Abstract
Psf1 (partner of sld five 1) forms a novel heterotetramer complex, GINS (Go, Ichi, Nii, and San; five, one, two, and three, respectively, in Japanese), with Sld5, Psf2, and Psf3. The formation of this complex is essential for the initiation of DNA replication in yeast and Xenopus laevis egg extracts. Although all of the components are well conserved in higher eukaryotes, the biological function in vivo is largely unknown. We originally cloned the mouse ortholog of PSF1 from a hematopoietic stem cell cDNA library and found that PSF1 is expressed in blastocysts, adult bone marrow, and testis, in which the stem cell system is active. Here we used the gene-targeting technique to determine the physiological function of PSF1 in vivo. Mice homozygous for a nonfunctional mutant of PSF1 died in utero around the time of implantation. PSF1−/− blastocysts failed to show outgrowth in culture and exhibited a cell proliferation defect. Our data clearly indicate that PSF1 is required for early embryogenesis.
Eukaryotic chromosomal replication is tightly regulated to maintain the integrity of genomic information. In Saccharomyces cerevisiae, Orc (origin recognition complex) is bound to replication origins throughout the cell cycle (1, 4, 21). From late M to G1 phase, MCM (minichromosome maintenance) protein is loaded onto the origin, marked by Orc, Cdc6, and Cdt1, and forms the prereplication complex (pre-RC) (2, 9). On activation and recruitment of additional factors, such as CDC45, the pre-RC is converted to the preinitiation complex, which is the complex essential for the transition to DNA replication (2, 23). In yeast, CDC45 is essential for the initiation and elongation of DNA replication (7, 20).
Recently, a novel multiprotein complex, GINS, was identified. The GINS complex contains Psf1 (partner of sld five 1), Psf2, Psf3, and Sld5 and forms a ring-like structure (8, 10, 19). During the S phase, the GINS complex is loaded onto chromatin after the formation of pre-RCs and then tightly associates with the replication origin. This binding is suppressed by p21 and geminin by inhibition of the loading of CDC45 onto chromatin (11) and the pre-RC formation by binding to Cdt1 (23). Moreover, the chromatin binding of GINS complex and that of CDC45 are mutually dependent processes, but they do not associate with each other. The association of PSF1 and Dpb11/Cut5 with the origins is also mutually dependent. All genes encoding GINS components are evolutionarily conserved and are essential for cell growth (22). However, the functions of the GINS complex in mammalian cells have not been reported.
In this study, we isolated the mouse ortholog of PSF1 and generated PSF1-deficient mice by a gene-targeting technique. In mice, PSF1 is specifically expressed in proliferating immature cells. Loss of PSF1 causes embryonic lethality around the implantation stage. PSF1−/− embryos revealed impaired proliferation of the inner cell mass (ICM) and trophoblasts. Our data suggest that PSF1 is essential for mouse embryogenesis.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were purchased from SLC. All animal studies were approved by the Animal Care Committee of Kanazawa University.
Construction and screening of a stem cell-specific cDNA library.
Preparation of bone marrow (BM) cells and fluorescence-activated cell sorting analysis were performed as described previously (17, 18). The antibodies used in flow cytometric analysis for lineage marker (Lin) were phycoerythrin-conjugated Mac-1 (M1/70), Gr-1 (RB6-8C5), B220 (RA3-6B2), TER119, anti-CD4 (GK1.5), and anti-CD8 (53-6.72). Allophycocyanin-conjugated anti-c-kit (ACK2) and biotin-conjugated anti-Sca-1 were also applied. Biotinylated anti-Sca-1 was visualized with peridinin chlorophyll protein-streptavidin. These antibodies were purchased from BD Pharmingen. Lin+ cells and Lin− c-kit+ Sca-1+ cells were purified from BM cells that had been obtained from an 8-week-old C57BL/6 mouse by cell sorting. The stained cells were analyzed with a FACSCalibur (Becton Dickinson) and sorted with an EPICS ALTRA (Beckman Coulter) instrument. Total RNAs were extracted from these cell populations and were subjected to cDNA synthesis and amplification using a SMART PCR synthesis kit (BD Clontech) according to the manufacturer's protocol. Subtractive cloning was performed using a PCR-Select cDNA subtraction kit (BD Clontech).
