Abstract
The essential, conserved yeast nucleolar protein Ytm1 is one of 17 proteins in ribosome assembly intermediates that contain WD40 protein-protein interaction motifs. Such proteins may play key roles in organizing other molecules necessary for ribosome biogenesis. Ytm1 is present in four consecutive 66S preribosomes containing 27SA2, 27SA3, 27SB, and 25.5S plus 7S pre-rRNAs plus ribosome assembly factors and ribosomal proteins. Ytm1 binds directly to Erb1 and is present in a heterotrimeric subcomplex together with Erb1 and Nop7, both within preribosomes and independently of preribosomes. However, Nop7 and Erb1 assemble into preribosomes prior to Ytm1. Mutations in the WD40 motifs of Ytm1 disrupt binding to Erb1, destabilize the heterotrimer, and delay pre-rRNA processing and nuclear export of preribosomes. Nevertheless, 66S preribosomes lacking Ytm1 remain otherwise intact.
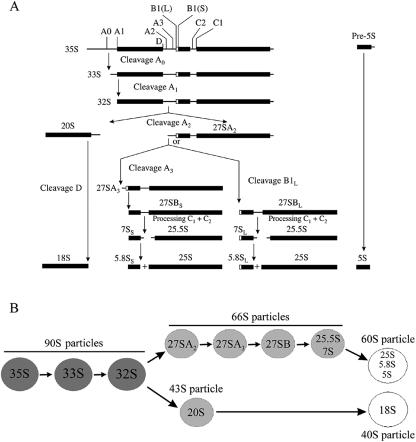
Biogenesis of eukaryotic ribosomes is a highly regulated and dynamic process that begins in the nucleolus with transcription of a precursor rRNA (pre-rRNA) that is rapidly packaged into the 90S ribonucleoprotein particle containing ribosomal proteins, nonribosomal proteins, and snoRNA-containing ribonucleoprotein particles (snoRNPs). The 90S pre-RNPs are converted into 43S and 66S ribosome assembly intermediates, which ultimately give rise to mature 40S and 60S ribosomal subunits (Fig. 1).
FIG. 1.
Pre-rRNA processing and pre-rRNP maturation pathway in Saccharomyces cerevisiae. (A) The 35S pre-rRNA contains sequences for mature 18S, 5.8S, and 25S rRNAs (represented as thick horizontal lines) along with additional internal and external spacer sequences (represented as thin horizontal lines). The 35S pre-rRNA is transcribed by RNA polymerase I and rapidly modified and processed to produce 33S pre-rRNA. Cleavage of 33S pre-rRNA at site A0 generates 32S pre-rRNA. The 20S and 27SA2 pre-rRNA processing intermediates are generated through internal cleavage of 32S pre-rRNA at the A2 site. Subsequent processing and cleavage of 20S and 27SA2 pre-rRNAs result in the production of the mature 18S, 25S, and 5.8 rRNAs, respectively. 5S rRNA is transcribed separately by RNA polymerase III. (B) Pre-rRNA processing occurs in preribosomal particles. The 35S primary transcript is found within the 90S pre-rRNP (dark gray circle). Cleavage at site A2 initiates subunit-specific maturation, generating the 43S and 66S pre-rRNPs (light gray circles). The 43S preribosome is exported to the cytoplasm, where final steps in 20S maturation take place. Multiple 66S preribosomes exist that contain each of the 27S or 25.5S plus 7S pre-rRNA processing intermediates. The mature 40S subunit contains 18S rRNA, whereas the 60S subunit contains 25S, 5.8S, and 5S rRNA (white circles).
Molecular genetic approaches in yeast identified more than 70 trans-acting factors required for ribosome assembly (12, 14, 46). Subsequent advances in proteomics enabled purification of pre-rRNPs from yeast and identification of an additional 80 assembly factors present in preribosomes, as well as most of those proteins previously discovered using genetic screens (3, 7, 11, 17, 20, 21, 24, 26, 37, 38, 41, 49-51). Metazoan homologues of most of the yeast ribosome assembly factors were discovered by proteomic analysis of purified nucleoli (2, 52).
Among the assembly factors found in yeast preribosomes are 17 proteins containing WD40 motifs (14). These motifs function as protein-protein interaction domains (53). Therefore, such WD40-containing proteins may nucleate assembly of preribosomes by interacting sequentially or simultaneously with other assembly factors or ribosomal proteins. Previously, we identified the WD40 protein Ytm1 as a constituent of purified 66S pre-rRNPs and showed that depletion of Ytm1 results in a deficiency of 60S ribosomal subunits (21).
In this study, we have further investigated the role of Ytm1 in ribosome biogenesis. Ytm1 is a constituent of multiple consecutive 66S preribosomes containing 27SA2, 27SA3, 27SB, 25.5S, and 7S pre-rRNAs plus a collection of ribosomal and nonribosomal proteins. Ytm1 is present in a heterotrimer with two other assembly factors, Nop7 and Erb1, both within 66S pre-rRNPs and as a subcomplex independent of preribosomes. Mutations in Ytm1 disrupt interactions between Ytm1 and Erb1, destabilize the heterotrimer, and significantly reduce association of these three proteins with 66S preribosomes. These 66S pre-rRNPs otherwise remain intact in the ytm1-1 mutant, but processing of 27SA3 pre-rRNAs is delayed and release of 66S preribosomes from the nucleolus is partially blocked. Thus, Ytm1 is necessary to nucleate the assembly of a heterotrimer that is important for intermediate-to-late steps in maturation of 66S preribosomes.
MATERIALS AND METHODS
Strains, plasmids, and media.
Yeast strains used in this work (Table 1) were grown in YEPD medium (2% dextrose, 2% peptone, and 1% yeast extract) or YEPGal medium (2% galactose, 2% peptone, and 1% yeast extract) at 30°C and harvested at 5 · 107 cells/ml unless otherwise indicated. The ytm1-1 mutant strain JWY7128 was generated by mutagenizing plasmid pRS317 containing wild-type YTM1 and LYS2 with hydroxylamine and transforming it into yeast strain SM412 cells which have GAL-YTM1 at the YTM1 locus. The ytm1-1 mutant plasmid that conferred temperature sensitivity to strains grown on selective medium containing 2% glucose was rescued, shuttled through Escherichia coli, and used to replace the chromosomal YTM1 gene in strain JWY7132 to generate ytm1-1 strain JWY7128. The GAL-YTM1 strain JWY6992 was described previously (21).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JWY3400 | MATaura3-52 lys2-801 trp1-1 leu2-1 his3-Δ200 pep4::HIS3 prb1-Δ1.6R can YTM1 | E. Jones |
| JWY7128 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ytm1-1 URA3 | S. Matsumoto |
| JWY7132 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 YTM1 | S. Matsumoto |
| JWY6149 | MATahis3-Δ1 leu2-Δ0 lys2-Δ0 MET15 ura3-Δ0 YTM1 | 26 |
| JWY6992 | MATα met15-Δ0 ura3-Δ0 his3-Δ1 leu2-Δ0 YTM1::kanMX4 pGAL-YTM1 URA3 | 21 |
| JWY6779 | MATaura3-52 trp1-Δ101 lys2-801 his3-Δ200 leu2-Δ1 YTM1 p1877 (RPL25eGFP URA3) | This study |
| JWY6790 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL1 ytm1-1 p1878 (RPL25-eGFP TRP1) | This study |
| JWY6147 | MATaura3-52 trp1-Δ101 lys2-801 his3-Δ200 leu2-Δ1 YTM1 | E. Jones |
| JWY6770 | MATaura3-52 trp1-Δ101 lys2-801 his3-Δ200 leu2-Δ1 YTM1-HA3 HIS3 | This study |
| JWY7124 | MATaura3-52 lys2-801 trp1-1 leu2-1 his3-Δ200 pep4::HIS3 prb1-Δ1.6R can YTM1-TAP TRP1 | This study |
| JWY7129 | MATα ura3-52 lys2-801 trp1 leu2-1 his3-Δ200 ade2-101 pep4::HIS3 prb1-Δ1.6R can 1 nop4::TRP1 [pRS315 LEU2 nop4-3] YTM1-TAP URA3 | This study |
| JWY6729 | MATα ura3-52 lys2-801 trp1 leu2-1 his3-Δ200 ade2-101 pep4::HIS3 prb1-Δ1.6R can1 nop4::TRP1 [pRS315 LEU2 nop4-3] NOP7-TAP TRP1 | 21 |
| JWY7131 | MATahis7 ade2 trp1 lys2 tyr1 gal1 gal2 rrp1-1 YTM1-TAP TRP1 | This study |
