Abstract
Caspase 9 is a critical component of the mitochondrial or intrinsic apoptotic pathway and is activated by Apaf-1 following release of cytochrome c from mitochondria in response to a variety of stimuli. Caspase 9 cleaves and activates effector caspases, mainly caspase 3, leading to the demise of the cell. Survival signaling pathways can impinge on this pathway to restrain apoptosis. Here, we have identified Ser144 of human caspase 9as an inhibitory site that is phosphorylated in a cell-free system and in cells in response to the protein phosphatase inhibitor okadaic acid. Inhibitor sensitivity and interactions with caspase 9 indicate that the predominant kinase that targets Ser144 is the atypical protein kinase C isoform zeta (PKCζ). Prevention of Ser144 phosphorylation by inhibition of PKCζ or mutation of caspase 9 promotes caspase 3 activation. Phosphorylation of serine 144 in cells is also induced by hyperosmotic stress, which activates PKCζ and regulates its interaction with caspase 9, but not by growth factors, phorbol ester, or other cellular stresses. These results indicate that phosphorylation and inhibition of caspase 9 by PKCζ restrain the intrinsic apoptotic pathway during hyperosmotic stress. This work provides further evidence that caspase 9 acts as a focal point for multiple protein kinase signaling pathways that regulate apoptosis.
Apoptosis is a controlled form of cell death that plays important roles during development and tissue homeostasis through the removal of damaged or unnecessary cells (22). A family of cysteine proteases, termed caspases, are key mediators of apoptosis and are present in cells as inactive or low-activity zymogens (45). Many apoptotic stimuli induce the release of cytochrome c from mitochondria, a step that is controlled by pro- and antiapoptotic proteins of the Bcl-2 family (13). In the cytosol, cytochrome c binds to Apaf-1, inducing its assembly into a high-molecular-weight complex, the apoptosome, which recruits and activates caspase 9, probably by enhancing the dimerization of procaspase 9 monomers (1, 29, 38). Active caspase 9 initiates a cascade of caspase activation by irreversibly cleaving and activating downstream effector caspases such as caspase 3 that are responsible for the demolition of the cell (6).
Induction of apoptosis must be tightly regulated to ensure that potentially dangerous cells are efficiently removed, for instance, those with severe genomic damage, while cells that are transiently stressed by environmental conditions can recover and survive. Aberrations in the balance between pro- and antiapoptotic controls are likely to underlie diseases that are characterized by inappropriate or insufficient apoptosis, such as degenerative diseases and cancer, respectively (44). The mitochondrial or intrinsic apoptotic pathway is regulated downstream of cytochrome c release by caspase inhibitor proteins such as XIAP (14). The activity of XIAP may be controlled by the release of other factors such as Smac/Diablo from mitochondria (41). Heat shock proteins can also bind to components of the pathway and prevent caspase activation during cellular stress (4). In addition, components of the pathway such as XIAP (11) and caspase 9 (3, 7, 32) are regulated posttranslationally through phosphorylation by protein kinases activated by signaling pathways. The ERK mitogen-activated protein (MAP) kinase pathway, which can suppress apoptosis in many cell types, phosphorylates caspase 9 in growth factor-stimulated cells at an inhibitory site, Thr125 (3). Caspase 9 may also be targeted by protein kinase B/Akt (7) and protein kinase A (32) and appears to act as a focal point for multiple signaling pathways that restrain apoptosis during mitogenesis and possibly also in response to cellular stresses. Phosphorylation of caspase 9 may contribute to the suppression of apoptosis in cancer cells in which inhibitory pathways such as those operating through ERK MAP kinase are constitutively activated. However, the regulation of caspase 9 phosphorylation is not fully characterized, particularly with respect to stress signaling.
A number of other protein kinases are thought to modulate apoptosis, including protein kinase C (PKC). The PKC family is composed of the classical α, β, and γ isoforms, which are activated by diacylglycerol in a Ca2+- and phospholipid-dependent manner; the novel δ, ɛ, θ, and η isoforms, which are also activated by diacylglycerol and phospholipids but are Ca2+ insensitive; and the atypical ζ and λ/i isoforms that are insensitive to both diacylglycerol and Ca2+ (33, 36). While some PKC isoforms have been implicated in apoptosis, loss of other isoforms can trigger this process, implying that they promote cell survival (47). In general, the δ and θ isoforms appear to play roles in the induction of apoptosis, whereas the classical α/β isoforms and atypical ζ/λ/i isoforms have been implicated in suppression of apoptosis (20). One mechanism by which PKC isoforms might regulate apoptosis is phosphorylation of caspase 9.
Cell-free systems that reproduce the intrinsic or mitochondrial apoptotic pathway have proved to be useful for dissecting the biochemical mechanisms controlling caspase activation, including regulation by signaling pathways (3, 9, 32). In the present study, we have used concentrated cell extracts to identify a novel inhibitory phosphorylation site in human caspase 9, Ser144. This site is targeted by the atypical PKC (aPKC) PKCζ both in vitro and in intact cells treated with the protein phosphatase inhibitor okadaic acid (OA). We show that phosphorylation of Ser144 in cells is stimulated by hyperosmotic stress, which activates PKCζ, but not by growth factors, phorbol ester, or other cellular stresses. Phosphorylation and inhibition of caspase 9 by PKCζ may therefore provide a mechanism to restrain apoptosis in osmotically stressed cells.
MATERIALS AND METHODS
Antibodies and reagents.
A sheep polyclonal antibody was generated against His6-caspase 9C287A and affinity purified on a glutathione S-transferase (GST)-caspase 9C287A column. An antibody that recognizes caspase 9 phosphorylated on Ser144 was raised in rabbits (Moravian Biotechnology, Brno, Czech Republic) inoculated with the phosphopeptide GALEpSLRGNAD, where pS represents phosphoserine (synthesized by G. Bloomberg, University of Bristol, Bristol, United Kingdom). Phosphorylation site-specific antibody was purified by two rounds of negative selection against nonphosphorylated peptide, followed by selection against the phosphopeptide. Commercial antibodies were purchased from Oncogene Research (pan-PKC), Santa Cruz Biotechnology (PKCζ/λ), and BD Transduction Laboratories (monoclonal PKC isoform antibody sampler kit). OA, 12-O-tetradecanoylphorbol 13-acetate (TPA), and epidermal growth factor were purchased from Affinity, Calbiochem, and Sigma, respectively. Myristoylated PKCζ pseudosubstrate (N-Myr-Ser-Ile-Tyr-Arg-Arg-Gly-Ala-Arg-Arg-Trp-Arg-Lys-Leu), myristoylated PKCα/β pseudosubstrate (N-Myr-Phe-Ala-Arg-Lys-Gly-Ala-Leu-Arg-Gln), and other protein kinase inhibitors were purchased from Calbiochem; acetyl Asp-Glu-Val-Asp 7-amido-4-methylcoumarin (AcDEVD-AMC) was from Biomol; and other reagents were from Sigma.
Plasmids and recombinant proteins.
pcDNA3.Caspase 9 and pcDNA3.Apaf-1(1-541) were generated as described previously (3). Wild-type and D386A mutant pcmv5.FLAG-PKCζ (mouse) were kind gifts from D. Alessi, University of Dundee, Dundee, Scotland, United Kingdom. Site-directed mutagenesis was carried out with the QuikChange kit (Stratagene). For recombinant protein production in Escherichia coli BLR(DE3), caspase 9 was subcloned into pGEX-4T1 (Amersham Pharmacia Biotech) or pET28a (Novagen). Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 30°C. Recombinant proteins tagged with GST or His6 were affinity purified on glutathione-Sepharose 4B beads (Amersham) or Ni-nitrilotriacetic acid-agarose beads (QIAGEN), respectively. Proteins were eluted from washed beads in HEPES-buffered saline (10 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 0.5 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride, 1 μg ml−1 each aprotinin, leupeptin, and pepstatin A) containing 15 to 50 mM glutathione or 25 to 250 mM imidazole, respectively. Glutathione or imidazole was removed by filtering through a PD10 desalting column (Amersham Pharmacia Biotech), and proteins were eluted in HEPES-buffered saline and stored in aliquots at −70°C. In vitro transcription and translation were carried out with the TNT Quick coupled transcription-translation kit according to the manufacturer's protocol (Promega), and proteins were labeled by including [35S]methionine (Amersham Pharmacia Biotech) in the reaction mixture.