Immunohistochemistry and fluorescence-activated cell sorting analysis.
Tissue fixation, preparation of tissue sections, and staining of sections with antibodies were performed as described previously (18). For immunohistochemistry, anti-PSF1 (see below) and antibromodeoxyuridine (anti-BrdU) antibodies (Zymed) were used. Cy3- or horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from Jackson ImmunoResearch.
To obtain a specific antibody against PSF1, a rabbit was immunized with a synthetic peptide (CEQLIRQGVLEH) derived from the C-terminal region of PSF1. Antisera were affinity purified with the same peptide. Preimmunized rabbit immunoglobulins were used as a negative control to confirm specific staining.
RT-PCR analysis.
Reverse transcription-PCR (RT-PCR) was performed as previously described (18). We used the following primer sets: 5′-TTA AGA AAT AGA CGC TGC ACG A-3′ and 5′-TGC CAT CAT CAA CTT CAA ATT C-3′ (for PSF1) and 5′-ACC ACA GTC CAT GCC ATC AC-3′ and 5′-TCC ACC ACC CTG TTG CTG TA-3′ (for glyceraldehyde-3-phosphate dehydrogenase [GAPDH]).
Gene targeting.
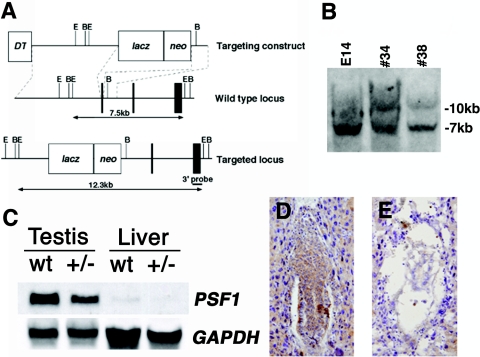
We isolated genomic clones encoding mouse PSF1 from the mouse 129Sv/J library (Stratagene) by using mouse PSF1 cDNA as a probe. In the targeting construct, exon 5 was replaced with the β-galactosidase gene and a pGK-neomycin resistance gene (see Fig. 2). We linearized this construct with NotI and electroporated it into 129Sv/J E14.1 embryonic stem (ES) cells. We selected G418-resistant clones and screened them by PCR and Southern blot analysis for the correct recombination (see Fig. 2). Chimera mice were generated by the aggregation method (12). We mated chimera males with C57BL/6J females and screened the offspring by Southern blotting and PCR analyses for mice bearing the PSF1+/− genotype.
FIG. 2.
Targeted disruption of mouse PSF1. Generation of PSF1-deficient mice. (A) Exons are represented by vertical bars and introns by intervening horizontal lines. Exon 5 of PSF1 was replaced by homologous recombination with a β-galactosidase gene (lacZ) and a neomycin resistance gene driven by the Pgk1 prompter (neo). E, EcoRI; B, BamHI. This increased the size of the EcoRI restriction fragment. (B) Southern blot analysis of wild-type (E14) and targeted (#34 and #38) ES cells demonstrated homologous recombination in PSF1. (C) Northern blot analysis of 30 μg of total RNA isolated from wild-type (wt) and heterozygous PSF1 mutants hybridized with a PSF1 cRNA probe. Heterozygous PSF1 mutants had decreased PSF1 mRNA in the testis (top). GAPDH mRNA signals were used as an internal standard (bottom). (D and E) PSF1 expression was analyzed by immunohistochemical staining using anti-PSF1 antibody in PSF1+/+ (D) and PSF1−/− (E) embryos at E6.5. Original magnification, ×100.
Northern blot.
Total RNA was extracted from testis and liver of 8-week-old mice. Thirty-microgram aliquots of total RNA were separated on 1% formaldehyde-agarose gels and transferred onto nylon membrane filters by capillary transfer using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer. Filters were hybridized overnight at 68°C in DIG Easy Hyb (Roche) with a digoxigenin-labeled PSF1 cRNA probe. Hybridized probe was detected with alkaline phosphatase-conjugated antidigoxigenin antibodies using the DIG luminescent detection kit (Roche), following the manufacturer's instructions. GAPDH mRNA was used as an internal standard to ensure that equal amounts of RNA were loaded in each lane.