| JWY6970 | MATahis7 ade2 trp1 lys2 tyr1 gal1 gal2 rrp1-1 NOP7-TAP TRP1 | This study |
| JWY6938 | MATaura3-52 lys2-801 trp1-1 leu2-1 his3-Δ200 pep4::HIS3 prb1-Δ1.6R can YTM1 NOP7-TAP TRP1 | 21 |
| JWY7019 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ytm1-1 NOP7-TAP TRP1 | This study |
| JWY7810 | MATapep4-3 his4-500 ura3-52 leu2-3,112 pEG(KG) (GST-ERB1 LEU2 URA3) | M. Snyder |
| JWY7808 | MATapep4-3 his4-500 ura3-52 leu2-3,112 pEG(KG) (GST-YTM1 LEU2 URA3) | M. Snyder |
| JWY7807 | MATapep4-3 his4-500 ura3-52 leu2-3,112 pEG(KG) (GST-NOP7 LEU2 URA3) | M. Snyder |
| JWY6300 | MATα trp1-901 leu2-3,112 ura3-52 his3-Δ200 gal4Δ gal80Δ LYS2-GAL1-HIS3 ade2-101GAL2-ADE2 met2::GAL7-LacZ | PJ69-4α (29) |
| JWY4340 | MATatrp1-901 leu2-3,112 ura3-52 his3-Δ200 gal4 gal80 LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | PJ69-4a (29) |
| JWY7138 | MATaura3-52 lys2-801 trp1-1 leu2-1 his3-Δ200 pep4::HIS3 prb1-Δ1.6R can BRX1-TAP TRP1 YTM1 | This study |
| JWY7140 | MATα ura3-52 lys2-801 ade2-101 trp1Δ63 his-3Δ200 leu2-Δ1 BRX1-TAP TRP1 ytm1-1 | This study |
Three-hemagglutinin epitope (HA3)- and tandem affinity purification (TAP)-tagged strains were generated as described in references 21 and 26. Integration of the HA3 tag or the TAP cassette in-frame with the last codon of each open reading frame was confirmed by genomic PCR and Western immunoblotting. Yeast strain JWY6790 expressing enhanced green fluorescent protein (eGFP)-tagged rpL25 was generated by transforming JWY7128 (ytm1-1) with a plasmid bearing rpL25eGFP (28). Transformants were screened by fluorescence microscopy for expression of rpL25eGFP. Yeast strains JWY7808 (GST-YTM1), JWY7810 (GST-ERB1) and JWY7807 (GST-NOP7) expressing glutathione S-transferase (GST) fusion proteins were obtained from Mike Snyder (Yale University).
Sucrose gradient analysis.
Ribosomes, preribosomes, and polyribosomes in yeast strains JWY3400 (YTM1), JWY7124 (YTM1-TAP), and JWY7128 (ytm1-1) were analyzed as described previously (26). JWY7128 was grown at 25°C and harvested or else shifted from 25 to 37°C for 3 h and harvested. Ytm1-HA3 or Ytm1-TAP were identified in gradient fractions by Western blotting using mouse monoclonal antibody 12CA5 or rabbit anti-mouse immunoglobulin G (Pierce), respectively.
Analysis of rRNA.
Steady-state levels of rRNAs were analyzed by Northern blotting (26) or primer extension assays (58). To carry out primer extension, radiolabeled oligonucleotide primers complementary to 35S, 27S, or 25.5S pre-rRNA were first annealed for 90 min at 46°C to total RNA or affinity-purified RNA. Primer extension reaction mixtures containing the annealed oligonucleotide primer/RNA hybrid, 10 mM deoxynucleoside triphosphates (dNTPs) (Amersham Biosciences), 12.5 U avian myeloblastosis virus reverse transcriptase (Promega), and 20 U RNasin (Promega) were incubated for 40 min at 46°C. To hydrolyze the RNA, 6 μl of 1 M NaOH and 1 μl of 0.5 M EDTA were added to each primer extension reaction mixture for 30 min to 1 h at 55°C. Next, 6 μl of 1 M HCl was added to each reaction mixture and DNA was precipitated with 4 μg glycogen, 30 μl 7.5 M NH4OAc, and 250 μl of 100% ethanol. DNA was suspended in DNA dye (95% formamide, 0.05% xylene cyanol, 0.05% bromophenol blue in 20 mM EDTA, pH 8.0) and subjected to electrophoresis on a 6% polyacrylamide-urea sequencing gel. Gels were dried and directly exposed to film for autoradiography. Oligonucleotide sequences are available upon request. Pulse-chase assays of pre-rRNA processing were carried out as previously described (26).
Ribosome export assays.
Release of preribosomes from nucleoli and export from the nucleoplasm to the cytoplasm were assayed as previously described (28), except that strains were grown overnight in C-Trp medium, washed and suspended in YEPD medium, and grown at 25°C or shifted to 37°C for 5 h.
Affinity purification and mass spectrometry.
Cell extracts were prepared, tandem affinity purification was carried out, and identification of copurifying proteins and RNAs was performed as previously described (26, 47).
Ribosome assembly subcomplexes were separated from 66S preribosomes and 60S ribosomal subunits by centrifugation on 7% to 47% sucrose gradients (26) or by differential centrifugation as performed previously (32) with the following modifications: whole-cell extracts were centrifuged for 2 h at 180,000 × g at 4°C, followed by a second centrifugation of supernatants for 30 to 45 min at 180,000 × g at 4°C. Subcomplexes were affinity purified from gradient fractions or from the 180,000 × g supernatant using TAP-tagged Nop7.
Assembly subcomplexes were isolated directly from whole-cell extracts by adding to the lysis buffer and calmodulin binding buffer a phosphatase inhibitor cocktail (20 mM pyrophosphate, 10 mM sodium azide, 20 mM sodium fluoride, 1 mM sodium orthovanadate, and 100 mM β-glycerophosphate) that disrupts pre-rRNPs.
Generation of anti-Ytm1 antibodies and Western immunoblotting.
Rabbit antibodies generated against the synthetic peptide ITREDKSVQKGVNDK (Alpha Diagnostics, Inc.) were used to detect Ytm1. Antibodies were concentrated by ammonium sulfate precipitation, dialyzed, and affinity purified using full-length filter-bound Ytm1 protein previously subjected to electrophoresis through a 10% polyacrylamide gel and electroblotted to nitrocellulose (Optitran; Schleicher and Schuell). Immunoblotting was carried out using standard protocols (26).
GST pull-down assays.
GST fusion proteins were harvested from yeast by glass bead lysis of frozen cell pellets suspended in 1.6 ml sorbitol buffer (300 mM sorbitol, 5 mM MgCl2, 100 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml leupeptin). One milliliter of protein extract was incubated with 50 μl glutathione-agarose beads overnight at 4°C. Beads were washed three times with 1 ml high-salt wash buffer (300 mM sorbitol, 5 mM MgCl2, 1 M NaCl, 10 mM Tris-HCl, pH 7.5), three times with 1 ml HKT buffer (10 mM HEPES, 100 mM KCl, 0.5% Triton X, 1% IGEPAL, 5% bovine serum albumin) and once with 1 ml sorbitol buffer lacking proteinase inhibitors. 35S-labeled Nop7, Erb1, and Ytm1 were synthesized in vitro using the TNT T7 Quick for PCR DNA kit (Promega Corporation) and oligonucleotides T7_NOP7_UP and NOP7_TRUC_2HY_GAP_DN, T7_ERB1_UP and ERB1_DN, or T7_YTM1_UP and YTM1-2HYB-GAP_REPR-DN. Labeled proteins (5 μl or 10% of the labeling reaction) were incubated with the glutathione bead-bound GST fusion proteins for 2 h at 4°C. GST beads were washed once with 0.5 ml of HKT buffer containing 1 M NaCl and three times with 1 ml HKT buffer. Protein complexes were eluted from GST beads by boiling in 35 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and subjected to electrophoresis on 10% polyacrylamide gels. Gels were fixed in a 50% methanol-15% acetic acid solution for 15 min, washed with water, treated with 1 M sodium salicylate for 1 h, dried, and subjected to autoradiography.
Yeast two-hybrid assays.