Cell culture, transfections, and extracts.
HeLa, HEK293, and U2OS cells were cultured in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) fetal bovine serum, 2 mM glutamine, 50 μg/ml streptomycin, and 50 U/ml penicillin G (Invitrogen). Plasmid DNA for transfections was purified on cesium chloride gradients, and transfections were carried out with Superfect reagent (QIAGEN) according to the manufacturer's protocol. At 24 h after transfection, U2OS or HEK293 cells were cultured in Dulbecco modified Eagle medium without serum for a further 24 h prior to treatment with 1 μM OA, 25 ng/ml epidermal growth factor, 1 μM TPA, 0.7 M NaCl, or 0.5 M sorbitol. Cells were lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and analyzed by Western blotting. HeLa cells were serum starved for 48 h prior to OA treatment and caspase 9 immunoprecipitation.
Preparation of cell extracts.
HeLa cell cytosolic (S100) extracts were purchased from Cilbiotech (Mons, Belgium) and were supplied in 10 mM HEPES-KOH (pH 7.5)-10 mM KCl-10 mM MgCl2-0.5 mM DTT at a protein concentration of 8 to 12 mg/ml. Extracts of NIH 3T3 and HEK293 cells were prepared as follows. Pelleted cells were washed and resuspended in an equal volume of cold cell extract buffer (20 mM HEPES-KOH, pH 7.5, 10 mM KCl, 2 mM MgCl2, 1 mM DTT), lysed by repeated passage through a 25-gauge needle, and then centrifuged at 16,000 × g, 30 min, 4°C. The supernatant (S16 extract) was snap-frozen and stored in aliquots in liquid nitrogen.
Phosphorylation of caspase 9 in HeLa cytosolic extract.
Caspase 9 was phosphorylated in HeLa cytosolic extract incubated with 1 μM OA and an ATP-regenerating system (1 mM ATP, 10 μg ml−1 creatine kinase, 5 mM creatine phosphate) for 4 h at 30°C. Reactions were stopped by boiling in SDS-PAGE sample buffer containing 5% (vol/vol) 2-mercaptoethanol and analyzed by Western blotting.
Coprecipitation of PKC and caspase 9.
GST-caspase 9 (5 to 10 μg) was phosphorylated in HeLa cytosolic extract containing 1 μM OA in a total volume of 100 μl for 2 h at 30°C. GST-tagged protein was affinity purified by incubation with 10 μl glutathione-Sepharose 4B beads for 1 h at 4°C in a total volume of 500 μl buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 1 μM OA). Beads were washed three times in the same buffer and boiled in SDS-PAGE sample buffer prior to separation of proteins by SDS-PAGE and detection by Western blotting.
Immunodepletion of caspase 9 from HeLa cytosolic extract.
A 10-μl volume of sheep anti-caspase 9 antibody was bound to 40 μl of protein A beads (Amersham Pharmacia Biotech) by incubation overnight at 4°C in 200 μl of cell extract buffer (20 mM HEPES-KOH, pH 7.5, 10 mM KCl, 2 mM MgCl2). Beads were washed five times in the same buffer and then incubated with 100 μl of HeLa cytosolic extract for 3 h at 4°C. A second round of depletion was carried out with 5 μl of caspase 9 antibody bound to 20 μl of protein A beads, as before, generating caspase 9-depleted extract. Successful immunodepletion was confirmed by Western blotting with a mouse monoclonal antibody to caspase 9.
Immunoprecipitations.
For precipitation from HeLa cytosolic extract, 200 μl extract was diluted to 500 μl with buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Igepal, 1 μM OA, 0.5 mM DTT). For precipitations from cultured cells, cells were first lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM Na3VO4, 50 mM NaF, 5 mM β-glycerophosphate, 2 mM EDTA, 1 μM OA, 0.5 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml each aprotinin, pepstatin A, and leupeptin). For caspase 9 immunoprecipitations, 200 μl lysate was incubated with 1 μg mouse monoclonal anti-caspase 9 antibody (Chemicon) at 4°C for 1 h and then incubated with 20 μl washed protein G beads for a further 1 h at 4°C. For immunoprecipitation of FLAG-PKCζ, 2 mg lysate was incubated with 10 μl washed mouse anti-FLAG M2-agarose conjugate (Sigma) for 1 h at 4°C. Beads were washed three times in respective buffer and, where necessary, boiled in SDS-PAGE sample buffer prior to electrophoresis and Western blotting.
Kinase assays.
Recombinant PKC isoforms were purchased from Upstate Biotechnology. Five hundred nanograms of recombinant His6-caspase 9 was incubated in a final volume of 10 μl of kinase assay buffer (20 mM MOPS, pH 7.2, 25 mM β-glycerophosphate, 1 mM Na3VO4) containing 0.1 mg/ml diglycerides, 0.01 mg/ml phosphatidylserine (PKC Lipid Activator; Upstate), 100 μM ATP, 10 mM MgCl2, 0.5 mM DTT, and 18.5 MBq [γ-32P]ATP, where necessary, for 30 min at 30°C. One millimolar CaCl2 was added to reaction mixtures with the α, β, and γ PKC isoforms. Reactions were stopped by boiling in SDS-PAGE sample buffer and analyzed by electrophoresis, followed by autoradiography or Western blotting. For immunoprecipitated FLAG-PKCζ kinase assays, anti-FLAG-agarose bead complexes were resuspended in a final volume of 25 μl kinase assay buffer, as described above, containing 50 μM PKCɛ substrate peptide (Upstate) and incubated for 10 min at 30°C. Reactions were stopped by spotting onto P81 phosphocellulose paper (Whatman), which was washed four times in 150 mM H3PO4, and incorporation of [32P]phosphate was determined by scintillation counting.
Caspase activation.
To activate caspases, HeLa cytosolic extract was incubated at 30°C with addition of an ATP-regenerating system (1 mM ATP, 10 μg/ml creatine kinase, 5 mM creatine phosphate) and 7.5 μM cytochrome c. For reconstitution of caspase 9-depleted HeLa cytosolic extract, 2 μl in vitro-translated caspase 9 was included. At the times shown, 4 μl of the reaction mixture was incubated for 15 min at 30°C with 16 μl assay buffer (50 mM HEPES, pH 7.5, 10 mM EDTA, 50 mM KCl) containing Ac-DEVD-AMC at a final concentration of 50 μM. Reactions were stopped by adding 20 μl 50 mM Na acetate (pH 4.0) and 160 μl H2O. Released AMC was measured with a fluorescence microtiter plate reader at excitation and emission wavelengths of 340 nm and 440 nm, respectively. For induction of caspase 9 processing with recombinant Apaf-1(1-541), 1 μl 35S-labeled, in vitro-translated caspase 9 was incubated with 5 ng recombinant His6-Apaf-1(1-541) in a final volume of 10 μl assay buffer (20 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM DTT) for 90 min at 30°C. Reactions were stopped by boiling in SDS-PAGE sample buffer.
RESULTS
Caspase 9 is phosphorylated on Ser144 in human cell extracts.
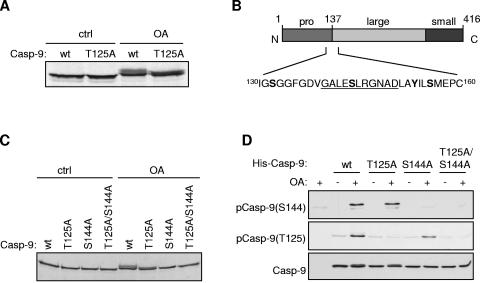
We have previously shown that OA, an inhibitor of protein phosphatases PP-1 and PP-2A, blocks cytochrome c-induced caspase 3 activation in HeLa cytosolic extracts by promoting the direct phosphorylation of caspase 9 at Thr125 by ERK MAP kinase. In HeLa cytosolic extract diluted fivefold (∼2 mg protein/ml), Thr125 was the only major site of phosphorylation in caspase 9 that was induced by OA and was responsible for inhibition of caspase 9 activation (3). In undiluted HeLa cytosolic extracts (∼10 mg protein/ml), we found that OA induced a decrease in electrophoretic mobility of in vitro-translated caspase 9, indicative of phosphorylation. However, this shift was altered, but not abolished, when Thr125 was mutated to nonphosphorylatable alanine (Fig. 1A), indicating that an additional site in caspase 9 was phosphorylated under these conditions. We therefore sought to identify this novel phosphorylation site and investigate its possible role in the regulation of caspase 9.