In vitro culture.
Embryos (E3.5) were flushed out from the uteri of pregnant mice and individually cultured in standard ES medium with leukemia inhibitory factor (Invitrogen) for 10 days. DNA synthesis was assessed by BrdU (10 μM; Sigma) incorporation in cultured blastocysts from days 4 to 5; on day 5 of culturing, the blastocysts were fixed with 4% paraformaldehyde in phosphate-buffered saline, stained with anti-BrdU antibody (1 μg/ml; Zymed), and counterstained with hematoxylin. The TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assay for detection of apoptotic cells was performed using the In Situ Cell Death Detection kit (Roche) according to the manufacturer's instructions.
RESULTS AND DISCUSSION
Cloning of mouse PSF1.
Although most terminally differentiated somatic cells are not able to proliferate, stem cells and immature progenitor cells are constitutively in cycle to produce mature cells. To elucidate the molecular mechanism regulating mammalian cell division, we constructed a subtraction library from the BM-derived Lin− c-kit+ Sca-1+ hematopoietic stem cell (KSL cells; stem/progenitor cells) fraction as a tester and the Lin+ mature hematopoietic cells as the driver in order to isolate genes encoding proteins that are involved in DNA replication and specifically expressed in immature cells. Among 521 clones that were abundantly expressed in KSL cells, one gene named #e11 was expressed in KSL cells and their progenitor cells (Lin− c-kit+ Sca-1−) but not in Lin+ mature hematopoietic cells, as confirmed by RT-PCR (Fig. 1A). This gene corresponded to a hypothetical gene in GenBank (accession no. AK013116) and was closely related to Psf1 in a budding yeast (partner of sld5-1), which was shown to encode a protein involved in DNA replication in yeast and in an in vitro model using Xenopus laevis egg extracts (8, 10). We identified the binding partner of mouse #e11 to be sld5 by the two-hybrid system (M. Ueno and N. Takakura, unpublished data). Therefore, we considered this #e11 gene to be a mouse ortholog and named it PSF1.
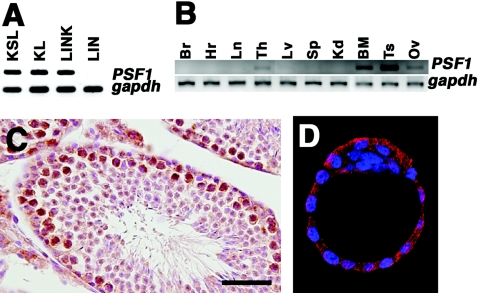
FIG. 1.
PSF1 expression in adult tissues. (A and B) RT-PCR of fractionated adult BM cells (A) and adult tissues (B). KSL, Lin− Sca-1+ c-kit+ cells; KL, Lin− Sca-1− c-kit+; LINK, Lin+ c-kit+; LIN, Lin+. Br, brain; Hr, heart; Ln, lung; Th, thymus; Lv, liver; Sp, spleen; Kd, kidney; Ts, testis; Ov, ovary. (C and D) Immunostaining of sections of the testis (C) and blastocysts (E3.5) (D) with anti-PSF1 antibody labeled with HRP (C) or Cy3 (D). Sections were counterstained with hematoxylin or 4′,6′-diamidino-2-phenylindole (DAPI). Color staining: HRP, brown; Cy3, red; DAPI, blue. Bar, 50 μm.
PSF1 is predominantly expressed in highly proliferative organs, especially in the immature cell population.
We analyzed PSF1 expression in several adult tissues. PSF1 expression was predominantly observed in hematopoietic tissues such as the adult BM and thymus on RT-PCR (Fig. 1B). Moreover, PSF1 expression was observed in reproductive tissues, i.e., the testis and ovary, which have an active stem cell system. In other adult tissues (brain, heart, lung, liver, spleen, and kidney), PSF1 expression was not detectable. These data suggested that PSF1 is expressed specifically in tissues with higher rates of proliferation.