The YTM1 and ERB1 open reading frames were amplified from the genome using primer pairs YTM1-2HYB-GAP_REPR-UP and YTM1-2HYB-GAP_REPR-DN or ERB1-2HYB-GAP_REPR-UP and ERB1-2HYB-GAP_REPR-DN, respectively. PCR products were cloned into the DNA binding domain vector pOBD-2 by gap repair and transformed into the two-hybrid host strain PJ69-4α (JWY6300). Yeast cells containing NOP7, ERB1, and YTM1 cloned into the DNA activation domain vector (pOAD) were obtained as a pool in the two-hybrid host strain PJ69-4a (JWY4340) from the Yeast Resource Center (University of Washington). The yeast two-hybrid assay was performed as previously described (5). Diploids containing activation domain and binding domain plasmids were screened on media lacking adenine or containing 2 to 50 mM 3-aminotriazole to assay the ADE3 and HIS3 reporter genes, respectively.
RESULTS
The ytm1-1 mutant contains mutations in two of the WD40 repeat motifs of Ytm1.
Ytm1 is an essential 51-kDa yeast nucleolar protein that is highly conserved from fungi to humans. Seven WD repeats are clustered at the C terminus of the protein (Fig. 2A). WD repeat-containing proteins are predicted to share a common circularized beta-propeller structure (Fig. 2B) based on the crystal structure of the Gβ subunit of heterotrimeric G proteins (16, 33, 54, 59). A paramount feature of WD repeat proteins is their ability to participate in multiple interactions either simultaneously or sequentially. The WD repeat motif is not known to have any enzymatic activity. Therefore, WD repeat proteins acting in disparate pathways may share a common function of establishing and regulating interactions within multiprotein complexes.
FIG. 2.
Ytm1 is a WD40 repeat-containing protein. (A) Predicted amino acid sequence of S. cerevisiae Ytm1. WD40 repeats are overlined. Amino acid residues altered in the ytm1-1 mutant are indicated by asterisks. (B) Ras Mol 2.6 was used to generate the top and side view of a model for amino acids 103 to 450 of Ytm1, based on the crystal structure of the WD repeat protein Gβ.
To investigate the importance of WD40 motifs in Ytm1, we used the ytm1-1 temperature-sensitive mutant that contains two mutations, G398D and S442N, in WD40 repeats 6 and 7 of Ytm1 (Fig. 2A). This mutant strain was generated by random mutagenesis of cloned YTM1 and replacement of chromosomal YTM1 with the mutant allele. The counterpart of Ytm1 residue G398 in the Gβ protein contacts phosducin (54). Thus, residues at this position may be important for mediating interactions with associated proteins. Within the conserved WD40 repeat consensus sequence is the structural tetrad, which contributes to local and global stability through inter- and intrablade hydrogen bonding (40, 53, 54). The G398D substitution occurs in the position adjacent to the highly conserved aspartate residue in the structural tetrad. Similarly, the S442N substitution occurs in a residue that is next to the serine/threonine residue of the structural tetrad. Thus, the ability of Ytm1 to bind to ligands may be compromised in the ytm1-1 mutant, which may result in defects in ribosome assembly.
The ytm1-1 mutant was unable to grow at 37°C and grew slower than the wild-type control strain at all temperatures below 37°C (data not shown). The G398D and S442N mutations might inactivate Ytm1 at 37°C, perhaps by distorting the structure of Ytm1 or by disrupting interactions with ligands of Ytm1. The ytm1-1 mutant protein is relatively stable: amounts of Ytm1-1 protein did not change drastically when compared to those of wild-type Ytm1 in strains grown at 25°C or shifted from 25°C to 37°C for 5 h (data not shown).
The ytm1-1 mutant is deficient in 60S ribosomal subunits.
To determine the effect of inactivation of Ytm1 on ribosome biogenesis, we assayed levels of ribosomal subunits, monoribosomes, and polyribosomes in ytm1-1 cells grown at 25°C or shifted from 25°C to 37°C for 3 h. Upon shifting the ytm1-1 mutant cells to 37°C, amounts of 60S ribosomal subunits and 80S monosomes were greatly reduced and half-mer polyribosomes were apparent. At 25°C, ytm1-1 mutant cells contained fewer free 60S subunits and 80S monoribosomes and accumulated half-mer polyribosomes, compared to wild-type YTM1 cells (Fig. 3). This suggests that the function of Ytm1-1 is compromised even at the permissive temperature and is consistent with the slow-growth phenotype at 25°C. These findings are consistent with previous results obtained when Ytm1 is metabolically depleted (21).
FIG. 3.
The ytm1-1 mutant is deficient in 60S ribosomal subunits. Free 40S and 60S ribosomal subunits, monoribosomes, and polyribosomes were assayed in yeast strains JWY3400 (YTM1) (left) or JWY7128 (ytm1-1) (center) grown at 25°C or JWY7128 grown at 25°C and shifted to 37°C for 3 h (right). Whole-cell extracts prepared from each strain were fractionated on 7 to 47% sucrose gradients. A260 peaks representing 40S and 60S ribosomal subunits and 80S monosomes are labeled. Half-mer polyribosomes are indicated by vertical arrows.
Pre-rRNA processing is slowed when Ytm1 is inactivated or depleted.
The kinetics of pre-rRNA processing were analyzed by pulse-labeling YTM1 wild-type and ytm1-1 mutant cells, grown at 25°C, and shifted to 37°C for 5 h, as well as the GAL-YTM1 cells shifted from galactose- to glucose-containing medium, in which Ytm1 is depleted. Processing of 35S pre-rRNA to mature 25S and 18S rRNAs occurred rapidly in the YTM1 strain. The 27S pre-rRNA processing intermediate was completely converted to 25S rRNA by the 60-min chase point. In the ytm1-1 mutant, however, 27S pre-rRNA was still present at the 60-min chase point; 27SA pre-rRNAs were converted to 27SB pre-rRNA less efficiently than in wild-type cells (Fig. 4B). Consequently, 3.6-fold less mature 25S rRNA was produced relative to 18S rRNA in the ytm1-1 mutant than in the wild-type cells (Fig. 4B, compare lanes 5 and 10). Both 35S pre-rRNA and 23S pre-rRNA accumulated in the ytm1-1 mutant. The effects on 35S and 23S are thought to be indirect and often are observed when 60S ribosomal subunit assembly is perturbed (4, 19). Effects on pre-rRNA processing observed in Ytm1-depleted cells are identical to those found in the ytm1-1 mutant (data not shown).
FIG. 4.
Processing of pre-rRNAs is altered in the ytm1-1 mutant. (A) Oligonucleotide probes or primers used to detect rRNAs and pre-rRNAs. (B) Yeast strains JWY3400 (YTM1) and JWY7128 (ytm1-1) were grown in YEPD medium at 25°C and shifted to 37°C for 5 h. Cells were pulse-labeled with [5,6 3H]uracil for 5 min and chased with an excess of unlabeled uracil for 2, 5, 10, and 60 min. Equal cpm of RNA isolated from cells at each time point were subjected to electrophoresis on agarose-formaldehyde gels to separate each pre-rRNA or rRNA and detected by autoradiography. (C) Primer extension was performed to determine steady-state levels of 27SA2, 27SA3, 27SBL plus 7SL, and 27SBS plus 7SS pre-rRNAs. RNA was extracted from whole-cell extracts from strains JWY3400 (YTM1) and JWY7128 (ytm1-1) grown in YEPD medium at 25°C or shifted from 25°C to 37°C for 3 h or 6 h or from strain JWY6149 (YTM1) or JWY6992 (GAL-YTM1) grown in galactose-containing medium and shifted to glucose-containing medium for 0, 10, 12, 15, or 18 h. (D) Northern blotting was used to determine steady-state levels of 25S, 18S, 5.8S, and 5S rRNA and 7S pre-rRNA. High-molecular-weight RNAs were subjected to electrophoresis on agarose-formaldehyde gels, whereas acrylamide-urea gels were used to separate low-molecular-weight RNAs. U3 snoRNA was used as a loading control. RNA was quantified by phosphorimaging and normalized to U3 snoRNA.
We analyzed the steady-state levels of rRNA intermediates and mature rRNAs by primer extension and Northern blotting (Fig. 4C and D). Ytm1 was depleted from GAL-YTM1 cells by shifting galactose-grown cells to glucose for 0 h, 10 h, 12 h, 15 h, or 18 h. The ytm1-1 cells were grown at 25°C and shifted to 37°C for 0 h, 3 h, or 6 h. Primer extension using an oligonucleotide that detects 27S pre-rRNAs (Fig. 4A) indicated that under nonpermissive conditions amounts of 27SA2 pre-rRNA decrease in ytm1-1 cells but increase in GAL-YTM1 cells (Fig. 4C). 27SA3 pre-rRNA strongly accumulated upon shifting GAL-YTM1 and ytm1-1 cells to the nonpermissive conditions (Fig. 4C). Amounts of 27SB pre-rRNAs were also affected: 27SBS pre-rRNA was drastically diminished relative to 27SBL pre-rRNA, making these species nearly equal in Ytm1-depleted or Ytm1-inactivated cells (Fig. 4C).