FIG. 1.
Phosphorylation of caspase 9 on Ser144 in a cell-free system. (A) Caspase 9 (Casp-9) is phosphorylated at a site other than Thr125 in response to OA. 35S-labeled, in vitro-translated wild-type (wt) or T125A caspase 9 was incubated in HeLa cytosolic extract with DMSO vehicle (ctrl [control]) or 1 μM OA. Reactions were analyzed for mobility-shifted caspase 9 by SDS-PAGE and autoradiography. (B) Diagram of caspase 9 protein with potential novel phosphorylation sites indicated in bold. The underlined sequence containing phosphorylated Ser144 was used to raise a specific antibody. (C) As for panel A, with wild-type and mutant caspase 9, as indicated. (D) Recombinant inactive His-caspase 9 (C287A) mutants were incubated as for panel A. Samples were immunoblotted with phospho-Ser144, phospho-Thr125, and caspase 9 antibodies.
The shift in electrophoretic mobility of fragments of caspase 9 incubated in OA-treated HeLa cytosolic extract indicated that the novel phosphorylation site was restricted to amino acids 130 to 160, which encompasses four potential phosphorylation sites, Ser133, Ser144, Tyr153, and Ser156 (data not shown; Fig. 1B). Mutation of Ser144 (but not any of the other three residues) to alanine abolished the mobility shift of full-length caspase 9T125A, indicating that Ser144 was the phosphorylation site (Fig. 1C and data not shown). To confirm that Ser144 was phosphorylated under these conditions, we raised an antibody against a peptide corresponding to residues 140 to 150 of caspase 9 in which the serine residue was phosphorylated. This antibody recognized recombinant human caspase 9 when incubated in HeLa cytosolic extract only after OA treatment, while mutation of Ser144 to alanine abolished recognition, confirming that Ser144 was phosphorylated under these conditions (Fig. 1D). Mutation of Thr125 to alanine did not prevent recognition of caspase 9 by the antibody directed against phospho-Ser144 and vice versa, showing that each site was phosphorylated independently. These data also show that the two antibodies are specific for the individual phosphorylation sites.
Ser144 phosphorylation inhibits caspase 9 activation.
Since Ser144 in caspase 9 was phosphorylated under conditions that antagonize cytochrome c-induced caspase 9/3 activation, i.e., when OA was added to cell extracts, we wished to test if phosphorylation at this site contributed to this inhibition. To this end, in vitro-translated caspase 9 was incubated with constitutively active Apaf-1(1-541), which, due to the lack of the regulatory WD-40 repeat region, does not require cytochrome c for oligomerization (21, 43). Autocatalytic processing of caspase 9 to a 35-kDa fragment, which is associated with its activation, was induced by Apaf-1(1-541). However mutation of Ser144 to a potentially phosphomimetic aspartic acid residue (S144D) abrogated processing, suggesting that phosphorylation at this site may inhibit caspase 9 activation (Fig. 2A).
FIG. 2.
Inhibition of caspase 9 activity by Ser144 phosphorylation. (A) 35S-labeled, in vitro-translated wild-type (wt) or S144D mutant caspase 9(Casp-9) was incubated with recombinant His-Apaf-11-541. Samples were analyzed for caspase 9 processing by SDS-PAGE and autoradiography. (B) HeLa cytosolic extract depleted of endogenous caspase 9 was incubated with in vitro-translated, 35S-labeled wild-type or S144A mutant caspase 9.Caspase activation was induced by incubation with cytochrome c (cyt c), where shown, for 4 h at 30°C. One micromolar OA was added as indicated. Samples were analyzed as described for panel A. (C) Caspase 9-depleted HeLa extract was incubated with unlabeled in vitro-translated wild-type or mutant caspase 9, and then caspase activation was induced as for panel B. Caspase 3 activity was assayed by measuring DEVD-AMC cleavage. The top part of panel C shows the removal of caspase 9 from the extract by two successive rounds of depletion (1, 2) compared to predepleted extract (Pre) and the addition of in vitro-translated wild-type or mutant caspase 9 to depleted extract.
To assess whether Ser144 phosphorylation affects caspase 9 activation, we used reconstituted HeLa cytosolic extract in which endogenous caspase 9 had been immunodepleted and replaced with in vitro-translated wild-type caspase 9 or caspase 9S144A. Addition of cytochrome c induced the processing of wild-type caspase 9 to its autocatalytic p35 and caspase 3-generated p37 proteolytic fragments, and this processing was strongly inhibited by OA, as expected (3) (Fig. 2B). However, although caspase 9S144A was processed to a similar extent as wild-type caspase 9 following the addition of cytochrome c, OA only partially inhibited the processing of this mutant. This result indicates that the phosphorylation of Ser144, induced by OA, attenuates caspase 9 activation and processing.
To confirm that the inhibition of caspase 9 activation through phosphorylation at Ser144 also regulated the subsequent activation of caspase 3, we carried out a similar experiment whereby the caspase 9-depleted HeLa cytosolic extract was reconstituted with in vitro-translated caspase 9, and cytochrome c-induced caspase 3 activation was assayed by measuring the cleavage of the fluorogenic tetrapeptide substrate AcDEVD-AMC (Fig. 2C). Depletion of caspase 9 from the extract abolished cytochrome c-induced caspase 3 activation, which was restored by the addition of in vitro-translated wild-type caspase 9 or its phosphorylation site mutants. As expected, OA strongly inhibited the activation of caspase 3 by wild-type caspase 9. This inhibition was partially attenuated by caspase 9S144A or caspase 9T125A, while the inhibitory effect of OA was further decreased by a double phosphorylation site mutant, caspase 9S144A/T125A, indicating that the inhibitory effect of OA on caspase 3 activation is due in part to the phosphorylation of caspase 9 at Ser144 and this phosphorylation is additive to the inhibitory effect of phosphorylation at T125A. The persistent partial inhibition of caspase 9S144A/T125A suggests that an additional inhibitory mechanism is also induced by OA.
Effects of protein kinase inhibitors on Ser144 phosphorylation in cytosolic extracts.
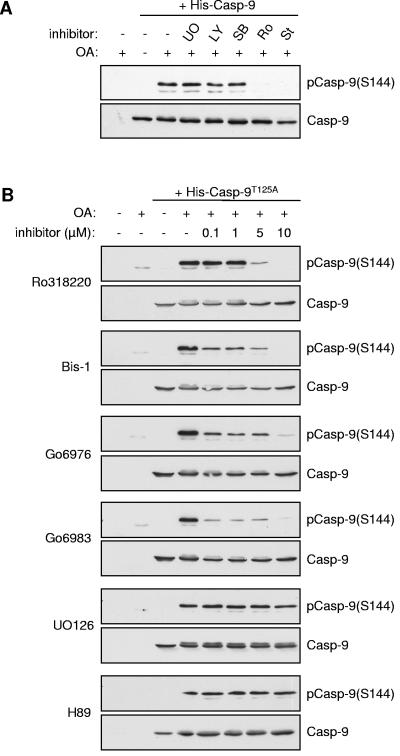
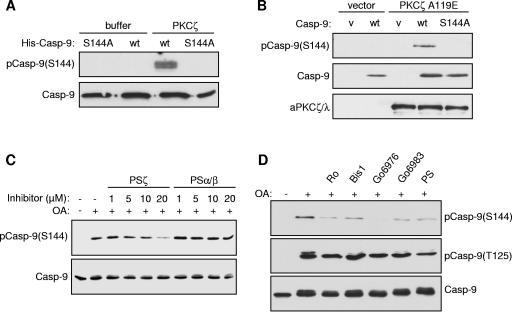
The Ser144 site occurs within a sequence that does not have strong similarity to known protein kinase consensus motifs, although the presence of a basic arginine residue at position +2 is suggestive of a PKC phosphorylation site (Fig. 1B). As a first step to identify the protein kinase that targets Ser144, we screened a number of protein kinase inhibitors for the ability to abrogate phosphorylation of this site induced by OA in HeLa cytosolic extract. Notably, UO126, which inhibits the activation of MEK1, the upstream activator of ERK1/2, had no effect on Ser144 phosphorylation (Fig. 3A and B), ruling out involvement of the ERK MAP kinase pathway that phosphorylates caspase 9 on Thr125 (3). Furthermore, LY294002, SB203580 (Fig. 3A), and H89 (Fig. 3B), inhibitors of phosphatidylinositol-3-OH kinase (which functions upstream of protein kinase B/Akt), p38 MAP kinase, and protein kinase A, respectively, were also without effect. By contrast, the PKC inhibitor Ro318220 and the broad-specificity protein kinase inhibitor staurosporine strongly inhibited Ser144 phosphorylation (Fig. 3A). These results suggest that phosphorylation of this site requires a kinase other than PKB/Akt (7), ERK1/2 MAP kinase (3), or PKA (32), which have been reported previously to phosphorylate other sites in caspase 9. The stress-activated p38 MAP kinase is also not required.