To determine the spatial distribution of PSF1 protein in the adult testis, we generated antibody against PSF1 peptide. Immunohistochemistry on mouse testis sections showed that PSF1 protein is present in immature cells, i.e., spermatogonia (Fig. 1C). PSF1 is expressed in other immature cell populations including blastocysts (Fig. 1D), adult thymic progenitor cells, and yolk sac-containing hematopoietic progenitor cells (data not shown). These data suggested that PSF1 is expressed specifically in the immature cell population.
Targeted disruption of the mouse PSF1 gene.
To analyze the function of PSF1, we generated mice lacking a functional PSF1 gene (Fig. 2). The mouse PSF1 gene contains seven putative coding exons. The targeting vector was designed by deleting exon 5 and inserting a lacZ-neo cassette. Among the 95 independent ES cell colonies examined, we found two homologous recombinants. Correct targeting was confirmed in these ES clones by Southern blot analyses with a 3′ probe (Fig. 2B). One ES clone was aggregated with C57BL/6 blastocysts to generate a chimera, which subsequently produced germ line transmission. The PSF1+/− line was established by backcrosses with C57BL/6 mice. To confirm the loss of PSF1 transcript in mutant mice by gene disruption, we performed Northern blot analysis. As expected, PSF1 mRNA was reduced in PSF1+/− testis (Fig. 2C). PSF1+/− mice were born at Mendelian frequency, and there were no apparent differences between PSF1+/− mice and wild-type mice.
PSF1 is required for cell proliferation, and loss of PSF1 leads to early embryonic lethality.
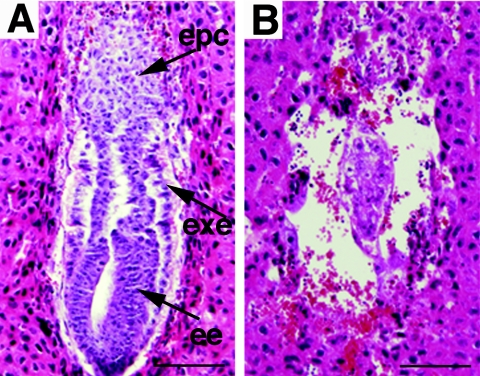
We analyzed the PSF1 gene in neonates resulting from PSF1+/− intercrosses and did not obtain any homozygous offspring (Table 1). In normal E6.5 embryos, a cylinder-like two-layered cellular structure was observed (Fig. 2D and 3A). However, PSF1-deficient embryos, which could be identified at this stage by the absence of PSF1 immunoreactivity (Fig. 2E), lacked such cylinder-like structures (Fig. 2E and 3B). These data suggested that the PSF1−/− embryos failed to develop past E5.5 and showed disorganized embryonic and extraembryonic structures.
TABLE 1.
Progeny from PSF1 heterozygotes
| Age | No. of offspring with genotype:
|
No. resolved | No. total | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| Neonate | 15 | 36 | 0 | 0 | 51 |
| E11.5 | 9 | 14 | 0 | 3 | 26 |
| E9.5 | 7 | 16 | 0 | 6 | 29 |
| E7.5 | 12 | 16 | 0 | 4 | 32 |
| E6.5 | NDa | ND | ND | 8 | 26 |
| E3.5 | 4 | 13 | 5 | NAb | 22 |
ND, not determined. The genotype of 16 E6.5 embryos was not determined.
NA, not available. This table shows the number of offspring obtained by mating PSF1 heterozygotes that were studied.
FIG. 3.
Histological analysis of PSF1−/− embryos. Hematoxylin- and-eosin-stained, longitudinal sections of E6.5 embryos in decidua. (A) Wild-type embryo; (B) mutant PSF1−/− embryo. epc, ectoplacental cone; exe, extraembryonic ectoderm; ee, embryonic endoderm. Bars, 100 μm.
The cellular proliferation and differentiation of mutant embryos were investigated in in vitro cultures of blastocysts. On light microscopy, there were no differences among individual blastocysts (data not shown). In PSF1+/+ and PSF1+/− embryos (total n = 104), trophoblasts started to spread over the culture dish after hatching and supported robust ICM outgrowths after 2 days. While the trophoblasts from PSF1−/− blastocysts (n = 32) also attached and spread over the dish, PSF1−/− ICM cells failed to form outgrowths (see Fig. 4). These data suggested that the lethality of PSF1−/− embryos in utero was caused at least by the death of ICM cells.