Phosphorimage analysis of Northern blots also indicated that 7S pre-rRNA and 5S rRNA decrease slightly upon shifting GAL-YTM1 and ytm1-1 cells to nonpermissive conditions (Fig. 4D). Despite the observed effects on processing of early and intermediate pre-rRNAs present in 66S pre-rRNPs, amounts of 25S and 5.8S rRNA were largely unaffected. Because preexisting 25S and 5.8S rRNAs are present in vast quantities, changes in steady-state amounts of 25S and 5.8S rRNAs may be masked and therefore difficult to observe by standard assays.
Taken together, these results indicate that in ytm1 mutants pre-rRNA processing delays begin early, at the step when 27SA2 pre-rRNA is converted to 27SA3 pre-rRNA. Subsequent steps in pre-rRNA processing are similarly slowed down, but no step in pre-rRNA processing is completely blocked. Mutation of Ytm1 through depletion or inactivation results in nearly identical phenotypes, suggesting that Ytm1-1 protein is largely inactive.
Ytm1 is necessary for release of 66S preribosomes from the nucleolus.
To further investigate the timing and role of Ytm1 in ribosome biogenesis, we assayed the ability of 60S preribosomes to exit the nucleolus and nucleus in the ytm1-1 mutant, using the ribosome export assay (28), in which eGFP-tagged rpL25 functions as a reporter. In ytm1-1 cells grown at the permissive temperature, 66S preribosomes were released to the cytoplasm (Fig. 5C). In wild-type cells grown at 25°C or shifted from 25°C to 37°C, rpL25eGFP signal was cytoplasmic (data not shown). When the ytm1-1 mutant strain was grown at 25°C and shifted to 37°C for 5 h, rpL25eGFP was strongly retained in the nucleolus in most cells (Fig. 5D, arrows), although in some cells signal was distributed throughout the nucleoplasm. Thus, Ytm1 is important for nucleolar release of 66S preribosomes and perhaps for subsequent nuclear export.
FIG. 5.
Inactivation of Ytm1 in the ytm1-1 mutant causes 66S preribosomes to accumulate in the nucleolus. The ytm1-1 mutant strain JWY6790 expressing eGFP-tagged rpL25 was grown in C-Trp medium at 25°C, washed and suspended in YEPD, and grown at 25°C (A and C) or shifted to 37°C for 5 h (B and D). Nuclei stained with 4′,6′-diamidino-2-phenylindole (DAPI) are shown in panels A and B (typically, nucleoli do not stain with DAPI). The signal from RpL25eGFP is shown in panels C and D. Arrows indicate nucleolar accumulation of rpL25eGFP (D) and corresponding DAPI staining (B).
Ytm1 is a component of 66S preribosomes.
Previously Ytm1 was identified in 66S pre-rRNPs purified using TAP-tagged ribosome assembly factors (3, 21, 26, 41). Consistent with this result, HA3-tagged Ytm1 peaks in sucrose gradient fractions 15 to 17 containing 66S preribosomes (21) (Fig. 6). A small amount of Ytm1 can be detected sedimenting at the top of the gradient in lighter fractions and larger amounts at the bottom of the gradient in heavier fractions. The significance of the sedimentation in heavier fractions is unclear.
FIG. 6.
Ytm1-HA3 cosediments on sucrose gradients with 66S preribosomes. Whole-cell extracts were prepared from yeast strain JWY6770 (YTM1-HA3) and fractionated on 7 to 47% sucrose velocity gradients. Fractions containing 40S and 60S ribosomal subunits and 80S monosomes are labeled. Proteins were trichloroacetic acid precipitated from gradient fractions and subjected to Western immunoblot analysis to detect Ytm1-HA3.
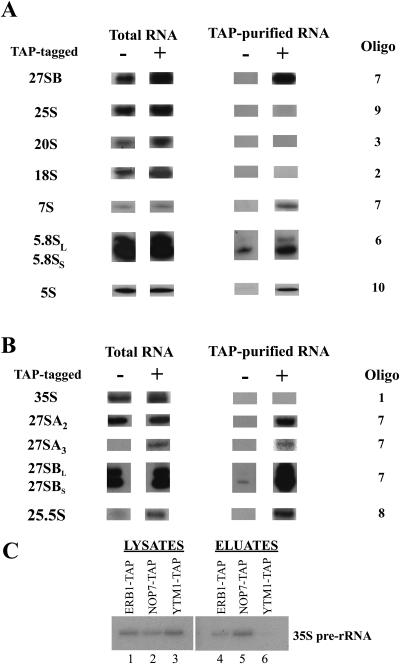
To examine in more detail ribosome assembly intermediates containing Ytm1, we identified the pre-rRNAs and proteins associated with affinity-purified Ytm1-TAP. TAP-tagged Ytm1 is fully functional: the tagged strain grows at wild-type rates and has a wild-type polysome profile, and Ytm1-TAP sediments on sucrose gradients with a peak at 66S (data not shown).
To determine in which preribosomes Ytm1 is present, we assayed which pre-rRNAs copurify with TAP-tagged Ytm1. The amounts of 27SA2, 27SA3, 27SB, 25.5S, and 7S pre-rRNAs recovered relative to each other were similar to those found in whole cells. Smaller relative amounts of 5.8S rRNA and no 35S or 20S pre-rRNA or 18S rRNA copurified with Ytm1-TAP (Fig. 7). Enrichment of Ytm1 with these RNA molecules is consistent with our finding that Ytm1 is important for assembly of 60S ribosomal subunits and for processing of 27S pre-rRNA (Fig. 3 and 4).
FIG. 7.
Ytm1 associates with pre-rRNAs in 66S preribosomes. (A) Whole-cell extracts were prepared from the YTM1-TAP strain JWY7124 and from untagged strain JWY3400 grown at 30°C in YEPD medium to 6 × 107 cells/ml. RNA was extracted from whole cells and from tandem affinity-purified samples, subjected to electrophoresis on agarose-formaldehyde or acrylamide-urea gels, blotted to nitrocellulose, and assayed by Northern blotting with specific oligonucleotide probes complementary to pre-rRNAs and mature rRNAs. Five percent of total RNA and 100% of tandem affinity-purified RNA were assayed. (B) Primer extension analysis was used to assay 35S, 27SA2, 27SA3, and 25.5S pre-rRNAs, as well as the BS and BL 5′ ends of 27S and 7S pre-rRNAs, using 32P-labeled oligonucleotides. Products of primer extension were resolved on sequencing gels, dried, and exposed to X-ray film for detection by autoradiography. (C) 35S pre-rRNA copurifies with TAP-tagged Erb1 and Nop7 but not Ytm1. RNA in whole-cell extracts and copurifying RNAs were assayed by primer extension as described above.
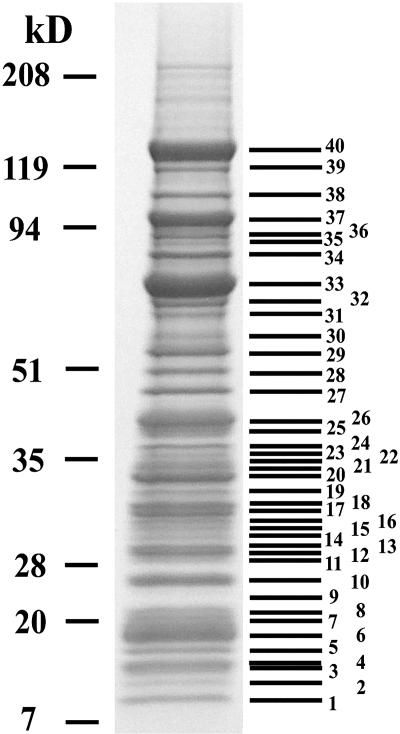
Fifty-three different proteins that copurify with TAP-tagged Ytm1 were identified by SDS-PAGE and mass spectrometry (Fig. 8). Among these are ribosomal proteins from both the large and small ribosomal subunits. Since these ribosomal proteins frequently contaminate TAPs (3, 11, 21), their significance cannot be assessed. Seventeen nonribosomal proteins specifically required for biogenesis of 60S ribosomal subunits are present in the Ytm1-containing particles (Fig. 8 and Table 2). Copurification of these pre-rRNAs and proteins with TAP-tagged Ytm1 indicates that Ytm1 first enters the ribosome assembly pathway by associating with 66S preribosomes and remains stably associated with 66S pre-rRNPs until late stages of maturation in the nucleoplasm.