FIG. 3.
Inhibition by PKC inhibitors of caspase 9 phosphorylation at Ser144. Recombinant T125A (C287A) His-caspase 9 (Casp-9) was incubated in HeLa cytosolic extract treated with 1 μM OA. Samples were analyzed by immunoblotting with phospho-Ser144 and caspase 9 antibodies where indicated. (A) Effect of kinase inhibitors on Ser144 phosphorylation. Ten micromolar UO126, 5 μM LY294002, 10 μM SB203580, 5 μM Ro318220, and 1 μM staurosporine were included as indicated. (B) Concentration-dependent effects on Ser144 phosphorylation of Ro318220, Bis-1, Go6976, Go6983, UO126, and H89.
To investigate further the potential involvement of PKC in Ser144 phosphorylation, additional inhibitors known to target this family of kinases were tested (Fig. 3B). Phosphorylation of Ser144 was strongly inhibited by Ro318220 at 5 μM and completely abolished at 10 μM. Other PKC inhibitors, bisindoylmaleimide 1 (Bis-1), Go6976, and Go6983, substantially reduced Ser144 phosphorylation at 0.1 μM, although some phosphorylation persisted with each inhibitor at up to 5 μM. By contrast, UO126 and H89 had no effect, even at 10 μM. While it should be noted that all of the PKC inhibitors tested are known to target kinases other than the PKC family (2, 12), these results are suggestive of a role for members of the PKC family in Ser144 phosphorylation.
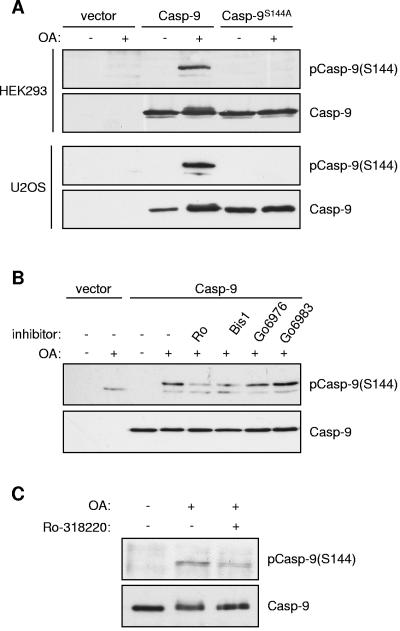
Phosphorylation of caspase 9 at Ser144 is induced by OA in cells.
To test if caspase 9 was phosphorylated at Ser144 in intact cells, we transfected HEK293 cells and U2OS cells with catalytically inactive caspase 9 containing Ser144 or a mutant in which this site was abolished (caspase 9S144A). In both cell types, treatment with OA induced strong phosphorylation at Ser144 that was absent in the S144A mutant (Fig. 4A). Phosphorylation of Ser144 induced by OA in U2OS cells was inhibited by Ro318220 and Bis-1, although Go6976 had less effect and Go6983 had none (Fig. 4B). Similarly, OA-induced phosphorylation of endogenous caspase 9 retrieved by immunoprecipitation was also partially inhibited by Ro318220 (Fig. 4C). These data confirm that Ser144 is indeed phosphorylated in cells treated with OA and are consistent with a role for PKC in this response.
FIG. 4.
Phosphorylation of caspase 9 (Casp-9) at Ser144 in cells. (A) HEK293 and U2OS cells were transfected with empty vector or wild-type or S144A caspase 9 (both C287A). At 24 h after transfection, cells were serum starved for 24 h and then treated with 1 μM OA for 60 min. Cell lysates were immunoblotted with phospho-Ser144 and caspase 9 antibodies as indicated. (B) Effects of PKC inhibitors on Ser144 phosphorylation. U2OS cells transfected with caspase 9 (C287A) were serum starved for 24 h and then treated with 5 μM Ro-318220, 10 μM Bis-1, 10 μM Go6976, or 10 μM Go6983 for 15 min prior to treatment with 1 μM OA for a further 60 min. Cell lysates were analyzed as before. (C) Phosphorylation of endogenous caspase 9 on Ser144. HeLa cells were serum starved for 48 h and then treated with 5 μM Ro-318220 for 15 min, followed by 1 μM OA for 60 min. Caspase 9 immunoprecipitates were Western blotted as in panel A.
Phosphorylation of caspase 9 by PKC isoforms and their interaction in cell extracts.
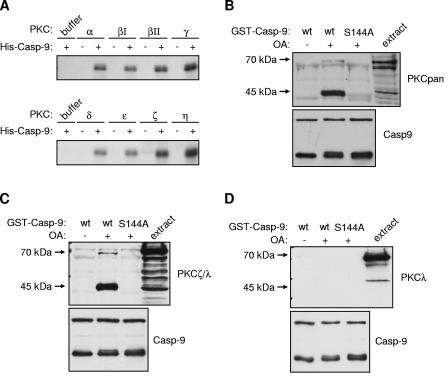
Eight recombinant PKC isoforms, spanning each of the three subfamilies (classical, novel, and atypical), phosphorylated recombinant His6-caspase 9 to similar extents in vitro, suggesting that caspase 9 could be a direct substrate for one or more PKC isoforms (Fig. 5A). The physiological selection of targets by protein kinases often involves the formation of complexes with their substrates. To determine if PKC isoforms interact stably with caspase 9, we incubated recombinant GST-caspase 9 in HeLa cytosolic extracts and analyzed coprecipitating proteins with specific antibodies. An antibody that recognizes all PKC isoforms detected a polypeptide of approximately 70 kDa (corresponding in mass to some PKC isoforms), which coprecipitated with GST-caspase 9 when the extract was treated with OA (Fig. 5B). In addition, a polypeptide of approximately 45 kDa, most likely a proteolytic fragment of PKC generated during preparation of the extracts (36), interacted strongly with GST-caspase 9 in the same manner. The requirement for OA suggests that either activation of the kinase by phosphorylation or phosphorylation of caspase 9 itself is required to stabilize the interaction. Consistent with the latter possibility, caspase 9S144A did not interact with PKC in the extracts, indicating that phosphorylation of Ser144 stabilizes the binding of PKC to caspase 9. Together, the observation that one or more PKC isoforms can phosphorylate caspase 9 and interact with the protease in a Ser144-dependent manner strongly suggests that this site is directly phosphorylated by PKC.
FIG. 5.
Phosphorylation of caspase 9 (Casp-9) at Ser144 by PKC isoforms. (A) 32P labeling of caspase 9 by PKC isoforms. Recombinant His-caspase 9 (C287A) was incubated with buffer control or recombinant purified PKC isoforms, normalized for activity, and [γ-32P]ATP before being analyzed by SDS-PAGE and autoradiography. (B) Coprecipitation of PKC from HeLa cytosolic extract with caspase 9. Wild-type (wt) or S144A GST-caspase 9 (C287A) was incubated in HeLa extract with 1 μM OA, as shown, before being recovered on glutathione-Sepharose beads. Samples were immunoblotted with an antibody which detects all PKC isoforms (PKCpan) or a caspase 9 antibody, as indicated. The upper band detected by the caspase 9 antibody corresponds to full-length GST-caspase 9, whereas the lower bands correspond to an N-terminal fragment tagged with GST that also contains the S144 site. (C and D) Coprecipitation of PKCζ with caspase 9 from HeLa cell extract. Wild-type or S144A (both C287A) GST-caspase 9 was incubated in OA-treated HeLa cytosolic extract and recovered on glutathione-Sepharose beads. Samples were analyzed by immunoblotting with antibodies to caspase 9 and PKCζ/λ (C) or PKCλ (D).