FIG. 4.
Defective growth of PSF1−/− blastocysts in vitro. Blastocysts (E3.5) were recovered from heterozygous intercrosses, cultured individually for a period of 6 days as indicated, and genotyped by PCR. Cultures of representative +/− (A to D) and −/− (E to H) blastocysts are shown. Arrow, ICM; arrowhead, trophoblast. (I and J) DNA synthesis in cultured blastocysts (BrdU incorporation). Immunostaining with anti-BrdU antibody was performed (brown). (I) PSF1+/− blastocyst; (J) PSF1−/− blastocyst. Inset in panel I shows a different field in this culture plate. Arrow, ICM; arrowhead, labeled trophoblast. (K and L) Apoptosis of cells in cultured ICM from blastocysts (TUNEL assay; green, apoptotic cells). (K) PSF1+/− blastocyst; (L) PSF1−/− blastocyst. Nuclei were counterstained with propidium iodide (red). Bars, 100 μm. (M to P) Blastocysts were cultured for 8 days (M and O) and 10 days (N and P). (M and N) PSF1+/− blastocyst; (O and P) PSF1−/− blastocyst.
To further delineate the proliferation defect of the PSF1−/− blastocysts, we carried out bromodeoxyuridine incorporation assays during blastocyst outgrowth (Fig. 4I and J). Vigorous DNA synthesis was observed in PSF1+/− ICM cells by immunostaining with anti-BrdU antibody (Fig. 4I). However, the presumed ICM cells from the mutants ceased to proliferate, while DNA synthesis was still observed in trophoblasts (Fig. 4J). Moreover, in the ICM of PSF1−/− blastocyst cultures, TUNEL-positive apoptotic cells appeared on day 5 (Fig. 4K and L). Therefore, the PSF1−/− ICM cells were unable to proliferate and underwent apoptosis in culture.
While BrdU+ cells were found in PSF1−/− trophoblasts after 5 days of culturing (Fig. 4J), PSF1−/− trophoblasts stopped proliferation after 8 days of culturing (Fig. 4O). BrdU was not incorporated in PSF1−/− trophoblasts beyond 8 days (data not shown), and the number of trophoblasts declined (Fig. 4P). By contrast, PSF1+/− embryo trophoblasts continuously proliferated after 10 days of culturing (Fig. 4M and N). These data indicated that PSF1 is essential for both ICM and trophoblast proliferation.
In this study, we examined the functions of PSF1 in vivo by gene targeting technology. Our results revealed impaired proliferation of the ICM and trophoblasts in PSF1−/− embryos. Recently, it was reported that Psf1 and CDC45 are involved cooperatively in the initiation of DNA replication in yeast (10, 19) and that both molecules are prerequisite for DNA replication in yeast (7, 20). In mice, the phenotype of CDC45-deficient embryos after uterine implantation (24) is quite similar to that of PSF1-null embryos. Mice deficient in the CDC45 ortholog of yeast show a defect in cell proliferation in blastocyst culture (24). Therefore, the molecular functions of PSF1 and CDC45 for DNA replication may be conserved in mammalian cells.
Although Psf1 is indispensable for cell proliferation in yeast (19), no obvious morphological abnormality was found in PSF1−/− embryos before implantation (data not shown). This observation raised the possibility of the existence of maternal PSF1 transcript stores and could account for the proliferation of PSF1−/− embryos through the early developmental stages. To ascertain this, we performed immunostaining on unfertilized eggs with anti-PSF1 antibody which revealed potent expression of PSF1 (data not shown). Because zygotic gene transcription initiates at the two-cell stage and paternal protein contributions are thought to be negligible (16), we conclude that the timing of PSF1−/− lethality is due to the loss and/or dilution of maternal PSF1 transcripts around implantation stage.
