Paules. 2000. Oxidative stress and cell cycle checkpoint function. Free Radic. Biol. Med. 28:1387-1404. [DOI] [PubMed] [Google Scholar]
- 48.Sherman, F., G. R. Fink, and C. W. Lawrence. 1978. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 49.Slekar, K. H., D. J. Kosman, and V. C. Culotta. 1996. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 271:28831-28836. [DOI] [PubMed] [Google Scholar]
- 50.Sturtz, L. A., and V. C. Culotta. 2002. Superoxide dismutase null mutants of baker's yeast, Saccharomyces cerevisiae. Methods Enzymol. 349:167-172. [DOI] [PubMed] [Google Scholar]
- 51.Sturtz, L. A., K. Diekert, L. T. Jensen, R. Lill, and V. C. Culotta. 2001. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 276:38084-38089. [DOI] [PubMed] [Google Scholar]
- 52.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 53.Sun, Z., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272-274. [DOI] [PubMed] [Google Scholar]
- 54.Sundstrom, M., Y. Lindqvist, G. Schneider, U. Hellman, and H. Ronne. 1993. Yeast TKL1 gene encodes a transketolase that is required for efficient glycolysis and biosynthesis of aromatic amino acids. J. Biol. Chem. 268:24346-24352. [PubMed] [Google Scholar]
- 55.Thompson, L. H., and D. Schild. 2002. Recombinational DNA repair and human disease. Mutat. Res. 509:49-78. [DOI] [PubMed] [Google Scholar]
- 56.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 57.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 58.Wallace, M. A., L. L. Liou, J. Martins, M. H. Clement, S. Bailey, V. D. Longo, J. S. Valentine, and E. B. Gralla. 2004. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis: cross-compartment protection by CuZnSOD. J. Biol. Chem. [DOI] [PubMed]
- 59.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 60.Yoon, S. J., Y. H. Koh, R. A. Floyd, and J. W. Park. 2000. Copper, zinc superoxide dismutase enhances DNA damage and mutagenicity induced by cysteine/iron. Mutat. Res. 448:97-104. [DOI] [PubMed] [Google Scholar]
- 61.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.