FIG. 8.
Nonribosomal proteins necessary for biogenesis of 60S ribosomal subunits, as well as ribosomal proteins, copurify with TAP-tagged Ytm1. Whole-cell extract was prepared from the YTM1-TAP strain JWY7124 grown at 30°C in YEPD medium to 6 × 107 cells/ml and subjected to tandem affinity purification. Proteins were trichloroacetic acid precipitated from column eluates and subjected to electrophoresis on 4 to 20% polyacrylamide gels. Proteins were stained with colloidal Coomassie blue, manually excised from the gel, digested with trypsin, and identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (Table 2).
TABLE 2.
Nonribosomal proteins that copurify with Ytm1-TAP
| Gel ID no.a | Protein | ORF | Characteristic
|
Pre-rRNA processingc | Localizationb | Reference | |||
|---|---|---|---|---|---|---|---|---|---|
| Metazoan homolog | Essential | Sediments at 66S | Decreased 60S Subunits | ||||||
| 33 | Ytm1 | YOR272w | + | + | + | + | 27SA3↑ 25.5S↓ 7S↓ | No/Nu | This work (21) |
| 10 | Nip7 | YPL211w | + | + | + | + | NDd | No/Nu/C | 61 |
| 24 | Nop16 | YER002c | − | − | + | + | ND | No/Nu | 21 |
| 26 | Brx1 | YOL077c | + | + | ND | + | 27SA↑ 27SB↓ | No | 31 |
| 28 | Rlp7 | YNL002c | + | + | ND | + | 27SA3↑ 27SB↓ 7S↓ | No | 9,15 |
| 28 | Rpf2 | YKR081c | + | + | + | + | 27SB↑ | No | 39,60 |
| 29 | Cic1/Nsa3 | YHR052c | − | + | + | ND | 27S↓ 7S↓ | No | 13, 21,41 |
| 31 | Nsa1 | YGL111w | − | + | ND | + | ND | ND | 21 |
| 32, 33 | Has1 | YMR290c | + | + | ND | +/− | 35S↑ 27SB↑ | No | 10 |
| 34 | Ebp2 | YKL172w | + | + | ND | + | 35S↑ 27SA3↑ | No | 27,57 |
| 35, 36 | Nog1 | YPL093w | + | + | + | + | 27S↓ 7S↓ | No | 30 |
| 37 | Nop7 | YGR103w | + | + | + | + | 27S→25S slow | No | 1 |
| 37 | Noc3 | YLR002c | + | + | + | + | 35S↑ 27SB↑ | No/Nu | 37 |
| 38 | Noc2 | YOR206w | + | + | + | + | 35S↑ 27SB↑ | No/Nu | 37 |
| 38 | Nop2 | YGL111w | + | + | + | + | 35S↑ 27S↑ | No | 6,25 |
| 39 | Drs1 | YLL008w | − | + | ND | + | 27SB↑ | No | 48 |
| 40 | Erb1 | YMR049c | + | + | + | + | 27S↑ | No | 44 |
Protein band 30 was identified as Tef2 and protein band 36 also contained Ssal. These proteins are common contaminants of affinity-purified complexes (17,24).
No, nucleolar; Nu, nuclear; C, cytoplasmic.
↑ and ↓, increased and decreased amounts of indicated RNAs, respectively.
ND, not determined.
Ribosome assembly factors Ytm1, Nop7, and Erb1 are present together in a subcomplex.
Several different experiments demonstrate that Ytm1, Erb1, and Nop7 form aheterotrimeric subcomplex that is present within 66S preribosomes and also exists independently of pre-rRNPs. (i) Affinity purification from whole-cell extracts using TAP-tagged Ytm1, Nop7, or Erb1 yielded 50 to 60 proteins present in 66S pre-rRNPs (Fig. 8) (21; data not shown). In each case, greater amounts of Ytm1, Nop7, and Erb1 (Fig. 8, bands 33, 37, and 40, respectively) were recovered than those of any of the other proteins. This suggests that a mixture of 66S pre-rRNPs and a heterotrimer of Ytm1, Nop7, and Erb1 copurify with each of these TAP-tagged proteins (Fig. 8 and see Fig. 11). (ii) Affinity purification using TAP-tagged Nop7 or Ytm1 from rrp1-1 or nop4-3 mutants in which 66S pre-rRNPs are unstable (21, 26) yielded mostly Ytm1, Nop7, and Erb1, and greatly diminished amounts of molecules comprising 66S pre-rRNPs (21) (Fig. 9A). Thus, under these conditions, many fewer 66S pre-rRNPs were recovered, but the heterotrimeric subcomplexes remained intact. (iii) The Ytm1/Nop7/Erb1 heterotrimer could be separated from 66S pre-rRNPs by sucrose gradient fractionation or differential centrifugation of whole-cell extracts. Affinity purification from such enriched fractions using TAP-tagged Ytm1, Nop7, or Erb1 yielded primarily Ytm1, Nop7, and Erb1 (8, 32) (Fig. 9B, lane 1, and C, lane 2). (iv) Treatment of whole-cell extracts with a cocktail of phosphatase inhibitors caused pre-rRNPs to be disrupted (P. Harnpicharnchai, unpublished), while the Nop7/Ytm1/Erb1 subcomplex remained intact. Nop7-TAP or Ytm1-TAP under these conditions resulted in the recovery of only the Ytm1/Nop7/Erb1 heterotrimer (Fig. 9D, lane 2) (5; data not shown). (v) Nop7-TAP from sucrose gradient fractions containing 66S preribosomes yielded most protein components of the 66S pre-rRNPs (Fig. 9E, lane 1). However, when gradient fractions containing 66S pre-rRNPs were treated with the phosphatase inhibitor cocktail prior to affinity purification with Nop7-TAP, mostly the heterotrimer was recovered (Fig. 9E, lane 2). The last result indicates that Ytm1, Erb1, and Nop7 form a stable complex within preribosomes sedimenting at 66S and can be released from the 66S pre-rRNPs by treatment with the phosphatase inhibitor cocktail. The molecular basis of the effect(s) of the phosphatase inhibitors is unknown.
FIG. 11.
66S preribosomes are largely intact but lack Ytm1 in the ytm1-1 mutant. Wild-type YTM1 cells and mutant ytm1-1 cells expressing Nop7-TAP or Brx1-TAP were grown in YEPD medium at 25°C and shifted to 37°C for 5 h. (A) Silver staining or (B) Western immunoblot analysis was performed on proteins trichloroacetic acid precipitated from samples affinity purified from extracts from these strains.
FIG. 9.
Ytm1, Erb1, and Nop7 form a heterotrimeric subcomplex both within 66S preribosomes and independently of these particles. (A) Ytm1, Erb1, and Nop7 are enriched (relative to other proteins found in 66S pre-rRNPs) among proteins copurifying with Ytm1-TAP or Nop7-TAP from rrp1-1 or nop4-3 mutants in which 66S preribosomes are unstable. Heterotrimer was purified (B) from sucrose gradient fractions, (C) by differential centrifugation, or (D) from whole-cell extracts treated with a cocktail of phosphatase inhibitors that disrupt 66S pre-rRNPs. Wild-type cells or mutant cells were grown at 25°C and shifted to 37°C for 5 h. Tandem affinity purification using Nop7-TAP or Ytm1-TAP was carried out from (A) whole-cell extracts from a 50-ml culture, (B) gradient fractions 5 to 7 (prepared from a 900-ml culture), (C) whole-cell extracts from 50 ml of cells (lanes 1 and 3) or 180,000 × g spin supernatants prepared from 500 ml of cells (lanes 2 and 4), or (D) untreated (−) or phosphatase inhibitor cocktail-treated extracts (+). Purified proteins were resolved by SDS-PAGE. Note that the heterotrimer is destabilized in the ytm1-1 mutant (B, lane 2; C, lane 4; D, lane 4). (E) Ytm1, Erb1, and Nop7 form a stable subcomplex within 66S preribosomes. Whole-cell extracts from YTM1 cells were subjected to centrifugation on 7 to 47% gradients. Fractions containing 66S preribosomes were pooled and subjected to tandem affinity purification in the presence (+) or absence (−) of phosphatase inhibitors, and proteins were resolved by SDS-PAGE. Bands indicated by asterisks in B and E are common contaminants that we observe upon TAP from any fractions of sucrose gradients (top, middle, or bottom) using any TAP-tagged protein.