Only the α, δ, ζ, and λ/i isoforms of PKC were detected in HeLa cytosolic extracts by Western blotting with isoform-specific antibodies (data not shown), indicating that it must be one or more of these isoforms that interact with caspase 9. Antibodies to PKCα or -δ failed to detect any proteins coprecipitating with GST-caspase 9 (data not shown). By contrast, an antibody that recognizes both aPKCs PKCζ and -λ/i (Fig. 5C) detected 70-kDa and 45-kDa proteins that coprecipitated with GST-caspase 9 from extracts treated with OA. Similar to the results obtained with the pan-PKC antibody, this interaction was dependent on the presence of Ser144. Furthermore, an antibody that recognizes PKCλ/i but not PKCζ did not detect these proteins in the GST-caspase 9 precipitate (Fig. 5D). This shows that caspase 9 interacts specifically with PKCζ, strongly suggesting that this isoform regulates caspase 9 phosphorylation.
Phosphorylation of Ser144 in caspase 9 by PKCζ.
We found that purified recombinant PKCζ did indeed phosphorylate caspase 9 at Ser144 in vitro, as detected by the site-specific antibody (Fig. 6A). Similarly, cotransfection of HEK293 cells with catalytically inactive caspase 9 and a constitutively active mutant of PKCζ with a mutation in the inhibitory pseudosubstrate region (PKCζA119E) resulted in strong phosphorylation of caspase 9 at Ser144 (Fig. 6B). Furthermore, an inhibitor peptide derived from the pseudosubstrate region of PKCζ, but not one derived from the pseudosubstrate region of PKCα/β (26), substantially inhibited phosphorylation of caspase 9 on Ser144 in OA-treated HeLa cytosolic extract, showing that PKCζ is the principal Ser144 kinase activity in the extract (Fig. 6C). Residual phosphorylation that was not inhibited by high concentrations of the pseudosubstrate inhibitor may be due to phosphorylation by another PKC isoform. This could account for the inhibition of Ser144 phosphorylation in HeLa cytosolic extracts by Go6976 (Fig. 3B and 6D), which inhibits classical PKC isoforms. Neither the PKCζ pseudosubstrate inhibitor nor the other PKC inhibitors tested inhibited caspase 9 phosphorylation in extracts at the other major inhibitory site induced by OA, Thr125 (Fig. 6D).
FIG. 6.
Phosphorylation of caspase 9 (Casp-9) at Ser144 by PKCζ. (A) Recombinant wild-type (wt) or S144A mutant His6-caspase 9 (C287A) was incubated with recombinant His-PKCζ. (B) HEK293 cells were transfected for 24 h with inactive (C287A) wild-type or S144A caspase 9 and constitutively active PKCζ (PKCζA119E) or empty vector (v). (C) Recombinant His-caspase 9 was incubated in HeLa cytosolic extract treated with 1 μM OA in the presence of PKCζ pseudosubstrate inhibitor (PSζ) or PKCα/β pseudosubstrate inhibitor (PSα/β) at the concentrations shown. (D) Effects of kinase inhibitors on Ser144 and Thr125 phosphorylation in HeLa cytosolic extract treated with 1 μM OA, 5 μM Ro318220, 10 μM Bis-1, 10 μM Go6976, 10 μM Go6983, or 10 μM pseudosubstrate inhibitor (PS). In each case, samples were analyzed by SDS-PAGE and Western blotting with the indicated antibodies.
Inhibition of PKCζ reverses the suppression of caspase activation by OA in cell extracts.
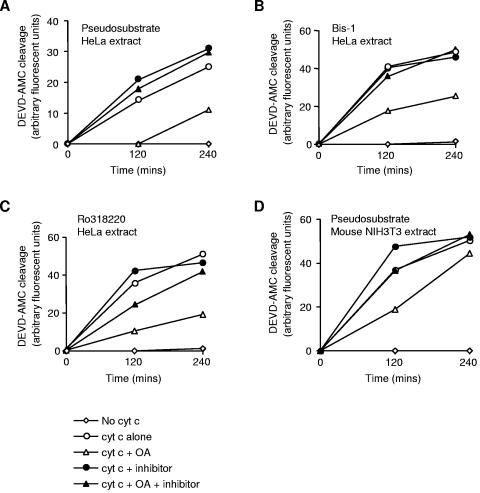
The PKCζ pseudosubstrate inhibitor that prevented caspase 9 phosphorylation at Ser144 also restored cytochrome c-induced caspase 3 activation in HeLa extract treated with OA (Fig. 7A), demonstrating that PKCζ suppresses caspase activation in this system. Similarly, Bis-1 (Fig. 7B) and Ro318220 (Fig. 7C), as well as other inhibitors of Ser144 phosphorylation (data not shown), also reversed suppression of caspase activation by OA. Restoration of caspase 3 activation in these experiments was not attributable to changes in phosphorylation at Thr125, since none of these inhibitors affected phosphorylation at this site (Fig. 6D). These results, together with those shown in Fig. 2C, are consistent with a significant role for phosphorylation of Ser144 in human caspase 9 in the suppression of caspase 3 activation. Nevertheless, OA also partially inhibited caspase activation in mouse NIH 3T3 cell extract, albeit less efficiently than in human HeLa cell extract, and this inhibition was reversed by the PKCζ pseudosubstrate inhibitor (Fig. 7D). Since the Ser144 site is apparently not present in rodent caspase 9 (data not shown), this suggests that PKCζ can also act through additional sites in caspase 9 or other targets.
FIG. 7.
Reversal of OA-induced inhibition of caspase activation by PKCζ inhibitors. Cytosolic extracts from human HeLa (A, B, and C) and mouse NIH 3T3 (D) cells were incubated with 7.5 μM (HeLa) or 1 μM (NIH 3T3) cytochrome c (cyt c), 1 μM OA, and 10 μM PKCζ pseudosubstrate, Bis-1, or Ro318220 as indicated. Caspase 3 activity was assayed by measuring DEVD-AMC cleavage.
Hyperosmotic stress induces caspase 9 phosphorylation.
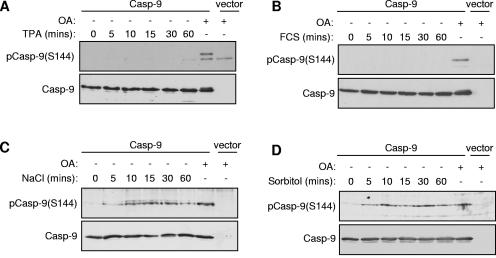
We were interested in determining the physiological conditions under which caspase 9 might be regulated by PKCζ. Unlike classical and novel PKC family members, which are characteristically activated by generation of diacylglycerol in response to growth factors or pharmacologically by phorbol esters (36), activators of aPKC signaling pathways are less well characterized. We therefore tested the abilities of a variety of stimuli to induce Ser144 phosphorylation in human cell lines transfected with catalytically inactive caspase 9. The phorbol ester TPA did not induce phosphorylation at this site (Fig. 8A), which is consistent with the finding that TPA-insensitive PKCζ, as opposed to the TPA-sensitive classical and novel PKCs, is the principal Ser144 kinase. In addition, we did not detect phosphorylation in response to serum stimulation, epidermal growth factor, or insulin (Fig. 8B and data not shown). By contrast, incubation of cells under conditions of acute hyperosmotic stress induced by treatment with 0.7 M sodium chloride or 0.5 M sorbitol, but not other stress stimuli such as heat shock or oxidative stress, promoted rapid and sustained phosphorylation of caspase 9 at Ser144 to a level similar to that induced by OA (Fig. 8C and D and data not shown).
FIG. 8.
Phosphorylation of caspase 9 (Casp-9) following hyperosmotic stress. U2OS (A and B) or HEK293 (C and D) cells were transfected with caspase 9 (C287A) or empty vector. At 24 h posttransfection, cells were serum starved for 24 h before treatment with 1 μM OA for 60 min or (A) 1 μM TPA, (B) 10% fetal calf serum (FCS), (C) 0.7 M NaCl, or (D) 0.5 M sorbitol. Cell lysates were analyzed by immunoblotting with phospho-Ser144 and caspase 9 antibodies as indicated.