Most of our knowledge on DNA replication has accumulated from studies on lower eukaryotes. However, it can be argued that DNA replication of higher eukaryotes is more complex. For example, DNA replication is linked to gene transcription in higher eukaryotes including Drosophila melanogaster (15), Xenopus (5), and mice (6) but not in yeast (14). Although origin recognition complex (Orc) binds to chromatin at replication origins throughout the cell cycle in yeast (1, 4, 21), a part of Orc2 localizes at the centrosomes and heterochromatin during the M phase and participates in chromosome segregation in human cells (13). Mcm10, which is a conserved protein and is involved in initiation of DNA replication in yeast, is required for chromosome condensation in fly cells (3). Interestingly, in nematodes, loss of PSF1 causes abnormality of chromatin segregation (M. Ueno and N. Takakura, unpublished data). Thus, PSF1 may have pivotal roles in other biological processes. Further analysis on the function of PSF1 will shed light on the complex mechanism of DNA replication in mammalian cells.
Acknowledgments
We thank Y. Shimizu, K. Ishida, M. Sato, and Y. Nakano for technical assistance.
This work was partly supported by the Japanese Ministry of Education, Culture, Sports, Science and Technology and the Japan Society for Promotion of Science.
REFERENCES
- 1.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 3.Christensen, T. W., and B. K. Tye. 2003. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol. Biol. Cell 14:2206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diffley, J. F., J. H. Cocker, S. J. Dowell, and A. Rowley. 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78:303-316. [DOI] [PubMed] [Google Scholar]
- 5.Fisher, D., and M. Mechali. 2003. Vertebrate HoxB gene expression requires DNA replication. EMBO J. 22:3737-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forlani, S., C. Bonnerot, S. Capgras, and J. F. Nicolas. 1998. Relief of a repressed gene expression state in the mouse 1-cell embryo requires DNA replication. Development 125:3153-3166. [DOI] [PubMed] [Google Scholar]
- 7.Hopwood, B., and S. Dalton. 1996. Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc. Natl. Acad. Sci. USA 93:12309-12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanemaki, M., A. Sanchez-Diaz, A. Gambus, and K. Labib. 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423:720-724. [DOI] [PubMed] [Google Scholar]
- 9.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 10.Kubota, Y., Y. Takase, Y. Komori, Y. Hashimoto, T. Arata, Y. Kamimura, H. Araki, and H. Takisawa. 2003. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 17:1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimura, S., and H. Takisawa. 1998. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 17:5699-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasanth, S. G., K. V. Prasanth, K. Siddiqui, D. L. Spector, and B. Stillman. 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23:2651-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 15.Schubeler, D., D. Scalzo, C. Kooperberg, B. van Steensel, J. Delrow, and M. Groudine. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 32:438-442. [DOI] [PubMed] [Google Scholar]
- 16.Schultz, R. M. 1993. Regulation of zygotic gene activation in the mouse. Bioessays 15:531-538. [DOI] [PubMed] [Google Scholar]
- 17.Takakura, N., T. Watanabe, S. Suenobu, Y. Yamada, T. Noda, Y. Ito, M. Satake, and T. Suda. 2000. A role for hematopoietic stem cells in promoting angiogenesis. Cell 102:199-209. [DOI] [PubMed] [Google Scholar]
- 18.Takakura, N., X. L. Huang, T. Naruse, I. Hamaguchi, D. J. Dumont, G. D. Yancopoulos, and T. Suda. 1998. Critical role of the TIE2 endothelial cell receptor in the development of definitive hematopoiesis. Immunity 9:677-686. [DOI] [PubMed] [Google Scholar]
- 19.Takayama, Y., Y. Kamimura, M. Okawa, S. Muramatsu, A. Sugino, and H. Araki. 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17:1153-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tercero, J. A., K. Labib, and J. F. Diffley. 2000. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 19:2082-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinreich, M., C. Liang, and B. Stillman. 1999. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc. Natl. Acad. Sci. USA 96:441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. E. Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. J. Hegemann, T. M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-McDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 23.Wohlschlegel, J. A., B. T. Dwyer, S. K. Dhar, C. Cvetic, J. C. Walter, and A. Dutta. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290:2309-2312. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida, K., F. Kuo, E. L. George, A. H. Sharpe, and A. Dutta. 2001. Requirement of CDC45 for postimplantation mouse development. Mol. Cell. Biol. 21:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]