Our current data suggest that the components of the Ytm1/Nop7/Erb1 heterotrimer join preribosomes separately since significant or small amounts of 35S pre-rRNA coprecipitate with Nop7 and Erb1, respectively, although no 35S pre-rRNA copurifies with Ytm1 (Fig. 7C). Thus, Nop7 likely joins nascent preribosomes first, followed by Erb1. Ytm1 later associates with 66S preribosomes containing 27SA2 pre-rRNA (Fig. 7B).
Ytm1 and Nop7 directly interact with Erb1.
To assay pairwise interactions between components of the heterotrimer and to determine whether the interactions are direct, we carried out GST pull-down assays. Ytm1 bound specifically to GST-Erb1, and Erb1 bound to GST-Ytm1, while Nop7 displayed strong binding to GST-Erb1 but not to GST-Ytm1 (Fig. 10A) (data not shown). Consistent with these observations, Pes1 and Bop1, the mammalian homologues of Nop7 and Erb1, bind to each other in vitro and interact in two-hybrid assays in vivo (34). The interactions between Erb1 and Ytm1 were corroborated by two-hybrid assays in vivo. Cells expressing AD-YTM1 and BD-ERB1 or AD-ERB1 and BD-YTM1 displayed strong expression of the GAL-HIS3 reporter gene (growth on 50 mM 3-aminotriazide) (data not shown). Thus, Ytm1 and Nop7 each bind directly to Erb1 but not to one another (Fig. 10C). Erb1, like Ytm1, contains WD40 repeats. Nop7 also contains a known protein-protein interaction motif, the BRCT domain (1). Further analysis is necessary to test whether these or other domains dictate the strong interactions among these three proteins.
FIG. 10.
Ytm1 and Nop7 directly interact with Erb1. (A) Synthetic radiolabeled proteins (*) were incubated with GST fusion proteins (lanes 2, 5, and 8). As negative controls, synthetic peptides were incubated with GST beads only (lanes 1, 4, and 7) or GST fusion proteins were incubated with the unrelated, radiolabeled 40S ribosome assembly factor Krr1 or ribosomal protein L11 (lanes 3, 6, and 9). (B) Radiolabeled wild-type Ytm1 protein was preincubated at 37°C for 15 min (lane 2). Mutant Ytm1-1 protein was preincubated at 37°C for 15 min (lane 3), 30 min (lane 4), or 60 min (lane 5). Following preincubation, wild-type Ytm1 or mutant Ytm1-1 radiolabeled protein was incubated with GST-Erb1. Mutant Ytm1-1 protein was incubated with GST beads only (lane 1) as a negative control. Complexes were eluted from glutathione beads, subjected to electrophoresis on 10% polyacrylamide gels, and detected by autoradiography. (C) Model for interactions between Ytm1, Erb1, and Nop7. Gray lines indicate interactions detected using GST pulldown assays, whereas the black line indicates interactions detected by two-hybrid assays.
Association of heterotrimer with 66S preribosomes is significantly weakened in the ytm1-1 mutant.
Since the two ytm1-1 mutations are in residues that may be important for interactions with ligands, we determined the effects of these mutations on the integrity of the heterotrimer and 66S preribosomes. We used TAP-tagged Nop7 or Brx1 to purify 66S preribosomes from YTM1 and ytm1-1 strains, since both of these proteins are present in all seven different 66S pre-rRNPs (21, 60) (Fig. 1B). SDS-PAGE, silver staining, and Western blotting revealed that most of the proteins present in wild-type 66S pre-rRNPs are also present in equivalent amounts in the particles isolated from the ytm1-1 mutant, indicating that the 66S preribosomes are largely intact in the ytm1-1 mutant (Fig. 11). However, Ytm1-1 mutant protein was greatly diminished or absent from the pool of purified 66S preribosomes (Fig. 11A, lanes 2 and 4, and B, lanes 2 and 4). Nop7 and Erb1 were still present, but in reduced amounts (Fig. 11A, lanes 2 and 4). Thus some of the mutant preribosomes contain Nop7 and/or Erb1 but not Ytm1, and others lack all three proteins. The relative amounts of each pre-rRNA copurifying with TAP-tagged Nop7 or Brx1 in 66S pre-rRNPs parallel those in whole-cell extracts (data not shown).
The Ytm1/Nop7/Erb1 heterotrimer is destabilized in the ytm1-1 mutant.
The effects of the ytm1-1 mutations on 66S pre-rRNPs and pre-rRNA processing could result from alterations of the heterotrimer containing Ytm1. Therefore, we purified the heterotrimer from the ytm1-1 mutant and examined its integrity, using three assays: sucrose gradient centrifugation, differential centrifugation, and treatment of whole-cell extracts with phosphatase inhibitors. In each case, only small amounts, if any, of Erb1 copurified with Nop7 and no Ytm1 could be detected (Fig. 9B, lane 2; C, lane 4; and D, lane 4). GST-pulldown assays confirmed that Ytm1-1 does not bind to Erb1 in vitro at 37°C (Fig. 10B). These results suggest that at the nonpermissive temperature, Ytm1-1 fails to interact with Erb1 and the Nop7-Erb1 association is significantly weakened, leading to destabilization of the heterotrimer and perturbations of preribosome maturation.
DISCUSSION
Although there has been much progress to identify yeast ribosome assembly intermediates and their protein and RNA constituents (reviewed in references 12 and 14), nothing is known about the architecture of these pre-rRNPs. For example, it is unclear which proteins are nearest neighbors within assembling ribosomes and to what extent neighboring molecules function together. Since Ytm1 contains multiple WD40 protein-protein interaction motifs, it is an excellent candidate for a molecule present in a multiprotein subcomplex comprising a neighborhood in assembling ribosomes.
Here we have shown that Ytm1 is a constituent of four consecutive 66S preribosomes and is necessary for steps in their maturation to 60S ribosomal subunits. Ytm1 associates with ribosome assembly factors Erb1 and Nop7 to form a stable subcomplex that is present within 66S preribosomes and that also exists separately from pre-rRNPs. Erb1 and Nop7 assemble into preribosomes prior to Ytm1. Mutations in WD40 motifs 6 and 7 of Ytm1 destabilize the heterotrimer and weaken association of each of the three proteins with 66S pre-rRNPs. Consequently, processing of pre-rRNAs and release of preribosomes from the nucleolus are delayed, resulting in production of fewer 60S ribosomal subunits. This is one of the first examples demonstrating the importance of protein-protein interactions and subcomplex integrity for assembly of eukaryotic ribosomes.
Ytm1 associates with 66S preribosomes.
The copurification of Ytm1 with the same relative proportions of 27SA2, 27SA3, 27SB, 25.5S, and 7S pre-rRNAs as found in whole-cell extracts (Fig. 7) indicates that Ytm1 is present in each of the four consecutive 66S pre-rRNPs containing these pre-rRNAs, throughout most or all of their lifetimes. No 20S pre-rRNA or 18S rRNA copurifies with Ytm1, consistent with Ytm1 participating in biogenesis of 60S ribosomal subunits but not 40S subunits. Although Ytm1-TAP particles contain moderate amounts of 5.8S rRNA, we failed to detect significant levels of 25S rRNA, consistent with a previous report indicating that conversion of 7S pre-rRNA to 5.8S rRNA occurs more rapidly than production of 25S rRNA from 25.5S pre-rRNA (18).
We also identified 53 proteins that copurify with TAP-tagged Ytm1 (Fig. 8, Table 2). Of the proteins isolated, 17 are ribosome assembly factors previously shown to be components of 66S preribosomes (Table 2). None of the proteins found specifically in late nucleoplasmic or cytoplasmic 66S pre-rRNPs (41) are present in preribosomes purified using Ytm1-TAP. This is consistent with our RNA and subcellular localization data indicating that Ytm1 dissociates from pre-rRNPs in the nucleoplasm prior to the latest nucleoplasmic and cytoplasmic stages of ribosome maturation. Thus 66S preribosomes purified using Ytm1-TAP may represent core complexes of molecules important for intermediate steps in 60S subunit ribosome biogenesis.
Ytm1 is present in a heterotrimeric complex with Nop7 and Erb1.