Activation of PKCζ and its role in the phosphorylation of caspase 9 in response to hyperosmotic stress of cells.
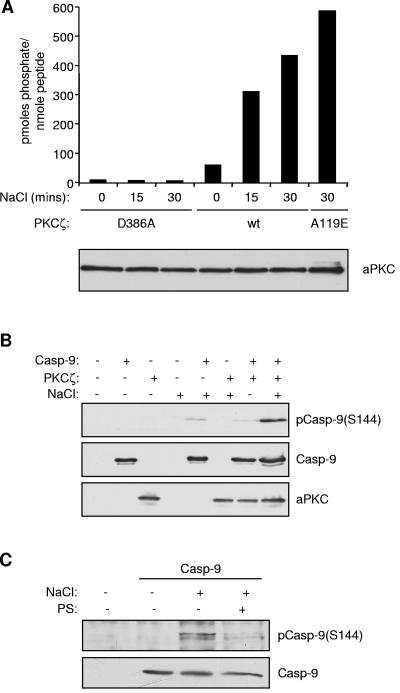
To investigate if Ser144 phosphorylation following osmotic stress occurs via a pathway involving PKCζ, we first assessed whether osmotic stress can increase the activity of this kinase (Fig. 9A). Wild-type FLAG-PKCζ was expressed in HEK293 cells and precipitated with an anti-FLAG antibody, and its activity was measured by assaying the incorporation of 32P from [γ-32P]ATP into a PKCε substrate peptide, which can also be used to assay the activity of the ζ isoform (35). A catalytically inactive mutant of PKCζ (D386A) was used to control for background precipitated kinase activity, while the constitutively active mutant of PKCζ (A119E) was used as a positive control for full activation. Treatment with 0.7 M NaCl resulted in three- and fourfold increases in the activity of wild-type PKCζ after 15 and 30 min, respectively, achieving about 80% of the activity of PKCζA119E, demonstrating that PKCζ is strongly activated by hyperosmotic stress in these cells.
FIG. 9.
Hyperosmotic stress induces PKCζ activation and caspase 9 (Casp-9) phosphorylation at Ser144. (A) Hyperosmotic stress increases PKCζ activity. HEK293 cells were transfected with wild-type (wt), constitutively active (A119E), or kinase-dead (D386A), FLAG-tagged PKCζ. At 24 h posttransfection, cells were serum starved for a further 24 h and then treated with 0.7 M NaCl for the times shown. FLAG-PKCζ was immunoprecipitated from cell lysates and assayed for kinase activity. The amount of aPKC (ζ/λ) recovered in each sample and detected by Western blotting is shown below. (B) HEK293 cells were cotransfected with caspase 9 (C287A) and PKCζ or empty vector. Cells were treated with 0.7 M NaCl for 30 min, and then cell lysates were analyzed by immunoblotting with phospho-Ser144, caspase 9, and aPKC antibodies, as indicated. (C) Phosphorylation of endogenous caspase 9 on Ser144 in response to osmotic stress. HeLa cells were serum starved for 48 h and then treated with 100 μM pseudosubstrate inhibitor (PS) for 45 min, followed by 0.7 M NaCl for 45 min. Caspase 9 immunoprecipitates were Western blotted as in panel A.
To determine whether PKCζ activated by hyperosmotic stress can phosphorylate caspase 9, HEK293 cells were cotransfected with wild-type PKCζ and catalytically inactive caspase 9. In the absence of a stimulus, increased expression of PKCζ caused only weak phosphorylation of caspase 9 at Ser144. However, the phosphorylation of this site in response to treatment with NaCl was markedly increased in cotransfected cells, showing that hyperosmotic stress can induce Ser144 phosphorylation via PKCζ (Fig. 9B). By contrast, cotransfection of caspase 9 with catalytically inactive PKCζD386A did not promote Ser144 phosphorylation (data not shown). Furthermore, pretreatment with a cell-permeable myristoylated PKCζ pseudosubstrate inhibitor substantially inhibited hyperosmotic stress-induced Ser144 phosphorylation in cells transfected with caspase 9 alone, providing compelling evidence that a PKCζ-dependent pathway phosphorylates this site under these conditions (Fig. 9C).
Interaction of PKCζ and caspase 9 in cells is regulated by hyperosmotic stress.
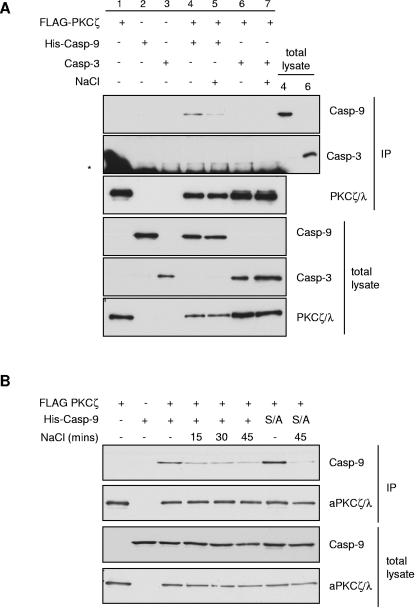
In cotransfected HEK293 cells, immunoprecipitation of FLAG-PKCζ resulted in coprecipitation of caspase 9 but not caspase 3 (Fig. 10A). When these cells were subjected to hyperosmotic shock with 0.7 M NaCl, the interaction between PKCζ and caspase 9 was disrupted within 15 min of NaCl treatment (Fig. 10A and B). These data show that a population of caspase 9 molecules interacts with PKCζ in nonstressed cells, and this interaction is regulated dynamically following hyperosmotic stress. However, phosphorylation of caspase 9 at Ser144, which occurs under similar conditions (Fig. 8), was not required for the change in interaction with PKCζ (Fig. 10B), suggesting that additional mechanisms regulate this interaction in cells.
FIG. 10.
Hyperosmotic stress regulates the interaction of PKCζ and caspase 9 (Casp-9) in cells. HEK293 cells were transfected with FLAG-PKCζ and His-caspase 9 (C287A) or caspase 3 (C163A). Cells were serum starved for 24 h prior to treatment with NaCl. FLAG-PKCζ immunoprecipitates (IP) and total lysates were analyzed by Western blotting with antibodies against caspase 9, caspase 3, and PKCζ/λ. (A) Coprecipitation of PKCζ with caspase 9 but not caspase 3 and disruption of the interaction between PKCζ and caspase 9 by treatment with 0.7 M NaCl for 40 min. Twenty micrograms of total lysate from samples 4 and 6 was also loaded to estimate the proportions of caspase 9 and caspase 3, respectively, that precipitated with PKCζ. The asterisk indicates mouse anti-FLAG immunoglobulin G light chains which react with the secondary antibody used to detect mouse anti-caspase 3. (B) Time course of the effect of 0.7 M NaCl on the interaction between PKCζ and caspase 9 and the effect of mutation of Ser144 to Ala (S/A).
DISCUSSION
Caspase 9 is a major point of control in the apoptotic pathway that is induced by a wide variety of stimuli that act through mitochondria. Caspase 9 activation also plays an important role in the amplification of caspase activation initiated through other pathways. In this report, we have identified a novel pathway that inhibits caspase 9 activation through the aPKC isoform PKCζ, which phosphorylates Ser144 in human caspase 9. This mechanism is activated pharmacologically both in a cell-free system and in cultured cells by the protein phosphatase inhibitor OA. We have also found that phosphorylation of caspase 9 in cells is induced by hyperosmotic stress, which activates PKCζ and regulates its interaction with caspase 9.
In a previous analysis (3), we identified Thr125 as the only major site in caspase 9 that was phosphorylated in diluted, OA-treated HeLa cytosolic extracts. Thr125 is targeted by ERK MAP kinase in these extracts, as well as in growth factor-stimulated cells. With concentrated HeLa cytosolic extracts, we have now found that caspase 9 is phosphorylated at an additional site, Ser144. The loss of phosphorylation at Ser144 following dilution of extracts is apparently due to the labile activity of the kinase acting on this site, which has so far prevented successful purification of the native enzyme or immunodepletion studies (data not shown). Nevertheless, inhibitor studies showed that PKCζ is the predominant kinase acting on Ser144, although residual phosphorylation of this site in cell extracts when PKCζ is inhibited may be due to another kinase, most likely another PKC isoform. In cells, however, the lack of phosphorylation of Ser144 in response to phorbol ester argues against a role for classical or novel PKC isoforms in the phosphorylation of this site in vivo, whereas strong inhibition by a specific pseudosubstrate inhibitor demonstrates a requirement for PKCζ.
Although both Ser144 and T125 are phosphorylated when caspase 9 activation is inhibited by OA in concentrated cytosolic extracts, we have found that selective inhibition of either PKCζ or the ERK MAP kinase pathway in these extracts is sufficient to reactivate caspase 9 and subsequently caspase 3. The apparent lack of redundancy between the two inhibitory pathways is most probably due to a requirement for only a fraction of the total caspase 9 molecules to be activated in response to cytochrome c in order to restore full caspase 3 activation. Thus, our analysis does not preclude other inhibitory sites being targeted by PKCζ or other kinases in OA-treated cell extracts. Indeed, PKCζ may phosphorylate both human and mouse caspase 9 at additional sites that could contribute to the suppression of caspase activation. It will therefore be of interest to identify such additional sites to determine what role they might also play in caspase 9 regulation. Nevertheless, we have shown that phosphorylation of Ser144 of human caspase 9 plays a significant inhibitory role in its regulation and this site acts as a marker for the activity of PKCζ toward caspase 9 in cells.
The results presented here provide a novel insight into the functions and mechanisms of action of PKCζ. Previous work supports a role for PKCζ in the promotion of cell survival. For example, expression of par-4 protein in NIH 3T3 cells inhibits aPKC and induces morphological changes typical of apoptosis that are antagonized by overexpression of PKCζ or PKCλ (16). Conversely, UV irradiation of NIH 3T3 cells causes a reduction in aPKC (PKCζ/λ/i) catalytic activity that precedes apoptosis (5). Furthermore, PKCζ can be cleaved by active caspases following etoposide treatment, and although this results in an initial increase in PKCζ activity due to the removal of the regulatory domain, the PKCζ fragments are subsequently degraded rapidly by the proteasome (42). The down-regulation of components of survival pathways or damage repair mechanisms is a common feature of apoptosis, and degradation of PKCζ therefore implies that this kinase otherwise plays a role in cell survival. One mechanism by which PKCζ might mediate antiapoptotic responses, at least in some cell types, is activation of the transcription factor NF-κB (15, 28, 34). We have now identified a direct mechanism for the regulation of apoptosis by PKCζ through inhibition of caspase 9 by phosphorylation.
We have observed phosphorylation of human caspase 9 at Ser144 in HEK293 cells following hyperosmotic shock, a stress that strongly activated PKCζ in those cells. This finding is consistent with a previous report of increased PKCζ activity in adipocytes treated with sorbitol (40). It will be of interest to investigate the mechanism of PKCζ activation during the osmotic stress response. One well-characterized pathway to PKCζ activation involves its recruitment to Cdc42 and Rac GTPases via the adaptor protein Par6, which plays a critical role in cell polarity (23, 30, 37). While this complex is clearly involved in aPKC activation in response to signals such as integrin ligation (18), it is interesting that both Cdc42 and Rac can also be activated by hyperosmotic stress (39, 46), and Cdc42 has been implicated in the regulation of apoptosis (17). In addition, we have shown that hyperosmotic stress regulates the interaction of PKCζ with caspase 9. Although the stable binding of PKCζ to caspase 9 in cytosolic extracts appears to require Ser144 phosphorylation, in cells this interaction occurred even when Ser144 phosphorylation was abolished by mutation and it was reduced following hyperosmotic stress. This suggests that the interaction of PKCζ and caspase 9 in cells is dynamic and is regulated by additional mechanisms inresponse to hyperosmotic stress, possibly through changes in the subcellular localization of the components. Interestingly, several PKC isoforms, including PKCζ, have been reported to undergo translocation to intracellular membrane compartments in response to osmotic and mechanical stresses (31).
In most cells within tissues, exposure to the extracellular fluid maintains a normal osmotic balance. However, particular cell types, such as those in the kidney medulla or mouth, can be exposed to severe changes in extracellular osmolarity. To survive in this environment, cells exposed to osmotic stress initiate an adaptive response involving modulation of ion transport and cell shrinkage, followed by accumulation of organic solutes, such as amino acids, to balance the osmotic pressure (8, 10). Such homeostatic responses to acute osmotic stress may require the restraint of apoptosis, and we propose that inhibition of caspase 9 through phosphorylation by PKCζ plays a role in this process. Nevertheless, abnormal osmotic stress can result in apoptosis in a number of cell types (19, 24). Embryonic fibroblasts and thymocytes derived from caspase 9-null mice are resistant to sorbitol-induced apoptosis, demonstrating that osmotic-stress-induced death in these cells requires caspase 9 (25). This strongly suggests that regulation of caspase 9 plays an important role in determining cell survival following osmotic stress. Pathologically, an inability to regulate apoptosis under such conditions may contribute to diseases like diabetes, in which hyperglycemia imposes an osmotic pressure on cells such as those of the lens epithelium, and this appears to be a prime factor in the development of diabetic cataracts (27). Conversely, constitutive activation of stress response pathways that restrain apoptosis may contribute to abnormal cell survival in diseases such as cancer.
Acknowledgments
This work was supported by a Cancer Research UK Studentship (S.C.B.) and grants from Cancer Research UK and the Medical Research Council. P.R.C. is a Royal Society-Wolfson Research Merit Awardee.
We are grateful to Dario Alessi (Dundee) for providing reagents and advice.
REFERENCES
- 1.Acehan, D., X. Jiang, D. G. Morgan, J. E. Heuser, X. Wang, and C. W. Akey. 2002. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell 9:423-432. [DOI] [PubMed] [Google Scholar]
- 2.Alessi, D. R. 1997. The protein kinase C inhibitors Ro 318220 and GF 109203X are equally potent inhibitors of MAPKAP kinase-1β (Rsk-2) and p70 S6 kinase. FEBS Lett. 402:121-123. [DOI] [PubMed] [Google Scholar]
- 3.Allan, L. A., N. Morrice, S. Brady, G. Magee, S. Pathak, and P. R. Clarke. 2003. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat. Cell Biol. 5:647-654. [DOI] [PubMed] [Google Scholar]
- 4.Beere, H. M. 2004. The stress of dying: the role of heat shock proteins in the regulation of apoptosis. J. Cell Sci. 117:2641-2651. [DOI] [PubMed] [Google Scholar]
- 5.Berra, E., M. M. Municio, L. Sanz, S. Frutos, M. T. Diaz-Meco, and J. Moscat. 1997. Positioning atypical protein kinase C isoforms in the UV-induced apoptotic signaling cascade. Mol. Cell. Biol. 17:4346-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budihardjo, I., H. Oliver, M. Lutter, X. Luo, and X. Wang. 1999. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15:269-290. [DOI] [PubMed] [Google Scholar]
- 7.Cardone, M. H., N. Roy, H. R. Stennicke, G. S. Salvesen, T. F. Franke, E. Stanbridge, S. Frisch, and J. C. Reed. 1998. Regulation of cell death protease caspase-9 by phosphorylation. Science 282:1318-1321. [DOI] [PubMed] [Google Scholar]
- 8.Chan, H. C., and D. J. Nelson. 1992. Chloride-dependent cation conductance activated during cellular shrinkage. Science 257:669-671. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, P. R. 2002. Apoptosis: lessons from cell-free systems, p. 176-199. In M. D. Jacobson and N. McCarthy (ed.), Apoptosis: the molecular biology of programmed cell death. Oxford University Press, Oxford, United Kingdom.