Our results and those of others indicate that Ytm1, Erb1, and Nop7 form a stable heterotrimeric complex (8, 21, 32) (Fig. 8, 9, and 10). The sedimentation of Nop7, Ytm1, and Erb1 on sucrose gradients indicates that in wild-type cells most of this microparticle exists within 66S preribosomes (21; our unpublished results) (Fig. 6 and 9E). Both Ytm1 and Nop7 bind tightly to Erb1; however, we find no evidence for direct interactions between Ytm1 and Nop7 (Fig. 10). The ability to purify the heterotrimer directly from 66S preribosomes using the phosphatase inhibitor cocktail that disrupts 66S pre-rRNPs (Fig. 9E) suggests that interactions among these three proteins are stronger than those with other molecules in the 66S pre-rRNP. Nevertheless, it is reasonable to assume that this subcomplex influences the assembly or function of a larger protein or RNP neighborhood within assembling ribosomes necessary for their efficient maturation (see below). Thus, it will be important to identify other molecules that are adjacent to Ytm1, Erb1, and Nop7 within preribosomes.
The heterotrimeric complex also exists independently of 66S preribosomes. It can be purified from fractions near the top of gradients or from high-speed supernatants (Fig. 9B and C). Our present data suggest that Nop7 and Erb1 may assemble into preribosomes before Ytm1. Consistent with this finding is the presence of Nop7 and Erb1, but not Ytm1, in 66S pre-rRNPs isolated from the ytm1-1 mutant. Nop7 and Erb1 may not require Ytm1 to assemble into or remain in preribosomes. Upon completion of their functions, Nop7, Erb1, and Ytm1 might dissociate from preribosomes together as a heterotrimer, prior to recycling into other nascent preribosomes. The substantial amount of heterotrimer that we purify from cells might also result from disassembly in vivo of unstable abortive assembly intermediates or upon dissociation from preribosomes in vitro during fractionation or purification.
A conserved network of protein interactions.
Interactions among Ytm1, Erb1, and Nop7 required for ribosome biogenesis are likely to be conserved. Interactions of Pes1, the mouse homologue of Nop7, with Bop1, the mouse homologue of Erb1, are required for assembly of Pes1 into preribosomes (34). Like their yeast counterparts, Pes1 and Bop1 are required for similar steps in processing of pre-rRNAs to mature 25S and 5.8S rRNA (1, 35, 44, 55, 56).
Mutations in the WD40 motifs of Ytm1 destabilize the heterotrimer and weaken its association with preribosomes.
The G398D and S442N mutations in WD40 repeats 6 and 7 of YTM1 prevent binding of Ytm1-1 to Erb1 in vitro and significantly weaken association between Nop7 and Erb1 (Fig. 9 and 10). Destabilization of the heterotrimer also weakens association of Ytm1, Erb1, and Nop7 with 66S pre-rRNPs, resulting in recovery of greatly diminished amounts of Ytm1 and slightly decreased amounts of Erb1, relative to Nop7, in the pool of purified 66S preribosomes (Fig. 11). The mixture of 66S pre-rRNPs purified from the ytm1-1 mutant using Brx1-TAP contains no detectable Ytm1-1 and less Nop7 and Erb1 compared to preribosomes isolated from wild-type cells (Fig. 11). (Note that no free heterotrimer could copurify with Brx1-TAP.) The greater decrease of Ytm1 compared to Erb1 in Nop7-TAP-purified ytm1-1 mutant particles suggests that the Ytm1-Erb1 interaction may be necessary to recruit or maintain Ytm1 in 66S pre-rRNPs. This interaction may also influence assembly of Nop7 and Erb1 into preribosomes or may be required to stabilize their association with preribosomes, since amounts of Nop7 are reduced slightly in the 66S preribosomes purified from ytm1-1 mutant cells expressing Brx1-TAP.
Ytm1 is required for pre-rRNA processing and trafficking of 66S preribosomes.
Our data show that pre-rRNA processing is slowed in the ytm1 mutants. The conversion of 27SA3 pre-rRNA to 27SB pre-rRNA is delayed, resulting in changes in amounts of 27SBS and 27SBL pre-rRNAs. Subsequent steps in pre-rRNA processing are also slowed, resulting in reduced amounts of 7S pre-rRNA and 5S rRNA (Fig. 4). Processing of 27SA3 to 27SB pre-rRNA involves rapid exonucleolytic trimming of the 5′ end of the 27SA3 pre-rRNA by Rat1 and Xrn1 (23). Ytm1 might function directly in all of these steps. Alternatively, the decreased rate of processing of 27SA3 pre-rRNA in the ytm1 mutants might indirectly affect subsequent processing of 27SB pre-rRNA, for example, by perturbing the architecture of pre-rRNPs. Depletion of Nop7 and Erb1 has effects on pre-rRNA processing similar but not completely identical to those of ytm1 mutants: increased levels of 27SA3 pre-rRNA and decreased amounts of 27SB and 7S pre-rRNAs (42, 44). Thus, the pre-rRNA processing phenotypes of the ytm1 mutants may reflect a combination of effects on the presence and/or functions of all three proteins in the Ytm1/Erb1/Nop7 heterotrimer.
Most, but not all steps of processing of precursors to 25S and 5.8S rRNA are thought to occur prior to nucleolar release and most likely require or are accompanied by many changes in the topology of these pre-rRNPs. Although the mechanism whereby preribosomes exit the nucleolus remains a mystery, it may require some changes in the composition and structure of preribosomes, including those resulting from pre-rRNA processing. Nuclear export of preribosomes requires binding of export factors to preribosomes, perhaps timed by the availability of ligands on the surface of preribosomes (22). Thus pre-rRNA processing, nucleolar release, and nuclear export may be intertwined to prevent premature exit of incompletely assembled ribosomes from the nucleolus and nucleus. Therefore, rather than binding directly to export factors, Ytm1, as well as Erb1 and Nop7, is likely to participate indirectly in nucleolar release and nuclear export of preribosomes, by functioning together to establish or maintain structures necessary for these processes. Indeed, as observed for the ytm1-1 mutant, preribosomes also accumulate in the nucleus upon depletion of Nop7 (42). The ytm1-1 mutations, however, cause 66S preribosomes to be retained in the nucleolus (Fig. 5).
Functions of the heterotrimer in ribosome biogenesis.
Taken together, our results suggest the following model for effects of ytm1-1 mutations on ribosome assembly. In wild-type cells, early in ribosome assembly during or after synthesis of 35S pre-rRNA, Nop7 and Erb1 join nascent preribosomes. Slightly later, many 60S ribosome assembly proteins, including Ytm1, associate with the 27SA2 pre-rRNA to form the earliest detectable 66S pre-rRNP. In the ytm1-1 mutant, Ytm1-1 may not efficiently assemble into this pre-RNP or may not remain stably associated with preribosomes, also resulting in destabilization of association of Nop7 and Erb1 with pre-60S ribosomes. Although it is also possible that Ytm1, Nop7, or Erb1 dissociates from the mutant particles during purification rather than in vivo, these mutant particles are likely otherwise intact. However, their overall architecture might be perturbed by the absence or weakened association of Ytm1, Erb1, and Nop7. When preribosomes lack these putative scaffolding proteins, they might be unable to establish or maintain structures necessary for efficient pre-rRNA processing, ribosome assembly, release of nascent ribosomes from the nucleolus, or export of preribosomes from the nucleus to the cytoplasm.
Moonlighting functions of the heterotrimeric proteins.
Clearly Ytm1 is necessary for efficient ribosome production. Depletion or inactivation of Ytm1 decreases the rate of 60S subunit biogenesis below levels necessary to sustain viability. However, Ytm1 may have a second “moonlighting” function, such as mitosis or chromosome transmission (36, 43; Matsumoto et al., personal communication). Interestingly, Nop7 and its metazoan homologue Pescadillo, and Bop1, the mammalian homologue of Erb1, also have independent functions. Nop7 and Pescadillo are implicated in DNA replication or cell proliferation (8, 35). Mutations in BOP1 induce p53-dependent cell cycle arrest (45). Thus, the heterotrimeric complex of Ytm1, Erb1, and Nop7 might be a depot for directing multiple functions of these proteins.
Acknowledgments
We thank Seiji Matsumoto for generously providing ytm1-1 and YTM1 strains and for communicating unpublished results. We thank Stan Fields and Tony Hazbun (Yeast Resource Center, University of Washington) for plasmids containing YTM1, ERB1, or NOP7 cloned into pOAD. We also thank Michael McAlear, Jonathan Warner, David Goldfarb, Janine Maddock, Jennifer Fuentes, and Bernard Trumpower for antibodies against ribosomal proteins or ribosome assembly factors. We are grateful to Susan Dowd and Mark Bier (Center for Molecular Analysis, Carnegie Mellon University) for assistance with mass spectrometry. We thank Jon Minden, Peter Berget, Jeff Brodsky, Elizabeth Jones, and members of our laboratory for fruitful discussions and for comments on the manuscript. We thank Brooke McCartney for use of her microscope.