- 10.Dall'Asta, V., P. A. Rossi, O. Bussolati, and G. C. Gazzola. 1994. Response of human fibroblasts to hypertonic stress. Cell shrinkage is counteracted by an enhanced active transport of neutral amino acids. J. Biol. Chem. 269:10485-10491. [PubMed] [Google Scholar]
- 11.Dan, H. C., M. Sun, S. Kaneko, R. I. Feldman, S. V. Nicosia, H. G. Wang, B. K. Tsang, and J. Q. Cheng. 2004. Akt phosphorylation and stabilization of X-linked inhibitor of apoptosis protein (XIAP). J. Biol. Chem. 279:5405-5412. [DOI] [PubMed] [Google Scholar]
- 12.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desagher, S., and J. C. Martinou. 2000. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10:369-377. [DOI] [PubMed] [Google Scholar]
- 14.Deveraux, Q. L., R. Takahashi, G. S. Salvesen, and J. C. Reed. 1997. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388:300-304. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Meco, M. T., I. Dominguez, L. Sanz, P. Dent, J. Lozano, M. M. Municio, E. Berra, R. T. Hay, T. W. Sturgill, and J. Moscat. 1994. ζ PKC induces phosphorylation and inactivation of IκBα in vitro. EMBO J. 13:2842-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz-Meco, M. T., M. M. Municio, S. Frutos, P. Sanchez, J. Lozano, L. Sanz, and J. Moscat. 1996. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell 86:777-786. [DOI] [PubMed] [Google Scholar]
- 17.Erickson, J. W., and R. A. Cerione. 2001. Multiple roles for Cdc42 in cell regulation. Curr. Opin. Cell Biol. 13:153-157. [DOI] [PubMed] [Google Scholar]
- 18.Etienne-Manneville, S., and A. Hall. 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCζ. Cell 106:489-498. [DOI] [PubMed] [Google Scholar]
- 19.Galvez, A. S., J. A. Ulloa, M. Chiong, A. Criollo, V. Eisner, L. F. Barros, and S. Lavandero. 2003. Aldose reductase induced by hyperosmotic stress mediates cardiomyocyte apoptosis: differential effects of sorbitol and mannitol. J. Biol. Chem. 278:38484-38494. [DOI] [PubMed] [Google Scholar]
- 20.Gutcher, I., P. R. Webb, and N. G. Anderson. 2003. The isoform-specific regulation of apoptosis by protein kinase C. Cell. Mol. Life Sci. 60:1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Y., L. Ding, D. M. Spencer, and G. Nunez. 1998. WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J. Biol. Chem. 273:33489-33494. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson, M. D., M. Weil, and M. C. Raff. 1997. Programmed cell death in animal development. Cell 88:347-354. [DOI] [PubMed] [Google Scholar]
- 23.Joberty, G., C. Petersen, L. Gao, and I. G. Macara. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531-539. [DOI] [PubMed] [Google Scholar]
- 24.Kubo, E., T. Urakami, N. Fatma, Y. Akagi, and D. P. Singh. 2004. Polyol pathway-dependent osmotic and oxidative stresses in aldose reductase-mediated apoptosis in human lens epithelial cells: role of AOP2. Biochem. Biophys. Res. Commun. 314:1050-1056. [DOI] [PubMed] [Google Scholar]
- 25.Kuida, K., T. F. Haydar, C. Y. Kuan, Y. Gu, C. Taya, H. Karasuyama, M. S. Su, P. Rakic, and R. A. Flavell. 1998. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase-9. Cell 94:325-337. [DOI] [PubMed] [Google Scholar]
- 26.Laudanna, C., D. Mochly-Rosen, T. Liron, G. Constantin, and E. C. Butcher. 1998. Evidence of zeta protein kinase C involvement in polymorphonuclear neutrophil integrin-dependent adhesion and chemotaxis. J. Biol. Chem. 273:30306-30315. [DOI] [PubMed] [Google Scholar]
- 27.Lee, A. Y., S. K. Chung, and S. S. Chung. 1995. Demonstration that polyol accumulation is responsible for diabetic cataract by the use of transgenic mice expressing the aldose reductase gene in the lens. Proc. Natl. Acad. Sci. USA 92:2780-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitges, M., L. Sanz, P. Martin, A. Duran, U. Braun, J. F. Garcia, F. Camacho, M. T. Diaz-Meco, P. D. Rennert, and J. Moscat. 2001. Targeted disruption of the ζPKC gene results in the impairment of the NF-κB pathway. Mol. Cell 8:771-780. [DOI] [PubMed] [Google Scholar]
- 29.Li, P., D. Nijhawan, I. Budihardjo, S. M. Srinivasula, M. Ahmad, E. S. Alnemri, and X. Wang. 1997. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479-489. [DOI] [PubMed] [Google Scholar]
- 30.Lin, D., A. S. Edwards, J. P. Fawcett, G. Mbamalu, J. D. Scott, and T. Pawson. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signaling and cell polarity. Nat. Cell Biol. 2:540-547. [DOI] [PubMed] [Google Scholar]
- 31.Liu, X., M. I. Zhang, L. B. Peterson, and R. G. O'Neil. 2003. Osmomechanical stress selectively regulates translocation of protein kinase C isoforms. FEBS Lett. 538:101-106. [DOI] [PubMed] [Google Scholar]
- 32.Martin, M. C., L. A. Allan, M. Lickrish, C. Sampson, N. Morrice, and P. R. Clarke. 2005. Protein kinase A regulates caspase-9 activation by Apaf-1 downstream of cytochrome c. J. Biol. Chem. 280:15449-15455. [DOI] [PubMed] [Google Scholar]
- 33.Mellor, H., and P. J. Parker. 1998. The extended protein kinase C superfamily. Biochem. J. 332:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moscat, J., M. T. Diaz-Meco, and P. Rennert. 2003. NF-κB activation by protein kinase C isoforms and B-cell function. EMBO Rep. 4:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakanishi, H., K. A. Brewer, and J. H. Exton. 1993. Activation of the zeta isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 268:13-16. [PubMed] [Google Scholar]
- 36.Nishizuka, Y. 1984. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature 308:693-698. [DOI] [PubMed] [Google Scholar]
- 37.Qiu, R. G., A. Abo, and G. S. Martin. 2000. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCζ signaling and cell transformation. Curr. Biol. 10:697-707. [DOI] [PubMed] [Google Scholar]
- 38.Renatus, M., H. R. Stennicke, F. L. Scott, R. C. Liddington, and G. S. Salvesen. 2001. Dimer formation drives the activation of the cell death protease caspase-9. Proc. Natl. Acad. Sci. USA 98:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roig, J., Z. Huang, C. Lytle, and J. A. Traugh. 2000. p21-activated protein kinase γ-PAK is translocated and activated in response to hyperosmolarity. Implication of Cdc42 and phosphoinositide 3-kinase in a two-step mechanism for γ-PAK activation. J. Biol. Chem. 275:16933-16940. [DOI] [PubMed] [Google Scholar]
- 40.Sajan, M. P., G. Bandyopadhyay, Y. Kanoh, M. L. Standaert, M. J. Quon, B. C. Reed, I. Dikic, and R. V. Farese. 2002. Sorbitol activates atypical protein kinase C and GLUT4 glucose transporter translocation/glucose transport through proline-rich tyrosine kinase-2, the extracellular signal-regulated kinase pathway and phospholipase D. Biochem. J. 362:665-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiozaki, E. N., and Y. Shi. 2004. Caspases, IAPs and Smac/DIABLO: mechanisms from structural biology. Trends Biochem. Sci. 29:486- 494. [DOI] [PubMed] [Google Scholar]
- 42.Smith, L., L. Chen, M. E. Reyland, T. A. DeVries, R. V. Talanian, S. Omura, and J. B. Smith. 2000. Activation of atypical protein kinase C zeta by caspase processing and degradation by the ubiquitin-proteasome system. J. Biol. Chem. 275:40620-406027. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasula, S. M., M. Ahmad, T. Fernandes-Alnemri, and E. S. Alnemri. 1998. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1:949-957. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, C. B. 1995. Apoptosis in the pathogenesis and treatment of disease. Science 267:1456-1462. [DOI] [PubMed] [Google Scholar]
- 45.Thornberry, N. A., and Y. Lazebnik. 1998. Caspases: enemies within. Science 281:1312-1316. [DOI] [PubMed] [Google Scholar]
- 46.Uhlik, M. T., A. N. Abell, N. L. Johnson, W. Sun, B. D. Cuevas, K. E. Lobel-Rice, E. A. Horne, M. L. Dell'Acqua, and G. L. Johnson. 2003. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell Biol. 5:1104-1110. [DOI] [PubMed] [Google Scholar]
- 47.Whelan, R. D., and P. J. Parker. 1998. Loss of protein kinase C function induces an apoptotic response. Oncogene 16:1939-1944. [DOI] [PubMed] [Google Scholar]