This work was supported by National Institutes of Health grants RO1 GM28301 to J.L.W., F31 GM65067 to T.D.R., and F31 GM19937 to E.W.H. P.H. was supported by the government of Thailand.
REFERENCES
- 1.Adams, C. C., J. Jakovljevic, J. Roman, P. Harnpicharnchai, and J. L. Woolford, Jr. 2002. Saccharomyces cerevisiae nucleolar protein Nop7 is necessary for biogenesis of 60S ribosomal subunits. RNA 8:150-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. L. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 3.Baβler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 4.Basu, U., K. Si, J. R. Warner, and U. Maitra. 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21:1453-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cagney, G., P. Uetz, and S. Fields. 2001. Two-hybrid analysis of the Saccharomyces cerevisiae 26S proteasome. Physiol. Genomics 7:27-34. [DOI] [PubMed] [Google Scholar]
- 6.de Beus, E., J. S. Brockenbrough, B. Hong, and J. P. Aris. 1994. Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J. Cell Biol. 127:1799-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large U3 snoRNP complex which is required for 18S rRNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du, Y. C., and B. Stillman. 2002. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109:835-848. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, D. A., F. Dragon, S. J. Lee, and S. J. Baserga. 2000. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA 97:13027-13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emery, B., J. de la Cruz, S. Rocak, O. Deloche, and P. Linder. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 52:141-158. [DOI] [PubMed] [Google Scholar]
- 11.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 12.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 13.Fatica, A., M. Oeffinger, D. Tollervey, and I. Bozzoni. 2003. Cic1p/Nsa3p is required for synthesis and nuclear export of 60S ribosomal subunits. RNA 9:1431-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 15.Gadal, O., D. Strauss, E. Petfalski, P. E. Gleizes, N. Gas, D. Tollervey, and E. Hurt. 2002. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 157:941-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudet, R., A. Bohm, and P. B. Sigler. 1996. Crystal structure at 2.4 Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell 87:577-588. [DOI] [PubMed] [Google Scholar]
- 17.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 18.Geerlings, T. H., J. C. Vos, and H. A. Raué. 2000. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA 6:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelperin, D., L. Horton, J. Beckman, J. Hensold, and S. K. Lemmon. 2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 9:1268-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 21.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt., and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 22.Hedges, J., M. West, and A. R. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry, Y., H. Wood, J. P. Morrissey, E. Petfalski, S. Kearsey, and D. Tollervey. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 10:2452-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 25.Hong, B., J. S. Brockenbrough, P. Wu, and J. P. Aris. 1997. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 17:378-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horsey, E. W., J. Jakovljevic, T. D. Miles, P. Harnpicharnchai, and J. L. Woolford, Jr. 2004. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA 10:813-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber, M. D., J. H. Dworet, K. Shire, L. Frappier, and M. A. McAlear. 2000. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem. 275:28764-28773. [DOI] [PubMed] [Google Scholar]
- 28.Hurt, E., S. Hannus, B. Schmeizl, D. Lau, D. Tollervey, and G. Simos. 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in Ran-cycle and nucleoporin mutants. J. Cell Biol. 144:389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James, P., J. Halladay, and E. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaser, A., E. Bogengruber, M. Hallegger, E. Doppler, G. Lepperdinger, M. Jantsch, M. Breitenbach, and G. Kreil. 2001. Brix from Xenopus laevis and Brx1p from yeast define a new family of proteins involved in the biogenesis of large ribosomal subunits. Biol. Chem. 382:1637-1647. [DOI] [PubMed] [Google Scholar]
- 32.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 33.Lambright, D. G., J. Sondek, A. Bohm, N. P. Skiba, H. E. Hamm, and P. B. Sigler. 1996. The 2Å crystal structure of a heterotrimeric G protein. Nature 379:311-319. [DOI] [PubMed] [Google Scholar]
- 34.Lapik, Y. R., C. J. Fernandes, L. F. Lau, and D. G. Pestov. 2004. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell 15:17-29. [DOI] [PubMed] [Google Scholar]
- 35.Lerch-Gaggl, A., J. Haque, J. Li, G. Ning, P. Traktman, and S. A. Duncan. 2002. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J. Biol. Chem. 277:45347-45355. [DOI] [PubMed] [Google Scholar]
- 36.Matsuzaki, F., S. Matsumoto, and I. Yahara. 1988. Truncation of the carboxy-terminal domain of yeast β-tubulin causes temperature-sensitive growth and hypersensitivity to antimitotic drugs. J. Cell Biol. 107:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 38.Milkereit, P., D. Strauss, J. Bassler, O. Gadal, H. Kuhn, S. Schutz, N. Gas, J. Lechner, E. Hurt, and H. Tschochner. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278:4072-4081. [DOI] [PubMed] [Google Scholar]
- 39.Morita, D., K. Miyoshi, Y. Matsui, A. Toh-E, H. Shinkawa, T. Miyakawa, and K. Mizuta. 2002. Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and is essential for the processing of 27SB pre-rRNA to 25S rRNA and the 60S ribosomal subunit assembly in Saccharomyces cerevisiae. J. Biol. Chem. 32:28780-28786. [DOI] [PubMed] [Google Scholar]
- 40.Neer, E. J., and T. F. Smith. 1996. G protein heterodimers: new structures propel new questions. Cell 84:175-178. [DOI] [PubMed] [Google Scholar]
- 41.Nissan, T. A., J. Baβler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. Pre-60S particles on the road to ribosomes. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oeffinger, M., A. Leung, A. Lamond, and D. Tollervey. 2002. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA 8:626-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouspenski, I., S. J. Elledge, and B. R. Brinkley. 1999. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 27:3001-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestov, D. G., M. G. Stockelman, Z. Strezoska, and L. F. Lau. 2001. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 29:3621-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pestov, D. G., Ž. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21:4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raué, H. A. 2003. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae. The machine that makes the machine, p. 1-24. In M. O. J. Olson (ed.), The nucleolus. Kluwer Academic/Plenum Publishers, Dordrecht, The Netherlands.
- 47.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 48.Ripmaster, T. L., G. P. Vaughn, and J. L. Woolford, Jr. 1992. A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl. Acad. Sci. USA 89:11131-11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saveanu, C., A. Namane, P.-E. Gleizes, A. Lebreton, J.-C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schäfer, T., D. Strauβ, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scherl, A., Y. Coute, C. Deon, A. Calle, K. Kindbeiter, J.-C. Sanchez, A. Greco, D. Hochstrasser, and J. J. Diza. 2002. Functional proteomic analysis of the human nucleolus. Mol. Biol. Cell 13:4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, T. F., C. Gaitatzes, K. Sacena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 54.Sondek, J., A. Bohm, D. G. Lambright, H. E. Hamm, and P. B. Sigler. 1996. Crystal structure of a G protein βγ dimer at 2.1Å resolution. Nature 379:369-374. [DOI] [PubMed] [Google Scholar]
- 55.Strezoska, Ž., D. G. Pestov, and L. F. Lau. 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 20:5516-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2002. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 277:29617-29625. [DOI] [PubMed] [Google Scholar]
- 57.Tsujii, R., K. Miyoshi, A. Tsuno, Y. Matsui, A. Toh-e, T. Miyakawa, and K. Mizuta. 2000. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells 5:543-553. [DOI] [PubMed] [Google Scholar]
- 58.Venema, J., R. J. Planta, and H. A. Raue. 1998. In vivo mutational analysis of ribosomal RNA in Saccharomyces cerevisiae. Methods Mol. Biol. 77:257-270. [DOI] [PubMed] [Google Scholar]
- 59.Wall, M. A., D. E. Coleman, E. Lee, J. A. Iniguez-Lluhi, B. A. Posner, A. G. Gilman, and S. R. Sprang. 1995. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83:1047-1058. [DOI] [PubMed] [Google Scholar]
- 60.Wehner, K. A., and S. J. Baserga. 2002. The σ70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9:329-339. [DOI] [PubMed] [Google Scholar]
- 61.Zanchin, N. I., P. Roberts, A. DeSilva, F. Sherman, and D. S. Goldfarb. 1997. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell. Biol. 17:5001-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]